Abstract
Background
Nontyphoidal Salmonella is the main cause of human salmonellosis. In order to study the prevalent serogroups and serovars of clinical isolates in Taiwan, 8931 Salmonellae isolates were collected from 19 medical centers and district hospitals throughout the country from 2004 to 2007. The pulsed-field eletrophoresis types (PFGE) and antibiotic resistance profiles of Salmonella enterica serovars Bareilly (S. Bareilly) and Braenderup (S. Braenderup) were compared, and multi-drug resistance (MDR) plasmids were characterized.
Results
Over 95% of human salmonellosis in Taiwan was caused by five Salmonella serogroups: B, C1, C2-C3, D1, and E1. S. Typhymurium, S. Enteritidis, S. Stanley and S. Newport were the four most prevalent serovars, accounting for about 64% of isolates. While only one or two major serovars from four of the most prevalent serogroups were represented, four predominant serovars were found in serogroup C1 Salmonellae. The prevalence was decreasing for S. Choleraeuis and S. Braenderup, and S. Virchow and increasing for S. Bareilly. S. Braenderup mainly caused gastroenteritis in children; in contrast, S. Bareiley infected children and elderly people. Both serovars differed by XbaI-PFGE patterns. Almost all S. Bareilly isolates were susceptible to antibiotics of interest, while all lacked plasmids and belonged to one clone. Two distinct major clones in S. Braenderup were cluster A, mainly including MDR isolates with large MDR plasmid from North Taiwan, and cluster B, mainly containing susceptible isolates without R plasmid from South Taiwan. In cluster A, there were two types of conjugative R plasmids with sizes ranging from 75 to 130 kb. Type 1 plasmids consisted of replicons F1A/F1B, blaTEM, IS26, and a class 1 integron with the genes dfrA12-orfF-aadA2-qacEΔ1-sulI. Type 2 plasmids belonged to incompatibility group IncI, contained tnpA-blaCMY-2-blc-sugE genetic structures and lacked both IS26 and class 1 integrons. Although type 2 plasmids showed higher conjugation capability, type 1 plasmids were the predominant plasmid.
Conclusions
Serogroups B, C1, C2-C3, D1, and E1 of Salmonella caused over 95% of human salmonellosis. Two prevalent serovars within serogroup C1, S. Bareilly and cluster B of S. Braenderup, were clonal and drug-susceptible. However, cluster A of S. Braenderup was MDR and probably derived from susceptible isolates by acquiring one of two distinct conjugative R plasmids.
Background
Non-typhoidal Salmonellae are major zoonotic pathogens that commonly cause salmonellosis outbreaks. Globally, salmonellosis caused by non-typhoidal salmonellae generally results in about 1.3 billion cases of acute gastroenteritis and 3 million deaths annually [1]. In the United States, Salmonellae cause an estimated 1.4 million cases of salmonellosis and over 500 deaths annually [2]. Multi-drug resistant (MDR) Salmonella, the global spread of which is mediated by international food trade and travel, is a global public health issue [3,4]. Often, clonal spread of MDR strains has been observed in particular serovars [4-6]. In most instances, resistance genes often associated with integrons and/or transposons are clustered within antimicrobial resistance islands that can be horizontally transferred by conjugative or mobilization plasmids [7].
In serogroup C1, S. Bareilly and S. Braenderup are closely related according to molecular analysis [8,9]. Both serovars have been highly susceptible to antimicrobials since 1971 [10,11] and are frequently isolated from feces of people with food-borne salmonellosis all over the world [12-16]. However, prevalence of both serovars differs between hosts and regions. In Denmark, S. Bareilly was isolated from diverse sources, including humans, animals and animal feed, while S. Braenderup was only found in humans [17]. In a study of a broiler-raising plant in the USA, S. Bareilly was often found in broilers and finished feed; however, S. Braenderup was only observed in hatcheries [18]. In addition, S. Braenderup was commonly isolated from cattle and turtles in Sweden [19], pigs [12] and chicken egg shells [20] in USA. These findings imply that animal reservoirs may be important sources of both serovars in human disease.
In this study, prevalent serogroups and serovars were determined for 8,931 Salmonella isolates collected from 2004 and 2007 in Taiwan. Because of the genetic similarity between S. Bareilly and S. Braenderup [8,9], the two serovars were compared with respect to antimicrobial resistance, resistance genes, PFGE and plasmid profiles. Both serovars disseminated clonally and varied in antimicrobial resistance patterns.
Results
Prevalent serogroups and serovars
Between 2004 and 2007, over 95% of 8,931 Salmonella isolates belonged to serogroups B, C1, C2-C3, D1 and E1 (Table 1). Prevalence differed between serogroups and across time within serogroups: prevalence decreased in serogroups B (46.9%→42.4%) and C1 (14.2%→9.1%) and increased in serogroups C2-C3 (9%→11.3%) and D1 (23.3%→30.2%) over the study period. Such changes were associated with the prevalence of major serovars in each serogroup and were due to only one or two main predominant serovars in each serogroup, except serogroup C1 with four prevalent serovars (Table 1). The top four serovars were S. Enteritidis (22.9-28.9%) of serogroup D1, S. Typhimurium (20.4-24.7%) and S. Stanley (8.2-11.4%) of serogroup B, and S. Newport of serogroup C2 (5.6 - 7.3%). In contrast to the decrease in prevalence of S. Typhimurium from 2005 to 2007, a gradual increase in prevalence was observed in S. Enteritidis.
Table 1.
Prevalence of Salmonella serogroups and their main serovars isolated from human from 2004 to 2007.
