Abstract
In 2003, a phase III placebo-controlled trial (VAX004) of a candidate HIV-1 vaccine (AIDSVAX B/B) was completed in 5,403 volunteers at high risk for HIV-1 infection from North America and the Netherlands. A total of 368 individuals became infected with HIV-1 during the trial. The envelope glycoprotein gene (gp120) from the HIV-1 subtype B viruses infecting 349 patients was sequenced from clinical samples taken as close as possible to the time of diagnosis, rendering a final data set of 1,047 sequences (1,032 from North America and 15 from the Netherlands). Here, we used these data in combination with other sequences available in public databases to assess HIV-1 variation as a function of vaccination treatment, geographic region, race, risk behavior, and viral load. Viral samples did not show any phylogenetic structure for any of these factors, but individuals with different viral loads showed significant differences (P = 0.009) in genetic diversity. The estimated time of emergence of HIV-1 subtype B was 1966–1970. Despite the fact that the number of AIDS cases has decreased in North America since the early 90s, HIV-1 genetic diversity seems to have remained almost constant over time. This study represents one of the largest molecular epidemiologic surveys of viruses responsible for new HIV-1 infections in North America and could help the selection of epidemiologically representative vaccine antigens to include in the next generation of candidate HIV-1 vaccines.
Keywords: America, demographics, gp120, HIV-1, vaccine trial
Introduction
Over 1.2 million people are currently living with AIDS in North America (Centers for Disease Control and Prevention 2007; UNAIDS 2008). Among these, 42% are African Americans, 40% non-Hispanic Whites, 17% Hispanic, and 1% Asian and other races. The main prevention strategy in America is to introduce widespread testing to identify HIV-positive people. This strategy has been successful in some areas, such as the prevention of mother-to-child transmission. In other areas, prevention efforts have been less effective, and although, combination antiretroviral treatment has helped to dramatically reduce the number of people developing and dying of AIDS, around 40,000 new AIDS cases are diagnosed every year. Indeed, in the last few years, there appears to be an increase again in the rate of HIV-1 infection (Centers for Disease Control and Prevention 2008).
Despite these statistics, there have been few comprehensive surveys of the viruses responsible for new (incident) infections in North America or to monitor their population dynamics (Flynn et al. 2005; Keele et al. 2008). Population genetic studies will help us understand the evolutionary history, origin, epidemiology, and population dynamics of pathogens and, ultimately, develop improved public health control strategies. Indeed, the emerging field of molecular epidemiology allows researchers to define the basic units of transmissible diseases and provides keen insights into the past history and future directions of infectious diseases (Tibayrenc 2005). A comprehensive survey of genetic diversity of HIV-1 across North America has never before been accomplished, yet such data are useful for the selection of representative antigens to include candidate vaccines and to understand the population dynamics of HIV-1 in this area.
In 2003, a phase III placebo-controlled trial (VAX004) of a candidate HIV-1 vaccine (AIDSVAX B/B) was completed in individuals at high risk for HIV-1 infection (Flynn et al. 2005). The study enrolled 5,403 volunteers from North America and the Netherlands of which 368 became infected with HIV-1 despite intensive risk-reduction counseling. Envelope glycoprotein sequences were generated for 349 HIV-1 subtype B–infected individuals using the plasma sample obtained closest to the time of diagnosis. A sample of three full-length gp120 clones with open reading frames were obtained per patient resulting in a final data set of 1,047 sequences.
Previously, Pérez-Losada et al. (2009) analyzed selective pressure variation across races in these data, finding significant differences. In this paper, we provide a much broader analysis of the VAX004 North American sequences in combination with other sequences available in public databases to document HIV-1 envelope glycoprotein sequence variation as a function of treatment status (vaccine or placebo), geography, race, risk group, and viral load. Here we studied potential differences in genetic diversity due to mutation and recombination and extend our previous analyses on selection across races to the four other factors. Finally, we tried to infer the demographic history of HIV-1 in North America and date the origin of the virus. The data analyzed in this paper represent the largest molecular epidemiologic survey of viruses responsible for new HIV-1 infections in North America and provide a unique opportunity to study HIV-1 evolution in an epidemiological context.
