The regenerative capacity of muscle is regulated by p38-γ, which phosphorylates MyoD and leads to formation of a complex that represses myogenin transcription.
Abstract
The mitogen-activated protein kinase p38-γ is highly expressed in skeletal muscle and is associated with the dystrophin glycoprotein complex; however, its function remains unclear. After induced damage, muscle in mice lacking p38-γ generated significantly fewer myofibers than wild-type muscle. Notably, p38-γ-deficient muscle contained 50% fewer satellite cells that exhibited premature Myogenin expression and markedly reduced proliferation. We determined that p38-γ directly phosphorylated MyoD on Ser199 and Ser200, which results in enhanced occupancy of MyoD on the promoter of myogenin together with markedly decreased transcriptional activity. This repression is associated with extensive methylation of histone H3K9 together with recruitment of the KMT1A methyltransferase to the myogenin promoter. Notably, a MyoD S199A/S200A mutant exhibits markedly reduced binding to KMT1A. Therefore, p38-γ signaling directly induces the assembly of a repressive MyoD transcriptional complex. Together, these results establish a hitherto unappreciated and essential role for p38-γ signaling in positively regulating the expansion of transient amplifying myogenic precursor cells during muscle growth and regeneration.
Introduction
Satellite cells are skeletal muscle stem cells responsible for postnatal growth and repair. These cells are activated from quiescence through a highly ordered program that governs their transient amplification and subsequent differentiation, both of which are regulated by the same transcription factor, MyoD (Rudnicki et al., 1992; Blais et al., 2005; Ishibashi et al., 2005; Tapscott, 2005). For instance, MyoD initiates the differentiation program by promoting cell cycle withdrawal and directly activating myogenin gene expression (Hollenberg et al., 1993; Halevy et al., 1995), the latter of which is mediated by the MyoD-dependent recruitment of cofactors responsible for achieving permissive chromatin architecture, including p300, PCAF, SWI/SNF, and p68/p72 (Puri et al., 1997a,b; Sartorelli et al., 1999; de la Serna et al., 2001, 2005; Dilworth et al., 2004; Simone et al., 2004; Caretti et al., 2006); and transcriptional initiation (Deato and Tjian, 2007; Deato et al., 2008).
The promyogenic kinase p38-α is also essential for differentiation (Cuenda and Cohen, 1999; Wu et al., 2000; Bergstrom et al., 2002; Perdiguero et al., 2007), as it indirectly regulates MyoD function through phosphorylation of the chromatin-modifying enzyme SWI/SNF (Simone et al., 2004; Serra et al., 2007). Moreover, p38-α also phosphorylates E47, an E protein that heterodimerizes with MyoD to promote DNA binding (Lluís et al., 2005); Mef2 proteins, which cooperate with MyoD as part of a feed-forward network (Molkentin et al., 1995; Zetser et al., 1999; Zhao et al., 1999; Wu et al., 2000; Penn et al., 2004; Rampalli et al., 2007); and KH-type splicing regulatory protein (KSRP), an mRNA decay factor that subsequently fails to bind to the 3′ untranslated regions of myogenic transcripts (Briata et al., 2005). Interestingly, p38-α signaling also promotes satellite cell activation (Jones et al., 2005); however, the specific targets remain to be identified.
Despite the expression of functional MyoD protein during myoblast proliferation (Blais et al., 2005; Ishibashi et al., 2005) and the presence of the MyoD coactivators noted earlier together with an accessible noncanonical MyoD binding site (Berkes et al., 2004), the chromatin structure of the myogenin promoter exists in a repressive state (Gerber et al., 1997), thereby ensuring the correct temporal patterning of myogenin gene expression. In fact, previous studies have documented the role of MyoD corepressors in preventing gene expression (Mal et al., 2001, 2006; Puri et al., 2001; Mal and Harter, 2003). However, the signals regulating these associations during the different stages of satellite cell activation remain to be established.
Although the promyogenic role of p38-α signaling has been well documented, the function and mechanism of action of p38-γ during satellite cell activation has remained elusive despite its more restricted expression and its activation in skeletal muscle (Lechner et al., 1996; Mertens et al., 1996; Boppart et al., 2000; Tortorella et al., 2003; Perdiguero et al., 2007; Ruiz-Bonilla et al., 2008; Wang et al., 2008). Our results define a novel function for p38-γ during adult myogenesis, whereby its direct phosphorylation of MyoD assembles a repressive transcriptional complex containing a MyoD corepressor. Through epigenetic modifications of chromatin within the promoter of an important myogenic regulator, p38-γ signaling ensures the correct temporal pattern of differentiation-specific gene expression during satellite cell–mediated muscle growth and regeneration.
Results
Impaired regeneration in p38-γ−/− muscle is caused by a satellite cell deficit
To investigate the role of the mitogen-activated protein kinase (MAPK) p38-γ in myogenesis in vivo, we began by examining the regeneration capacity of muscle lacking the p38-γ gene (Sabio et al., 2005). These mice are viable, grow at a similar rate and produce comparable litter sizes to wild-type controls, and display no overt muscle phenotype or pathology. Importantly, there is no apparent compensation for the loss of p38-γ by up-regulation of the expression or activity of the promyogenic p38-α MAPK (Sabio et al., 2005).
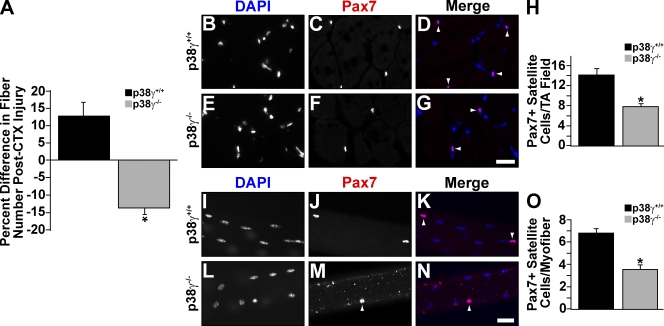
Acute injury was induced by injection of cardiotoxin (CTX) into the belly of the tibialis anterior (TA) muscle. Importantly, we detected activation of p38-γ after injury of wild-type muscle (Fig. S1, A and B). Histological examination of sections of regenerating muscle did not reveal an overt phenotype such as delayed or impaired regeneration. However, we detected a small but statistically significant decrease in mutant fiber size 21 d after injury (Fig. S1, C–H). Enumeration of total fiber number 21 d after injury revealed a significant regeneration deficit in p38-γ−/− muscle (Fig. 1 A). Wild-type TA muscle displayed a 13% increase in the total number of fibers, whereas TA muscle lacking p38-γ displayed a 14% decrease, relative to uninjured contralateral TA controls. Therefore, the reduced numbers and sizes of fibers formed after acute injury of p38-γ−/− muscle suggested that the function of satellite cells was perturbed.
Figure 1.
Impaired regeneration resulting from a satellite cell deficit in p38-γ−/− muscle. (A) Enumeration of total fiber number from wild-type and p38-γ−/− TA muscle 21 d after CTX injury. Numbers were normalized to the contralateral TA muscle. Error bars represent ±SEM for n = 5–6. Asterisk denotes significance (P < 0.0003). (B–H) Immunofluorescent staining for Pax7 (C and F) from wild-type and p38-γ−/− TA muscle. Nuclei were counterstained with DAPI (B and E). (D and G) Pax7-DAPI merged pictures. Representative pictures are shown, with arrowheads denoting Pax7-positive nuclei. Pax7-positive satellite cells were enumerated (H). Error bars represent standard error of the means (±SEM) for n = 10. Asterisk denotes significance (P < 0.0001). (I–O) Immunofluorescent staining for Pax7 (J and M) on freshly isolated single fibers from wild-type and p38-γ−/− EDL muscle. Nuclei were counterstained with DAPI (I and L). (K and N) Pax7-DAPI merged pictures are shown. Representative pictures are shown, with arrowheads indicating Pax7-positive nuclei. Pax7-positive satellite cells were enumerated (O). Error bars represent ±SEM for n = 84 myofibers. Asterisk denotes significance (P < 0.0001). Bars, 25 µm.