| Serogroup/Serovar | Number of isolates | Prevalence (%)2 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2004 | 2005 | 2006 | 2007 | Total | 2004 | 2005 | 2006 | 2007 | Total | |
| Serogroup B | 1133 | 1045 | 938 | 854 | 3970 | 44.3 | 46.9 | 44.0 | 42.4 | 44.5 |
| S. Typhimurium | 571 | 551 | 441 | 412 | 1975 | 22.3ab | 24.7a | 20.7b | 20.4b | 22.1ab |
| S. Stanley | 287 | 183 | 242 | 168 | 880 | 11.2 | 8.2 | 11.4 | 8.3 | 9.9 |
| Serogroup C1 | 364 | 229 | 234 | 184 | 1101 | 14.2 | 10.3 | 11.0 | 9.1 | 11.3 |
| S. Choleraesuis | 111 | 65 | 30 | 17 | 223 | 4.3 (30.5) |
2.9 (28.4) |
1.41 (12.8) |
0.84 (9.23) |
2.50 (22.6) |
| S. Braenderup | 96 | 46 | 66 | 32 | 240 | 3.8 (26.4) |
2.1 (20.9) |
3.1 (28.2) |
1.6 (17.4) |
2.7 (23.7) |
| S. Bareilly | 54 | 41 | 47 | 54 | 196 | 2.1 (14.8) |
1.8 (17.9) |
2.2 (20.1) |
2.7 (29.4) |
2.2 (19.4) |
| S. Virchow | 43 | 34 | 33 | 19 | 129 | 1.7 (11.8) |
1.5 (14.8) |
1.6 (14.1) |
0.9 (10.3) |
1.4 (12.8) |
| Other serovars1 | 60 | 43 | 58 | 62 | 223 | 2.3 (16.5) |
1.9 (18.8) |
2.7 (24.8) |
3.1 (33.7) |
2.5 (22.1) |
| Serogroup C2-C3 | 231 | 246 | 239 | 228 | 944 | 9.0 | 11.0 | 11.2 | 11.3 | 10.6 |
| S. Newport | 144 | 137 | 135 | 147 | 563 | 5.6 | 6.1 | 6.3 | 7.3 | 6.3 |
| S. Albany | 87 | 109 | 104 | 81 | 381 | 3.4 | 4.9 | 4.9 | 4.0 | 4.3 |
| Serogroup D | 597 | 550 | 583 | 609 | 2339 | 23.3 | 24.7 | 27.4 | 30.2 | 26.2 |
| S. Enteritidis | 586 | 543 | 567 | 582 | 2278 | 22.9c | 24.4bc | 26.6ab | 28.9a | 25.5 |
| Serogroup E1 | 122 | 76 | 64 | 70 | 332 | 4.8 | 3.4 | 3.0 | 3.5 | 3.7 |
| S. Weltevreden | 94 | 61 | 556 | 62 | 273 | 3.7 | 2.7 | 2.6 | 3.1 | 3.1 |
| Sum3 | 2447 | 2147 | 2058 | 1954 | 8736 | 95.6 | 96.3 | 96.6 | 96.5 | 96.3 |
| Total Salmonellae | 2,557 | 2,228 | 2,131 | 2,015 | 8,931 | |||||
1Other serogroup C1 serovars include are mainly S. Infantis, S. Potsdam, S. Mbandaka, and S. Montevideo.
2Numbers in parenthesis indicate the percentage of isolates of a C1 serovar over total serogroup C1 isolates.
3Sum is the total number of serogroup B, C1, C2-C3, D, and E isolates.
abcDifferent letters indicate significant difference between years.
Prevalence of serogroup C1 serovars
S. Braenderup, S. Choleraesuis, S. Bareilly and S. Virchow were the predominant serovars in serogroup C1 and consisted of 66 - 84% of total serogroup C1 isolates from 2004 to 2007 (Table 1). Other serovars, including S. Infantis, S. Potsdam, S. Mbandaka, and S. Montevideo, were occasionally isolated with prevalence less than 1% for each serovar. Over the study period, the prevalence of S. Choleraesuis declined dramatically, and S. Braenderup prevalence declined mildly. In contrast, the prevalence of S. Bareilly and other serovars gradually increased from 2004 to 2007. Since S. Braenderup and S. Bareilly were the two main serogroup C1 serovars in 2006-2007 and differed in prevalence trends, 45 S. Braenderup and 51 S. Bareilly isolates were analyzed for their antimicrobial resistance profiles and genetic characteristics.
Age distribution of patients
Patients infected with S. Braenderup and S. Bareilly were separated into four age groups. Although, both serovars were found primarily to infect children (age ≤ 4 years), S. Bareilly was isolated far more frequently from the elderly (age ≥ 50 years) (8.9% for S. Braenderup vs. 31.4% for S. Bareilly, p < 0.05) (Table 2). However, S. Braenderup was predominantly isolated from children (68.9% for S. Braenderup vs. 49% for S. Bareilly, p < 0.05).
Table 2.
Age prevalence of patient infected by S. Bareilly and S. Braenderup
| Rate (%) of each age group | ||||
|---|---|---|---|---|
| Serovar | 0 ~ 4 | 5 ~ 12 | 13 ~ 50 | > 50 |
| S. Bareilly | 49.0b (25/51) |
9.8 (5/51) |
9.8 (5/51) |
31.4b (16/51) |
| S. Braenderup | 68.9a (31/45) |
8.9 (4/45) |
13.3 (6/45) |
8.9a (4/45) |
abDifferent letters indicate significant difference between S. Bareilly and S. Braenderup (Pp<0.05).
PFGE phylogenetic analysis
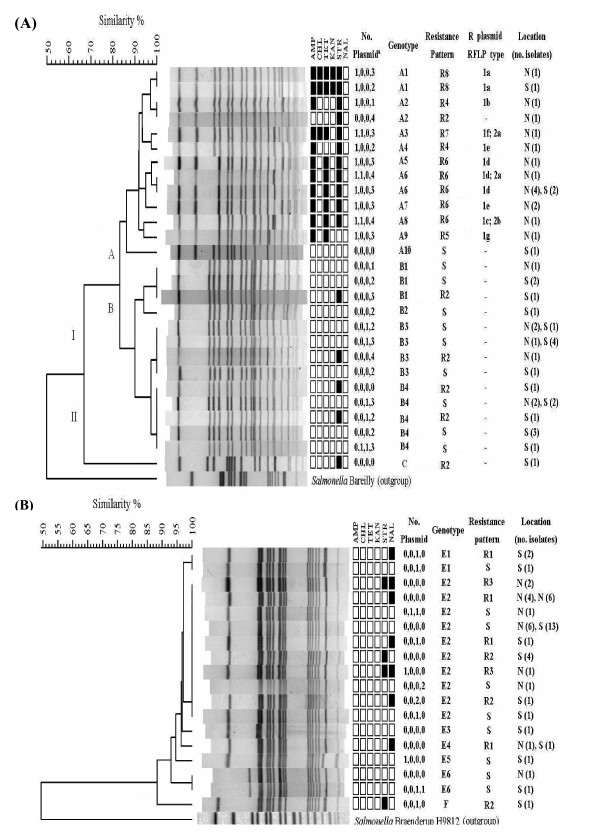
The clustering analysis of XbaI-digested PFGE patterns demonstrated genetic differences between S. Braenderup and S. Bareilly and within each serovar (Figure 1). In S. Braenderup, all isolates were separated into 2 clusters (I and II) at S = 0.68. Most isolates belonged to cluster I, which was further separated into two subgroups (A and B) at S = 0.84 (Figure 1A). In cluster A, 19 isolates were separated into 9 PFGE patterns, and 78.9% (15/19) of the isolates were from northern Taiwan (Figure 1A). In cluster B, 25 isolates were grouped into 4 PFGE patterns, and 72% (18/25) of the isolates were from southern Taiwan (Figure 1A). S. Bareilly isolates were highly genetically homogenous and shared more than 90% pattern similarity (Figure 1B).
Figure 1.
Dendrograms were constructed by PFGE-XbaI patterns to determine the genotypes for S. Braenderup (A) and S. Bareilly (B) with corresponding information including the number and size of plasmids, PFGE subtypes, antimicrobial resistance patterns and collection location of each isolate. The dendrograms were generated by the unweighted pair group method with arithmetic mean (UPGMA) using the Dice-predicted similarity value of two patterns. The BioNumerics version 4.5 statistics program was used with settings of 1.0% optimization and 0.7% tolerance. Symbols of black square and white square represent resistant and susceptible respectively. Plasmids were separated into four groups by size. Ex, 1, 1, 1, 3 indicates that this strain harbored 6 plasmids, one is >90 kb, one is from >50 to <90 kb, one is from >6.6 to <50 kb, and three are <6.6 kb.
