Abstract
Eukaryotic initiation factor (eIF) eIF1 maintains the fidelity of initiation codon selection by enabling 43S complexes to reject codon–anticodon mismatches, to recognize the initiation codon context, and to discriminate against establishing a codon–anticodon interaction with AUGs located <8 nt from the 5′-end of mRNA. To understand how eIF1 plays its discriminatory role, we determined its position on the 40S ribosomal subunit using directed hydroxyl radical cleavage. The cleavage of 18S rRNA in helices 23b, 24a, and 44 by hydroxyl radicals generated from Fe(II) tethered to seven positions on the surface of eIF1 places eIF1 on the interface surface of the platform of the 40S subunit in the proximity of the ribosomal P-site. The position of eIF1 on the 40S subunit suggests that although eIF1 is unable to inspect the region of initiation codon directly, its position close to the P-site is very favorable for an indirect mechanism of eIF1's action by influencing the conformation of the platform of the 40S subunit and the positions of mRNA and initiator tRNA in initiation complexes. Unexpectedly, the position of eIF1 on the 40S subunit was strikingly similar to the position on the 30S ribosomal subunit of the sequence and structurally unrelated C-terminal domain of prokaryotic initiation factor IF3, which also participates in initiation codon selection in prokaryotes.
Keywords: eIF1, mRNA, ribosome, translation, directed hydroxyl radical cleavage
The ribosomal scanning model (Kozak 1989) describes translation initiation on most eukaryotic mRNAs. First, initiator tRNA (Met-tRNAiMet), eukaryotic initiation factor (eIF) eIF2, and GTP form a ternary complex, which together with the multisubunit eIF3 binds to a 40S ribosomal subunit to form a 43S preinitiation complex (Dever 2002). Formation of 43S complexes is stimulated by eIF1A (Chaudhuri et al. 1997). It is homologous to prokaryotic initiation factor IF1 (Sette et al. 1997; Battiste et al. 2000), which occupies the ribosomal A-site of the 30S subunit (Carter et al. 2001). The initiation factors eIF4F, eIF4A, and eIF4B cooperatively bind to the 5′-proximal region of mRNA and unwind its secondary structure, which allows attachment of 43S complexes to this region, most likely via the eIF4G–eIF3 interaction (Gingras et al. 1999). Indirect evidence suggests that after attachment to the cap-proximal region, 43S complexes scan the 5′-untranslated region (5′-UTR) in a 3′-direction until they encounter an AUG triplet in a favorable nucleotide context GCC(A/G)CCAUGG, in which the presence of A/G in the -3 position and G in the +4 position relative to the A of the initiation codon is the most important (Kozak 1986, 1987a,b, 1989), stop at it, and form 48S initiation complexes with an established codon–anticodon interaction.
The scanning process and AUG codon recognition require another factor, eIF1 (Pestova et al. 1998; Pestova and Kolupaeva 2002). In the absence of eIF1, 40S subunits do not reach the initiation codon of native capped β-globin mRNA in an in vitro reconstituted translation system and form only aberrant ribosomal complexes near the 5′-end of the mRNA (Pestova et al. 1998). eIF1 plays a principal role in initiation codon selection, enabling 43S complexes to discriminate between cognate and near-cognate initiation codons and to sense the nucleotide context of initiation codons (Pestova and Kolupaeva 2002). eIF1 also discriminates against assembly of 48S complexes on AUG triplets located only 1–4 nt from the 5′-end of mRNA (Pestova and Kolupaeva 2002). Genetic analysis in yeast shows that mutations in eIF1 allow initiation at a UUG codon in vivo (Yoon and Donahue 1992). The activities of eIF1 are very similar to some of the activities of the prokaryotic initiation factor IF3: IF3 also discriminates against initiation at non-AUG codons and at the 5′-proximal AUG codons of leaderless mRNAs (Hartz et al. 1990; Tedin et al. 1999).
The mechanism by which eIF1 maintains the fidelity of initiation codon selection is not known. eIF1 could influence initiation codon selection either directly by inspection of the codon–anticodon interaction and the region of mRNA surrounding the initiation codon, or indirectly by inducing conformational changes in 43S complexes that enable them to recognize and reject codon–anticodon mismatches.
To provide the foundation for understanding the mechanism of action of eIF1, we determined the position and orientation of eIF1 on the 40S ribosomal subunit by directed hydroxyl radical cleavage (Culver and Noller 2000) using Fe(II)-BABE-derivatized single-cysteine eIF1 mutants. We consider that the high degree of homology between 18S and 16S rRNA justifies the use of the crystal structure of the 30S ribosomal subunit from Thermus thermophilus (Yusupov et al. 2001) for modeling the eIF1/40S subunit interaction because the crystal structure of a 40S subunit is not available. Sites of cleavage of 18S rRNA and tRNA from Fe(II) tethered to seven positions on the surface of eIF1 provided a sufficient number of constraints to model the eIF1/40S subunit interaction. Here we show that eIF1 binds to the interface surface of the platform of the 40S subunit close to the ribosomal P-site. The position of eIF1 relative to mRNA and to the anticodon loop of initiator Met-tRNAiMet on the 40S subunit favors an indirect mechanism of action of eIF1 in maintaining the fidelity of initiation codon selection.
Results
Binding of eIF1 to the 40S ribosomal subunit
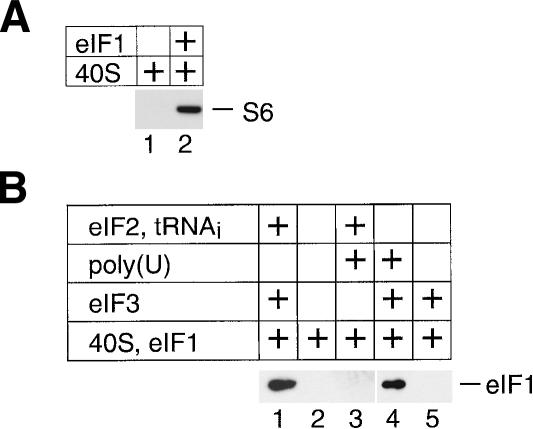
Binding of eIF1 to 40S subunits can be detected by a pull-down assay at high concentrations of eIF1 (Fig. 1A, lane 2), but eIF1/40S subunit binary complexes are not stable enough to withstand sucrose density gradient centrifugation (Fig. 1B, lane 2). eIF1 binds stably to 40S subunits in the presence of eIF2–ternary complex, eIF3, and eIF1A (Pestova et al. 1998), or of eIF2–ternary complex and eIF3 (Fig. 1B, lane 1), but not in the presence of eIF2–ternary complex alone (Fig. 1B, lane 3). eIF1 binds to the p110 subunit of eIF3 (Asano et al. 1998; Phan et al. 1998; Fletcher et al. 1999). We therefore studied the influence of eIF3 alone on binding of eIF1 to 40S subunits. We recently found that in the absence of eIF2–ternary complex, stable binding of eIF3 to 40S subunits requires the presence of a U-rich oligonucleotide cofactor (V.G. Kolupaeva, I.B. Lomakin, C.U.T. Hellen, and T.V. Pestova, in prep.). Here we report that in the presence of poly(U) but not in its absence (Fig. 1B, lanes 4,5), eIF3 also promoted stable binding of eIF1 to the 40S subunit. The ability of eIF3/poly(U)/40S subunit complexes to interact normally with eIF2–ternary complexes (V.G. Kolupaeva, I.B. Lomakin, C.U.T. Hellen, and T.V. Pestova, in prep.) points to the functional nature of the eIF3/40S subunit interaction in these stable complexes and indicates that they are suitable for analysis of the eIF1/40S subunit interaction.
Figure 1.
