Abstract
We have investigated human male demographic history using 590 males from 51 populations in the Human Genome Diversity Project - Centre d’Étude du Polymorphisme Humain worldwide panel, typed with 37 Y-chromosomal Single Nucleotide Polymorphisms and 65 Y-chromosomal Short Tandem Repeats and analyzed with the program Bayesian Analysis of Trees With Internal Node Generation. The general patterns we observe show a gradient from the oldest population time to the most recent common ancestors (TMRCAs) and expansion times together with the largest effective population sizes in Africa, to the youngest times and smallest effective population sizes in the Americas. These parameters are significantly negatively correlated with distance from East Africa, and the patterns are consistent with most other studies of human variation and history. In contrast, growth rate showed a weaker correlation in the opposite direction. Y-lineage diversity and TMRCA also decrease with distance from East Africa, supporting a model of expansion with serial founder events starting from this source. A number of individual populations diverge from these general patterns, including previously documented examples such as recent expansions of the Yoruba in Africa, Basques in Europe, and Yakut in Northern Asia. However, some unexpected demographic histories were also found, including low growth rates in the Hazara and Kalash from Pakistan and recent expansion of the Mozabites in North Africa.
Keywords: Y-STR, Y-SNP, HGDP–CEPH, male demographic history, BATWING, serial founder model
Introduction
Current models of human evolution differ in detail, but all include a recent origin in Africa and an expansion, both geographical and demographic, of fully modern humans from a small African population to the current large worldwide population within the last ∼100,000 years (KY) (Jobling et al. 2004). The timing and rate of this expansion and its variation in different parts of the world are, however, unclear. The patterns of DNA variation in modern populations carry powerful information about their evolutionary history, including demographic information (Cavalli-Sforza 2007). Many analyses of worldwide DNA data sets support the hypothesis of serial founder events starting from a single origin in sub-Saharan Africa and leading to the Americas as the last continents to be inhabited (e.g., Prugnolle et al. 2005; Hellenthal et al. 2008; Li et al. 2008). However, the demographic changes accompanying these events merit further investigation.
The haploid Y chromosome can provide unique insights into the human past. Its long nonrecombining segment carries the most informative stable haplotypes in the genome, whereas its permanent location in the male genome links these to male-specific history (Jobling and Tyler-Smith 2003). Consequently, it has been an attractive target for demographic inference. Previous studies have usually been carried out at worldwide or continentwide resolution and have suggested demographic expansion beginning in the Paleolithic ∼18 (7–41) KYA (Pritchard et al. 1999) or ∼22 (8.5–50) KYA (Macpherson et al. 2004) but have sometimes focused on smaller areas. Such detailed studies have revealed that there can be significant variation between neighboring regions, for example, expansion in the northern part of East Asia beginning before the last glacial maximum 18–21 KYA contrasted with expansion in the southern part beginning afterward (Xue, Zerjal, et al. 2006) or, within Europe, a much later start to expansion in Armenia (Weale et al. 2001).
Two factors have led us to reinvestigate this subject. First, the Human Genome Diversity Project - Centre d’Étude du Polymorphisme Humain (HGDP-CEPH) panel of 1,064 DNAs (Cann et al. 2002) has become a standard resource for many evolutionary genetic studies (e.g., Rosenberg et al. 2002; Hellenthal et al. 2008; Li et al. 2008; Pickrell et al. 2009), so it would be useful to have detailed information about male demographic history in this sample set that can then be compared with the results of other analyses. Second, the number of useful Y-chromosomal markers available has increased by more than an order of magnitude (Kayser et al. 2004; Lim et al. 2007) since the initial exploration of Y-chromosomal variation in this panel (Macpherson et al. 2004), providing greatly increased haplotype resolution (Vermeulen et al. 2009). We therefore set out to investigate three areas: the influence of marker number and type (simple or complex Y-chromosomal Short Tandem Repeats [Y-STR]) on the conclusions that could be drawn, the demographic inferences that could be obtained at the individual population level, and the support (or lack of it) that the Y data would provide for the serial founder model.
Materials and Methods
Data
Haplotypes of 590 HGDP–CEPH male samples chosen from the H952 subset (Cann et al. 2002; Rosenberg 2006) based on a total of 67 Y-STRs have been determined (Vermeulen et al. 2009), but the two DYS385 loci were excluded from the current analyses because they could not be distinguished using the typing method employed, and all work was based on 65 Y-STRs or subsets of them. Duplicated or fractional alleles were treated as missing data. Thirty-three Y-SNPs identifying major branches in the Y-chromosomal phylogeny were genotyped using a standard multiplex amplification and minisequencing protocol modified from that of Sanchez et al. (2003). Further details and genotypes are available on request from PdeK. The populations were assigned to seven geographical regions for some analyses as shown in supplementary table S1, Supplementary Material Online. Some individual population sample sizes were very small, and this raised the question of what minimum size should be used. Because of the particular interest of the San, we did not want to exclude this sample, so we provisionally accepted n ≥ 4 and investigated the constraints imposed by such a small sample as described in the Results section. The Colombian and Mayan samples (two individuals each) fell below this threshold and were combined.