To enumerate the number of satellite cells in mutant muscle, we performed immunostaining on sections of undamaged TA muscle (Fig. 1, B–G). In addition, the number of satellite cells on single muscle fibers freshly isolated from the extensor digitorum longus (EDL) muscle was examined (Fig. 1, I–N). Notably, we observed a 50% reduction in satellite cell number in both p38-γ−/− TA (Fig. 1 H) and EDL (Fig. 1 O) muscles.
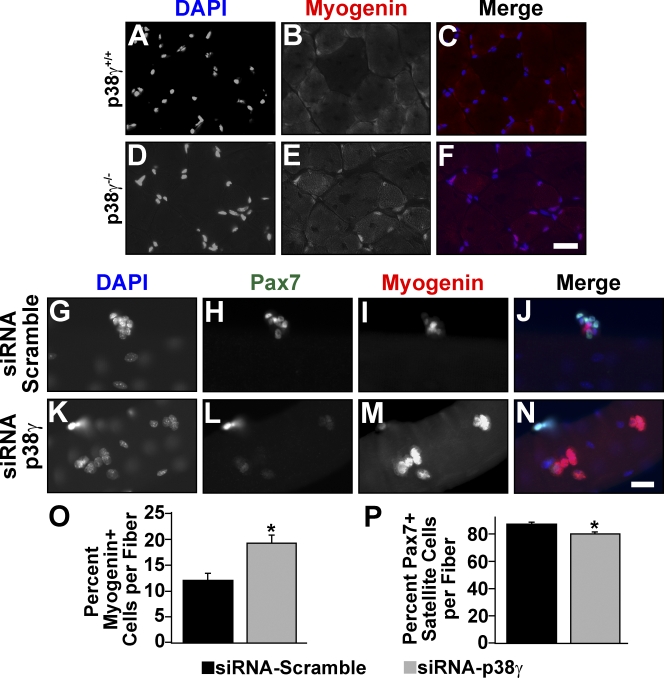
Strikingly, Myogenin protein was detected in large numbers of nuclei in p38-γ−/− TA muscle, whereas it was absent in wild-type muscle (Fig. 2, A–F). Myogenin is typically highly expressed in myogenic cells entering the terminal differentiation program and is down-regulated in mature fibers. Together, these results suggest that p38-γ–deficient satellite cells are defective in satellite cell maintenance and that satellite cell–derived myogenic precursors lacking p38-γ are undergoing premature differentiation.
Figure 2.
Precocious up-regulation of Myogenin in p38-γ–deficient muscle. (A–F) Immunofluorescent staining for Myogenin (B and E) from wild-type and p38-γ−/− TA muscle. Nuclei were counterstained with DAPI (A and D). (C and F) Myogenin-DAPI merged pictures. Representative pictures are shown (n = 9). (G–P) Immunofluorescent staining for Pax7 (H and L) and Myogenin (I and M) in single EDL fibers from wild-type mice transfected with siRNA against p38-γ or control scrambled siRNA immediately after their isolation. Nuclei were counterstained with DAPI (G and K). (J and N) Pax7-Myogenin-DAPI merged pictures. Representative pictures are shown. The percentage of Myogenin-expressing and Pax7-expressing myogenic precursor cells was determined 72 h after transfection (O and P). Error bars represent ±SEM for n = 28. Asterisks denote significance (P < 0.001). Bars, 25 µm.
Both differentiated myofibers and satellite cells express p38-γ. To assess the role of p38-γ specifically in satellite cells, we performed siRNA knockdown of p38-γ in satellite cells cultured on single fibers isolated from wild-type EDL muscle (Fig. 2, G–N). Importantly, we detected a 40% increase in the percentage of Myogenin-expressing myogenic precursor cells in fibers treated with a p38-γ–specific siRNA compared with a nonspecific scrambled siRNA control (Fig. 2 O). Furthermore, we observed a 10% decrease in the percentage of Pax7-expressing cells in fibers treated with p38-γ siRNA (Fig. 2 P).
Collectively, the reduced numbers of satellite cells, the elevated numbers of nuclei containing the differentiation marker Myogenin, and the reduced numbers of fibers generated after acute injury all support the assertion that satellite cells lacking p38-γ display an increased propensity to initiate the differentiation program.
Reduced growth and precocious Myogenin activation in p38-γ−/− satellite cells
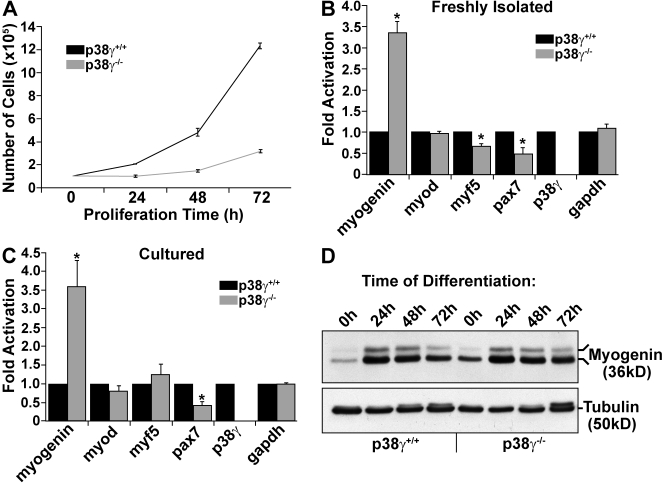
To further investigate the role of p38-γ in satellite cells, we isolated them from p38-γ−/− muscle by FACS and examined their growth and differentiation characteristics. The growth rate of established primary cultures of wild-type and p38-γ−/− myoblasts was examined by direct enumeration of cells over several days under growth conditions. Strikingly, cultured p38-γ−/− myoblasts displayed a severe reduction in their growth rate (Fig. 3 A).
Figure 3.
Impaired proliferation and precocious activation of Myogenin in p38-γ–deficient satellite cells. (A) Proliferation analysis of wild-type and p38-γ−/− satellite cells. Cells were seeded at the same density (105), and cells on separate plates were dissociated and counted every 24 h for 3 d. Error bars represent ±SEM for n = 3. (B and C) Real-time RT-PCR for myogenin, Myod, Myf5, Pax7, and p38-γ transcripts from wild-type and p38-γ−/− satellite cells. RNA was extracted from satellite cells immediately after isolation (B) or after several days in culture (C). Error bars represent ±SEM for n = 7–9. Asterisks denote significance (P < 0.008). (D) Immunoblot analysis of Myogenin protein expression from proliferating and differentiating wild-type and p38-γ−/− satellite cell–derived myoblasts. Tubulin was used as a loading control.
To confirm this proliferation defect, we performed Ki67 immunostaining on these cells and observed a significant reduction in Ki67-positive p38-γ–deficient satellite cell–derived myoblasts over 72 h of culture under growth conditions (Fig. S2 A). In addition, we also detected elevated apoptosis in mutant myoblasts under growth conditions (Fig. S2 B), which could be attributed to the increase in apoptosis normally observed upon entry into the differentiation program.