Binding of eIF1 to the 40S ribosomal subunit. (A) Interaction of 40S subunits with T7-Tag antibody agarose-immobilized eIF1 in an in vitro binding assay. Ribosomal protein S6 was visualized by Western blotting. (B) Presence of eIF1 in ribosomal complexes isolated from sucrose density gradients. Prior to centrifugation, 40S subunits were incubated with eIF1 and different combinations of eIF3, eIF2–ternary complex, and poly(U) RNA as indicated. eIF1 in fractions containing 40S subunits was detected by Western blotting using anti-T7 tag antibodies.
Analysis of the eIF1/40S subunit interaction by enzymatic footprinting
For footprinting analysis of the eIF1/40S subunit interaction, 18S rRNA was digested by RNase VI, which is specific for double-stranded RNA. Binding of eIF1 to 40S subunits in the presence of eIF3 and poly(U) protected nucleotides 1811–1813 in helix 44 from cleavage (Fig. 2, lane 2). Binding of eIF3 alone to the 40S subunit did not protect these nucleotides from cleavage (Fig. 2, lane 3). At high concentrations, eIF1 alone was also able to protect nucleotides 1811–1813 of 18S rRNA from RNase VI cleavage (Fig. 2, lane 4). This confirms that the interaction of eIF1 with the 40S subunit is similar in the presence and in the absence of eIF3.
Figure 2.
Enzymatic footprinting of ribosomal complexes containing eIF1. cDNA products obtained by primer extension show protection of nucleotides 1811–1813 of 18S rRNA from RNase VI cleavage in the presence of eIF1. Reference lanes G, A, C, and T depict 18S rRNA sequence generated using the same primer.
Construction and activity of single-cysteine eIF1 mutants
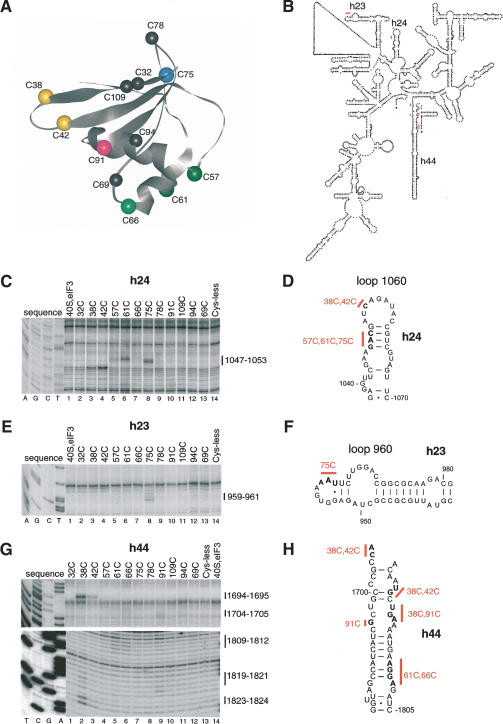
To gain insights into the mechanism by which eIF1 maintains the fidelity of initiation, we investigated the position and orientation of eIF1 on the 40S subunit by directed hydroxyl radical probing (Culver and Noller 2000). In this approach, locally generated hydroxyl radicals are used to cleave 18S rRNA in the vicinity of Fe(II) specifically tethered to a unique cysteine residue on the surface of eIF1 via the linker 1-(p-bromoacetamidobenzyl)-EDTA (BABE). eIF1 consists of a tightly folded domain with two α-helices on one side of a five-stranded β-sheet (residues 29–113; Fig. 4A, below) and 28 unstructured N-terminal amino acids (Fletcher et al. 1999). The unstructured N-terminal regions of human and yeast eIF1 contain conserved sequences (Fletcher et al. 1999). We therefore investigated the influence of this region on the activity of eIF1 in 48S complex formation on β-globin mRNA. In an in vitro reconstituted translation system, 48S complexes do not form on the initiation codon of native capped β-globin mRNA in the absence of eIF1 but instead an aberrant ribosomal complex (complex I) forms near the 5′-end of the mRNA (Fig. 3A, lane 2; Pestova et al. 1998). Like wild-type eIF1, mutant eIF1Δ29 (lacking 29 N-terminal amino acids) dissociated the aberrant complex I and promoted efficient 48S complex formation on β-globin mRNA (Fig. 3A, lanes 3,4). Because the unstructured region of eIF1 was not essential for its activity, we concentrated on mapping the position of the structured domain of eIF1 on the 40S subunit. eIF1 contains cysteine residues at positions 69 and 94 (Fig. 4A). Cysteineless mutant eIF1 containing Cys69Ala and Cys94Ser substitutions bound stably to 40S subunits in the presence eIF3 and poly(U) (data not shown). Like wild-type eIF1, cysteineless mutant eIF1 mock-conjugated with Fe(II)-BABE dissociated complex I and promoted formation of 48S complexes on the initiation codon of β-globin mRNA (Fig. 3B, lanes 3,16). Cysteineless mutant eIF1 also allowed formation of a very small amount of initiation complex on a GUG triplet in the β-globin 5′-UTR (Fig. 3B, lane 16). We had previously observed formation of this complex on β-globin mRNA in the absence of eIF1 in a reaction mixture that contained eIF4G and eIF4A instead of eIF4F (Pestova and Kolupaeva 2002).
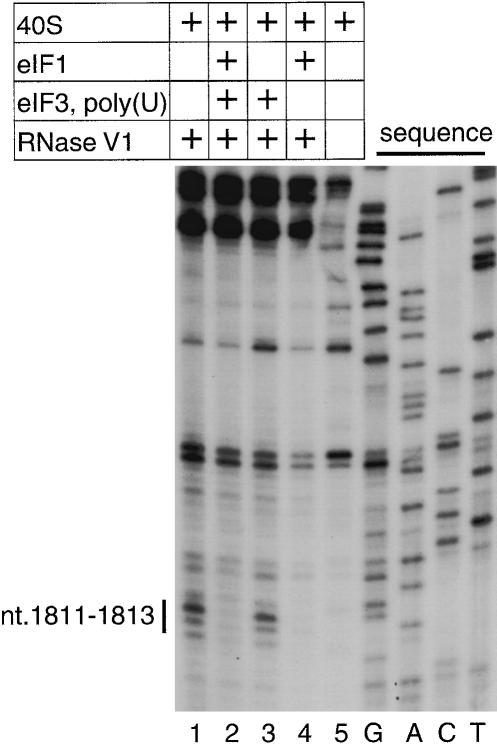
Figure 4.
Directed hydroxyl radical cleavage of 18S rRNA in 40S/eIF3/eIF1 complexes from Fe(II) tethered to different positions on the surface of eIF1. (A) Ribbon diagram of the structured domain of human eIF1. Spheres indicate positions of cysteines introduced on the surface of eIF1 for tethering of Fe(II)-BABE. The positions of cysteines from which hydroxyl radicals were able to cleave 18S rRNA are shown in colors (C38 and C42, yellow; C57 and C61, green; C66, light green; C75, light blue; C91, magenta). (B) Secondary structure of 18S rRNA with sites of directed hydroxyl radical cleavage shown as red bars. (C,E,G) Primer extension analysis of directed hydroxyl radical cleavage of 18S rRNA (in helices 24, 23, and 44, respectively) in 40S/eIF3/eIF1 complexes from Fe(II) tethered to positions on eIF1 as indicated. Reaction mixtures corresponding to the lanes marked “40S, eIF3” did not contain eIF1. Reaction mixtures corresponding to the lanes marked “Cys-less” contained the cysteineless eIF1 mutant. Reference lanes G, A, C, and T depict 18S rRNA sequence generated from the same primer. The positions of cleaved nucleotides are shown as black bars on the right. (D,F,H) Elements of helices 24, 23, and 44 of 18S rRNA with hydroxyl radical cleavage sites shown as red bars.