Demographic Inferences
We used the program BATWING (Bayesian Analysis of Trees With Internal Node Generation) (Wilson et al. 2003). BATWING uses a Markov Chain Monte Carlo (MCMC) procedure to generate a series of genealogical trees with associated parameter values consistent with the data. After equilibration, posterior estimates of these parameters can be obtained, along with their confidence intervals (CIs). BATWING makes a number of assumptions, including single-step STR mutations and no mutation at SNPs (treated as unique event polymorphisms), recombination, or selection. In each run, the input data set consisted of Y-STR allele sizes and all the Y-SNPs that showed a variant in >1 individual in the sample, except that phylogenetically equivalent duplicate SNPs were omitted. We used a population model of exponential growth from an initially constant-sized population with the settings and priors described previously (Xue, Zerjal, et al. 2006), except for the mutation rate. Three sets of mutation rates were compared: 1) an “observed” mutation rate (OMR) for each Y-STR compiled from previously described mutation counts in father–son pairs (Dupuy et al. 2004; Gusmao et al. 2005; Lee et al. 2007; Shi et al. 2007; Decker et al. 2008; Padilla-Gutierrez et al. 2008; Toscanini et al. 2008; Goedbloed et al. 2009; Kim et al. 2009) or in deep-rooted pedigrees (Vermeulen et al. 2009) tabulated in supplementary table S2, Supplementary Material Online. 2) A widely used calibrated “evolutionary” mutation rate (EMR) based on well-dated historical events (Zhivotovsky et al. 2004). 3) A recalibration of the EMR that corrected for the difference in variance between the Y-STRs used by Zhivitovsky et al. and some of those used here, the recalibrated evolutionary mutation rate (rEMR). BATWING convergence was assessed by extending runs to at least x MCMC cycles such that x and 10x cycles gave similar results (Xue et al. 2008).
BATWING was run on the Sanger Institute “computer farm,” using the Platform LSF job schedular. The farm consisted of a mixture of Intel Xeon EMT64 and AMD Opteron processors with 8–16 GB of RAM each. From each run, we recorded 1) time to the most recent common ancestor (TMRCA) of the population and individual Y-SNPs, 2) effective population size before population growth (Ne), 3) time when growth began, and 4) growth rate. All runs were carried out three times starting with different random seeds, and values given are means of the three.
A reduced-median network was constructed from a worldwide data set consisting of the Y-SNPs plus 11 standard complex Y-STRs using Network 4.10 (http://www.fluxus-engineering.com/netwinfo.htm) (Bandelt et al. 1995). Time estimates for branches defined by SNPs, and their standard errors, were determined using the rho statistic implemented in the Network program and calibrated using the EMR/rEMR, which were equivalent for these Y-STRs.
Geographical distributions of demographic parameters were displayed as contour plots on a world map using Surfer 9 from Golden Software (http://www.goldensoftware.com/). Interpolation between populations was carried out using the default Kriging method; some large regions of the map that lack data (e.g., Australia and Greenland) were omitted and appear white. Pearson's correlation coefficient (R2), Spearman's rank correlation coefficient (ρ), and their significance were calculated using SPSS (version 16.0) for Windows.
Results
We wished to investigate the information about male historical demography contained in the HGDP–CEPH data set. In order to do this, we first needed to explore some features of the data: whether or not the choice of Y-STRs was important, whether or not small sample sizes could be used, and which mutation rate to adopt.
Y-STR and Sample Properties
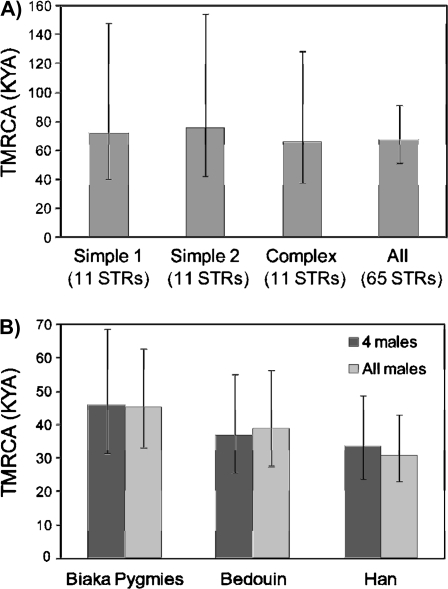
We began by exploiting the large number of Y-STRs for which data were available in order to investigate the effect of STR type and number on demographic inferences. Among the 65 Y-STRs were 11 with complex structures (i.e., more than one repeat unit sequence), which include most of the commonly used loci, and we matched these with two sets of 11 simple-structure Y-STRs (a single repeat-unit sequence) with similar variance (supplementary table S3, Supplementary Material Online) to determine the reproducibility of the outcome and the effect of simple or complex structure. We needed to establish the number of MCMC cycles required for convergence of the program to a stable state and found that with sample sizes of up to 77 males and 65 Y-STRs, convergence had occurred after 107 cycles (supplementary fig. 1, Supplementary Material online). We therefore used this number of cycles, or more, in subsequent analyses with sample sizes that were usually smaller. The three different 11-Y-STR sets gave very similar demographic inferences in 77 males from sub-Saharan Africa (fig. 1A and supplementary fig. 2, Supplementary Material online), illustrating the independence of these inferences from the particular set of markers used. Similar inferences were again obtained using all 65 Y-STRs (fig. 1A and supplementary fig. 2, Supplementary Material online). Here, it was notable that the increased number of Y-STRs narrowed the 95% CI for the TMRCA from 41 to 148, 42 to 155, and 38 to 129 KYA for 11 simple or complex Y-STRs to 51 to 91 KYA for 65 Y-STRs but not the CI for the expansion time or Ne. A larger number of Y-STRs therefore has some advantages, and all 65 loci were used subsequently.