Materials and Methods
VAX004 Vaccine Trial Participants
Five thousand four hundred and three volunteers were enrolled in the VAX004 vaccine trial and randomly assigned to vaccine or placebo groups according to a 2:1 ratio. Epidemiologic data (e.g., self-reported race, risk behavior, and geographic location) were collected upon enrollment and at various times during the course of the study. All subjects were injected with AIDSVAX B/B, a bivalent vaccine prepared by combining purified recombinant gp120s from two different strains of virus (MN and GNE8) and alum (aluminum hydroxide) adjuvant. All subjects were immunized according to a 0, 1, 6, 12, 18, 24, and 30-month schedule. Serum samples were collected immediately prior to each injection and 2 weeks after each injection, with a final blood sample taken at 6 months following the final injection. The specimen taken prior to each injection was used to calculate preboost anti-gp120 titer values and submitted for HIV-1 testing (enzyme-linked immunosorbent assay). The tests selected for HIV-1 testing were unaffected by antibodies to the AIDSVAX B/B antigens. If evidence of HIV-1 infection was obtained, confirmatory testing was carried out by immunoblot. Once HIV-1 infections were confirmed, HIV-1+ subjects were enrolled in a separate protocol (Step B) where plasma and cells were collected at regular intervals for up to 2 years postinfection. Plasma samples were used for viral load testing and envelope glycoprotein sequencing. Frozen lymphocytes were cryopreserved for immunologic and genetic testing. The date of infection was defined as the midpoint between the last seronegative specimen and the first seropositive specimen.
The volunteers participating in the VAX004 trial were recruited from 58 clinical sites distributed throughout the United States, Canada, Puerto Rico, and one site in the Netherlands. The North American participants were distributed in five groups: Northeast, South, Southwest, Midwest, and West (table 1). All regions included individuals from the United States; the Northeast, Midwest, and West regions included also individuals from Montreal, Toronto, and Vancouver (Canada), respectively, and the South region from Puerto Rico.
Table 1.
Mean intrapatient genetic diversity (θ), population recombination rate (ρ), and selection (ω) estimates.
| HIV-1 subtype B | θ | ρ | ω |
| Treatment | |||
| Placebo (122) | 0.0046 | 5.53 | 1.06 |
| Vaccine (228) | 0.0042 | 4.55 | 0.90 |
| Racea | |||
| Asian (5) | 0.0030 | 5.2 | 0.72 |
| Black (12) | 0.0052 | 2.15 | 1.45 |
| Hispanic (22) | 0.0046 | 3.61 | 0.73 |
| Other (15) | 0.0041 | 2.57 | 1.20 |
| White (291) | 0.0043 | 5.22 | 0.95 |
| North American region | |||
| Midwest (44) | 0.0039 | 6.26 | 1.06 |
| Northeast (82) | 0.0038 | 4.90 | 0.85 |
| South (63) | 0.0049 | 3.91 | 0.89 |
| Southwest (66) | 0.0046 | 4.97 | 1.11 |
| West Coast (90) | 0.0044 | 4.85 | 0.93 |
| Risk | |||
| Low (43) | 0.0045 | 8.54 | 0.99 |
| Medium (267) | 0.0044 | 4.23 | 0.98 |
| High (58) | 0.0038 | 5.60 | 0.90 |
| Viral load categories (virions/ml) | |||
| 1: <0.1 × 104 (16) | 0.0034 | 2.81 | 0.86 |
| 2: 0.1 × 104 to 0.5 × 104 (51) | 0.0031 | 6.82 | 0.81 |
| 3: 0.5 × 104 to 1 × 104 (28) | 0.0038 | 5.40 | 0.94 |
| 4: 1 × 104 to 5 × 104 (75) | 0.0046 | 4.31 | 0.82 |
| 5: 5 × 104 to 10 × 104 (34) | 0.0040 | 4.71 | 0.90 |
| 6: 10 × 104 to 50 × 104 (69) | 0.0054 | 4.05 | 1.06 |
| 7: 50 × 104–100 × 104 (15) | 0.0043 | 4.27 | 1.43 |
| 8: >100 × 104 (12) | 0.0048 | 4.28 | 0.85 |
NOTE.—Individuals analyzed are indicated between parentheses.
Selection estimates were taken from Pérez-Losada et al. (2009).