Figure 3.
Activities of eIF1 mutants in 48S complex formation. Toeprinting analysis of 48S complexes assembled on β-globin mRNA in the presence of 40S subunits, eIF2, eIF3, eIF4A, eIF4B, eIF4F, eIF1A, aminoacylated initiator tRNA, and the N-terminally truncated eIF1Δ29 mutant (A) or Fe(II)-BABE-derivatized cysteineless and single-cysteine eIF1 mutants (B) as indicated. Reaction mixtures containing only β-globin mRNA are shown in lane 1 of each panel. Reaction mixtures corresponding to lane 2 of each panel contained all translation components except eIF1. Reference lanes A and G depict 18S rRNA sequence generated using the same primer. Complex I is arrested 21–24 nt from the 5′-end of β-globin mRNA. The 48S complex assembled on the AUG triplet is designated as “complex II (+15–17 nt from AUG).” The 48S complex assembled on the GUG triplet located 23 nt from the 5′-end of β-globin mRNA is designated as “+16 nt from GUG.”
On the basis of the cysteineless mutant, we constructed 10 other mutants containing single surface-exposed Cys residues at different well-distributed positions on the structured part of eIF1 (Fig. 4A). Two mutants (C69 and C94) each contained one native cysteine: in mutant C69, cysteine 94 was substituted by Ser, and in mutant C94, cysteine 69 was substituted by Ala. All eIF1 mutants bound to 40S subunits in the presence of eIF3 and poly(U) RNA, although with a reduced efficiency of 30%–40% of that of the wild-type protein (data not shown). Taking into account the reduced ribosomal binding activity of eIF1 mutants, a 10:1 molar excess of eIF1 over 40S subunits was used for 48S complex formation to enhance eIF1/40S subunit binding. All eIF1 mutants to various extents dissociated the aberrant complex I and promoted 48S complex formation at the initiation codon of β-globin mRNA (Fig. 3B, lanes 4–15). Fe(II)BABE-derivatized C32, C38, and C42 mutants were least active in 48S complex formation on the AUG codon of β-globin mRNA and had ∼50% of the activity of wild-type eIF1. Conjugation of eIF1 mutants with Fe(II)-BABE could not account for the reduced proofreading activity of some mutants because underivatized proteins also showed reduced activity in this assay (data not shown). Except for mutant C69 (Fig. 3B, lane 15), all other eIF1 mutants, and in particular C32, C38, and C42, also allowed formation of initiation complexes at the GUG triplet in the β-globin 5′-UTR (Fig. 3B, lanes 4–14). These mutants all contained the Cys69Ala substitution that by itself in mutant C94 allowed formation of low levels of 48S complexes on the GUG triplet (Fig. 3B, lane 14). However, the fact that all eIF1 mutants were able to bind to 40S subunits and promote formation of 48S complexes on β-globin mRNA with at least 50% of the activity of wild-type eIF1 made them suitable for mapping the position of eIF1 on the 40S subunit.
Directed hydroxyl radical cleavage of 18S rRNA in eIF1/40S subunit complexes
To ensure stable binding of eIF1 to 40S subunits, which is required for directed hydroxyl radical probing, we investigated complexes of eIF1 with the 40S subunit formed in the presence of eIF3 and poly(U). We used these complexes rather than complexes that contained eIF2to avoid potential shielding of 18S rRNA from hydroxyl radical cleavage by components of the eIF2–ternary complex. Mock Fe(II)-BABE-conjugated cysteineless mutant eIF1 was used as a negative control for hydroxyl radical cleavage. Hydroxyl radicals generated from Fe(II) tethered to seven of the 12positions (shown in color in Fig. 4A) cleaved 18S rRNA in helices 23, 24, and 44 with different intensities (Fig. 4C,E,G).
The most prominent cleavage occurred in the 1060 loop at the top of helix 24 (nucleotides 1047–1053; Fig. 4C,D). Hydroxyl radicals generated from Fe(II) tethered to positions C38, C42, C57, C61, and C75 cleaved this region with different intensities. Hydroxyl radicals originating from Fe(II) tethered to position 61 in α-helix 1 cleaved strongly at nucleotides 1047–1049. Strong cleavage also occurred at nucleotides 1048–1049 when Fe(II) was tethered to position 75 in β-strand 3. Hydroxyl radicals generated from C38 and C42in the loop between β-strands 1 and 2cleaved at nucleotide 1053 with medium and strong intensities, respectively. Weak cleavage was also observed at nucleotide 1048 when Fe(II) was tethered to C57 in α-helix 1.
Hydroxyl radicals generated from Fe(II) tethered to C75 also cleaved 18S rRNA in the 960 loop at the top of helix 23 (nucleotides 959–961) with medium intensity (Fig. 4E,F).
The most intense cleavage in helix 44 was observed when Fe(II) was tethered to C38 and C42(Fig. 4G,H). Strong cleavage at nucleotide 1695 and nucleotides 1823–1824 occurred from Fe(II) tethered to C38 and medium cleavage at the same nucleotides from Fe(II) tethered to C42. Weak cleavage at nucleotides 1809–1812 of helix 44 was induced by Fe(II) tethered to C61 and C66 in α-helix 1 and at nucleotides 1819–1821 by Fe(II) tethered to C38, and C91 in α-helix 2. Hydroxyl radicals generated from Fe(II) tethered to C91 also cleaved weakly at nucleotides 1704–1705. No cleavage of 18S rRNA was observed by hydroxyl radicals generated from positions 32, 69, 78, 94, or 109.
No additional cleavage sites in 18S rRNA were detected in binary eIF1/40S subunit complexes compared with eIF1/eIF3/poly(U)/40S subunit complexes (data not shown). Cleavage sites that were of strong or medium intensity in eIF1/eIF3/poly(U)/40S subunit complexes were also observed in binary eIF1/40S subunit complexes, but their intensity was weak. Cleavage sites that were weak in eIF1/eIF3/poly(U)/40S subunit complexes were not detected in binary complexes.
All cleavage sites occur in regions of 18S rRNA (Fig. 4B) that are highly conserved between it and prokaryotic 16S rRNA. In 16S rRNA, the 790 loop at the top of helix 24 (analog of the 1060 loop in 18S rRNA), the 690 loop on top of helix 23 (analog of the 960 loop in 18S rRNA), nucleotide 1399 (analog of nucleotide 1695 in 18S rRNA), and nucleotides 1497–1498 (analog of nucleotides 1823–1824 in 18S rRNA) in helix 44 surround the ribosomal P-site of the 30S subunit (Yusupov et al. 2001).
Directed hydroxyl radical cleavage of initiator tRNA in 43S preinitiation complexes
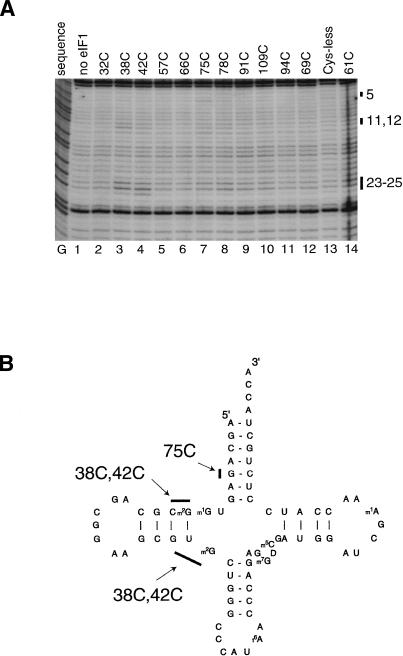
Hydroxyl radical cleavage of regions of 18S rRNA that are analogous to regions that surround the ribosomal P-site in 16S rRNA prompted us to investigate the orientation of eIF1 relative to initiator tRNA in 43S complexes that contained 40S subunits, eIF3, eIF2, and initiator tRNA. We used a primer complementary to the 3′-terminal 16 nt of initiator tRNA to map hydroxyl radical cleavage sites on it by primer extension. Although this method does not permit inspection of ∼20 3′-terminal nucleotides of tRNA and requires synthetic in vitro transcribed unmodified tRNA (because some modified nucleotides in native tRNA strongly arrest reverse transcription), we nevertheless were able to detect cleavage of initiator tRNA induced by hydroxyl radicals generated from Fe(II) tethered to three positions on eIF1 (Fig. 5A,B). Hydroxyl radicals generated from Fe(II) tethered to C38 and C42 cleaved tRNA at nucleotides 23–25 and nucleotides 11–12in the D-stem. Hydroxyl radicals generated from Fe(II) tethered to C75 caused very weak cleavage at nucleotide 5 in the A-stem.