FIG. 1.
Properties of STRs and sample size. (A) Effect of simple or complex Y-STR structure and Y-STR number. All Y-STR sets produce similar median estimates of TMRCA, but the larger number of Y-STRs led to a reduced 95% CI. (B) Effect of sample size. Similar median estimates of TMRCA were obtained, but the 95% CIs of the TMRCA were slightly reduced for the larger sample sizes.
Because some population sample sizes were as small as four, we needed to determine whether useful demographic information could be obtained from such a small sample and decide whether to include such samples, merge them with others, or omit them entirely. We therefore randomly subsampled four males from three larger samples—Biaka Pygmies (n = 20), Bedouin (n = 24), and Han Chinese (n = 23)—and compared the posteriors from these subsamples with those from the whole sample. Similar median posterior estimates were obtained (fig. 1B and supplementary fig. S3, Supplementary Material Online), although the 95% CI for the TMRCA and Ne (but not for expansion time) were wider for the small samples. We therefore concluded, somewhat to our surprise, that a sample size of four was often sufficient to provide useful information and that we did not need to omit or combine such samples.
The final methodological issue that we needed to address was the choice of mutation rate. For all the Y-STRs, information about the OMR was available from either direct counts in father–son pairs or deep-rooting family data (supplementary table S2, Supplementary Material online), and a general EMR (Zhivotovsky et al. 2004) is commonly applied to all Y-STRs. We used both of these but were concerned that although the EMR was appropriate for Y-STRs with similar levels of variability to the eight loci used by Zhivitovsky et al. to estimate it, it would not be appropriate for significantly more or less variable Y-STRs that are expected to have different mutation rates. We therefore devised the following strategy to overcome this problem. We first compared the OMR of each marker with its variance in the 590 individuals and found them to be highly correlated (ρ = 0.444, P = 0.001; supplementary fig. S4, Supplementary Material online). We could thus use variance to guide the choice of appropriate mutation rate priors. For Y-STRs with variances within the range of variances of the eight Y-STRs used by Zhivitovsky et al., we used the Zhivitovsky et al. rate. The 15 Y-STRs with variances above or below this range were assigned to four additional classes as shown in table 1. These rEMRs provided a third set of mutation rates.
Table 1.
Recalibrated Mutation Rates for five subsets of Y-STRs Grouped by Repeat Count Variance in the HGDP–CEPH Data Set.
| Subset* | Mean Variance | Recalibrated Mutation Rate | Prior Distribution for Recalibrated Mutation Rate | 95% Interval of Gamma Distribution |
| 1 | 1.70 × 10−3 | 1.35 × 10−6 | Gamma (0.1; 75,000) | (3.38 × 10−8)–(4.92 × 10−6) |
| 2 | 6.00 × 10−2 | 4.59 × 10−5 | Gamma (1; 22,000) | (1.15 × 10−6)–(1.68 × 10−4) |
| 3 | 2.01 × 10−1 | 1.56 × 10−4 | Gamma (1; 6,400) | (3.96 × 10−6)–(5.76 × 10−4) |
| 4 | 9.06 × 10−1 | 6.90 × 10−4 | Gamma (1.47; 2,130) | (4.76 × 10−5)–(2.17 × 10−3) |
| 5 | 4.61 | 3.51 × 10−3 | Gamma (1; 2,85) | (8.88 × 10−5)–(1.29 × 10−2) |
*Subset 1: DYS472. Subset 2: DYS579, DYS480, DYS583, DYS530, DYS590, and DYS569. Subset 3: DYS575, DYS580, DYS554, DYS476, DYS636, DYS494, and DYS640. Subset 4: DYS391, DYS488, DYS491, DYS567, DYS540, DYS617, DYS618, DYS568, DYS638, DYS578, DYS437, DYS492, DYS537, DYS497, DYS594, DYS531, GATA_H4, DYS389CD, DYS565, DYS573, DYS511, DYS572, DYS490, DYS556, DYS456, DYS393, DYS438, DYS525, DYS549, DYS439, DYS533, DYS389AB, DYS522, DYS589, DYS495, DYS508, DYS505, DYS485, DYS19, DYS448, DYS388, DYS641, DYS487, DYS390, DYS458, DYS643, DYS635, DYS576, DYS392, and DYS570. Subset 5: DYS481.
Posterior estimates of TMRCA, expansion time, Ne, and growth rate were then calculated for the 51 HGDP–CEPH populations using each of the three mutation rates. For the first three parameters, median values followed the order rEMR > EMR > OMR, whereas for growth rate, the opposite order was usually seen (supplementary table S1; supplementary fig. S5, Supplementary Material Online). In the following section, we present results from the rEMR calculations.