Immediately after the isolation of p38-γ−/− satellite cells by FACS, we observed an almost 3.5-fold increase in the levels of myogenin mRNA, with no change in MyoD mRNA (Fig. 3 B). Similarly, p38-γ−/− satellite cell–derived myoblasts cultured for 1–2 wk continued to express elevated levels of myogenin transcript, with no significant alterations in MyoD expression (Fig. 3 C). We also observed decreased Pax7 and Myf5 expression in the p38-γ−/− satellite cells analyzed immediately after their isolation (Fig. 3 B), and depressed levels of Pax7 in cultured p38-γ−/− myoblasts (Fig. 3 C). Moreover, we also detected elevated Myogenin protein in proliferating p38-γ−/− satellite cell–derived myoblasts (Fig. 3 D).
In addition, we observed slightly elevated myosin heavy chain (MyHC) protein expression by 24 h of differentiation in p38-γ–deficient myoblasts (Fig. S2 C). Interestingly, this trend fails to continue throughout differentiation, as by 48 h, MyHC expression was enhanced in wild-type myocytes/myotubes.
Both Myf5 and Pax7 are normally down-regulated upon differentiation (Megeney et al., 1996; Sabourin et al., 1999; Seale et al., 2000). Myogenin is normally up-regulated in myogenic cells after cell cycle withdrawal as the cells enter the terminal differentiation program (Wright et al., 1989). Therefore, the markedly decreased rate of proliferation together with the premature expression of myogenin in growth conditions in p38-γ–deficient myoblasts supports the hypothesis that p38-γ signaling acts to inhibit the premature initiation of differentiation in myogenic precursor cells.
Myogenin transcription is repressed by p38-γ signaling
To further investigate the mechanism through which p38-γ represses myogenic differentiation, we examined the effects of p38-γ signaling on myogenin transcription in differentiating C2C12 myoblasts. Our analysis was performed at 24 h after induction of differentiation, as this represents the time of maximal expression of Myogenin protein (Fig. 4 A). To ensure that we specifically induced activation of p38-γ, we also forced expression of a constitutively active mutant of its upstream activator MKK6 (MKK6EE; Raingeaud et al., 1996; Cuenda et al., 1997) together with p38-γ. Importantly, activation of p38-α, a positive regulator of myogenic differentiation, was not altered under these conditions (Fig. S2 D), which indicates that any functional effects on myogenic differentiation were specific to p38-γ.
Figure 4.
Activation of p38-γ signaling represses MyoD activation of Myogenin transcription. (A) Schematic representation of myoblast differentiation, with MyoD binding to the myogenin promoter within 6–12 h of growth factor removal, and maximal MyoD transcriptional activity achieved between 12 and 48 h. (B–J) Immunofluorescent staining for the Myc (C and I) or Flag epitopes (F), and Myogenin (D, G, and J) in C2C12 myoblasts expressing empty vector (B–D), MKK6EE and p38-α (E–G), or MKK6EE and p38-γ (H–J) after 24 h of differentiation. Note the Myogenin-positive nuclei in cells containing activated p38-α (arrows in F and G). Conversely, note the lack of Myogenin staining in cells containing activated p38-γ (arrowheads in I and J). Nuclei were counterstained with DAPI (B, E, and H). (K) Luciferase assays on 10T1/2 fibroblast extracts expressing MKK6EE and p38-γ, or empty vector. MyoDwt or MyoDmut were expressed as indicated. A diagram of the myogenin-luciferase reporter, shown above the graph, contains two canonical E-boxes (E1 and E2), as well as the PME site, which contains the binding site for Pbx-Meis along with a noncanonical E box. Arrows indicate the location of primers for ChIP assays. Error bars represent ±SEM for n = 9. Asterisk denotes significance (P < 0.00002). (L) Quantification of a Myogenin immunoblot, normalized to MyoD protein levels, from 10T1/2 fibroblast extracts expressing MKK6EE, p38-γ, or empty vector. MyoDwt and MyoDmut were expressed as indicated. Note that the original immunoblots are shown in Fig. S2 G. MyoDmut, MyoD [S199A/S200A] mutant; MyoDwt, wild-type MyoD. Bar, 25 µm.
Normally, ∼20% of C2C12 cells contain nuclear-localized Myogenin protein as assessed by immunostaining at 24 h after induction of differentiation (Fig. 4, B–D). To prove we could stimulate differentiation, we coexpressed MKK6EE with the promyogenic p38-α MAPK. In agreement with the published role for p38-α (Cuenda and Cohen, 1999; Wu et al., 2000; Bergstrom et al., 2002), we detected Myogenin protein in 46% of these nuclei (Fig. 4, E–G). In contrast, Myogenin protein was detected in only 10.9% of C2C12 cells that were cotransfected with p38-γ and MKK6EE (n = 329; Fig. 4, H–J).
To further demonstrate that our results are caused by specific activation of p38-γ, we expressed only MKK6EE in C2C12 myoblasts. After 24 h of differentiation, we observed phosphorylation of p38-α together with increased Myogenin protein (Fig. S2 E), which is consistent with the known role of p38-α signaling in promoting myogenesis. In contrast, specific activation of p38-γ, in response to coexpression of MKK6EE and p38-γ, resulted in diminished Myogenin protein despite the invariant expression of MyoD (Fig. S2 F). Therefore, these results support the assertion that p38-γ signaling represses Myogenin expression.
MyoD transcriptional activity is repressed by p38-γ signaling
The activation of myogenin transcription during cell cycle withdrawal and entrance into the differentiation program is mediated by MyoD binding and transactivation (Hollenberg et al., 1993). To investigate the molecular mechanism through which p38-γ signaling represses Myogenin expression, we assessed the ability of MyoD to activate myogenin transcription in 10T1/2 fibroblasts in response to p38-γ activation. These cells do not intrinsically express MyoD, but are converted to the skeletal muscle lineage upon overexpression of MyoD (Davis et al., 1987), which makes them a useful model for studying MyoD-dependent gene regulation. Once again, our analysis was performed after 24 h of differentiation, which represents the time of maximal MyoD binding to the myogenin promoter and maximal activation of myogenin gene expression (Fig. 4 A).
We used a luciferase reporter under the control of a 224-bp fragment of the myogenin promoter previously shown to be sufficient for expression in vivo (Cheng et al., 1993; Berkes et al., 2004). On its own, MyoD transactivated this reporter ∼10-fold in 10T1/2 fibroblasts (Fig. 4 K). Strikingly, activation of p38-γ signaling resulted in a fourfold down-regulation of MyoD transactivation of the myogenin promoter (Fig. 4 K).