The vaccine trial protocol specified the following racial categories: white, black, non-white Hispanic, Asian, and “other” (table 1). All of the ethnic data were self-reported, and no effort was made to distinguish linguistic and geographic groups within major racial groups. The major risk factor for infection in this trial was male homosexual transmission. However, a small subgroup of subjects with other risk factors (exchanged drugs for sex) were also included. Although all of the trial participants were considered to be at high risk for transmission, the protocol defined relative risk as low, medium, or high based primarily on the frequency of predefined high-risk behaviors. Specifically, risk score was defined as the total number of risk factors reported from the following: 1) unprotected receptive anal sex with an HIV-1–infected male partner; 2) unprotected insertive anal sex with an HIV-1–infected male partner; 3) unprotected receptive anal sex with an HIV-1–uninfected male partner; 4) five acts of unprotected receptive anal sex with a male partner of unknown HIV-1 status; 5) 10 sex partners; 6) anal herpes; 7) hepatitis A; 8) use of poppers; and 9) use of amphetamines. Behavioral risk scores ranged from 0 to 7; 0 was categorized as low, 1–3 was categorized as medium, and 4–7 was categorized as high (Flynn et al. 2005).
Viral load measurements were subdivided into eight categories for genetic analyses (table 1) ranging from ≤0.1 × 104 virions/ml (category 1) to ≥100 × 104 (category 8). Because of possible differences in the sequence and antigenic structure of viruses from early infections and late infections, it was of interest to characterize the data set with respect to time after infection. To accomplish this, we compared the estimated time postinfection with viral load measurements determined for the first postdiagnosis blood draw that was collected at the same time as the specimen used for gp120 sequence determination.
Molecular Data Sets
Three hundred and sixty-eight individuals became infected during the course of this study, and HIV-1 subtype B viruses from 349 of them were sequenced for the gp120 gene. Three clones per individual were collected from the same earliest postinfection plasma sample. A listing of the sequence data used for this analysis has recently been released online and can be accessed at http://www.gsid.org. All gp120 sequences were determined using an ABI 3100 sequencer and assembled using Sequencher (www.genecodes.com).
Because this study is focused on North American HIV-1 dynamics, the 15 VAX004 gp120 sequences from Dutch individuals were excluded from the analyses. From now on, we will refer to the VAX004 North American data (1,032 sequences) as the VAX004NA data set. For the population dynamic analyses, the VAX004NA data set was combined with 1,010 full-length gp120 sequences from Los Alamos (LA) (http://hiv-web.lanl.gov/content/index) and GenBank (GB) (http://www.ncbi.nlm.nih.gov/) databases as of May 2007 to generate the final data set of 2,060 sequences, hereafter the VAX004NA-LAGB data set. All these sequences corresponded to 1,218 haplotypes unevenly distributed between 1981 and 2006. Some years, such as 1985, 2004, and 2005, were overrepresented, with at least 240 haplotypes each, but others, such as 1982, 1992, 1999, and 2001, were represented by less than five haplotypes each. On average, most years were represented by 10–20 haplotypes. Due to excessive computational burden, a coalescent analysis of population dynamics inference on the whole data set was not attempted; therefore, we selected up to 20 haplotypes per year that were randomly chosen three times. This rendered three data sets of 292 haplotypes that were used to study the past population dynamics of HIV-1 in North America (see below).
Sequence Alignment
VAX004NA and VAX004NA-LAGB nucleotide sequences were translated into amino acids and aligned with MAFFT v5.7 (Katoh et al. 2005) using the global algorithm (G-INS-i) and amino acid–specific frequencies. Questionable regions in our amino acid alignment were identified and removed using GBlocks v0.91b (Castresana 2000). Conserved amino acid regions were backtranslated to nucleotides generating an alignment of 1,401 sites for VAX004NA and 1,491 sites for VAX004NA-LAGB. Intrapatient alignments were trivial, so the full gp120 sequences (1,341–1,509 bp) were analyzed for each patient.
Phylogenetic Analysis
The best-fit model of DNA was selected with the Akaike Information Criterion (Akaike 1974) as implemented in Modeltest 3.6 (Posada and Crandall 1998). Maximum likelihood phylogenetic trees were inferred using GARLI (Zwickl 2006). Nodal support was assessed using the bootstrap procedure (Felsenstein 1985) with 1,000 replicates. Heuristic searches were performed under the best-fit model. In addition, Bayesian inference was performed in MrBayes 3.0 (Ronquist and Huelsenbeck 2003) using three codon-position partitions. We ran four chains (one cold and three heated) for 107 generation, sampling every 1,000 steps. Each run was repeated four times. Convergence and mixing of the Markov chains was assessed using Tracer v1.3 (Rambaut and Drummond 2003).