Demographic Inferences in 51 Populations
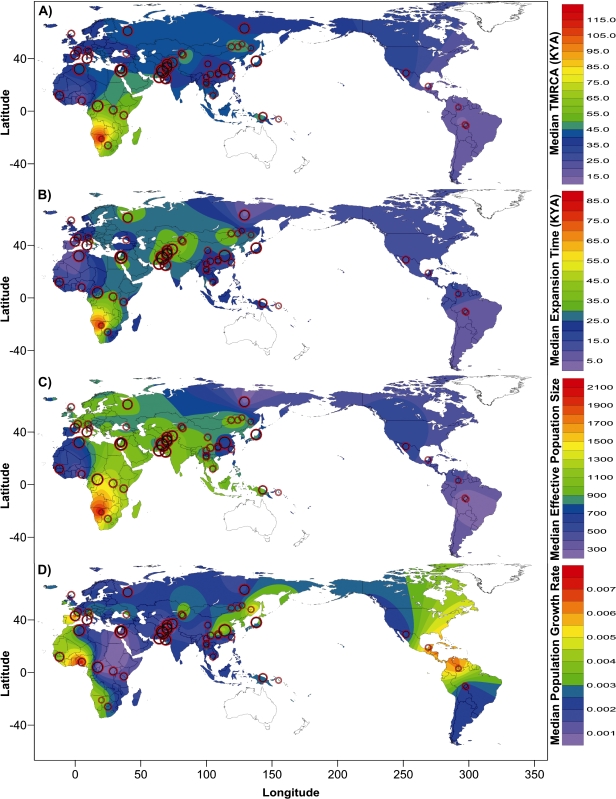
Median values of the four demographic parameters were plotted according to the geographical location of the sample site in Figure 2A–D. A number of general features are apparent in the data. First, the parameters are correlated, with a tendency for populations with an older TMRCA to have an older expansion time and larger effective population size but a lower growth rate and vice versa.
FIG. 2.
Contour plot showing the posterior distribution of (A) TMRCA, (B) Expansion time, (C) Initial effective population size, and (D) Population growth rate. Each population is marked by a circle, centered on the sampling site and with a diameter proportional to its sample size. The sample sizes of different populations are shown in supplementary table S1, Supplementary Material online.
Second, these correlations are not perfect. Although all pairwise comparisons between TMRCA, expansion time, and Ne were highly significant (P < 0.001, table 2), the ρ values ranged from 0.61 to 0.75. In contrast, these three parameters were all negatively correlated with growth rate, but only the correlation between Ne and growth rate was significant (ρ = −0.39, P = 0.005, table 2). For example, the population with the highest values for TMRCA, expansion time, and Ne (the San) showed an intermediate value for growth rate rather than the lowest.
Table 2.
Correlations of the Four Demographic Parameters in the 51 Populations.
| TMRCA | Expansion Time | Ne | Growth Rate | |
| TMRCA | – | 0.611 | 0.750 | −0.104 |
| Expansion time | <0.001 | – | 0.685 | −0.211 |
| Ne | <0.001 | <0.001 | – | −0.385 |
| Growth rate | 0.468 | 0.137 | 0.005 | – |
Correlation coefficients are shown above the diagonal. Corresponding P values are shown in italics below the diagonal.
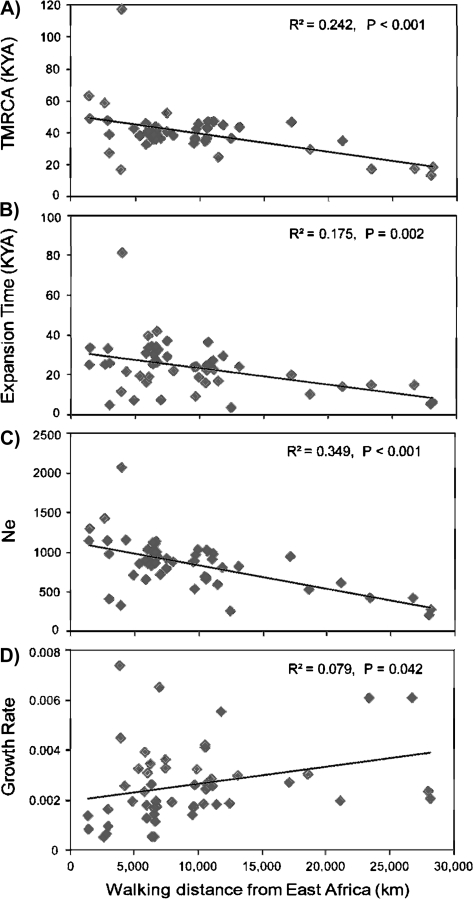
Third, strong geographical patterns are seen on a continental scale. Sub-Saharan African populations tend to have the oldest TMRCAs, the largest Nes and the earliest expansion times, whereas the American populations have some of the most recent TMRCAs and expansion times and the smallest Nes (fig. 2A–C). The other continental populations fall in between. Walking distance from an origin in East Africa (conventionally set at Addis Ababa) has been found to correlate with several characteristics of human populations, for example, negatively with mean STR diversity (Prugnolle et al. 2005), so we tested the correlation of male demographic parameters with these distances. TMRCA, expansion time, and Ne were negatively correlated, and these correlations were highly significant (fig. 3A–C). In contrast, the correlation with growth rate was much weaker and positive but still reached significance (fig. 3D).
FIG. 3.
Correlation between distance from East Africa with four demographic parameters in 51 populations. Strong negative correlations are seen between distance and (A) TMRCA, (B) expansion time, and (C) effective population size; a weak but significant positive correlation between distance and (D) population growth rate is also seen.