Antimicrobial resistance profiles
Among six traditional antibiotics tested, S. Braenderup and S. Bareilly isolates were almost all susceptible to chloramphenicol (CHL; 6.7% for S. Braenderup vs 0% for S. Bareilly) and kanamycin (KAN; 4.4% vs 0%) and differed significantly in resistance to ampicillin (AMP, 37.7% for S. Braenderup vs 0% for S. Bareilly), nalidixic acid (NAL; 0% vs 15.7%), streptomycin (STR, 37.7% vs 15.7%), and tetracycline (TET; 33.3% vs 0%) (Figure 1). Additionally, nine resistance patterns were determined, ranging from susceptibility to all antimicrobials to resistance to four antimicrobials. In S. Braenderup, 7 resistance patterns (S, R2, R4 to R8) were found, and significant differences were observed between cluster A (patterns R2, R4-R8) and B (patterns S and R2) for AMP (77.3% vs 0%), STR (63.6% vs 13%) and TET (54.5% vs 13%). In addition, most isolates in cluster A were MDR (73.7%) while most isolates in cluster B were susceptible (84%). In cluster A, pattern R6 (AMP, TET, and STR) was the predominant and was found in four genotypes (A3, A5, A6, and A7). In S. Bareilly, most isolates were either susceptible (S pattern; 52.9%) or resistant to one (pattern R1 and R2; 31.4% and 9.8%, respectively) or two (pattern R3; 5.9%) antimicrobials. NAL resistant isolates were found in S. Bareilly (patterns R2 and R3) but not in S. Braenderup. Since there were susceptible to levofloxacin (LEV) and moxifloxacin (MOX), NAL resistance may result from a mutation in the gyrA gene, which encodes a subunit of the enzyme DNA gyrase.
Characterization of MDR plasmids
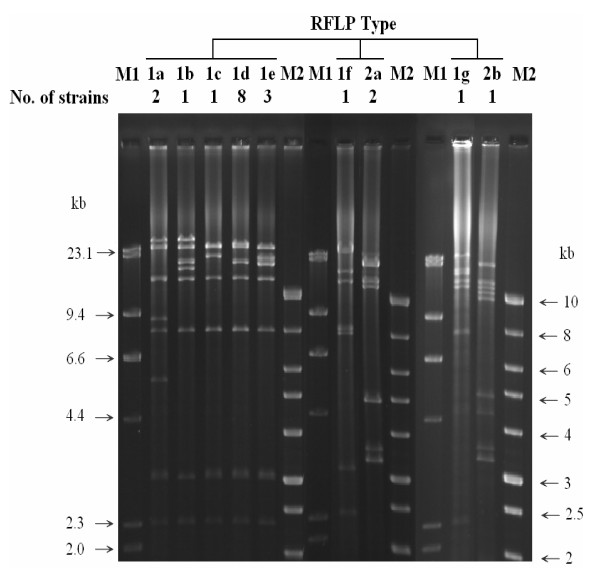
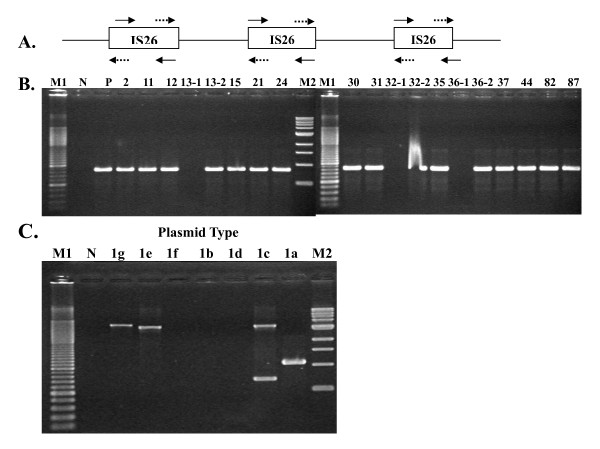
The prevalence of plasmid profile determined by plasmid number and size differed between these two serovars. Most S. Braenderup isolates [93.3%, (42/45)] carried plasmids, while few S. Bareilly isolates [23.5 % (12/51)] did (Figure 1). Plasmids larger than ca.75 kb were only found in resistance isolates of cluster A with the R4 to R8 patterns. Cluster B S. Braenderup isolates and S. Bareilly isolates carried smaller plasmids with the size smaller than 6.6 kb or lacked plasmids. Larger plasmids were further identified as R plasmids by analysis of the antimicrobial resistance profiles of E. coli pir116 transformants, and assigned to type 1 and 2 based on HindIII-restriction patterns (Table 3, Figure 2). Further conjugation, antibiotic resistance and PCR characterization of incompatibility and oriT types, mobile element IS26, class 1 integron, and AMP resistance genes blaTEM and blaCMY-2 were performed for these two plasmid types. Type 1 plasmids were separated into 7 subtypes (1a ~1g) based on differences in plasmid size ranging from 99.1 kb to 137.4 kb and restriction pattern. All plasmids carried blaTEM, replicons F1A and F1B, IS26, and a class 1 integron (Additional files 1 and 2: Figure S1 and S2) with a gene cluster of dfrA12-orfF-aadA2-qacEΔ1-sulI, conferring resistance to trimethoprim-sulfamethoxazole (Sxt) and disappearing in plasmid 1 g (Table 3), which apparently coincides with that in the plasmid of S. Typhimurium (Accession number AB365868). The size of R plasmid was associated with antimicrobial resistance and conjugation capability (Table 3). Only type 1a plasmids, with a size of 137.4 kb and conferring resistance to AMP, CHL, KAN, Sxt and TET, and 1b plasmids, with a size of 122.6 kb and encoding resistance to AMP and Sxt, were capable of conjugation, with efficiencies ranging 4.22 ~ 8.25 × 10-6. The other smaller plasmids, with sizes ranging from 99.1 kb to 104.8 kb and encoding resistance to AMP and Sxt for 1c-1e and 1g, and to AMP, CHL, Sxt and TET for 1f, were not capable of conjugation. Due to differences in plasmid size and since IS26 could be involved in plasmid transposition and recombination, we performed PCR amplification with the IS26 in primers and IS26out primers for all type 1 plasmids (Figure 3). In contrast to a 1.1-kb PCR product in the largest 1a plasmid, 1b, 1d, and 1e plasmids lacked any PCR products; 1e and 1g plasmids presented 3.1 kb PCR products; and 1c plasmid yielded two PCR products with sizes of 3.1 kb and 0.7 kb. These results suggest that the number of IS26 and/or distance between two IS26 elements differed among these type 1 plasmids. In contrast to type 1 plasmids, type 2 plasmids were much smaller in size (77.5 kb and 85 kb) and had higher conjugation efficiencies, ranging from 8.41 × 10-2 to 1.28 × 10-1 (Table 3). In addition, type 2 plasmids were the IncI1 plasmid and contained oriT as well as tnpA-blaCMY-2-blc-sugE (Table 3, Additional files 3: Figure S3).
Table 3.