In addition, reporter assays were recapitulated by immunoblotting experiments, which revealed that Myogenin protein expression was severely diminished after activation of p38-γ signaling (Figs. 4 L and S2 G). Importantly, MyoD protein levels remained unchanged by MKK6EE and p38-γ (Fig. S2 G), which confirms that the effects we observed were not a result of alterations in MyoD expression. Therefore, these data demonstrate that p38-γ signaling represses MyoD transcriptional activity, thereby inhibiting myogenin gene expression, which provides an explanation for the precocious myogenin expression observed in p38-γ−/− satellite cells.
p38-γ directly phosphorylates MyoD
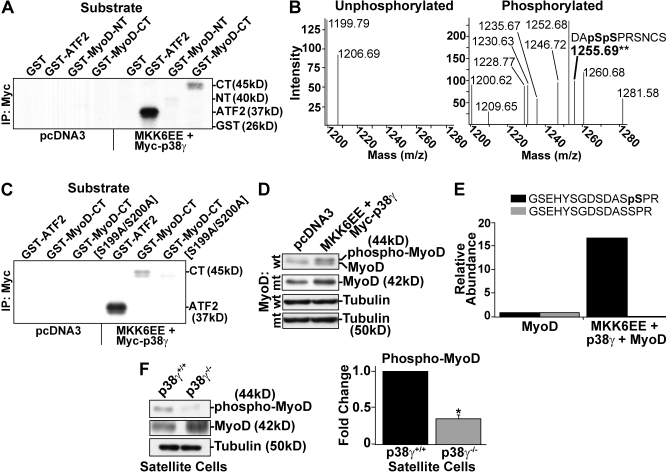
To examine whether MyoD was a substrate for p38-γ phosphorylation, we immunoprecipitated activated p38-γ from cell extracts and incubated it with recombinant MyoD in the presence of [γ32P]ATP. Incubation with the known p38-γ substrate ATF2 demonstrated that p38-γ was indeed activated by MKK6EE (Fig. 5 A), which is in agreement with our phospho–p38-γ immunoblots. More importantly, incubation of full-length MyoD protein with immunoprecipitated, activated p38-γ suggested that MyoD was phosphorylated by p38-γ (Fig. S3 A).
Figure 5.
p38-γ directly phosphorylates Ser199 and Ser200 within the C terminus of MyoD. (A) IP-kinase assays from 10T1/2 fibroblasts expressing MKK6EE and p38-γ, or empty vector. Extracts were immunoprecipitated with antibody reactive to the Myc tag on p38-γ, and incubated with the recombinant substrates GST–MyoD–N terminal (GST-MyoD-NT) and GST–MyoD–C terminal (GST-MyoD-CT), along with GST (negative) and GST-ATF2 (positive) as controls. (B) MALDI-TOF MS spectra of IP-kinase assays from 10T1/2 fibroblasts expressing empty vector or MKK6EE and p38-γ. The peak with an m/z ratio of 1,255.69 (indicated by the asterisks) represents the phosphorylated MyoD peptide. (C) IP-kinase assay from 10T1/2 fibroblasts expressing MKK6EE and p38-γ, or empty vector. Myc–p38-γ immunoprecipitates were incubated with the recombinant substrates GST-MyoD-CT or GST-MyoD-CT [S199A/S200A] containing mutated p38-γ phosphorylation sites. GST and GST-ATF2 were used as controls. (D) Immunoblot analysis of 293T extracts expressing MKK6EE and p38-γ, or empty vector. Wild type (wt) and mutant (mt) MyoD S199A/S200A were expressed as indicated. (E) LC-MS/MS analysis of 293T nuclear extracts expressing MyoD alone or together with MKK6EE and p38-γ. For relative abundance calculations, indicated MyoD peptides were normalized against additional MyoD peptides and α-actin peptides. Representative data are shown. (F) Immunoblot analysis of S200-phosphorylated MyoD and total MyoD in proliferating p38-γ+/+ and p38-γ−/− satellite cell–derived myoblasts. Tubulin was used as a loading control. Fold change is shown relative to p38-γ+/+ satellite cells after quantification and normalization to tubulin. Error bars represent ±SEM for n = 3. Asterisk denotes significance (P < 0.004). CT, C terminus; NT, N terminus.
To delineate the region of MyoD phosphorylated by p38-γ, we incubated immunoprecipitated, activated p38-γ with N- and C-terminal fragments of MyoD. Notably, only the C terminus of MyoD was specifically phosphorylated by activated p38-γ (Fig. 5 A). In fact, we observed similar levels of phosphorylation compared with full-length MyoD (104% ± 2% relative to full length). Moreover, a dominant-negative mutant of p38-γ (T183A/Y185F) blocked phosphorylation of MyoD (not depicted). Together, these results indicate that p38-γ directly phosphorylates the C terminus of MyoD.
To identify amino acids that are phosphorylated by p38-γ, we analyzed the C terminus of MyoD by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry after kinase reactions. Spectra from unphosphorylated (from empty vector control cells) and phosphorylated reactions are shown in Fig. 5 B. In the phosphorylated spectra, we observed a specific peak with the mass/charge (m/z) ratio of 1,255.69, which corresponded to a MyoD peptide containing phosphorylated Ser199 and Ser200. The additional peaks observed are likely a result of background contaminants due to other components in the reactions or inefficient immobilized metal ion affinity chromatography enrichment of phosphopeptides.
To confirm that these residues were targets for phosphorylation by p38-γ, we mutated them to alanine (S199A/S200A) and incubated them with immunoprecipitated, activated p38-γ. Importantly, this mutant abolished any phosphorylation (Fig. 5 C). To confirm the specificity of phosphorylation, we also mutated other potential MAPK phosphorylation sites within the C terminus to alanine and failed to alter the phosphorylation of MyoD by p38-γ using immunoprecipitation (IP)-kinase assays (not depicted). Collectively, these results indicate that p38-γ directly phosphorylates MyoD on Ser199 and Ser200.
To analyze MyoD phosphorylation in vivo, we expressed MyoD in the presence or absence of activated p38-γ and analyzed its mobility on immunoblots. We detected a slower migrating band in the presence of p38-γ signaling, which represents phosphorylated MyoD (Fig. 5 D; Tapscott et al., 1988). Importantly, expression of the MyoD S199A/S200A mutant completely abolished detection of this slower migrating band (Fig. 5 D), which confirms that MyoD is phosphorylated by p38-γ in vivo.
To further validate MyoD phosphorylation in vivo, we analyzed MyoD expressed in the presence or absence of activated p38-γ by liquid chromatography tandem mass spectrometry (LC-MS/MS). After MyoD IP, we observed a 17-fold increase in detection of the MyoD phosphopeptide in the presence of activated p38-γ, with a sixfold decrease in the corresponding unphosphorylated peptide (Fig. 5 E). However, this phosphopeptide represented phosphorylation of Ser200, and we were unable to detect phosphorylation of both Ser199 and Ser200 together. This result is not surprising given the extreme technical difficulties in identifying a dually phosphorylated peptide from a more complex sample by MS/MS, such as poor column retention, reduced ionization efficiency, and poor fragmentation (Aebersold and Goodlett, 2001). Therefore, we conclude that Ser200 likely represents the major phosphorylated residue in vivo, with phosphorylation of Ser199 a functionally significant but less frequent event.
To examine MyoD phosphorylation in p38-γ–deficient satellite cells, we took advantage of our LC-MS/MS observations and used a phospho-Ser200–specific antibody. Notably, we detected a 65% reduction in MyoD phosphorylation in mutant versus wild-type satellite cells (Fig. 5 F). Together, these results indicate that MyoD represents a physiological target of p38-γ phosphorylation in vivo.
Phosphorylation of MyoD by p38-γ enhances promoter occupancy
To determine the role of MyoD phosphorylation, we examined the transcriptional activation properties of MyoD during myogenic differentiation. Although the MyoD S199A/S200A mutant transactivated the myogenin-luciferase reporter 18-fold in 10T1/2 fibroblasts, p38-γ signaling failed to significantly down-regulate its activity (Fig. 4 K), a result recapitulated by Western analysis (Fig. 4 L). These results suggest that the down-regulation of MyoD transcriptional activity is dependent on its phosphorylation.