Fourth, some individual populations stand out from this general pattern. The Yoruba showed low TMRCA and Ne compared with the rest of the African populations. Both Palestinians and Mozabite from the Middle East and North Africa, respectively, showed extremely recent expansion times (5.0 and 7.4 KYA) but not very high population growth rates, whereas the Basques in Europe showed a very recent expansion time (7.5 KYA) coupled with a very high population growth rate. In northern Asia, the Yakut showed a very recent expansion time of 3.7 KYA, a small Ne, and average growth rate. On the other hand, a number of populations from several parts of the world showed relatively low population growth rates: Biaka Pygmies and Mbuti Pygmies in Africa, Bedouin in the Middle East, and Hazara and Kalash in Central/South Asia.
Haplotype Patterns outside Africa
We next estimated TMRCAs for individual Y-SNPs in the worldwide data set using both BATWING and, for comparison, the rho statistic in Network; we also estimated TMRCAs in individual population samples from the BATWING data (supplementary table S4, Supplementary Material online). The BATWING and Network estimates were highly correlated (R2 = 0.43, P = <0.001), but the Network times were, on average, 1.2-fold older than those from BATWING. Furthermore, the differences were systematic in that their ratio was correlated with the TMRCA (R2 = 0.16, P = 0.028): TMRCAs below ∼40 KYA were in general similar between the two methods, whereas older TMRCA estimates tended to be higher with Network. We ascribe a substantial part of this difference to the difficulty in specifying the correct location of the root required by the Network calculation (Xue, Daly, et al. 2006) and use mainly the BATWING TMRCAs.
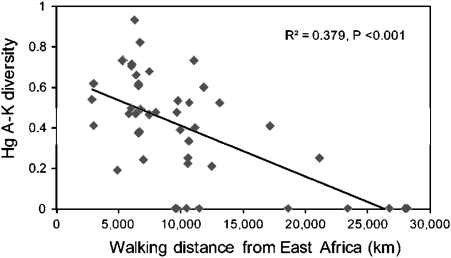
Both sets of estimates were consistent in their suggestions that the major branches of the haplogroup tree, A–K, had originated by ∼40 KYA, soon after the migration out of Africa, and could therefore be used to explore the applicability of a serial founder model to the Y data. According to this model, progressive loss of lineages would be expected as humans migrated further from Africa. This is indeed seen in the Y data, with a significant decrease in both the number of these lineages with distance (R2 = 0.36, P <0.001) and the haplogroup diversity (R2 = 0.38, P <0.001; fig. 4).
FIG. 4.
Relationship between diversity of Y haplogroups A–K in populations outside Africa with distance from East Africa. A strong negative correlation is seen, consistent with serial founder models.
In addition, the TMRCAs of the lineages are predicted by the model to be more recent at greater distances from Africa. A test of this prediction requires lineages that are both geographically widespread and abundant enough to give meaningful TMRCA estimates. These requirements are potentially met by the nested set of lineages C–K (defined by marker M168), F–K (M213), and K (M9). Their TMRCAs do decrease with distance from East Africa (table 3, supplementary fig. S6, Supplementary Material online), and these correlations are significant for all except haplogroup K where the reduced numbers of samples (median 6 per population sample; 10 samples ≤3 individuals) introduces noise into several of the TMRCA estimates. Overall, however, there is strong support for the hypothesis of fewer and more recent Y lineages at increased distances outside Africa.
Table 3.
Correlations of Lineage TMRCAs with Walking Distance from East Africa.
| Lineage | Frequency (%) | R2 Value | P Value |
| All | 100 | 0.382 | <0.001 |
| C–K (M168) | 99 | 0.335 | <0.001 |
| F–K (M213) | 85 | 0.216 | 0.002 |
| K (M9) | 61 | 0.011 | 0.506 |
Discussion
In this study, we made decisions about a number of technical issues arising from the markers, the small sample sizes available from some populations, and the range of mutation rates that might be chosen and now discuss these before considering the broader conclusions from our data.
The use of simple versus complex Y-STRs had no apparent influence on the demographic inferences, and the increase in number of Y-STRs from 11 to 65 had only a small effect but did lead to a decrease in the CI in some parameters (fig. 1A): reassuring findings in view of the small numbers of predominantly complex Y-STRs that have been used in all previous studies of this kind. Subsampling showed that random subsets of four Y chromosomes typically produced similar inferences to larger sets (fig. 1B). This outcome must depend on the phylogenetic structure of a sample. If, for example, a sample contained a common haplogroup and a rare highly divergent haplogroup, an old TMRCA would be observed in the few subsamples that contained the rare haplogroup and a more recent one in the majority of subsamples that lacked it. From the general correspondence found between large and small samples, we conclude that demographic inferences from these samples are not dominated by rare lineages. Further support for the representative nature of small samples comes from a comparison of the current results with those of Xue, Zerjal, et al. (2006): TMRCAs were compared using EMRs in the three populations Daur, Hezhen, and Oroqen where the sample sizes (previous:current) were 39:6, 45:5, and 31:5, respectively. The TMRCAs were very similar (supplementary fig. S7, Supplementary Material online).