Figure 5.
Directed hydroxyl radical cleavage of initiator tRNA in 43S complexes from Fe(II) tethered to different positions on the surface of eIF1. (A) Primer extension analysis of directed hydroxyl radical cleavage of initiator tRNA in 43S complexes that contained 40S subunits, eIF2, initiator tRNA, eIF3, and eIF1 from Fe(II) tethered to different positions on eIF1 as indicated. The reaction mixture corresponding to the lane marked “Cys-less” contained the cysteineless eIF1 mutant. The reaction mixture corresponding to the lane marked “no eIF1” did not contain eIF1. Reference lane G depicts tRNA sequence generated using the same primer. The positions of cleaved nucleotides are shown as black bars on the right. (B) Secondary structure of initiator tRNA with sites of directed hydroxyl radical cleavage shown as black bars.
Modeling of the interaction between eIF1 and the 40S subunit: eIF1 binds to the interface surface of the platform of the small ribosomal subunit
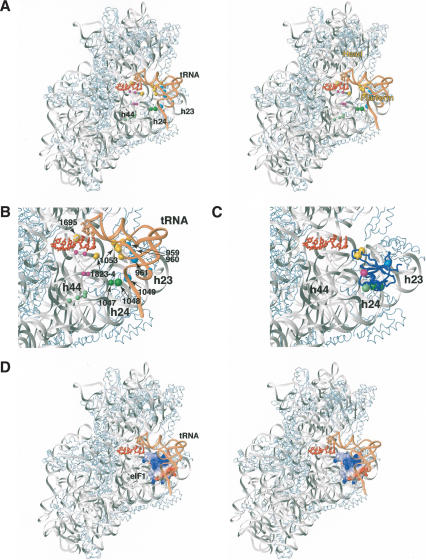
Although the crystal structure of the 40S subunit is not available, we considered that the high degree of homology between 18S and 16S rRNA (particular at the sites of cleavage) justifies using the crystal structure of the T. thermophilus 30S subunit (Yusupov et al. 2001) for modeling the eIF1/40S subunit interaction. The sites in the 16S rRNA and in tRNA bound to the ribosomal P-site of the 30S subunit that correspond to cleavage sites in 18S rRNA and initiator tRNA are shown in Figure 6A. R38 and K42are located on the opposite face of eIF1 to K61 and E75 (Fig. 4A; Fletcher et al. 1999). Consistently, Fe(II)-derivatized C38 and C42mutants yielded similar cleavage patterns, distinct from those obtained with Fe(II)-derivatized C61 and C75 mutants (Fig. 4). Considering the strong cleavage at the top of helix 44 and in the 1060 loop (analog of the 790 loop in 16S rRNA) and also the cleavage in the D-stem of initiator tRNA (Fig. 6A,B), we placed eIF1 directly on the interface surface of the 40S subunit between the platform and initiator tRNA (Fig. 6C). The orientation of eIF1 was optimized to maximally satisfy the strong hydroxyl radical cleavage of 18S rRNA from Fe(II) tethered to positions 38, 42, 61, and 75.
Figure 6.
Stereo view showing the position of eIF1 on the small ribosomal subunit. (A) Positions of directed hydroxyl radical cleavage in 18S rRNA and initiator tRNA (shown as colored spheres) mapped onto corresponding regions of 16S rRNA (gray) and tRNA (light brown) bound to the ribosomal P-site in a ribbon diagram of the crystal structure of the 30S ribosomal subunit from T. thermophilus (Yusupov et al. 2001). Colors of cleavage sites correspond to colors of cysteines on the surface of eIF1 in panel B and Figure 4A (C38 and C42, yellow; C57 and C61, green; C66, light green; C75, light blue; C91, magenta) from which hydroxyl radical cleavage occurs. The radius of the spheres is proportional to the efficiency of the cleavage: weak, medium, and strong. Ribosomal proteins are shown as light-blue ribbons. mRNA in the A- and P-sites is shown in red. (B) Close-up view of the region around the ribosomal P-site from panel A. The strongest cleavage sites in 18S rRNA modeled onto 16S rRNA structure are numbered and indicated by arrows. (C) Position of eIF1 (blue ribbon) on the 30S subunit from T. thermophilus (Yusupov et al. 2001) modeled using data from hydroxyl radical cleavage of 18S rRNA by Fe(II) tethered to different positions on the surface of eIF1. Colored spheres represent positions of cysteines on eIF1 from which hydroxyl radicals were able to cleave 18S rRNA. (D) Stereo view showing a model of the eIF1/30S/[P-site tRNA]/mRNA complex based on hydroxyl radical cleavage data and using the structure of the T. Thermophilus 30S subunit (Yusupov et al. 2001). eIF1 is in surface representation, painted by electrostatic potential (positive charge is blue and negative is red). The coloring of 16S rRNA, mRNA, tRNA, and ribosomal proteins in panels C and D is as in panels A and B.
The resulting model indicates that the loop between strands β1 and β2(containing R38 and K42) is oriented toward the codon–anticodon base pairs in the P-site but does not contact them directly, whereas α-helix 1 (containing K61) would be able to interact with helix 24 (Fig. 6B). This model is also consistent with our footprinting data. Although in our model, eIF1 does not directly contact nucleotides 1811–1813 of 18S rRNA, which are protected by eIF1 from RNase VI cleavage, binding of eIF1 to the 40S subunit would sterically block access of the enzyme to these nucleotides. Whereas most restraints were consistent with each other, it was impossible to reconcile the strong cleavage at nucleotide 1695 (equivalent to nucleotide 1399 in 16S rRNA) from K42with the rest of the restraints. Therefore, the distance restraint to nucleotide 1399 was not used in refining eIF1's position. Cleavage data for Met-tRNAi in 43S complexes was not used for docking, because the orientation of the P-site tRNA may differ between the 70S ribosome (the structure of which was used for modeling) and 43S complexes. Adding the P-site tRNA as a steric constraint did not significantly affect the final orientation of eIF1, indicating that the proposed binding mode of eIF1 was compatible with the orientation of the P-site tRNA in the X-ray structure of the 70S ribosome.
In the final model (Fig. 6B,C), the distances from the Cβ atoms of residues R38, K42, K61, and E75 to their corresponding strong and medium cleavage sites were 17 Å or less, except for the distance from K42of eIF1 to nucleotide 1695 (equivalent to nucleotide 1399 in 16S rRNA), which was ∼27 Å. The long distance (>30 Å) between nucleotide 5 in the acceptor stem of tRNA and E75 of eIF1 (from which cleavage in nucleotide 5 of tRNA occurred) indicates that Met-tRNAi in 43S complexes might be slightly rotated toward the E-site, compared with the P-site tRNA in the 70S ribosome, bringing the tRNA acceptor stem closer to the platform of the small subunit.