Characteristics of MDR plasmids from 17 S. Braenderup isolates.
| Antimicrobial resistance gene | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Strains | Plasmid RFLP profile |
Antibiogram1 | aadA2 | blaTEM | blaCMY-2 | Plasmid size (kb) |
Conjugation rate | Inc3 | Class I integron | IS26 | Month of isolation | Number of isolates |
| S. Braenderup 2 | 1a | ACKTSSxt | + | + | - | 137.4 | 4.22 × 10-6 | F1A/1B | + | ND | 2004.8 | 2 |
| E. coli/p2 | ACKSxtT | + | ||||||||||
| S. Braenderup 96 | 1a | ACKSSxtT | + | + | - | 137.4 | 6.04 × 10-6 | F1A/1B | + | ND | 2004.8 | |
| E. coli/p96 | ACKSxtT | + | ||||||||||
| S. Braenderup 24 | 1b | ASSxt | + | + | - | 122.6 | 8.25 × 10-6 | F1A/1B | + | ND | 2004.8 | 1 |
| E. coli/p24 | ASxt | + | ||||||||||
| S. Braenderup 874 | 1d | ASSxtT | + | + | - | 102.5 | -- | F1A/1B | + | ND | 2004.7 | 7 |
| E. coli/p30 | ASxt | + | ||||||||||
| S. Braenderup 12 | 1e | ASSxtT | + | + | - | 99.1 | - | F1A/1B | + | ND | 2005.4 | 3 |
| E. coli/p12 | ASxt | + | ||||||||||
| S. Braenderup 11 | 1g | ASxtT | - | + | - | 104.4 | - | F1A/1B | - | ND | 2005.1 | 1 |
| E. coli/p11 | ASxt | + | ||||||||||
| S. Braenderup 13 | ACSSxtT | + | + | + | + | ND | 2004.7 | |||||
| E. coli/p13-1 | A | - | - | + | 75.5 | 8.41 × 10-2 | IncI1 | - | - | 1 | ||
| E. coli/p13-2 | 1f | ACSxtT | + | + | - | 127.8 | - | F1A/1B | + | + | 1 | |
| S. Braenderup 32 | ASSxtT | + | + | + | + | ND | 2005.9 | |||||
| E. coli/p32-1 | 2a | A | - | - | + | 75.5 | 8.66 × 10-2 | IncI1 | - | - | 1 | |
| E. coli/p32-2 | 1d | ASxt | + | + | - | 102.5 | ND | F1A/1B | + | + | 1 | |
| S. Braenderup 36 | ASSxtT | + | + | + | + | ND | 2005.5 | |||||
| E. coli/36-1 | 2b | A | - | - | + | 85 | 1.28 × 10-1 | IncI1 | - | - | 1 | |
| E. coli/p36-2 | 1c | ASxt | + | + | - | 104.8 | - | F1A/1B | + | + | 1 | |
1Abbreviation: A, ampicillin; C, chloramphenicol; K, kanamycin; S, streptomycin; Sxt, trimethoprim-sulfamethoxazole; T, tetracycline.
2ND, not determined; +, conjugative; -, .non-conjugative.
3Inc, plasmid incompatibility group.
4Other 6 isolates 30 from 2005/2, 31 from 2004/10, 35 from 2005/7, 37 from 2005/3, 44 from 2004/6, and 82 from 2004/7 were not tested for conjugation.
5Other 2 isolates 15 from 2005/5 and 21 from 2004/9 were not tested for conjugation.
Figure 2.
HindIII-digested RFLP profiles of ampicillin resistance plasmids in S. Braenderup isolates. M1: HindIII-digested lambda DNA size marker. M2: 1 kb size marker.
Figure 3.
PCR amplification of IS26 and IS26-associated DNA fragments. (A) Primer design. Symbols of arrow and dashed arrow represent IS26in primers and IS26out primers, respectively. (B) PCR products amplified by IS26in primers. (C) PCR products amplified by IS26out primers. M1: 100-bp size marker. N: negative control. M2: 1-kb size marker.
Discussion
Human salmonellosis was limited to five Salmonella serogroups: B, C1, C2-C3, D1, and E1 (Table 1). Despite the decrease in prevalence of S. Typhimurium and the increase in the prevalence of S. Enteritidis from 2005 to 2007, serogroups B and D Salmonellae were the major pathogens for foodborne salmonellosis in Taiwan due to S. Typhimurium, S. Stanley, and S. Enteritidis of serogroup D1 being the three most prevalent serovars overall. Although the prevalence of serogroups C1 and C2-C3 were similar (around 11%), 4 prevalent serovars and 2 main serovars were found in serogroup C1 and serogroup C2-C3, respectively. In the present study, a shift in prevalence was observed in these four prevalent serogroup C1 serovars: a rapidly decrease in the prevalence of S. Choleresuis, mainly due to enhancement of sanitation and control of swine in Taiwan, and an increase in prevalence of S. Bareilly and other serovars (Table 1). Compared to the 1.6% increase in the prevalence of S. Braenderup from 1978 to 1987 in southern Taiwan [21], the change in the prevalence of isolates in this study ranged from 1.6% to 3.8%, with a trend of decrease from 2004 to 2007, except an increase of S. Braenderup infection in 2006 (Table 1), suggesting possibly occurrence of outbreaks in this year.
Contrary to earlier reports that S. Bareilly and S. Braenderup are closely related genetically [8,9], resistant to 10 Salmonella bacteriophages [22], and infect immuno-compromised patients, differences between S. Braenderup and S. Bareilly were found in the prevalence trend from 2004 to 2007 (Table 1), patients' age group (Table 2), and plasmid profile as well as antimicrobial resistance groups and XbaI-PFGE patterns (Figure 1A). In addition to genetic differences between these two serovars, differences in animal hosts were also observed in both serovars based on the geographic regions from which they were isolated [13,17,18,23]. In this study, we found that S. Bareilley isolates were highly homogeneous genetically and that S. Braenderup isolates were much diverse in our PFGE and plasmid analysis (Figure 1). This may explain why S. Braenderup, but not S. Bareilly, has been frequently reported [19,20,24]. To differentiate S. Braenderup, several molecular methods have been developed, including phage typing [25] and plasmid analysis as performed in this study (Table 1, Figure 1 and 2).
Unlike MDR S. Choleraesuis isolated from pigs and humans [5,6], S. Braenderup and S. Bareilly isolated from pigs were highly susceptible to antibiotics in 1971 [10]. In addition, in a study of resistance to 11 antibiotics for Salmonella isolated from turtles, S. Bareilly was still susceptible to all antibiotics, and, in contrast, few S. Braenderup isolates were resistant to gentamycin (6/15), sulfisoxazole (6/15) and TET (2/15) [11]. In our study, almost all of the cluster A isolates of S. Braenderup were MDR and associated with large MDR plasmids (Table 3, Figure 1). Although RFLP analysis separated type 1 plasmids into 7 subtypes, based on antimicrobial resistance encoded by these plasmids, 3 subtypes were observed, conferring resistance to AMP and Sxt (1b-1e and 1g), AMP, CHL, Sxt, and TET (1f) and AMP, CHL, KAN, Sxt and TET (1a), respectively (Table 3). Apparently, the dfrA12-orfF-aadA2-qacEΔ1-sulI region of class 1 integrons, which is frequently found in MDR Salmonella [26-28], was located on MDR plasmid and conferred resistance to Sxt (Table 3). Insertion sequence IS26 existed in all (Table 3) and differed from plasmids in S. Braenderburg found in Spain [29]. The size change in type 1 plasmids may be due to presence of multiple IS26 elements that may be involved in plasmid rearrangement (Figure 3).