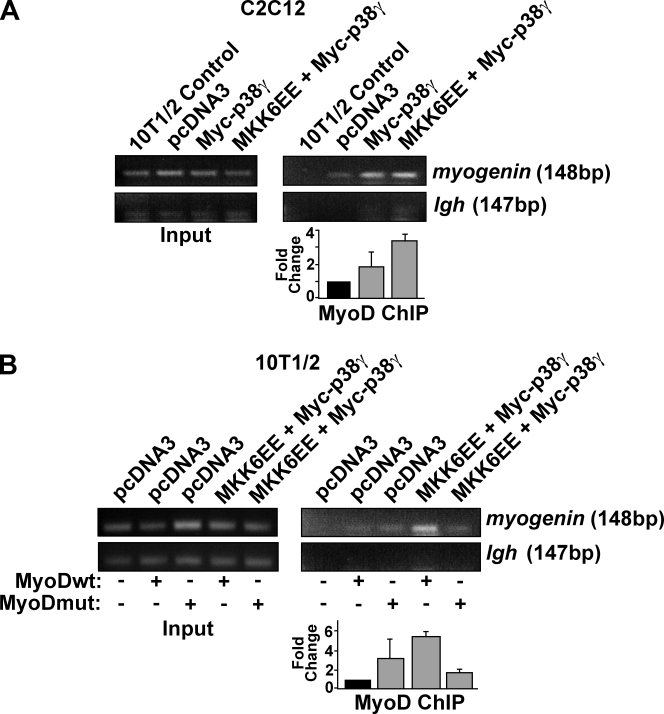
To examine the DNA-binding properties of p38-γ-phosphorylated MyoD, we performed chromatin IP (ChIP) assays from C2C12 myoblasts 24 h after induction of differentiation. Using PCR primers spanning the regulatory region of the myogenin promoter (Fig. 4 K; Cheng et al., 1993), we detected binding of endogenous MyoD to the endogenous myogenin promoter (Fig. 6 A), as has been described previously (Bergstrom et al., 2002; Berkes et al., 2004; Simone et al., 2004; de la Serna et al., 2005; Parker et al., 2006).
Figure 6.
Phosphorylation of MyoD by p38-γ enhances its occupancy on the Myogenin promoter. (A) Endogenous MyoD ChIP assay on C2C12 myoblasts expressing p38-γ, MKK6EE and p38-γ, or empty vector after 24 h of differentiation. 10T1/2 fibroblast extracts were used as control. (B) MyoD ChIP assay from 10T1/2 fibroblasts expressing MKK6EE and p38-γ, or empty vector after 24 h of differentiation. Wild-type (wt) and mutant (mut) MyoD S199A/S200A were expressed as indicated. PCR was performed using primers spanning the Igh enhancer as a control. Fold changes for each condition relative to empty vector (C2C12) or wild-type MyoD (10T1/2) are shown below representative gels.
Importantly, in the presence of either p38-γ alone, or MKK6EE and p38-γ, the occupancy of MyoD on the myogenin promoter was markedly increased (Fig. 6 A), which suggests that p38-γ signaling enhanced MyoD DNA binding. To confirm the specificity of the ChIP, we performed PCR for the Igh enhancer, which contains E boxes that do not bind MyoD in vivo (Weintraub et al., 1994; Bergstrom et al., 2002), and detected no MyoD binding (Fig. 6 A).
To ensure our results were specific to activation of p38-γ, we analyzed MyoD DNA binding by ChIP after expression of MKK6EE, which results in activation of p38-α but not p38-γ under our conditions. We failed to detect any differences in the association between MyoD and the myogenin promoter as a result of MKK6–p38-α signaling (Fig. S3 B). Furthermore, expression of a dominant-negative mutant of p38-γ prevented the p38-γ–dependent increase in MyoD occupancy of the myogenin promoter (Fig. S3 C). These results indicate that enhanced occupancy of MyoD on the promoter of myogenin is a specific result of p38-γ activation.
Similar to C2C12 differentiating myoblasts, ChIP assays from 10T1/2 fibroblasts expressing MKK6EE, p38-γ, and MyoD showed increased MyoD occupancy on the endogenous myogenin promoter compared with cells expressing MyoD alone (Fig. 6 B). However, the MyoD mutant lacking p38-γ phosphorylation sites (S199A/S200A) abolished the p38-γ–dependent increase in MyoD occupancy of the myogenin promoter (Fig. 6 B).
We also detected increased interaction between MyoD and the E protein HeLa E box binding factor (HEB; Parker et al., 2006) in vivo after activation of p38-γ in C2C12 myoblasts (Fig. S3 D). Importantly, this enhanced interaction occurred even though total protein levels for both MyoD and HEB remained unchanged (Fig. S3 E). This result implies that the observed increase in MyoD DNA binding may be a result of an increased association of MyoD with HEB. Together, these results demonstrate that phosphorylation of MyoD by p38-γ promotes MyoD association with the myogenin promoter.
Phosphorylation of MyoD by p38-γ directs histone H3K9 methylation of the myogenin promoter
The myogenin promoter is subject to regulation by histone modifications, which are dependent on the presence of MyoD (Puri et al., 1997a,b, 2001; Sartorelli et al., 1999; Mal et al., 2001; Mal and Harter, 2003; Dilworth et al., 2004; Sims et al., 2004). The only repressive lysine methylation modification described for the myogenin promoter has been the methylation of lysine 9 on histone H3 (H3K9; Zhang et al., 2002; Mal and Harter, 2003). To determine if methylation of H3K9 was associated with the negative effect of p38-γ on myogenin gene expression, we performed ChIP assays for dimethylated H3K9 in differentiating C2C12 myoblasts. H3K9 methylation was detected on this promoter during the differentiation of control cells, likely a result of unsynchronized, incomplete induction of C2C12 differentiation. To control for any effects of p38-α signaling, we expressed MKK6EE on its own and failed to alter the methylation state of this promoter (Fig. S3 F). However, levels of H3K9 methylation were substantially increased in the presence of MKK6EE and p38-γ together (Fig. 7 A).
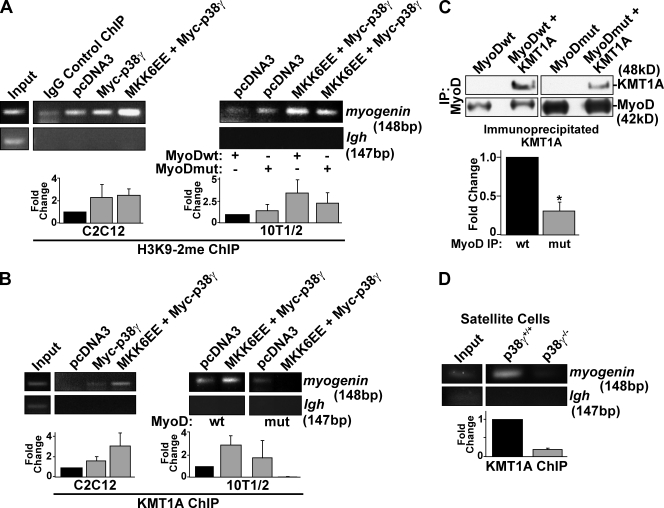
Figure 7.