Genetic Estimators and Patient Factors
The VAX004 trial gathered individuals from different geographic, biologic, social, and racial groups, but none of them constitute natural populations, so the genetic estimates correspond always to intrapatient data, unless otherwise stated. Genetic diversity (θ) was estimated for each patient using a version of Watterson's (1975) estimator that relaxes the assumption of an infinite-sites model and is implemented in LDhat v2.0 (McVean et al. 2002). The population recombination rate (ρ) was also estimated for each patient using LDhat. Here, each analysis was repeated 10 times, and the ρ mean estimate was used for subsequent analyses. Adaptive selection was assessed using the ratio of nonsynonymous (dN) to synonymous (dS) substitution rates (ω = dN/dS) and estimated in HYPHY (Kosakovsky Pond et al. 2005) using the fixed-effects likelihood (Kosakovsky Pond and Frost 2005) method with tree branch correction. Single ρ and θ estimates were also calculated for the entire VAX004NA data set.
Intrapatient ρ, θ, and ω estimates were pooled into factors (e.g., race, geographic region; table 1). Average estimates were then compared across factors using the Kruskal–Wallis test in R (R Development CoreTeam 2008). Tests based on linear models (e.g., analysis of variance) were not applied because their underlying assumptions were not met by some of the data sets.
Population Dynamics
Past population dynamics of HIV-1 in North America was inferred in BEAST v1.4.2 (Drummond and Rambaut 2007) using the Bayesian skyline plot model (Drummond et al. 2005) and a relaxed clock (lognormal) model of rate of substitution (Drummond et al. 2006). Such a model was clearly superior to a strict clock model as indicated by Bayes factors. Relative genetic diversity through time (Neτ) was estimated directly from serial samples under the best-fit model of nucleotide substitution. The hyperparameter m (number of grouped intervals) was set up to 1/4 of the sequences in each data set. All output generated by BEAST was analyzed in Tracer v1.3 to test for convergence and mixing. We performed six preliminary runs for 107 generations under the relaxed model using the three reduced versions (292 haploypes each) of the VAX004NA-LAGB data set (see Molecular Data Sets). Because no noticeable differences in parameter estimates and Bayesian skyline plots were observed among them, we chose one data set to perform the final runs (2 × 107 generations).
Results
Characterization of the VAX004 HIV-1 Subtype B gp120 Data Set
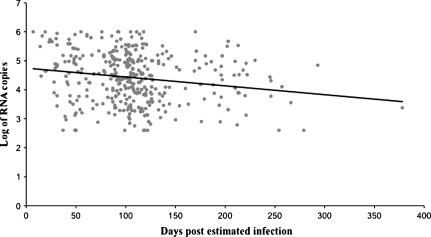
The estimated time of HIV-1 infection to first sampling ranged from 0 to 13 months with a mean time of infection of 109 ± 58 days. Thus, by the criteria of Fiebig et al. (2003), most of the sequences described in this report appear to represent stage V and VI infections. Time-course analysis of viral loads after HIV-1 infection (Clark et al. 1991; Daar et al. 1991) indicate the virus replicates to a very high level (105–106 virions/ml) within a few weeks of HIV-1 infection and then declines to a stable setpoint value. As expected, there was a significant decline in mean viral loads over time (fig. 1). However, we noted a significant compression of the data around 3 months postinfection. This appears to be an artifact of the 6-month interval scheduled between most clinic visits and the convention used to define the estimated day of infection (described in VAX004 Vaccine Trial Participants). Therefore, the estimated date of infection represents an overestimation, and the actual date of infection is earlier than indicated. Because there were approximately 20 infections with estimated dates of infection of 30 days or less, we suspect that these truly represent new and acute infections and should properly fall into Fieberg stage II–IV infections (Fiebig et al. 2003). It is likely that other early-stage infections occur in this cohort, but other assay methodologies will be required to identify them.
FIG. 1.
Characterization of gp120 sequence data set with respect to viral load and estimated time after infection. Line indicates the mean viral load as a function of time. Plasma used for viral load testing was collected as soon as possible following diagnostic tests confirming HIV-1 infection. The date of infection was defined as the midpoint between the last seronegative specimen and the first seropositive specimen.
Phylogenetic Analyses
GTR+Γ+I (Tavaré 1986) was chosen as the best-fit model for both the VAX004NA and VAX004NA-LAGB data sets and for all their corresponding codon-position partitions. The initial phylogenetic analyses focused on investigating the quality of the VAX004NA data set. As expected, the three clone sequences collected per patient clustered together and were not interdispersed with sequences from other individuals. Although, in general, both ML and Bayesian phylogenetic analyses did not reveal population structuring, 17 well-supported small clades (bootstrap proportions ≥70% or posterior probabilities ≥0.95) were observed (fig. 2). These could be transmission linkages where individuals infected each other or were infected by a common partner. Eleven of these clades grouped patients from different geographic areas, but the other six clustered individuals from the same area and gender of which five were also from the same race. Because HIV-1 infection occurs primarily in well-defined risk groups, these types of linkages are likely to be observed in studies of this nature. Unfortunately, because information on partner relationships was restricted, we could not confirm the epidemiological history of these potential transmission chains. The phylogenetic trees did not show any obvious clustering of individuals based on treatment, geographic region, race, risk, or viral load (fig. 2). In fact, in all cases, individuals within each factor seemed to be randomly distributed across the gene trees.