Although conjugation capability of type 2 plasmids was higher than that of type 1 plasmids, we only identified coexistence of type 1 and 2 plasmids in three S. Braenderup isolates, which differed in isolation day and PFGE pattern (Table 3). Isolate 13 with type 1f and 2a plasmids was collected in July of 2004 from Taipei. Isolate 32 with type 1d and 2a plasmids and isolate 36 with 1c and 2b plasmids were collected in March and May of 2005, respectively, from Taichung (Table 3). Only one isolate 44 with a type 1d plasmid was collected before those three isolates, in June of 2004 from Taichung. These results suggest possibly that isolate 32 with A6 genotype and R6 resistance pattern may be derived from isolate 44 with a type 1 plasmid, A4 genotype and R6 resistance pattern by introduction of a type 2 plasmid. Interestingly, type 2 plasmids are IncI1 plasmids, carrying the tnpA-blaCMY-2-blc-sugE structure (Table 3). AmpC β-lactamases are broadly distributed among the Enteribacteriaceae, and plasmid-mediated AmpC β-lactamases include ACC, ACT, CFE, CMY, DHA, FOX, LAT, MIR, and MOX [30]. At least three transposase associated genetic structures for blaCMY include ISEcp1-blaCMY-2-blc-sugE, ISCR1-blaCMY-9-yqgF-yqgE and IS26-frdC-frdD-ampR-blaCMY-13-blc-sugE-IS26 [30]. Recently, blaCMY has been shown to be broadly spread in Salmonella worldwide [29,31,32] and to be present in S. Braenderup [33]. In Taiwan, since we reported the tnpA-blaCMY-2-blc-sugE structure in S. Choleraesuis in 2004 [34], this transposon-like element has been found in other Salmonella serovars and Enterobacteriaceae [32]. In the present study, we first reported that S. Braenderup harbors tnpA-blaCMY-2-blc-sugE on a type 2 plasmid. Comparing this plasmid with the 138-kb plasmid pSC138 (accession no. NC_006856) of S. Choleraesuis, both are IncI1 plasmids with the tnpA-blaCMY-2-blc-sugE structure. However, type 2 plasmids were conjugative and much smaller in size due to lack of a 60-kb DNA region with multiple integrons and transposons, which carry MDR genes [35-37].
Conclusions
Over 95% cases of human salmonellosis surveyed in this study were caused by 5 Salmonella serogroups: B, C1, C2, D1, and E1. As two prevalent serogroup C1 serovars, S. Braenderup and S. Bareiley differed in patients' age groups and XbaI-PFGE patterns. Both serovars were clonally disseminated and drug-susceptible. However, in S. Braenderup, cluster A MDR isolates were derived from susceptible isolates by sequential introduction of two distinct R plasmids. Type 1 plasmids carry blaTEM, F1A/F1B replicons, insertion sequence IS26, and a class 1 integron with a gene cluster comprised of dfrA12-orfF-aadA2-qacEΔ1-sulI. In contrast, type 2 plasmids consist of IncI1 replicon and tnpA-blaCMY-2-blc-sugE. Although type 2 plasmids showed higher conjugation capability, type 1 plasmids were the predominant plasmids responsible for MDR dissemination in S. Braenderup.
Methods
Bacterial isolates
Salmonella isolates were collected from 19 medical centers and district hospitals located throughout Taiwan from 2004 to 2007. Serotypes of the isolates were determined in the Salmonella Reference Laboratory of Centers for Disease Control (CDC), Department of Health, Taiwan, with antisera purchased from S&A Reagents Lab (Bangkok, Thailand), Denka Seiken (Tokyo, Japan), Statens Serum Institut (Copenhagen, Denmark), and a local biotech company, LTK Biolaboratories (Taoyuan, Taiwan). Phase induction was performed using a paper-bridged method developed by the Taiwan CDC [38]. In total, 51 S. Bareilly isolates and 45 S. Braenderup isolates collected in 2004 and 2005 were selected for further characterization. Isolates were separated into two groups based on their geographic origin: the north Taiwan group, consisting of isolates collected from north of Taichung county (including Taichung county), and the south Taiwan group, consisting of isolates collected from south of Taichung county.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing was performed using the disc diffusion method in accordance with the guidelines of the CLSI standards [39] with 7 antibiotics: ampicillin (AMP, 50 μg), chloramphenicol (CHL, 20 μg), kanamycin (KAN, 30 μg), streptomycin (STR, 10 μg), tetracycline (TET, 12 μg), trimethoprim-sulfamethoxazole (Sxt, 23.75/1.25 μg), and quinolone antibiotics including nalidixic acid (NAL, 30 μg), levofloxacin (LEV, 5 μg) and moxifloxacin (MOX, 5 μg). The antimicrobials were purchased from BD (Becton Dickinson and Company, Sparks, Maryland, USA). Escherichia coli ATCC 25922 was used as the reference strain. An MDR isolate was defined as having resistance to three or more antibiotics belonging to different antibiotic classes.
Pulsed-field gel electrophoresis (PFGE)
The PulseNet Standardized Laboratory PFGE Protocol for Molecular Subtyping of Echerichia coli O157:H7, non-typhoidal Salmonella serotypes, and Shigella sonnei [40] was used for analysis of the Salmonella isolates: 10 U of XbaI were used for the restriction digestion. PFGE images were analyzed by using the fingerprint analysis software BioNumerics version 4.5 (Applied Maths). A unique PFGE pattern was defined as one or two DNA bands differing between PFGE patterns of two isolates. A dendrogram was generated by the unweighted pairgroup method with arithmetic mean (UPGMA) algorithm using the Dice-predicted similarity value of two Xbal-digested PFGE patterns.
Plasmid profile analysis
Plasmid profiles of each isolate were determined by the Kado and Liu method [41], and plasmid size was estimated by comparison with the plasmids of two S. Choleraesuis strains: OU7085 (50 kb and 6.6 kb) and OU7526 (50 kb and 90 kb).