The choice of mutation rate was perhaps the most difficult issue. Although it influences only the temporal scale of the conclusions, this scale is crucial for comparisons with other genetic and archaeological or climatic data. The OMR is based on a large number of observed transmissions for some loci (Goedbloed et al. 2009), but far fewer for others (Vermeulen et al. 2009), so the uncertainties differ between loci. These uncertainties were accommodated by using narrower or wider CIs in the BATWING priors. Overall, the rEMR provided the best combination of insights from both OMR and EMR for prior values, and the posterior times obtained were generally consistent with other data sets. For example, the oldest TMRCA was found in the San population (117 KYA, 95% CI 83–168 KYA) who show highly divergent patterns of classical markers (Cavalli-Sforza et al. 1994) and ancient mitochondrial DNA (mtDNA) lineages (Behar et al. 2008). Two other African populations show TMRCAs older than 55 KYA, but no non-African population has such an ancient TMRCA, consistent with the idea of a single Y lineage surviving from a migration out of Africa ∼50–60 KYA and demonstrating that the ancient African TMRCAs are not dependent on the small San sample. More recent TMRCAs in Asia and even more recent ones in the Americas are consistent with serial founder effects and loss of lineages by drift (Prugnolle et al. 2005; Li et al. 2008).
Estimates of the time when demographic expansion began lie within the Paleolithic period for most populations, as in most previous studies using the Y chromosome (Pritchard et al. 1999; Macpherson et al. 2004; Xue, Zerjal, et al. 2006), genomewide autosomal STRs in the HGDP-CEPH panel (Zhivotovsky et al. 2003), and mtDNA coding-region sequences (Atkinson et al. 2008). The estimates from mtDNA show growth in sub-Saharan Africa beginning 143–193 KYA, in South Asia at 52 KYA, followed by Northern, Central, and East Asia (49 KYA); Europe (42 KYA); the Middle East and North Africa (40 KYA); New Guinea (39 KYA); and the Americas (18 KYA) (Atkinson et al. 2008). Earliest expansion times from similar geographical regions in our Y-chromosomal study give the same order: The earliest expansion begins at 81 (40–130) KYA for the San in sub-Saharan Africa, followed by Burusho and Brahui of Pakistan, South Asia (42 and 40 KYA), Xibe and Mongolians of East Asia (37 and 36 KYA), Tuscans and Russians of Europe (35 and 33 KYA), Bedouin and Druze of Middle East and North Africa (33 and 26 KYA), Papuans of Oceania (20 KYA), and Pima of America (14 KYA). The times are very highly correlated (R2 = 0.97, P < 0.001), but the Y-chromosomal times are on average 76% of the mtDNA times. Although this raises the question of whether there might be sex-specific influences or selection on one or both loci, the most obvious explanation would be a calibration difference; we note that the alternative mutation rates considered would lead to even more recent Y-chromosomal times and so less agreement with the mtDNA inferences.
The largest male effective population sizes are found in Africa among the San and Pygmies (median values ∼1,300–2,100), but sizes ≥1,000 are found among 11 populations outside Africa in many parts of Asia and Europe, and only in the Americas are sizes consistently small (380 on average). The lowest growth rates are found not only among the Pygmy populations, as might be expected from their hunter-gatherer lifestyles and generally stable population sizes, but also among Bedouin and Druze (both Middle East), and Hazara and Kalash (both Pakistan), and point to features worth investigating further in these populations.
A number of populations stand out as exceptions to these general patterns. In Africa, the Yoruba have a particularly low TMRCA (17 KYA) and expansion time (12 KYA), perhaps linked to the expansion of Bantu-speaking farmers beginning ∼5 KYA, which erased much of the ancient Y-chromosomal diversity from this region (Jobling et al. 2004; Berniell-Lee et al. 2009). In Europe, the Basques, a well-studied linguistic isolate (Cavalli-Sforza et al. 1994) do not have an unusual TMRCA or effective population size but do have a strikingly recent expansion time and high growth rate consistent with rapid expansion after a bottleneck. The oldest TMRCA outside Africa (52 KYA) is found in the Xibe from Northern China, but this somewhat surprising finding may reflect their history of recent migration and admixture (Powell et al. 2007) and thus a poor fit to the population model assumed by BATWING. In contrast, the youngest expansion time and smallest effective population size in Asia occur in the Yakut from Siberia, consistent with previous observations of their low male lineage diversity (Zerjal et al. 1997) and likely origin through recent migration of a small population into a new area (Pakendorf et al. 2006). Intriguing observations that suggest areas for additional study are the low TMRCA (25 KYA) in the She from Southern China, and the particularly recent expansion times in the Mozabites (7.4 KYA, North Africa) and Palestinians (5.0 KYA, Middle East).
The serial founder model for the origin of populations outside Africa proposes a single-source population, the formation of a new population by a subset of individuals from this source (a bottleneck), followed by expansion of the new population that then becomes the source of the next population, and so on. Several features of autosomal data from the HGDP–CEPH populations fit the model well, including a decrease in diversity and increase in linkage disequilibrium with distance from East Africa (Prugnolle et al. 2005; Hellenthal et al. 2008; Jakobsson et al. 2008; Li et al. 2008). It is therefore expected, but still encouraging, to see a corresponding decrease in the diversity of Y-chromosomal lineages (fig. 4). The detailed haplotypes provided by the Y chromosome, however, allow an additional test to be performed: The TMRCAs of lineages are predicted to decrease with distance from East Africa, and this decrease is indeed seen in the Y data (table 3). The model therefore gains additional support.