FIG. 2.
Maximum likelihood phylogenetic inference of North American HIV-1 subtype B population structure based on the VAX004NA data set. Branch lengths are shown proportional to the amount of change along the branches. Clades supported by bootstrap proportions ≥70% or posterior probabilities ≥0.95 in the Bayesian tree are shown in bold. Only one clone per patient is represented for simplicity.
Viral Evolution and Patient Factors
The estimates of the population mutation (θ = 0.1049) and recombination (ρ = 100) rates were extremely high for the whole VAX004NA data set. Indeed, the average θ and ρ intrapatient estimates within the VAX004NA categories were much lower (table 1). Purifying selection (ω < 1) was operating in most data sets, but evidence of adaptive evolution (ω > 1) was also found in a few cases (table 1). The nonparametric Kruskal–Wallis tests only indicated significant differences in θ for different viral load groups (P = 0.009). Here, gp120 sequences from individuals belonging to the lower viral load categories (categories 1–3) showed on average lower θ values (0.0034) than individuals belonging to higher categories (categories 6–8; θ = 0.0048).
Population Dynamics and Dating
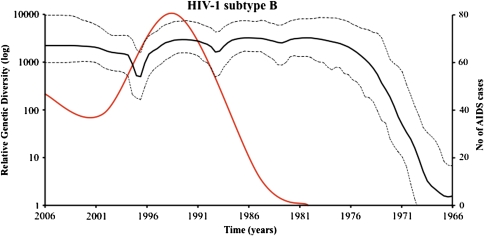
Substitution rates per site for gp120 ranged between 0.0041 and 0.0047 (mean = 0.0044), and the most recent common ancestor was dated in 1968 (1966–1970). The skyline demographic plot (fig. 3) suggested a rapid increase (exponential growth) in Neτ since the late 60s to the late 70s followed by a horizontal phase (constant growth) since the early 80s with three successive low inflections (population decrease) 6–10 years apart. This expansion preceded by at least 10 years the subsequent explosion of detected HIV/AIDS cases in United States.
FIG. 3.
Bayesian skyline plots of the past population dynamics of HIV-1 subtype B in North America. Solid lines show the median estimate and dashed lines the 95% highest posterior density limits. Number of AIDS cases (×1000) since 1980–2006 are indicated in red.
Discussion
Relevance of the VAX004 Data Set
The sequence data from the VAX004 trial (Flynn et al. 2005; GSID HIV-1 Data Browser; www.gsid.org/gsid_hiv_browser.html) represents the largest survey of gp120 envelope sequences of clade B viruses from recent infections yet assembled (Flynn et al. 2005). Although the VAX004 sequences were not from acute infections, they represent a period of time where virus loads are high and where individuals are particularly infectious (Quinn et al. 2000). These data, along with the acute infection data of Keele et al. (2008), provide the basis for the selection of epidemiologically representative vaccine antigens to include in the next generation of HIV-1 vaccines. Because sequence data from new and recent infections have not previously been available, the selection of vaccine antigens depended on consensus sequences from viruses from chronic infections. It is possible that the lack of success of the HIV-1 vaccines tested to date may, in part, be due to the fact that envelope proteins derived from chronic infections may have different antigenic structures than envelope proteins from transmission viruses. Several studies have reported differences in neutralization sensitivity (Derdeyn et al. 2004) and structure (Chohan et al. 2005; Jobes et al. 2006) between viruses from new infections compared with viruses from chronic infections. Such differences may reflect the fact that viruses from new infections are selected primarily for infectivity in the absence of an effective immune response, whereas viruses from chronic infections have undergone years of selection to evade the immune response. Indeed, many studies have shown that the immune response to HIV-1 needs to mature for a year or more before antibodies capable of broad cross-neutralization are detected (Richman et al. 2003; Wei et al. 2003; Gray et al. 2007). The availability of sequence data from new and recent infections will enable investigations to determine whether fundamental differences exist in antigenic structure between viruses from recent and chronic infections. The sequences will also enable the assembly of panels of viruses representative of new and acute infections that can be used to evaluate the immunogenicity and potency of candidate HIV-1 vaccines.