Restriction fragment length polymorphism (RFLP) and antibiotic susceptibility analysis of the MDR-plasmid
Large plasmids (> 50 kb) of 17 AMP and STR-resistant S. Braenderup isolates were characterized. Plasmid DNA was purified from resistant wild-type isolates by the alkaline lysis method [42] and then transformed into the competent E. coli strain pir116 (STRR), which was prepared by the CaCl2 method. Transformants were selectively grown on LB agar plates supplemented with AMP (100 μg/ml) and further tested for resistance to CHL, TET, and KAN, but not for resistance to STR, since the recipient strain was inherently resistant to streptomycin. The antibiotic resistance genes blaTEM, aadA, and blaCMY-2, class 1 integron as well as the insertion sequence IS26 and its related DNA fragments were amplified using the primers listed in Table 4. The genes blaSHV and blaCTX-M3 and M14 were also detected by the multiplex method [43]. The R-plasmids of each transformant were purified by use of the Geneaid Plasmid Midi Kit (Geneaid, Taiwan) and were digested with HindIII (New England Biolabs, USA) to determine similarity. Plasmid DNA fragments were separated by electrophoresis through a 0.6 % SeaKem GTG agarose gel (Cambrex Bio Science Rockland, Inc., Rockland, ME, USA) at 25 V for 16 h. The PCR product of class 1 integron was purified by DNA Clean/Extraction kit (GeneMark, Taiwan) and sequenced by Mission Biotech co. (Taiwan).
Table 4.
The PCR primers for PCR and size of PCR products
| Primer | Target | DNA sequence (5' to 3') | Product Sizesize | Note |
|---|---|---|---|---|
| Tem-F | blaTEM | GAAGATCAGTTGGGTGCACGAGT | 550 bp | This study |
| Tem-R | CAACTTTATCCGCCTCCATCCAGT | |||
| STR-F1 | aadA2 | AGACGCTCCGCGCTATAGAAGT | 203 bp | (46) |
| STR-R1 | CGGACCTACCAAGGCAACGCT | |||
| CS-F | CS region | GGCATCCAAGCAGCAAG | Variable | (47) |
| CS-R | AAGCAGACTTGACCTGA | |||
| 1.9CS-F | Flanking region of CS region | CTGCTGCGTAACATCGTTGCT | Variable | This study |
| 1.9CS-R | GGCGAGATCATCAAGTCAGT | |||
| ColE1-F | ColE1 oriT | CAAATGCTGTCCTTCCAGTGT | 225 bp | This study |
| ColE1-R | CTCAGTTCGGTGTAGGTCGT | |||
| F-F | IncFI oriT | CAACAACGCGCCGACACCGT | 288 bp | This study |
| F-R | CCCTTCCTGTCGACGCTTCT | |||
| R100-F | IncF2 oriT | CCACCAAAAGCACCACACACT | 266 bp | This study |
| R100-R | AGACACTCCTAGCAGCGCCT | |||
| pSC138-F | IncI oriT | TGTCACGAACATCTGCCAGT | 193 bp | This study |
| pSC138-R | GAGAGAAAGTGCCCATGGCT | |||
| IS26in-F | IS26 | GGCACTGTTGCAAAGTTAGC | 820 bp | DQ390455.1 |
| IS26in-R | GGCACTGTTGCAAATAGTCG | |||
| IS26out-F | Variable | GCTAACTTTGCAACAGTGCC | Variable | DQ390455.1 |
| IS26out-R | CGACTATTTGCAACAGTGCC | |||
| Tn-F | Tn | ACCTAGATTCTACGTCAGTAC | Variable | (35) |
| AmpC-F | AmpC | CAAGTTTGATTCCTTGGACTCT | AY253913 | |
| AmpC-R | CTCATCGTCAGTTATTGCAGCT | |||
| SugE-R | sugE | GCCTGATATGTCCTGGATCGT |
Plasmid conjugation and incompatibility group
Transferability of R plasmids from each RFLP group was determined by performing the conjugation test following a previously described method [44] with NAL-resistant S. Typhimurium LBNP4417 as the recipient strain. Briefly, 0.6 ml of overnight culture of donor strain was mixed with 1 ml of the overnight recipient strain. Then 1 ml fresh LB broth was added, and the mixture was incubated at 37°C with shaking at 100 rpm for 4 h. The bacterial solution was diluted at 101, 103, and 105 times with LB broth, and then 100 μl of the diluted solution was plated on MacConkey agar supplemented with AMP (100 μg/ml) and/or NAL (15 μg/ml). Conjugation efficiency was calculated by determining the number of transconjugants relative to the total number of recipients. Four primer sets were used to amplify the oriT regions of the ColE1, F (IncFI), R100 (IncFII), and pSC138 (IncI1-like) plasmids (Table 1). In addition, replicon types of these resistant plasmids were determined as described by Carattoli et al. [45].
Statistical analysis
The difference in the antimicrobial resistance rates between two serovars was analyzed by the independent t test. P values of < 0.05 were considered significant.
List of abbreviations
AMP: ampicillin; CDC: Center for Disease Control; CHL: cloramphenicol; KAN: kanamycin; LEV: levofloxacin; MDR: multi-drug resistance; MOX: moxifloxacin; NAL: nalidixic acid; PCR: polymerase chain reaction; PFGE: pulsed-field gel electrophoresis; QRDR: quinolone resistance determining region; RFLP: restriction fragment length polymorphism; STR: streptomycin; Sxt: trimethoprim-sulfamethoxazole; TET: tetramycin; UPGMA: unweighted pairgroup method with arithmetic mean.
Authors' contributions
CC designed, instructed and supervised most aspects of this project. CSC did PFGE analysis and prepared the manuscript. JML and SWC performed the experiments and data analysis. CHC, BCW and JGT assisted in the design of the study and helped to prepare the manuscript. CLC, CHC, and CHL gave useful comments and critically read the manuscript. YFC edited and revised the manuscript. All authors read and approved the final manuscript.
Authors' information
Chien-Shun Chiou is Chief Investigator of The Central Region Laboratory, Center of Research and Diagnostics, Centers for Disease Control, Taichung, Taiwan. Jui-Ming Lin and Shu-Wun Chen are research assistants, Bor-Chun Weng is an assistant professor, Jwu-Guh Tsay is a professor, and Chishih Chu is the chairman of Department of Microbiology and Immunology, National Chiayi University, Chiayi, Taiwan. Cheng-Hsun Chiu is a professor in the Department of Pediatrics, Chang Gung Children's Hospital and Chang Gung University College of Medicine, Taoyuan, Taiwan, Chi-Hong Chu is the superintendent of the National Defense Medical Center, Taipei, Taiwan. Yung-Fu Chang is a professor in the Department of Population Medicine and Diagnostic Sciences, College of Veterinary Medicine, Cornell University, Ithaca, NY 14853, USA. Chyi-Liang Chen is an assistant professor at the Molecular Infectious Diseases Research Center, Chang Gung Memorial Hospital, Taoyuan, Taiwan. Chien-Hsing Liu is the director of the Laboratory Department, Tainan Hospital, Taiwan, ROC.
Supplementary Material
Electrophoretic pattern of 1.9 kb PCR products of CS region amplified from type 1 plasmids. All type 1 plasmids consisted of CS region, except type 1 g and 2 plasmids.
Electrophoretic profile of inverted PCR products of CS-flanking region amplified from type 1 plasmids. Inversed PCR of CS flanking region amplified same PCR products from all type 1 plasmids, except those plasmid that did not show any PCR product of CS region.
PCR amplification of plasmid-mediated tnpA-blaCMY-2-blc-sugE genetic structure of type 2 plasmids. All type 2 plasmids consisted of tnpA-blaCMY-2-blc-sugE genetic structure.
Contributor Information
Chien-Shun Chiou, Email: nipmcsc@cdc.gov.tw.