In conclusion, this survey provides the most detailed view of human male demographic history thus far available. It is based on a number of simplifications, such as the demographic model in BATWING that assumes a constant size followed by exponential expansion and does not permit more complex size changes, mixing, or other characteristics of real populations. Nevertheless, it appears to capture key features of human history. In both broad outline and also in many of the exceptions to the general patterns, the conclusions fit the findings from other genetic and nongenetic studies. But some unusual patterns remain unexplained and provide directions for future work.
Supplementary Material
Supplementary figures S1–S7 and supplementary tables S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank the HGDP–CEPH donors for their contribution to this panel and Howard Cann for providing DNAs. Our work was funded by The Wellcome Trust, the Netherlands Forensic Institute, and received additional support by a grant from the Netherlands Genomics Initiative (NGI)/Netherlands Organization for Scientific Research (NWO) within the framework of the Forensic Genomics Consortium Netherlands (FGCN) to M.K. and P.dK.
References
- Atkinson QD, Gray RD, Drummond AJ. mtDNA variation predicts population size in humans and reveals a major Southern Asian chapter in human prehistory. Mol Biol Evol. 2008;25:468–474. doi: 10.1093/molbev/msm277. [DOI] [PubMed] [Google Scholar]
- Bandelt HJ, Forster P, Sykes BC, Richards MB. Mitochondrial portraits of human populations using median networks. Genetics. 1995;141:743–753. doi: 10.1093/genetics/141.2.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Villems R, Soodyall H, et al. (16 co-authors) The dawn of human matrilineal diversity. Am J Hum Genet. 2008;82:1130–1140. doi: 10.1016/j.ajhg.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berniell-Lee G, Calafell F, Bosch E, Heyer E, Sica L, Mouguiama-Daouda P, van der Veen L, Hombert JM, Quintana-Murci L, Comas D. Genetic and demographic implications of the Bantu expansion: insights from human paternal lineages. Mol Biol Evol. 2009;26:1581–1589. doi: 10.1093/molbev/msp069. [DOI] [PubMed] [Google Scholar]
- Cann HM, de Toma C, Cazes L, et al. (41 co-authors) A human genome diversity cell line panel. Science. 2002;296:261–262. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. Human evolution and its relevance for genetic epidemiology. Annu Rev Genomics Hum Genet. 2007;8:1–15. doi: 10.1146/annurev.genom.8.080706.092403. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. The history and geography of human genes. Princeton University Press: Princeton (NJ); 1994. [Google Scholar]
- Decker AE, Kline MC, Redman JW, Reid TM, Butler JM. Analysis of mutations in father-son pairs with 17 Y-STR loci. Forensic Sci Int Genet. 2008;2:e31–e35. doi: 10.1016/j.fsigen.2007.08.016. [DOI] [PubMed] [Google Scholar]
- Dupuy BM, Stenersen M, Egeland T, Olaisen B. Y-chromosomal microsatellite mutation rates: differences in mutation rate between and within loci. Hum Mutat. 2004;23:117–124. doi: 10.1002/humu.10294. [DOI] [PubMed] [Google Scholar]
- Goedbloed M, Vermeulen M, Fang RN, et al. (17 co-authors) Comprehensive mutation analysis of 17 Y-chromosomal short tandem repeat polymorphisms included in the AmpFlSTR® Yfiler® PCR amplification kit. Int J Legal Med. 2009;123:471–482. doi: 10.1007/s00414-009-0342-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusmao L, Sanchez-Diz P, Calafell F, et al. (42 co-authors) Mutation rates at Y chromosome specific microsatellites. Hum Mutat. 2005;26:520–528. doi: 10.1002/humu.20254. [DOI] [PubMed] [Google Scholar]
- Hellenthal G, Auton A, Falush D. Inferring human colonization history using a copying model. PLoS Genet. 2008;4:e1000078. doi: 10.1371/journal.pgen.1000078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsson M, Scholz SW, Scheet P, et al. (24 co-authors) Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451:998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- Jobling MA, Hurles ME, Tyler-Smith C. Human evolutionary genetics. New York and Abingdon. Garland Science; 2004. [Google Scholar]
- Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- Kayser M, Kittler R, Erler A, et al. (12 co-authors) A comprehensive survey of human Y-chromosomal microsatellites. Am J Hum Genet. 2004;74:1183–1197. doi: 10.1086/421531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Kim NY, Kim KS, Kim JJ, Park JT, Chung KW, Han MS, Kim W. Population genetics and mutational events at 6 Y-STRs in Korean population. Forensic Sci Int Genet. 2009;3:e53–e54. doi: 10.1016/j.fsigen.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Lee HY, Park MJ, Chung U, Lee HY, Yang WI, Cho SH, Shin KJ. Haplotypes and mutation analysis of 22 Y-chromosomal STRs in Korean father–son pairs. Int J Legal Med. 2007;121:128–135. doi: 10.1007/s00414-006-0130-x. [DOI] [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. (11 co-authors) Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Lim SK, Xue Y, Parkin EJ, Tyler-Smith C. Variation of 52 new Y-STR loci in the Y Chromosome Consortium worldwide panel of 76 diverse individuals. Int J Legal Med. 2007;121:124–127. doi: 10.1007/s00414-006-0124-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macpherson JM, Ramachandran S, Diamond L, Feldman MW. Demographic estimates from Y chromosome microsatellite polymorphisms: analysis of a worldwide sample. Hum Genomics. 