Phylogenetic Structure of HIV-1 in North America
Our phylogenetic trees indicated that intrapatient variation was restricted compared with interpatient variation and gave no signal of cross-contamination. According to the gp120 phylogeny, the North American HIV-1 subtype B population is not structured by any of the epidemiological and clinical factors studied. This agrees with previous results reported by Keele et al. (2008), who also showed that viral env genes evolving from individual transmitted or founder HIV-1 subtype B viruses generally exhibited a star-like phylogeny. This lack of phylogenetic structuring based on treatment is also consistent with the overall outcome of the VAX004 trial where immunization with AIDSVAX B/B did not significantly affect the rate of infection, the viral load, the CD4 count, or the clinical outcome of vaccine recipients compared with placebo recipients (Flynn et al. 2005). Given the age of the HIV-1 epidemic in North America (AIDS was recognized in United States in 1981) and the fact that the virus is thought to mutate at a rate of 1% per year (Shankarappa et al. 1999; Korber et al. 2000), the possibility existed that different clades would have emerged in different parts of North America. But again, our trees indicate that, contrary to what was observed in other continents such as Africa (Tessier et al. 1993; Papathanasopoulos et al. 2003; Peeters et al. 2003; Ferrante et al. 2005) and Asia (Weniger et al. 1994; Oelrichs and Crowe 2003), the North American population was homogeneous across the entire area of study. Although unexpected, this is not totally surprising considering that individuals in developed countries travel much more than individuals in undeveloped ones. Homogeneity across North American HIV-1 populations was also detected by Gilbert et al. (2007) in their study of the emergence of HIV-1 in the Americas. Phylogenetic differences based on risk factors have been observed in Thailand where the prevalent viruses among people infected through heterosexual transmission are predominately Clade A/E, whereas in intravenous drug users, both Clade A/E and Clade B viruses are in cocirculation (Ou et al. 1993; Weniger et al. 1994). This does not seem to be the case in North America, where the predominant strain among homosexuals and those who exchanged drugs for sex is HIV-1 subtype B (Centers for Disease Control and Prevention 2007; UNAIDS 2008).
HIV-1 Evolution and Patient Factors
No significant differences in recombination, mutation, and selection rates were observed among vaccinated and placebo individuals, risk behavior, or samples from different geographic regions; but lower viral load was associated with lower mutation rates. Because genetic diversity is positively correlated with Ne, one could expect that greater viral loads (census size) would also cause an increase on the number of effective virions. Significant differences in selection pressure (ω) among races were detected by Pérez-Losada et al. (2009) using these same data, suggesting that immune response induced by HIV-1 is stronger in black than in white volunteers. If additional data confirm that HIV-1 evolution is impacted by ethnic background or viral load, investigating the genetic determinants of these differences should become a public health priority. Future HIV-1 vaccine studies and trials need to make an effort to include a broader representation of volunteers so these factors can be all considered.
Demographics and Dating of HIV-1 in North America
Our coalescent estimate of the time of emergence of HIV-1 subtype B using North American samples was 1968 (1966–1970). This estimate corresponds to that of the “pandemic” clade in Gilbert et al. (2007), which encompasses the vast majority of non-Haitian subtype B infections in the United States and elsewhere around the world and that was dated as 1969 (1966–1972). Both studies then suggest that HIV-1 was circulating cryptically in North America for 11–15 years before the first cases of HIV/AIDS were recognized in United States in 1981. As indicated in Gilbert et al. (2007), this dating is supported by serological evidence from studies performed in 1978 in New York (Stevens et al. 1986) and San Francisco (Jaffe et al. 1985), which suggest that the virus had been spreading in the men who have sex with men (MSM) population for several years before this point. Even more, our analysis of subtype B past dynamics also suggests that viral populations have already greatly expanded before the number of detected AIDS cases exploded in the 80s when the virus entered the highest risk MSM population, and that, despite the fact that the number of AIDS cases has decreased by about 50% since the early 90s (http://www.cdc.gov/hiv/), HIV-1 relative genetic diversity has remained almost constant and invariably high (fig. 3). This is particularly important because under circumstances of high HIV-1 diversity and low surveillance, new (recombinant or mutant) or existing infective strains could expand exponentially. Coincidentally with this result, in the last few years, an increase in the rate of HIV-1 infection has been observed (Centers for Disease Control and Prevention 2008). These two observations then suggest that North America needs to strengthen prevention efforts, especially among high-risk groups such as MSM and injecting drug users, but also among the general population to avoid AIDS resurgence.