Jui-Ming Lin, Email: s0933365@mail.ncyu.edu.tw.
Cheng-Hsun Chiu, Email: chchiu@adm.cgmh.org.tw.
Chi-Hong Chu, Email: chu12450@ms18.hinet.net.
Shu-Wun Chen, Email: mabo3032@yahoo.com.tw.
Yung-Fu Chang, Email: yc42@cornell.edu.
Bor-Chun Weng, Email: brian@mail.ncyu.edu.tw.
Jwu-Guh Tsay, Email: jgtsay@mail.ncyu.edu.tw.
Chyi-Liang Chen, Email: dinoschen@mail.cgu.edu.tw.
Chien-Hsing Liu, Email: c32037695@hotmail.com.
Chishih Chu, Email: cschu@mail.ncyu.edu.tw.
Acknowledgements
This work was funded by grants from the Council of Agriculture 97
AS-14.6.1-BQ-B4(9), the National Science Council NSC96-2314-B415-001 (C. C.), and Tainan Hospital, Department of Health 93037 (C. L.) Executive Yuan, Taiwan.
References
- Pang T, Bhutta ZA, Finlay BB, Altwegg M. Typhoid fever and other salmonellosis: a continuing challenge. Trends Microbiol. 1995;3:253–255. doi: 10.1016/S0966-842X(00)88937-4. [DOI] [PubMed] [Google Scholar]
- Mead PS, Slutsker L, Dietz V, McCaig LF, Bresee JS, Shapiro C, Griffin PM, Tauxe RV. Food-related illness and death in the United States. Emerg Infect Dis. 1999;5:607–625. doi: 10.3201/eid0505.990502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aarestrup FM, Hendriksen RS, Lockett J, Gay K, Teates K, McDermott PF, White DG, Hasman H, Sorensen G, Bangtrakulnonth A, Pornreongwong S, Pulsrikarn C, Angulo FJ, Gerner-Smidt P. International spread of multidrug-resistant Salmonella Schwarzengrund in food products. Emerg Infect Dis. 2007;13:726–731. doi: 10.3201/eid1305.061489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butaye P, Michael GB, Schwarz S, Barrett TJ, Brisabois A, White DG. The clonal spread of multidrug-resistant non-typhi Salmonella serotypes. Microbes Infect. 2006;8:1891–1897. doi: 10.1016/j.micinf.2005.12.020. [DOI] [PubMed] [Google Scholar]
- Chang CC, Lin YH, Chang CF, Yeh KS, Chiu CH, Chu C, Chien MS, Hsu YM, Tsai LS, Chiou CS. Epidemiologic relationship between fluoroquinolone-resistant Salmonella enterica Serovar Choleraesuis strains isolated from humans and pigs in Taiwan (1997 to 2002) J Clin Microbiol. 2005;43:2798–2804. doi: 10.1128/JCM.43.6.2798-2804.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, Su LH, Chu C, Chia JH, Wu TL, Lin TY, Lee YS, Ou JT. The emergence in Taiwan of fluoroquinolone resistance in Salmonella enterica serotype Choleraesuis. N Engl J Med. 2002;346:413–419. doi: 10.1056/NEJMoa012261. [DOI] [PubMed] [Google Scholar]
- Miriagou V, Carattoli A, Fanning S. Antimicrobial resistance islands: resistance gene clusters in Salmonella chromosome and plasmids. Microbes Infect. 2006;8:1923–1930. doi: 10.1016/j.micinf.2005.12.027. [DOI] [PubMed] [Google Scholar]
- Rasschaert G, Houf K, Imberechts H, Grijspeerdt K, De Zutter L, Heyndrickx M. Comparison of five repetitive-sequence-based PCR typing methods for molecular discrimination of Salmonellaenterica isolates. J Clin Microbiol. 2005;43:3615–3623. doi: 10.1128/JCM.43.8.3615-3623.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Park SH, Kim HY. Comparison of Salmonella enterica Serovar Typhimurium LT2 and non-LT2 Salmonella genomic sequences, and genotyping of Salmonellae by using PCR. Appl Environ Microbio. 2006;72:6142–6151. doi: 10.1128/AEM.00138-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pocurull DW, Gaines SA, Mercer HD. Survey of infectious multiple drug resistance among Salmonella isolated from animals in the United States. Appl Microbiol. 1971;21:358–362. doi: 10.1128/am.21.2.358-362.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppe C, Kolar JJ, Demczuk WH, Harris JE. Drug resistance and biochemical characteristics of Salmonella from turkeys. Can J Vet Res. 1995;59:241–248. [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) PHLIS Salmonella Surveillance Annual Summary, 2005. US Department of Health and Human Services, CDC. 2007.
- Martin WJ, Ewing WH. Prevalence of Serotypes of Salmonella. Appl Microbiol. 1969;17:111–117. doi: 10.1128/am.17.1.111-117.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong B, Oberg J, Svenungsson B. Outbreak of salmonellosis in a restaurant in Stockholm, Sweden, September - October 2006. Euro Surveill. 2007;12:E13–14. doi: 10.2807/esm.12.11.00749-en. [DOI] [PubMed] [Google Scholar]
- Gupta SK, Nalluswami K, Snider C, Perch M, Balasegaram M, Burmeister D, Lockett J, Sandt C, Hoekstra RM, Montgomery S. Outbreak of Salmonella Braenderup infections associated with Roma tomatoes, northeastern United States, 2004: a useful method for subtyping exposures in field investigations. Epidemiol Infect. 2007;135:1165–1173. doi: 10.1017/S0950268807007911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urfer E, Rossier P, Mean F, Krending MJ, Burnens A, Bille J, Francioli P, Zwahlen A. Outbreak of Salmonella Braenderup gastroenteritis due to contaminated meat pies: clinical and molecular epidemiology. Clin Microbiol Infect. 2000;6:536–542. doi: 10.1046/j.1469-0691.2000.00148.x. [DOI] [PubMed] [Google Scholar]
- Grunnet K, Nielsen B. Salmonella Types Isolated from the Gulf of Aarhus Compared with Types from Infected Human Beings, Animals, and Feed Products in Denmark. Appl Microbiol. 1969;18:985–990. doi: 10.1128/am.18.6.985-990.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann AF, Feeley JC. Culture survey of Salmonella at a broiler-raising plant. Public Health Rep. 1968;83:417–422. [PMC free article] [PubMed] [Google Scholar]
- Boqvist S, Hansson I, Bjerselius UN, Hamilton C, Wahlström H, Noll B, Tysen E, Engvall A. Salmonella Isolated from Animals and Feed Production in Sweden Between 1993 and 1997. Acta Vet Scand. 2003;44:181–197. doi: 10.1186/1751-0147-44-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ching-Lee MR, Katz AR, Sasaki DM, Minette HP. Salmonella egg survey in Hawaii: evidence for routine bacterial surveillance. Am J Public Health. 1991;81:764–766. doi: 10.2105/AJPH.81.6.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CF. Incidence and antimicrobial resistance of Salmonella serotypes in southern Taiwan from 1978 through 1987. Gaoxiong Yi Xue Ke Xue Za Zhi. 1992;8:247–54. [PubMed] [Google Scholar]