Similarity of the interactions of eIF1 and prokaryotic IF3-CTD with the small ribosomal subunit
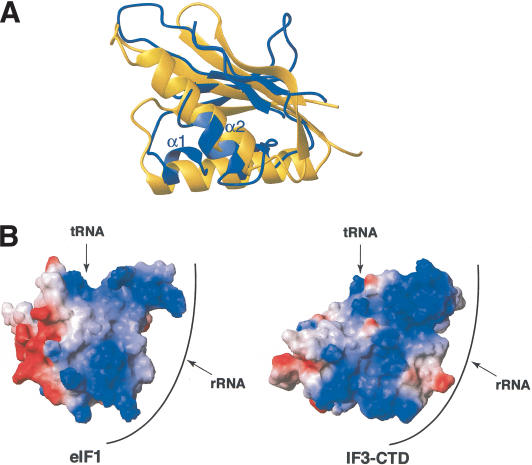
The directed hydroxyl radical cleavage data presented here indicate that eIF1 binds to the same surface of the small ribosomal subunit as the C-terminal domain (CTD) of IF3 (Dallas and Noller 2001). Regarding also the functional similarity between IF3 and eIF1, we therefore compared their interactions with rRNA at the structural level. Although eIF1 (Fletcher et al. 1999) and IF3-CTD (Biou et al. 1995) share a similar fold, consisting of a single β-sheet with two α-helices packed against it, the topology is completely different and no structural homology is found between these proteins using DALI/FSSP (Holm and Sander 1993). It appears, however, that not only do eIF1 and IF3-CTD bind to the same surface on the small subunit, but also that the orientations of the β-sheet and the α-helices described here and in studies performed in H. Noller's laboratory (Dallas and Noller 2001) are similar (Fig. 7A). Furthermore, the overall shapes and charge distributions of the two proteins are similar: the two structures loosely resemble a cone, whose sides are mainly positively charged and in proximity to rRNA and/or P-site tRNA, whereas the base is negatively charged and solvent-exposed (Figs. 6C, 7B).
Figure 7.
Superposition of eIF1 and the C-terminal domain of IF3 (IF3-CTD). (A) Superposition of structural elements of eIF1 (blue ribbon) and the IF3-CTD (yellow ribbon; Biou et al. 1995), based on their positions on the small ribosomal subunit (Dallas and Noller 2001; this study). The orientation of eIF1 is as in Figure 6. (B) Comparison of the surface charge distribution of eIF1 and IF3-CTD based on their positions on the small ribosomal subunit (Dallas and Noller 2001; this study). Positively and negatively charged surfaces are blue and red, respectively. Both eIF1 and IF3-CTD are rotated 180° compared with panel A. The extensive rRNA-binding surface (front) and the tRNA contact surface (top) are indicated. The solvent-exposed surface facing the ribosomal E-site is on the left.
Two Fe(II)-BABE derivatives of IF3-CTD efficiently cleaved nucleotide 1399, even though it was expected to be distant from the IF3 binding site (Dallas and Noller 2001). Therefore, although cleavage at nucleotide 1695 (eukaryotic equivalent of nucleotide 1399) by eIF1 could be explained by significant differences in the P-site structures of 40S and 30S subunits, it may equally indicate that IF3 and eIF1 induce similar conformational changes around the P-site. Cleavage by both IF3 and eIF1 at nucleotide 1399/1695 is of particular interest, because: (1) this nucleotide is located at the junction between the 3′-major and 3′-minor domains; (2) it is in close proximity to mRNA in the P-site; and (3) it is on the opposite side of mRNA with respect to the IF3/eIF1 binding site. Therefore, a conformational change bringing nucleotide 1399/1695 closer to IF3/eIF1 would probably affect the position of mRNA in the P-site and could also be part of long-range domain rearrangements in rRNA. Thus, based on hydroxyl radical cleavage in the 3′-major domain of 16S RNA and cryo-EM data, it was proposed that IF3 induces significant conformational changes in the small ribosomal subunit, including tilting of the head toward the P-site (Dallas and Noller 2001).
Discussion
The position of eIF1 near the P site on the interface surface of the 40S subunit platform is very similar to the position of the IF3-CTD on the 30S subunit, also as determined by directed hydroxyl radical probing (Dallas and Noller 2001). Taking into account the similarities in function of eIF1 and IF3 and in their positions on the ribosome, and also because of the paucity of biochemical data concerning eIF1's mechanism of action, we found it most useful to discuss eIF1's activities in parallel with those of IF3. Like eIF1, IF3 discriminates against initiation at non-AUG codons and at the 5′-proximal AUG codons of leaderless mRNAs (Hartz et al. 1990; Tedin et al. 1999). It is also involved in initiator tRNA selection (Hartz et al. 1989, 1990) and antagonizes ribosomal subunit association (Subramanian and Davis 1970). The IF3-CTD alone can fulfill all of these functions (Petrelli et al. 2001).
A physical basis for eIF1's involvement in antiassociation of ribosomal subunits
IF3 interferes with subunit joining directly by binding to an area on the 30S subunit that forms intersubunit bridges B2b, B2c, and B7a (Dallas and Noller 2001). In eukaryotes, eIF3 mediates ribosome dissociation and eIF1 has not previously been directly implicated in this process (Trachsel and Staehelin 1979). According to our model (Fig. 6B), binding of eIF1 to the 40S subunit would block access of the 60S subunit to elements of helices 23, 24, and 45 of 18S rRNA that participate in forming the B2b and B2d intersubunit bridges (Spahn et al. 2001). Although eIF1 alone does not dissociate ribosomes, we found that it strongly enhances eIF3's ribosomal dissociation/antiassociation activities (V.G. Kolupaeva, I.B. Lomakin, C.U.T. Hellen, and T.V. Pestova, in prep.), consistent with its position on the 40S subunit reported here. It is also worth mentioning that the presence of eIF1 in 43S preinitiation complexes increased their resistance to dissociation by 60S subunits compared with 43S complexes assembled in the presence of only eIF2, eIF3, and eIF1A (Majumdar et al. 2003). The mechanism of action of eIF1 in this process is likely similar to that proposed for IF3. A clearer understanding of the synergy of eIF1 and eIF3 in this process and of the role of the eIF1/eIF3 interaction in mediating this synergy will require accurate determination of eIF3's position on the 40S subunit.
Initiator tRNA selection in prokaryotes and eukaryotes
Three GC base pairs in the anticodon stem of prokaryotic initiator tRNA are important for its discrimination by the translation apparatus (Hartz et al. 1989, 1990). A mechanism for initiator tRNA selection has been proposed in which binding of IF3 to the 30S subunit causes the head to tilt toward the platform (Dallas and Noller 2001). This rearrangement indicated by hydroxyl radical cleavage in the 3′-major domain of 16S rRNA would enable nucleotides 1338–1339 of 16S rRNA in the head to inspect the minor groove of the anticodon stem directly. In contrast to prokaryotes, eukaryotic initiator tRNA is stringently selected (by eIF2) before it binds to the 40S subunit. If the mechanisms of initiator tRNA and initiation codon selection rely on different conformational changes caused by binding of IF3 to the 30S subunit, then eIF1 need not be able to participate in initiator tRNA selection. However, if these mechanisms are simply different manifestations of the same conformational changes in the 30S subunit caused by binding of IF3, then eIF1 may have the potential to play this role even though it does not usually need to perform it. Although eukaryotic initiator tRNA also has three GC pairs in the anticodon stem and mutation of them slightly reduces tRNA activity (Drabkin et al. 1993), we did not detect hydroxyl radical cleavage from eIF1 in the 3′-major domain of 18S rRNA in the 40S subunit's head that would have indicated tilting in a manner like that observed for 30S subunits. Although this observation suggests that eIF1 is not involved in initiator tRNA selection (at least by the mechanism proposed for IF3 by Dallas and Noller [2001]), we cannot exclude the possibility that the locations of cysteines in our eIF1 mutants do not allow cleavage of the 3′-major domain of 18S rRNA.