Phosphorylation of MyoD by p38-γ directs the assembly of a repressive transcriptional complex at the Myogenin promoter. (A and B) Endogenous H3K9-2me (A) and endogenous KMT1A (B) ChIP assays on a differentiating (24 h) C2C12 myoblast and 10T1/2 fibroblast extracts expressing p38-γ, MKK6EE and p38-γ, or empty vector. MyoDwt and MyoDmut were expressed as indicated for 10T1/2 fibroblasts ChIPs. (C) Nuclear extracts from proliferating 293T cells expressing wild-type or mutant MyoD in the presence or absence of Myc-KMT1A (or Flag-KMT1A; not depicted) were immunoprecipitated for MyoD and immunoblotted for KMT1A. Equal exposure times are shown. Input immunoblots are found in Fig. S3 H. Fold change in the relative association of KMT1A with wild-type versus mutant MyoD is also shown after quantification and normalization to immunoprecipitated MyoD. Error bars represent ±SEM for n = 3. Asterisk denotes significance (P < 0.02). (D) Endogenous KMT1A ChIP assays from proliferating p38-γ+/+ and p38-γ−/− satellite cell–derived myoblasts. PCR was performed on the Igh enhancer as a control. Fold changes for each condition relative to empty vector (C2C12), wild-type MyoD (10T1/2), or p38-γ+/+ (satellite cells) are shown below representative gels. Error bars represent ±SEM for n = 3. H3K9-2me, dimethylated histone H3 Lys9; MyoDwt, wild-type MyoD; MyoDmut, MyoD [S199A/S200A] mutant.
Furthermore, expression of MKK6EE and p38-γ, together with MyoD, in 10T1/2 fibroblasts similarly resulted in markedly increased H3K9 methylation of the myogenin promoter compared with cells expressing MyoD alone (Fig. 7 A). Importantly, there was little increase in H3K9 methylation when the MyoD S199A/S200A mutant was used (Fig. 7 A). These data indicate that p38-γ signaling induces formation of heterochromatin at the myogenin promoter, which correlates with its ability to repress gene expression after MyoD phosphorylation.
MyoD phosphorylation-dependent assembly of a repressive transcriptional complex
To identify the enzymatic activity responsible for methylation of H3K9, we examined the association of the K-methyltransferase KMT1A (Rea et al., 2000), which was recently shown to interact with and negatively regulate MyoD (Mal, 2006), with the myogenin locus. ChIP assays on differentiating C2C12 myoblasts revealed increased recruitment of endogenous KMT1A to the myogenin promoter after p38-γ activation (Fig. 7 B). Importantly, this result was specific to activation of p38-γ, as expression of only MKK6EE, which results in p38-α activation, precluded detection of KMT1A on the myogenin promoter (Fig. S3 G).
Notably, we observed increased KMT1A detection on the myogenin promoter after ChIP assays from 10T1/2 fibroblasts expressing MKK6EE, p38-γ, and MyoD, compared with MyoD on its own (Fig. 7 B). Strikingly, KMT1A recruitment was severely inhibited by expression of the MyoD S199A/S200A mutant together with MKK6EE and p38-γ (Fig. 7 B), which confirms that its recruitment is dependent on MyoD phosphorylation.
To investigate whether MyoD interacts with its corepressor KMT1A, as has previously been described (Mal, 2006), we coexpressed MyoD and KMT1A in 293T cells, and were able to detect KMT1A by immunoblotting after MyoD IP from nuclear extracts (Figs. 7 C and S3 H). Moreover, we observed an almost 70% decrease in the intensity of this interaction when the MyoD S199A/S200A mutant was expressed (Fig. 7 C).
To further ascertain the significance of p38-γ signaling in assembling a repressive transcriptional complex, we examined KMT1A recruitment to the myogenin promoter in proliferating satellite cell–derived myoblasts. Notably, KMT1A binding was severely attenuated in p38-γ–deficient myoblasts (Fig. 7 D), establishing a physiological role for p38-γ signaling in KMT1A recruitment during myogenic precursor cell proliferation.
Collectively, these results establish that p38-γ phosphorylation of MyoD results in the assembly of a repressive transcriptional complex for the purpose of temporally regulating myogenin gene expression during the myogenic differentiation program. Furthermore, this suppression allows for appropriate expansion of the transient amplifying population of myogenic precursor cells during regenerative myogenesis.
Discussion
For skeletal muscle differentiation to proceed, MyoD must directly activate transcription of myogenin (Hollenberg et al., 1993). However, MyoD is also required for myogenic precursor cell specification (Rudnicki et al., 1992), and therefore binds and regulates target genes in proliferating myoblasts (Blais et al., 2005; Ishibashi et al., 2005). As a result, the signals regulating MyoD–cofactor interactions are crucial in defining its regulation of these two distinct processes.
We have established for the first time that p38-γ signaling is responsible for assembling both MyoD and KMT1A on the myogenin promoter in a MyoD phosphorylation–dependent manner, which results in methylation of H3K9 and maintenance of a transcriptionally nonpermissive chromatin state. These results are supported by the observation that deletion of a region of MyoD containing the p38-γ–targeted serine residues results in a sevenfold increase in endogenous myogenin expression (Gerber et al., 1997). Moreover, KMT1A was also shown to interact with and repress the transcriptional activity of MyoD during myoblast proliferation (Mal, 2006).
Although our results show that p38-γ signaling promotes H3K9 methylation of the myogenin promoter, we cannot ascertain whether this represents the initiating event in myogenin repression or if it is involved in maintenance of the repressive state. In Schizosaccharomyces pombe, it has been documented that transcription can influence local chromatin structure through an RNAi-dependent pathway (Cam et al., 2009); however, we are unaware of any studies of this mechanism outside of centromeric heterochromatin in mammalian cells. Nevertheless, our data indicate that MyoD phosphorylation-dependent H3K9 methylation is still an important event in myogenin regulation, and it will be very interesting to determine if similar pathways to those described in yeast are active in muscle to control gene expression through facultative heterochromatin.
To equate our results with a biological role in vivo, we established that in the absence of p38-γ, premature myogenin expression severely impairs expansion of transient amplifying myogenic precursors by forcing these cells to initiate the differentiation program (Fig. S4). Subsequently, mutant cells become stalled as a result of low myocyte numbers available for fusion. In fact, we detected diminished MyHC expression in p38-γ−/− myocytes, which is in agreement with a recent paper by Perdiguero et al., (2007).
We recently demonstrated that the satellite cell pool was composed of two hierarchical subpopulations: satellite stem cells and satellite myogenic cells (Kuang et al., 2007). It is interesting that the reduced numbers of satellite cells observed in p38-γ−/− muscle are proposed to be caused by the premature initiation of differentiation in the transient amplifying satellite myogenic cell population. It will be interesting for future studies to address whether a subset of satellite stem cells can actually differentiate directly into myocytes independently of satellite myogenic cells in p38-γ–deficient muscle.
Although Myogenin is essential for embryonic and fetal myogenesis (Hasty et al., 1993; Nabeshima et al., 1993; Meadows et al., 2008), Knapp et al., (2006) found that it is not required for adult myogenesis. However, in the absence of myogenin, fiber size is dramatically reduced, and differentiating myoblasts display aberrant gene expression patterns (Meadows et al., 2008), which suggests that differentiation is not proceeding optimally. Moreover, the contribution of the remaining myogenic regulatory factors to differentiation in this mutant is unclear (Meadows et al., 2008). Our data, together with papers documenting the role of Myogenin in patterning differentiation-specific gene expression through feed-forward networks with MyoD (Bergstrom et al., 2002; Penn et al., 2004; Blais et al., 2005; Cao et al., 2006) and repression of specification factors (Olguin et al., 2007), suggest that Myogenin is indeed an important regulator of differentiation, and that its suppression during proliferation is critical to allow for the appropriate expansion of myogenic precursor cells.