- Atterbury RJ, Van Bergen MAP, Ortiz F, Lovell MA, Harris JA, De Boer A, Wagenaar JA, Allen VM, Barrow PA. Bacteriophage Therapy To Reduce Salmonella Colonization of Broiler Chickens. Appl Environ Microbiol. 2007;73:4543–4549. doi: 10.1128/AEM.00049-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langeland G. Salmonella spp. in the working environment of sewage treatment plants in Oslo, Norway. Appl Environ Microbiol. 1982;43:1111–1115. doi: 10.1128/aem.43.5.1111-1115.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savage W. Problems of Salmonella Food-poisoning. Br Med J. 1956;2:317–323. doi: 10.1136/bmj.2.4988.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sechter I, Gerichter CB. Phage Typing Scheme for Salmonella braenderup. Appl Microbiol. 1968;16:1708–1712. doi: 10.1128/am.16.11.1708-1712.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes P, Machado J, Sousa JC, Peixe L. Dissemination amongst humans and food products of animal origin of a Salmonella Typhimurium clone expressing an integron-borne OXA-30 beta-lactamase. J Antimicrob Chemother. 2004;54:429–34. doi: 10.1093/jac/dkh333. [DOI] [PubMed] [Google Scholar]
- Hsu SC, Chiu TH, Pang JC, Hsuan-Yuan CH, Chang GN, Tsen HY. Characterisation of antimicrobial resistance patterns and class 1 integrons among Escherichia coli and Salmonella enterica serovar Choleraesuis strains isolated from humans and swine in Taiwan. Int J Antimicrob Agents. 2006;27:383–391. doi: 10.1016/j.ijantimicag.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Molla B, Miko A, Pries K, Hildebrandt G, Kleer J, Schroeter A, Helmuth R. Class 1 integrons and resistance gene cassettes among multidrug resistant Salmonella serovars isolated from slaughter animals and foods of animal origin in Ethiopia. Acta Trop. 2007;103:142–149. doi: 10.1016/j.actatropica.2007.05.018. [DOI] [PubMed] [Google Scholar]
- Martínez N, Mendoza MC, Rodríguez I, Soto S, Bances M, Rodicio MR, Martínez N. Detailed structure of integrons and transposons carried by large conjugative plasmids responsible for multidrug resistance in diverse genomic types of Salmonella enterica serovar Brandenburg. J Antimicrob Chemothe. 2007;60:1227–1234. doi: 10.1093/jac/dkm336. [DOI] [PubMed] [Google Scholar]
- Jacoby GA. AmpC beta-lactamases. Clin Microbiol Rev. 2009;22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlet G, Barrett TJ, Butaye P, Cloeckaert A, Mulvey MR, White DG. Salmonella resistant to extended-spectrum cephalosporins: prevalence and epidemiology. Microbes Infect. 2006;8:1945–1954. doi: 10.1016/j.micinf.2005.12.029. [DOI] [PubMed] [Google Scholar]
- Su LH, Chen HL, Chia JH, Liu SY, Chu C, Wu TL, Chiu CH. Distribution of a transposon-like element carrying bla (CMY-2) among Salmonella and other Enterobacteriaceae. J Antimicrob Chemother. 2006;57:424–9. doi: 10.1093/jac/dki478. [DOI] [PubMed] [Google Scholar]
- Gray JT, Hungerford LL, Fedorka-Cray PJ, Headrick ML. Extended-Spectrum-Cephalosporin Resistance in Salmonella enterica Isolates of Animal Origin. Antimicrob Agents Chemother. 2004;48:3179–3181. doi: 10.1128/AAC.48.8.3179-3181.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu CH, Su LH, Chu C, Chia JH, Wu TL, Lin TY, Lee YS, Ou JT. Isolation of Salmonella enterica serotype choleraesuis resistant to ceftriaxone and ciprofloxacin. Lancet. 2004;363:1285–126. doi: 10.1016/S0140-6736(04)16003-0. [DOI] [PubMed] [Google Scholar]
- Chiou CS, Jones AL. Nucleotide sequence analysis of a transposon (Tn5393) carrying streptomycin resistance genes in Erwinia amylovora and other gram-negative bacteria. J Bacteriol. 1993;175:732–40. doi: 10.1128/jb.175.3.732-740.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali F, Kehrenberg C, Manfreda G, Schwarz S. Physical linkage of Tn3 and part of Tn1721 in a tetracycline and ampicillin resistance plasmid from Salmonella Typhimurium. J Antimicrob Chemother. 2005;55:562–5. doi: 10.1093/jac/dkh553. [DOI] [PubMed] [Google Scholar]
- Rao S, Maddox CW, Hoien-Dalen P, Lanka S, Weigel RM. Diagnostic accuracy of class 1 integron PCR method in detection of antibiotic resistance in Salmonella isolates from swine production systems. J Clin Microbiol. 2008;46:916–920. doi: 10.1128/JCM.01597-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiou CS, Huang JF, Tsai LH, Hsu KM, Liao CS, Chang HL. A simple and low-cost paper-bridged method for Salmonella phase reversal. Diagn Microbiol Infect Dis. 2006;54:315–317. doi: 10.1016/j.diagmicrobio.2005.10.009. [DOI] [PubMed] [Google Scholar]
- Clinical and Laboratory Standards Institute. M100-S17. Performance standards for antimicrobial susceptibility testing; 16th informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA; 2007. [Google Scholar]
- Ribot EM, Fair MA, Gautom R, Cameron DN, Hunter SB, Swaminathan B, Barrett TJ. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- Kado CI, Liu ST. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birnboim HC, Doly J. rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia JH, Chu C, Su LH, Chiu CH, Kuo AJ, Sun CF, Wu TL. Development of a multiplex PCR and SHV melting-curve mutation detection system for detection of some SHV and CTX-M beta-lactamases of Escherichia coli, Klebsiella pneumoniae, and Enterobacter cloacae in Taiwan. J Clin Microbiol. 2005;43:4486–4491. doi: 10.1128/JCM.43.9.4486-4491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C, Chiu CH, Chu CH, Ou JT. Nucleotide and amino acid sequences of oriT-traM-traJ-traY-traA-traL regions and mobilization of virulence plasmids of Salmonella enterica serovars enteritidis, gallinarum-pullorum, and typhimurium. J Bacteriol. 2002;184:2857–2862. doi: 10.1128/JB.184.11.2857-2862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. 2005;63:219–228. doi: 10.1016/j.mimet.2005.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Electrophoretic pattern of 1.9 kb PCR products of CS region amplified from type 1 plasmids. All type 1 plasmids consisted of CS region, except type 1 g and 2 plasmids.
Electrophoretic profile of inverted PCR products of CS-flanking region amplified from type 1 plasmids. Inversed PCR of CS flanking region amplified same PCR products from all type 1 plasmids, except those plasmid that did not show any PCR product of CS region.
PCR amplification of plasmid-mediated tnpA-blaCMY-2-blc-sugE genetic structure of type 2 plasmids. All type 2 plasmids consisted of tnpA-blaCMY-2-blc-sugE genetic structure.