Initiation codon selection by eIF1
Based on the mechanism of eukaryotic translation initiation, the necessity for eIF1's participation in initiator tRNA selection is not apparent, whereas eIF1's role in initiation codon selection is critical. The scanning mechanism of initiation requires continuous monitoring and protection against premature, partial base-pairing of triplets in the 5′-UTR with the anticodon of initiator tRNA, a requirement that is fulfilled by eIF1 (Pestova and Kolupaeva 2002). The position of eIF1 on the 40S subunit (Fig. 6B,C) suggests that eIF1 is unable to inspect the initiation codon directly and therefore monitors it indirectly by influencing the conformation of the 40S subunit and the positions of bound mRNA and initiator tRNA. The strong cleavage at nucleotide 1695 of 18S rRNA could be a manifestation of this hypothetical conformational change in the 40S subunit. Prokaryotic IF3 does induce conformational changes in the decoding site of the ribosome causing rearrangement of mRNA on the 30S subunit (La Teana et al. 1995; Shapkina et al. 2000; Petrelli et al. 2001). However, we make the conclusion about an indirect mechanism of eIF1's action very cautiously because eIF1 binds very close to the site of the codon–anticodon interaction and because the eIF1/40S subunit complex was modeled using the structure of the 30S subunit.
We propose that in the presence of eIF1, the position of initiator tRNA in 43S complexes is favorable for ribosomal scanning, but not to establish a stable codon–anticodon interaction, so that these complexes would have to undergo conformational change upon base-pairing. The necessity for reorientation of initiator tRNA and possibly also of mRNA on the 40S subunit either for or following establishment of codon–anticodon base-pairing in 48S complexes provides the most probable mechanism for initiation codon selection by eIF1. The presence of eIF1 would antagonize this hypothetical rearrangement and only those complexes with correct codon–anticodon base-pairing would be able to undergo such changes to form 48S complexes. The initiation codon would therefore be selected at this stage. This process could also be influenced by the context of the initiation codon: the important -3 and +4 nucleotides could interact with elements of the 40S subunit to stabilize conformational changes in the initiation complex that occur upon base-pairing. In support of this proposed stabilizing effect of “context” nucleotides, we found that 48S complexes assembled on AUG triplets in “bad” context in the absence of eIF1 were dissociated following its addition even though the codon and anticodon were properly base-paired (Pestova and Kolupaeva 2002). As a corollary of this conformational switch, it is possible that codon–anticodon base-pairing weakens binding of eIF1 to 40S subunits or even leads to displacement of eIF1 from 40S subunits.
In the absence of eIF1, because there would be no challenge to the stability of the 48S complex, complexes with partial base-pairing could form and participate in subsequent steps in the translation pathway. However, complexes with codon–anticodon mismatches assembled in the absence of eIF1 would likely not be able to keep their conformation upon subsequent binding of eIF1, thus, a mispaired tRNA would either be ejected or returned to the position that it occupied in 43S complexes, in which case they can resume scanning.
Relationships between the structure and function of eIF1
The position of eIF1 on 40S subunits suggests that mutations in its ribosome binding surface (which would weaken the eIF1/40S subunit interaction) or mutations on its opposite face (which would impair simultaneous binding of eIF1 and tRNA to 40S subunits) should affect eIF1's activity. eIF1 C38 and C42mutants with substitutions in the loop between β-strands 1 and 2on the predicted 40S subunit binding surface and the C32mutant (with a substitution on the surface that faces initiator tRNA) were least able to dissociate aberrant complex I and to promote 48S complex formation (Fig. 3B). The activities of our eIF1 mutants are therefore consistent with the proposed model for the eIF1/40S subunit interaction.
D88, Q89, and G112suppressor mutations in yeast eIF1 allow initiation at a UUG codon (Yoon and Donahue 1992; Cui et al. 1998). These residues are not located directly on eIF1's 40S subunit-binding surface but slightly to the side that is closer to the ribosomal A-site. These suppressor mutations most likely represent a compromise between the necessity for the essential eIF1 to monitor initiation codon selection adequately for cell viability and the necessary mild dysfunction that occasionally allows 48S complexes to form on the mutated (UUG) initiation codon of the selection marker.
Structure–function relationship between eIF1 and IF3
The prokaryotic initiation factors IF1 and IF2are closely related to the eukaryotic factors eIF1A and eIF5B, respectively (Sette et al. 1997; Lee et al. 1999; Battiste et al. 2000). The functions of the third prokaryotic factor, IF3, and its position on the small ribosomal subunit are very similar to those of eIF1, although IF3 and eIF1 have unrelated primary and tertiary structures. Therefore to perform the evolutionarily conserved function of maintaining the fidelity of initiation codon selection, a factor (eIF1 or IF3) may optimally have to be sandwiched between initiator tRNA and the platform. The requirement for eIF1 and IF3 to bind to conserved ribosomal regions imposes common demands and can account for similarities in their shape, size, and surface charge distributions (Fig. 7). However, given the sequence and structural similarities between IF1/eIF1A and IF2/eIF5B, the question arises why the AUG-fidelity monitoring function in prokaryotes and eukaryotes is not carried out by homologous factors. A satisfying explanation is that the differences between eIF1 and the IF3-CTD may reflect the necessity for different conformational changes in the small subunit to accommodate the fundamental differences between prokaryotic and eukaryotic initiation mechanisms. Most significantly, initiation in eukaryotes involves ribosomal scanning, which has no parallel in prokaryotes, and as a result the conformation of the platform and the mRNA-binding channel of the 40S subunit, which are most likely to be influenced by eIF1, must be able to accommodate this process. In addition, the protein environments of eIF1 and IF3 on the small subunit differ: prokaryotic initiator tRNA is bound by IF2, whereas eukaryotic initiator tRNA is bound by the unrelated eIF2. It is, in turn, stabilized on the 40S subunit by eIF3, which interacts with eIF1 and has no apparent prokaryotic counterpart. Our observations indicate that despite these major differences, prokaryotes and eukaryotes use similar mechanisms to ensure the fidelity of initiation codon selection, which is mediated by binding of unrelated factors to very similar positions on the small ribosomal subunit. Notably, some bacteria also contain a homolog of eIF1, YciH, the function of which is unknown. Its structure and surface charge distribution are very similar to those of eIF1 (Cort et al. 1999), therefore, it is tempting to speculate that in some circumstances YciH may play an analogous role to IF3 in bacteria, and that its homolog eIF1 has completely replaced IF3 in eukaryotes.
Materials and methods
Construction and purification of eIF1 mutants
eIF1 substitution mutants and the N-terminally truncated eIF1Δ29 deletion mutant were generated by PCR using the pET28b-eIF1 plasmid (Pestova et al. 1998). C69A and C94S mutations were combined to obtain a cysteineless eIF1 mutant. To create single-cysteine eIF1 mutants, cysteine residues were introduced at positions 32, 38, 42, 57, 61, 66, 75, 78, 91, or 109 on the surface of the cysteineless mutant. Two other single-cysteine eIF1 mutants contained one native cysteine each. Recombinant wild-type eIF1 and eIF1 mutants were expressed in Escherichia coli BL21(DE3) and purified on Ni2+-NTA (QIAGEN) and heparin-Sepharose (Roche) as described (Pestova et al. 1998). All recombinant eIF1 proteins had an N-terminal T7-Tag for immunological detection.