Acknowledgments
We thank Antonio Carvajal and Miguel Arenas for their help with the LDhat and fixed-effects likelihood analyses, respectively. This study was supported by a Bill & Melinda Gates Foundation grant to Global Solutions for Infectious Diseases and by the Spanish Ministry of Science and Education (grant number BIO2007-61411 to DP). We also thank the reviewers for their excellent suggestions.
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. HIV/AIDS surveillance report. 2007. Atlanta (GA): Centers for Disease Control and Prevention (CDC) [Google Scholar]
- Centers for Disease Control and Prevention. Centers for Disease Control and Prevention (CDC) 2008. [cited 2009 Dec 4] Available from: http://www.cdc.gov. [Google Scholar]
- Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005;79:6528–6531. doi: 10.1128/JVI.79.10.6528-6531.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark SJ, Saag MS, Decker WD, Campbell-Hill S, Roberson JL, Veldkamp PJ, Kappes JC, Hahn BH, Shaw GM. High titers of cytopathic virus in plasma of patients with symptomatic primary HIV-1 infection. New Engl J Med. 1991;324:954–960. doi: 10.1056/NEJM199104043241404. [DOI] [PubMed] [Google Scholar]
- Daar ES, Moudgil T, Meyer RD, Ho DD. Transient high levels of viremia in patients with primary human immunodeficiency virus type 1 infection. New Engl J Med. 1991;324:961–964. doi: 10.1056/NEJM199104043241405. [DOI] [PubMed] [Google Scholar]
- Derdeyn CA, Decker JM, Bibollet-Ruche F, et al. (14 co-authors) Envelope-constrained neutralization-sensitive HIV-1 after heterosexual transmission. Science. 2004;303:2019–2022. doi: 10.1126/science.1093137. [DOI] [PubMed] [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- Ferrante P, Delbue S, Mancuso R. The manifestation of AIDS in Africa: an epidemiological overview. J Neurovirol. 2005;11(Suppl 1):50–57. [PubMed] [Google Scholar]
- Fiebig EW, Wright DJ, Rawal BD, et al. (11 co-authors) Dynamics of HIV viremia and antibody seroconversion in plasma donors: implications for diagnosis and staging of primary HIV infection. AIDS. 2003;17:1871–1879. doi: 10.1097/00002030-200309050-00005. [DOI] [PubMed] [Google Scholar]
- Flynn NM, Forthal DN, Harro CD, Judson FN, Mayer KH, Para MF. Placebo-controlled phase 3 trial of a recombinant glycoprotein 120 vaccine to prevent HIV-1 infection. J Infect Dis. 2005;191:654–665. doi: 10.1086/428404. [DOI] [PubMed] [Google Scholar]
- Gilbert MT, Rambaut A, Wlasiuk G, Spira TJ, Pitchenik AE, Worobey M. The emergence of HIV/AIDS in the Americas and beyond. Proc Natl Acad Sci USA. 2007;104:18566–18570. doi: 10.1073/pnas.0705329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ES, Moore PL, Choge IA, et al. (13 co-authors) Neutralizing antibody responses in acute human immunodeficiency virus type 1 subtype C infection. J Virol. 2007;81:6187–6196. doi: 10.1128/JVI.00239-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe HW, Darrow WW, Echenberg DF, et al. (11 co-authors) The acquired immunodeficiency syndrome in a cohort of homosexual men. A six-year follow-up study. Ann Intern Med. 1985;103:210–214. doi: 10.7326/0003-4819-103-2-210. [DOI] [PubMed] [Google Scholar]
- Jobes DV, Daoust M, Nguyen V, Padua A, Michele S, Lock MD, Chen A, Sinangil F, Berman PW. High incidence of unusual cysteine variants in gp120 envelope proteins from early HIV type 1 infections from a phase 3 vaccine efficacy trial. AIDS Res Hum Retrovir. 2006;22:1014–1021. doi: 10.1089/aid.2006.22.1014. [DOI] [PubMed] [Google Scholar]
- Katoh K, Kuma K, Toh H, Miyata T. MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Res. 2005;33:511–518. doi: 10.1093/nar/gki198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, et al. (37 co-authors) Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci USA. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber B, Muldoon M, Theiler J, Gao F, Gupta R, Lapedes A, Hahn BH, Wolinsky S, Bhattacharya T. Timing the ancestor of the HIV-1 pandemic strains. Science. 2000;288:1789–1796. doi: 10.1126/science.288.5472.1789. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- McVean G, Awadalla P, Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics. 2002;160:1231–1241. doi: 10.1093/genetics/160.3.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oelrichs RB, Crowe SM. The molecular epidemiology of HIV-1 in South and East Asia. Curr HIV Res. 2003;1:239–248. doi: 10.2174/1570162033485302. [DOI] [PubMed] [Google Scholar]