2004;1:345–354. doi: 10.1186/1479-7364-1-5-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padilla-Gutierrez JR, Valle Y, Quintero-Ramos A, Hernandez G, Rodarte K, Ortiz RO, Olivares N, Rivas F. Population data and mutation rate of nine Y-STRs in a mestizo Mexican population from Guadalajara, Jalisco, Mexico. Leg Med (Tokyo) 2008;10:319–320. doi: 10.1016/j.legalmed.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Pakendorf B, Novgorodov IN, Osakovskij VL, Danilova AP, Protod'jakonov AP, Stoneking M. Investigating the effects of prehistoric migrations in Siberia: genetic variation and the origins of Yakuts. Hum Genet. 2006;120:334–353. doi: 10.1007/s00439-006-0213-2. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, et al. (11 co-authors) Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell GT, Yang H, Tyler-Smith C, Xue Y. The population history of the Xibe in northern China: a comparison of autosomal, mtDNA and Y-chromosomal analyses of migration and gene flow. FSI Genetics. 2007;1:115–119. doi: 10.1016/j.fsigen.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Seielstad MT, Perez-Lezaun A, Feldman MW. Population growth of human Y chromosomes: a study of Y chromosome microsatellites. Mol Biol Evol. 1999;16:1791–1798. doi: 10.1093/oxfordjournals.molbev.a026091. [DOI] [PubMed] [Google Scholar]
- Prugnolle F, Manica A, Balloux F. Geography predicts neutral genetic diversity of human populations. Curr Biol. 2005;15:R159–R160. doi: 10.1016/j.cub.2005.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg NA. Standardized subsets of the HGDP–CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann Hum Genet. 2006;70:841–847. doi: 10.1111/j.1469-1809.2006.00285.x. [DOI] [PubMed] [Google Scholar]
- Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW. Genetic structure of human populations. Science. 2002;298:2381–2385. doi: 10.1126/science.1078311. [DOI] [PubMed] [Google Scholar]
- Sanchez JJ, Borsting C, Hallenberg C, Buchard A, Hernandez A, Morling N. Multiplex PCR and minisequencing of SNPs—a model with 35 Y chromosome SNPs. Forensic Sci Int. 2003;137:74–84. doi: 10.1016/s0379-0738(03)00299-8. [DOI] [PubMed] [Google Scholar]
- Shi MS, Tang JP, Bai RF, Yu XJ, Lv JY, Hu B. Haplotypes of 20 Y-chromosomal STRs in a population sample from southeast China (Chaoshan area) Int J Legal Med. 2007;121:455–462. doi: 10.1007/s00414-007-0186-2. [DOI] [PubMed] [Google Scholar]
- Toscanini U, Gusmao L, Berardi G, Amorim A, Carracedo A, Salas A, Raimondi E. Y chromosome microsatellite genetic variation in two Native American populations from Argentina: population stratification and mutation data. Forensic Sci Int Genet. 2008;2:274–280. doi: 10.1016/j.fsigen.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Wollstein A, Gaag Kvd, et al. (11 co-authors) Improving global and regional resolution of male lineage differentiation by simple single-copy Y-chromosomal short tandem repeat polymorphisms. Forensic Sci Int Genet. 2009;3:205–213. doi: 10.1016/j.fsigen.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weale ME, Yepiskoposyan L, Jager RF, Hovhannisyan N, Khudoyan A, Burbage-Hall O, Bradman N, Thomas MG. Armenian Y chromosome haplotypes reveal strong regional structure within a single ethno-national group. Hum Genet. 2001;109:659–674. doi: 10.1007/s00439-001-0627-9. [DOI] [PubMed] [Google Scholar]
- Wilson IJ, Weale ME, Balding DJ. Inferences from DNA data: population histories, evolutionary processes and forensic match probabilities. J R Stat Soc Series A (Stat Soc) 2003;166:155–188. [Google Scholar]
- Xue Y, Daly A, Yngvadottir B, et al. (14 co-authors) Spread of an inactive form of caspase-12 in humans is due to recent positive selection. Am J Hum Genet. 2006;78:659–670. doi: 10.1086/503116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zerjal T, Bao W, et al. (11 co-authors) Male demography in East Asia: a north-south contrast in human population expansion times. Genetics. 2006;172:2431–2439. doi: 10.1534/genetics.105.054270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zerjal T, Bao W, et al. (13 co-authors) Modelling male prehistory in East Asia using BATWING. In: Matsumura S, Forster P, Renfrew C, editors. Simulations, genetics and prehistory: a focus on islands. McDonald Institute: Cambridge; 2008. pp. 81–90. [Google Scholar]
- Zerjal T, Dashnyam B, Pandya A, et al. (18 co-authors) Genetic relationships of Asians and Northern Europeans, revealed by Y-chromosomal DNA analysis. Am J Hum Genet. 1997;60:1174–1183. [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Rosenberg NA, Feldman MW. Features of evolution and expansion of modern humans, inferred from genomewide microsatellite markers. Am J Hum Genet. 2003;72:1171–1186. doi: 10.1086/375120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Cinnioglu C, et al. (17 co-authors) The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.