Purification of initiation factors, 40S ribosomal subunits, and aminoacylation of initiator tRNA
40S subunits, eIF3, eIF2, and eIF4F were purified from rabbit reticulocyte lysate (Green Hectares), and recombinant eIF1A, eIF4A, and eIF4B were expressed in E. coli and purified as described (Pestova et al. 1996, 1998). Aminoacylation of in vitro transcribed initiator tRNA by aminoacyl-tRNA synthetases from E. coli strain MRE 600 was done as described (Pestova and Hellen 2001).
Assembly and analysis of ribosomal complexes
In vitro eIF1–40S subunit binding assay was done essentially as described (Lomakin et al. 2000). eIF1 (10 μg) was immobilized on 20 μL of T7-Tag antibody-agarose (Novagen Inc.) suspension by incubation in 60 μL of buffer (20 mM Tris-HCl at pH 7.5, 200 mM KCl, 3 mM MgAc, 1 mM DTT, 10% glycerol, 0.5% Triton X-100) at 25°C for 30 min. Then 25 pmole of 40S subunits and 40 μg of BSA (New England Biolabs) were added to the immobilized eIF1 in 40 μL of final volume of the same buffer and incubated at 25°C for 30 min and then on ice for 90 min. After incubation, beads were washed four times with 400 μL of the same buffer. Bound material was resolved by electrophoresis using NuPAGE 12% gel (Invitrogen) and analyzed by Western blotting using antibodies against ribosomal protein S6 (Cell Signaling Technology).
For analysis of the eIF1–40S subunit interaction by sucrose density gradient centrifugation, 30 pmole of 40S subunits, 50 pmole of eIF3, 60 pmole of eIF2, 40 pmole of Met-tRNAiMet, 200 pmole of wild-type or mutant eIF1, and 8 μg of poly(U) (Sigma) in different combinations were incubated at 37°C for 10 min in 200 μL of buffer (20 mM Tris-HCl at pH 7.5, 100 mM KCl, 2.5 mM MgAc, 1 mM DTT) and analyzed by centrifugation through 10%–30% sucrose density gradients as described (Pestova et al. 1996). Fractions that corresponded to 40S subunits by optical density were analyzed for the presence of eIF1 by Western blotting using antibodies against T7-tag (QIAGEN).
For toeprinting analysis of 48S initiation complexes, reaction mixtures that contained 5 pmole of 40S subunits, 2μg of eIF2, 6 μg of eIF3, 0.5 μg of eIF4F, 2μg of eIF4A, 1 μg of eIF4B, 0.3 μg of eIF1A, 6 pmole of Met-tRNAiMet, 0.3 μg of native globin mRNA (Invitrogen), and 15 pmole of wild-type eIF1 or N-terminally truncated eIF1Δ29 mutant or 50 pmole of Fe(II)-BABE-derivatized eIF1 substitution mutants in 40 μL of buffer (20 mM Tris-HCl at pH 7.5, 100 mM KAc, 2.5 mM MgAc, 2 mM DTT, 1 mM ATP, 0.4 mM GTP, 0.25 mM spermidine) were incubated at 37°C for 10 min. 48S complex formation was analyzed by primer extension as described (Pestova et al. 1996, 1998).
Footprinting analysis of eIF1/40S subunit complexes
For footprinting analysis, 25 pmole of 40S subunits were incubated with either 250 pmole of eIF1 alone or with 50 pmole of eIF1 in the presence of 50 pmole of eIF3 and 3 μg of poly(U) RNA at 37°C for 10 min in 40 μL of buffer (20 mM Tris-HCl at pH 7.5, 100 mM KAc, 2.5 mM MgAc, and 2 mM DTT) and after that enzymatically digested by incubation with RNase VI (Pierce; final concentration 0.9 units/mL) in the same buffer at 37°C for 15 min as described (Kolupaeva et al. 1996). Ribosomal 18S RNA was phenol-extracted and cleavage sites were identified by primer extension using AMV reverse transcriptase (AMV RT; Promega) and primers complementary to different regions of rRNA.
Fe(II)-BABE modification of eIF1 mutants
Derivatization of single-cysteine eIF1 mutants with Fe(II)-BABE (Dojindo Molecular Technologies) was done using a previously described procedure (Culver and Noller 2000). Then 100 μg of single-cysteine eIF1 mutants and (as a negative control) cysteineless eIF1 mutants was incubated with 1 mM Fe(II)-BABE at 37°C for 30 min in 60 μL of buffer (80 mM HEPES at pH 7.5, 300 mM KCl, 10% glycerol). Fe(II)-BABE-derivatized eIF1 was separated from unincorporated Fe(II)-BABE on Microcon-10 microconcentrators (Millipore). Derivatized eIF1 proteins (0.3–0.4 mg/mL) were stored at -80°C.
Directed hydroxyl radical probing
For hydroxyl radical cleavage of 18S rRNA, 40S/eIF3/[Fe(II)-BABE]-eIF1 complexes were formed by incubating 7 pmole of 40S subunits, 10 pmole of eIF3, 3 μg of poly(U) RNA, and 50 pmole of derivatized eIF1 in 20 μL of buffer (80 mM HEPES at pH 7.5, 100 mM KCl, 2.5 mM MgAc, 10% glycerol) at 37°C for 10 min and chilled on ice. To generate hydroxyl radicals, the reaction mixture was supplemented with 0.025% H2O2 and 5 mM ascorbic acid (Culver and Noller 2000) and incubated on ice for 10 min. Reactions were quenched by addition of 10 mM thiourea. 18S rRNA was phenol-extracted, ethanol-precipitated, and analyzed by primer extension using AMV RT and primers complementary to different regions of 18S rRNA.
For hydroxyl radical cleavage of initiator tRNA, 43S complexes were formed by incubating 12pmole of 40S subunits, 12 pmole of eIF3, 10 pmole of eIF2, and 2 pmole of Met-tRNAiMet transcript in 35 μL of buffer (80 mM HEPES at pH 7.5, 100 mM KCl, 3 mM MgAc, 1 mM GMPPNP, 10% glycerol) at 37°C for 10 min and chilled on ice. After hydroxyl radical cleavage, per-formed as described above, initiator tRNA was analyzed by primer extension using primer 5′-TGGTAGCAGAGGATGG3′ complementary to its 3′-terminal 16 nt.
Modeling of eIF1/40S subunit interaction
The eIF1/40S subunit interaction was modeled using the structure of the T. thermophilus ribosome (Yusupov et al. 2001). MOLMOL (Koradi et al. 1996) was used for analysis of the hydroxyl radical cleavage data in the context of the ribosome structure and for manual docking. Regarding the radius of action of hydroxyl radicals (Culver and Noller 2000), 15-Å restraints for strong cleavage from the side chains of mutated amino acids of eIF1 to the corresponding cleavage sites on rRNA were assigned for initial modeling. The position of eIF1 was slightly adjusted to avoid backbone clashes between eIF1 and tRNA bound to the ribosomal P-site.
Acknowledgments
We thank Christopher Hellen for helpful discussions and great help in preparation of this manuscript. T.V.P. acknowledges support from National Institutes of Health Grant GM59660. G.W. acknowledges support by National Institutes of Health Grant CA68282 and National Science Foundation Grant MCB9816072. A.M. was funded by postdoctoral fellowship PF-02-117-01-GMC from the American Cancer Society.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1141803.