- Ou CY, Takebe Y, Weniger BG, Luo CC, Kalish ML, Auwanit W, Yamazaki S, Gayle HD, Young NL, Schochetman G. Independent introduction of two major HIV-1 genotypes into distinct high-risk populations in Thailand. Lancet. 1993;341:1171–1174. doi: 10.1016/0140-6736(93)91001-3. [DOI] [PubMed] [Google Scholar]
- Papathanasopoulos MA, Hunt GM, Tiemessen CT. Evolution and diversity of HIV-1 in Africa—a review. Virus Genes. 2003;26:151–163. doi: 10.1023/a:1023435429841. [DOI] [PubMed] [Google Scholar]
- Peeters M, Toure-Kane C, Nkengasong JN. Genetic diversity of HIV in Africa: impact on diagnosis, treatment, vaccine development and trials. AIDS. 2003;17:2547–2560. doi: 10.1097/01.aids.0000096895.73209.89. [DOI] [PubMed] [Google Scholar]
- Pérez-Losada M, Posada D, Arenas M, Jobes DV, Sinangil F, Berman PW, Crandall KA. Ethnic differences in the adaptation rate of HIV gp120 from a vaccine trial. Retrovirology. 2009;6:67. doi: 10.1186/1742-4690-6-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Quinn TC, Wawer MJ, Sewankambo N, Serwadda D, Li C, Wabwire-Mangen F, Meehan MO, Lutalo T, Gray RH. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. New Engl. J Med. 2000;342:921–929. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Drummond AJ. Tracer: MCMC trace analysis tool. Oxford: University of Oxford; 2003. Available from: http://evolve.zoo.ox.ac.uk. [Google Scholar]
- R Development Core Team. R Foundation for Statistical Computing. Vienna: Austria; 2008. R: a language and environment for statistical computing. Available from: http://www.R-project.org. [Google Scholar]
- Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proc Natl Acad Sci USA. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Shankarappa R, Margolick JB, Gange SJ, et al. (12 co-authors) Consistent viral evolutionary changes associated with the progression of human immunodeficiency virus type 1 infection. J Virol. 1999;73:10489–10502. doi: 10.1128/jvi.73.12.10489-10502.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens CE, Taylor PE, Zang EA, Morrison JM, Harley EJ, Rodriguez de Cordoba S, Bacino C, Ting RC, Bodner AJ, Sarngadharan MG. Human T-cell lymphotropic virus type III infection in a cohort of homosexual men in New York City. JAMA. 1986;255:2167–2172. [PubMed] [Google Scholar]
- Tavaré S. Some probabilistic and statistical problems in the analysis of DNA sequences. In: Miura RM, editor. Some mathematical questions in biology—DNA sequence analysis. Vol. 17. Providence (RI): American Mathematical Society; 1986. pp. 57–86. [Google Scholar]
- Tessier SF, Remy G, Louis JP, Trebucq A. The frontline of HIV1 diffusion in the Central African region: a geographical and epidemiological perspective. Int J Epidemiol. 1993;22:127–134. doi: 10.1093/ije/22.1.127. [DOI] [PubMed] [Google Scholar]
- Tibayrenc M. Bridging the gap between molecular epidemiologists and evolutionists. Trends Microbiol. 2005;13:575–580. doi: 10.1016/j.tim.2005.09.004. [DOI] [PubMed] [Google Scholar]
- UNAIDS. Report on the global HIV/AIDS epidemic. Joint United Nations programme on HIV/AIDS (UNAIDS) 2008. Geneva (Switzerland): WHO Library Cataloguing-in-Publication Data; 2008. p. 362. [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- Wei X, Decker JM, Wang S, et al. (15 co-authors) Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- Weniger BG, Takebe Y, Ou CY, Yamazaki S. The molecular epidemiology of HIV in Asia. AIDS. 1994;8(Suppl 2):S13–S28. [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Austin (TX): The University of Texas at Austin; 2006. Available from: http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html. [Google Scholar]