We observed that p38-γ is expressed and active during both proliferation and differentiation (Fig. S5, A and B). We therefore hypothesize that myogenin expression is not the only target of p38-γ–mediated repression, and this may aid in the temporal patterning of myogenic gene expression. Moreover, there must be additional mechanisms to specifically target p38-γ to MyoD, and for MKK6 to activate p38-γ and not p38-α.
For differentiation, the association between p38-α and its scaffold JLP controls its specific activation downstream of the cell surface receptor Cdo (Takaesu et al., 2006; Kang et al., 2008). We propose the existence of a p38-γ–specific scaffold, which functions during myogenic precursor cell proliferation to specifically activate p38-γ by promoting its association with MKK6. Moreover, this p38-γ activation mechanism could either occur independently of MyoD or be a common factor that binds MyoD, p38-γ, and MKK6, such as a MyoD cofactor. Future studies aimed at identifying these factors will be of the utmost interest.
Recently, Perdiguero et al. (2007) and Ruiz-Bonilla et al. (2008) analyzed p38-γ–deficient muscle and obtained differing conclusions compared with our results. For instance, Ruiz-Bonilla et al., (2008) concluded that regeneration occurs normally based on fiber size, whereas our conclusions about impaired regeneration were based on fiber number. Moreover, when we examined fiber size, we observed only a very small decrease in p38-γ mutant muscle. In addition, Perdiguero et al. (2007) noted a fusion defect in cultured p38-γ–deficient myoblasts, which we also detected. However, Ruiz-Bonilla et al., (2008) reported no difference in the growth, differentiation, or gene expression of these myoblasts. Although the exact reasons for these discrepancies is not clear, we speculate that this could be caused by the differences in satellite cell isolation, concentration of growth factors used in culture, or the use of wild-type versus heterozygous mice as baseline controls.
Combining IP-kinase assays with mass spectrometry, we identified Ser199 and Ser200 of MyoD as novel targets of p38-γ phosphorylation. Ser200, but not Ser199, is also a phosphorylation site for the cyclin-dependent kinases Cdk1 and Cdk2 during myoblast proliferation (Song et al., 1998; Kitzmann et al., 1999; Tintignac et al., 2004). Moreover, Ser5 in the N terminus of MyoD is also a target of Cdk phosphorylation, a modification we do not detect in response to p38-γ activation. This suggests that our p38-γ phosphorylation results are unique and not simply caused by p38-γ or MKK6 activation of Cdk1 or Cdk2, followed by their phosphorylation of MyoD.
Notably, p38-γ can be localized to the membrane of differentiated muscle fibers by interacting with α1-syntrophin (Hasegawa et al., 1999), a component of the dystrophin glycoprotein complex. In the absence of the dystrophin glycoprotein complex, such as that seen with Duchenne muscular dystrophy, α1-syntrophin fails to localize to the sarcolemma. We would therefore predict that this would also result in mislocalization of p38-γ in dystrophic muscle, which could affect its activation state, and have detrimental consequences on the pathology of this disease. To what extent dysregulated p38-γ signaling contributes to the loss of muscle fibers in Duchenne muscular dystrophy is a question of significant therapeutic relevance.
Materials and methods
Mice and satellite cell isolation
All experiments were performed in accordance with the University of Ottawa Animal Care Committee regulations. Mice lacking p38-γ have been described previously (Sabio et al., 2005). To induce activation of satellite cells in vivo, 4–5-wk-old age- and sex-matched p38-γ+/+ and p38-γ−/− mice (C57BL/6 background) were injected with 25 µl of 10 µM CTX (Latoxan; Seale et al., 2004) into the TA muscle and examined 21 d later, both in a blind manner. TA muscles from both the injected and contralateral control leg from each mouse were fixed in 4% PFA and subsequently processed by a professional pathology laboratory at the University of Ottawa in a blind manner. Specifically, TA muscles were lined up and bisected at the same site between muscles, embedded in paraffin, cross sectioned away from the cut site, and stained with hematoxylin and eosin according to standard pathology protocols. Slides were returned to us, and images were acquired at room temperature using a microscope (Axioskop) with a 10× 0.3 NA Plan-Neofluar (0.17 mm coverslip) objective (Carl Zeiss, Inc.), and full TA cross sections were reconstructed using Photoshop (Adobe), all in a blind manner. For fiber number counts, fibers across the entire cross section were counted in a blind manner. For fiber area measurements, three random fields from each muscle were analyzed using ImageJ software (National Institutes of Health), also in a blind manner. All injured muscles were compared directly with their respective uninjured contralateral TA. Error bars were calculated using ±SEM, and significance was measured using the Student's t test for fiber number analysis and Welch's t test for fiber size data. Satellite cell–derived primary myoblasts were isolated from 4–6-wk-old age- and sex-matched p38-γ+/+ and p38-γ−/− mice by FACS, gating on the α7-integrin–positive, CD31/Sca1/CD45-negative population as described previously (Kuang et al., 2007). Isolation of satellite cells on intact myofibers from EDL muscle was performed as described previously (Rosenblatt et al., 1995). In brief, EDL muscle was partially dissociated with 0.2% collagenase I in DME at 35°C for 30–60 min, and single fibers were isolated by trituration using flame-polished Pasteur pipettes of varying bore widths, then immediately fixed in 4% PFA (Kuang et al., 2007).
Cell culture and transfections
C2C12 myoblasts and C3H10T1/2 fibroblasts were maintained in DME with 10% FCS and 1× penicillin/streptomycin. Cells were transfected using the calcium phosphate protocol as described previously (Perry et al., 2001). 24 h after transfection, cells were switched to differentiation media (DME, 2% horse serum, and 1× penicillin/streptomycin) for another 24 h, and subsequently harvested. Satellite cell–derived primary myoblasts were cultured in Ham's F10 media supplemented with 20% FCS, 1× penicillin/streptomycin, and 2.5 ng/ml basic FGF, and were differentiated in DME supplemented with 5% horse serum and 1× penicillin/streptomycin. siRNA knockdown experiments were performed on intact EDL myofibers as described previously (Holterman et al., 2007).
Plasmids
MKK6EE (S207E/T211E) was generated by overlapping PCR and cloned into an HA-pcDNA3 vector. Mouse p38-γ was isolated and cloned into a Myc-pcDNA3 vector. This cloning was performed by R. Perry (York University, Toronto, Canada). MyoD cDNA (Davis et al., 1987) was expressed under control of the phosphoglycerate kinase promoter. MyoD S199A/S200A was generated by overlapping PCR and expressed from the phosphoglycerate kinase promoter. Recombinant GST proteins were generated by cloning respective cDNAs into the pGEX4T plasmid. The Myogenin-luciferase reporter was a gift from S. Tapscott (Fred Hutchinson Cancer Research Center, Seattle, WA; Berkes et al., 2004). Flag–p38-α was a gift from J. Han (The Scripps Research Institute, La Jolla, CA; Han et al., 1994). Myc- and Flag-KMT1A vectors were gifts from M. Horwitz (University of Washington, Seattle, WA)/T. Jenuwein (Max Planck Institute of Immunology, Freiburg, Germany) and A. Mal (Roswell Park Cancer Institute, Buffalo, NY), respectively (Rea et al., 2000; Mal, 2006).