References
- Asano K., Phan, L., Anderson, J., and Hinnebusch, A.G. 1998. Complex formation by all five homologues of mammalian translation initiation factor 3 subunits from yeast Saccharomyces cerevisiae. J. Biol. Chem. 273: 18573-18585. [DOI] [PubMed] [Google Scholar]
- Battiste J.L., Pestova, T.V., Hellen, C.U.T., and Wagner, G. 2000. The eIF1A solution structure reveals a large RNA-binding surface important for scanning function. Mol. Cell 5: 109-119. [DOI] [PubMed] [Google Scholar]
- Biou V., Shu, F., and Ramakrishnan, V. 1995. X-Ray crystallography shows that translational initiation factor IF3 consists of two compact α/β domains linked by an α-helix. EMBO J. 14: 4056-4064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Clemons Jr., W.M., Brodersen, D.E., Morgan-Warren, R.J., Hartsch, T., Wimberly, B.T., and Ramakrishnan, V. 2001. Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498-501. [DOI] [PubMed] [Google Scholar]
- Chaudhuri J., Si, K., and Maitra, U. 1997. Function of eukaryotic translation initiation factor 1A (eIF1A) (formerly called eIF-4C) in initiation of protein synthesis. J. Biol. Chem. 272: 7883-7891. [DOI] [PubMed] [Google Scholar]
- Cort J.R., Koonin, E.V., Bash, P.A., and Kennedy, M.A. 1999. A phylogenetic approach to target selection for structural genomics: Solution structure of YciH. Nucleic Acids Res. 15: 4018-4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Dinman, J.D., Kinzy, T.G., and Peltz, S.W. 1998. The Mof2/Sui1 protein is a general monitor of translational accuracy. Mol. Cell. Biol. 18: 1506-1516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver G.M. and Noller, H.F. 2000. Directed hydroxyl radical probing of RNA from iron(II) tethered to proteins in ribonucleoprotein complexes. Methods Enzymol. 318: 461-475. [DOI] [PubMed] [Google Scholar]
- Dallas A. and Noller, H.F. 2001. Interaction of translation initiation factor 3 with the 30S ribosomal subunit. Mol. Cell 8: 855-864. [DOI] [PubMed] [Google Scholar]
- Dever T.E. 2002. Gene-specific regulation by general translation factors. Cell 108: 545-556. [DOI] [PubMed] [Google Scholar]
- Drabkin H.J., Helk, B., and RajBhandary, U.L. 1993. The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J. Biol. Chem. 268: 25221-25228. [PubMed] [Google Scholar]
- Fletcher C.M., Pestova, T.V., Hellen, C.U., and Wagner G. 1999. Structure and interactions of the translation initiation factor eIF1. EMBO J. 18: 2631-2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras A.-C., Raught, B., and Sonenberg, N. 1999. eIF4 initiation factors: Effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 68: 913-963. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters, D.S, and Gold, L. 1989. Selection of initiator tRNA by Escherichia coli initiation factors. Genes & Dev. 3: 1899-1912. [DOI] [PubMed] [Google Scholar]
- Hartz D., Binkley, J., Hollingworth, T., and Gold L. 1990. Domains of initiator tRNA and initiation codon crucial for initiator tRNA selection by Escherichia coli IF3. Genes & Dev. 4: 1790-1800. [DOI] [PubMed] [Google Scholar]
- Holm L. and Sander, C. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233: 123-138. [DOI] [PubMed] [Google Scholar]
- Kolupaeva V.G., Hellen, C.U., and Shatsky, I.N. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2: 1199-1212. [PMC free article] [PubMed] [Google Scholar]
- Koradi R., Billeter, M., and Wuthrich, K. 1996. MOLMOL: A program for display and analysis of macromolecular structures. J. Mol. Graph. 14: 51-55. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1986. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell 44: 283-292. [DOI] [PubMed] [Google Scholar]
- ____. 1987a. At least six nucleotides preceding the AUG initiator codon enhance translation in mammalian cells. J. Mol. Biol. 196: 947-950. [DOI] [PubMed] [Google Scholar]
- ____. 1987b. Analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 15: 8125-8148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 1989. The scanning model for translation: An update. J. Cell. Biol. 108: 229-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Teana A., Gualerzi, C., and Brimacombe, R. 1995. From stand-by to decoding site. Adjustment of the mRNA on the 30S ribosomal subunit under the influence of the initiation factors. RNA 1: 722-782. [PMC free article] [PubMed] [Google Scholar]
- Lee J.H., Choi, S.K., Roll-Mecak, A., Burley, S.K., and Dever, T.E. 1999. Universal conservation in translation initiation revealed by human and archaeal homologs of bacterial translation initiation factor IF2. Proc. Natl. Acad. Sci. 96: 4342-4347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin I.B., Hellen, C.U., and Pestova T.V. 2000. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internalribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 20: 6019-6029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R., Bandyopadhyay, A., and Maitra, U. 2003. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40S preinitiation complex. J. Biol. Chem. 278: 6580-6587. [DOI] [PubMed] [Google Scholar]
- Pestova T.V. and Hellen, C.U.T. 2001. Preparation and activity of synthetic unmodified mammalian tRNAiMet in initiation of translation in vitro. RNA 7: 1496-1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V. and Kolupaeva, V.G. 2002. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 16: 2906-2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Hellen, C.U.T., and Shatsky, I.N. 1996. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol. Cell. Biol. 16: 6859-6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Borukhov, S.I., and Hellen, C.U.T. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854-859. [DOI] [PubMed] [Google Scholar]
- Petrelli D., LaTeana, A., Garofalo, C., Spurio, R., Pon, C.L., and Gualerzi, C.O. 2001. Translation initiation factor IF3: Two domains, five functions, one mechanism? EMBO J. 20: 4560-4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phan L., Zhang, X., Asano, K., Anderson, J., Vornlocher, H.P., Greenberg, J.R., Qin, J., and Hinnebusch, A.G. 1998. Identification of a translation initiation factor 3 (eIF3) core complex, conserved in yeast and mammals, that interacts with eIF5. Mol. Cell. Biol. 18: 4935-4946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sette M., van Tilborg, P., Spurio, R., Kaptein, R., Paci, M., Gualerzi, C.O., and Boelens, R. 1997. The structure of translation initiation factor IF1 from E. coli contains an oligomer-binding motif. EMBO J. 16: 1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapkina T.G., Dolan, M.A., Babin, P., and Wollenzien, P. 2000. Initiation factor 3-induced structural changes in the 30S ribosomal subunit and in complexes containing tRNAfMet and mRNA. J. Mol. Biol. 299: 615-628. [DOI] [PubMed] [Google Scholar]
- Spahn C.M.T., Beckmann, R., Eswar, N., Penczek, P.A., Sali, A., Blobel, G., and Frank, J. 2001. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell 107: 373-386. [DOI] [PubMed] [Google Scholar]
- Subramanian A.R. and Davis, B.D. 1970. Activity of initiation factor F3 in dissociating Escherichia coli ribosomes. Nature 228: 1273-1275. [DOI] [PubMed] [Google Scholar]
- Tedin K., Moll, I., Grill, S., Resch, A., Graschopf, A., Gualerzi, C.O., and Blasi, U. 1999. Translation initiation factor 3 antagonizes authentic start codon selection on leaderless mRNAs. Mol. Microbiol. 31: 67-77. [DOI] [PubMed] [Google Scholar]
- Trachsel H. and Staehelin, T. 1979. Initiation of mammalian protein synthesis. The multiple functions of the initiation factor eIF-3. Biochim. Biophys. Acta 565: 305-314. [DOI] [PubMed] [Google Scholar]
- Yoon H. and Donahue, T.F. 1992. The sui1 suppressor locus in Saccharomyces cerevisiae encodes a translation factor that functions during tRNAiMet recognition of the start codon. Mol. Cell. Biol. 12: 248-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusupov M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883-896. [DOI] [PubMed] [Google Scholar]