IP-kinase assays
To extract protein from 10T1/2 fibroblasts, cells were washed twice with PBS containing 100 mM NaF and 1 mM Na3VO4, scraped, pelleted, lysed with NP-40 buffer (50 mM Tris, pH 8.0, 150 mM NaCl, 2 mM EDTA, 100 mM NaF, 10 mM sodium pyrophosphate, and 0.5% NP-40) supplemented with 10 µg/ml aprotinin, 10 µg/ml leupeptin, 10 µg/ml pepstatin A, 1 mM PMSF, and 1 mM Na3VO4, then clarified by centrifugation as described previously (Perry et al., 2001). Myc 9E10 immunoprecipitates were washed with NP-40 buffer and kinase buffer (25 mM Hepes, pH 7.6, 20 mM MgCl2, 10 mM β-glycerophosphate, 2 mM DTT, 0.1 mM Na3VO4, and 1 mM NaF). Reactions were performed in kinase buffer containing 25 µM cold ATP, 2.5 µg of recombinant substrate, and 5 µCi [γ32P]ATP for 30 min at 30°C. Reactions were stopped with Laemmli protein sample buffer (100 mM Tris, pH 6.8, 20% glycerol, 4% SDS, 2% bromophenol blue, and 200 mM DTT). Samples were separated by SDS-PAGE, stained with Coomassie blue, destained, washed with 10% PEG, dried, and visualized by autoradiography. Coomassie blue–stained blots are shown in Fig. S5 (C and D).
Mass spectrometry
IP-kinase assays were performed as described in “IP-kinase assays,” proteins were digested in solution with AspN, and phosphopeptides were enriched by immobilized metal ion affinity chromatography and analyzed by MALDI-TOF MS (all steps including and following digestion were performed by Custom Biologics). Resulting m/z peaks were compared with theoretical digest of GST–MyoD–C terminal (GST-MyoD-CT) containing all possible posttranslational modifications. For in vivo studies, nuclear extracts were prepared as described previously (Dignam et al., 1983; McKinnell et al., 2008). In brief, cells were swollen in Hypotonic buffer (10 mM Hepes, pH 7.6, 10 mM KCl, 1.5 mM MgCl2, 0.5 mM DTT, and 50 mM NaF) containing protease inhibitors and lysed with a 27-gauge needle. After centrifugation to collect intact nuclei, nuclear contents were extracted in 20 mM Hepes, pH 7.6, 420 mM NaCl, 25% glycerol, 1.5 mM MgCl2, 2 mM DTT, 0.2 mM EDTA, 50 mM NaF, and protease inhibitors using a 28-gauge insulin syringe, then agitated for 60 min at 4°C. Nuclear extracts were clarified by centrifugation and dialyzed to reduce the concentrations of NaCl to 150 mM, glycerol to 20%, and Hepes to 15 mM. MyoD 5.8A antibody (BD) was cross-linked to protein G Dynabeads (Invitrogen) using dimethyl pimelimidate (Thermo Fisher Scientific) according to the manufacturer's instructions. MyoD immunoprecipitates were separated by SDS-PAGE, stained with Silver (SilverQuest; Invitrogen) or Coomassie Blue (SimplyBlue SafeStain; Invitrogen), excised, digested with Trypsin (Promega), separated by online reversed-phase high-performance liquid chromatography with an increasing gradient of 2–35% acetonitrile over 90 min, and analyzed using a mass spectrometer (LTQ-Orbitrap; Thermo Fisher Scientific). Peptides were identified by searching MS/MS data against the IPI murine database (v3.38; downloaded January 31, 2008) using X!Tandem (Thermo Fisher Scientific), and probabilities were determined by PeptideProphet (Keller et al., 2002) and ProteinProphet (Nesvizhskii et al., 2003). Peptide abundances were normalized to four different peptides on both MyoD and α-actin.
Co-IP
MyoD-KMT1A co-IPs were performed as described for in vivo mass spectrometry, except that gels were transferred for Western blotting rather than stained.
ChIP assays
Protein–DNA complexes were cross-linked with a final concentration of 1% formaldehyde using a 10× fixation buffer (50 mM Hepes, pH 8.0, 1 mM EDTA, 0.5 mM EGTA, 100 mM NaCl, and 11% formaldehyde) for 10 min at room temperature. Reactions were quenched with 125 mM glycine for 4–5 min, and cells were harvested as described earlier (see “IP-kinase assays”) and resuspended in buffer to lyse nuclei (40 mM Tris, pH 8.0, 1% Triton X-100, 4 mM EDTA, and 300 mM NaCl). Lysates were sonicated using a tip sonicator and clarified by centrifugation, then the NaCl and Triton X-100 concentrations were diluted to 150 mM and 0.5%, respectively. 1,000–2,000 µg of protein–DNA complexes were immunoprecipitated with 4–5 µg of antibody overnight. Antibody–protein–DNA complexes were collected, washed, and eluted, and cross links were reversed according to manufacturer's instructions (Millipore). DNA was purified using PCR purification columns (QIAGEN, Invitrogen), and PCR was performed using primers for the myogenin promoter (forward, 5′-CAGGTTTCTGTGGCGTTGGC-3′; reverse, 5′-CATACAGCTCCATCAGGTCGC-3′) and Igh enhancer (Bergstrom et al., 2002). Input DNA was amplified for 25 cycles, whereas experimental DNA was amplified between 30–35 cycles. Quantifications were performed using densitometry and represent averages from three independent experiments, with error bars representing ±SEM.
Reporter assays
Transfected cells were resuspended in 1× reporter lysis buffer (Promega) and subjected to three rounds of freeze thaw lysis (dry ice/EtOH and 37°C water bath). Luciferase assays were performed according to the manufacturer's instructions (Promega) using 5–10 µl of protein extract per reaction, and analyzed on a luminometer (Lumat LB9507; EG & G Berthold). Protein concentrations were determined using the Bradford assay, and relative light units were normalized to this value as described previously (Parker et al., 2006). Fold activation was calculated relative to the reporter-only transfected cells.
Western blotting and antibodies
Western blotting was performed as described previously (Perry et al., 2001). We used the following antibodies for immunoblotting, immunofluorescence, IP, and ChIP: anti-phospho-p38 (Cell Signaling Technology), anti-MKK6 (Assay Designs), anti–p38-γ (Millipore), anti-MyoD (C20; Santa Cruz Biotechnology, Inc.), anti-MyoD (5.2F; Sigma-Aldrich), anti-MyoD (5.8A; BD), anti-MyoD phospho Ser200 (Novus Biologicals), anti-Myogenin (F5D; Developmental Studies Hybridoma Bank), anti-Myogenin (M225; Santa Cruz Biotechnology, Inc.), anti-Pax7 (Developmental Studies Hybridoma Bank), anti-tubulin (DM1A; Sigma-Aldrich), anti-Myc (9E10; Santa Cruz Biotechnology, Inc.), anti-Ki67 (Abcam), anti-H3K9-2me (Millipore), and anti-KMT1A/Suv39h1 (Millipore). For phospho–p38-γ detection, the phospho-p38 antibody was found to cross-react with phosphorylated p38-γ only upon overexpression.
Immunochemistry
Immunocytochemistry was performed essentially as described previously (Parker et al., 2006). In brief, cells were fixed with 4% PFA for 5–10 min, quenched with 125 mM glycine for 5 min, permeabilized with 0.3% Triton X-100 in PBS, and blocked in 3% BSA in PBS before the addition of primary (2 h) and secondary antibody (45 min). Nuclei were counterstained with DAPI (Sigma-Aldrich) and mounted with fluorescence mounting media (Dako). TUNEL staining was performed using the in situ Cell Death Detection kit (Roche) according to the manufacturer's instructions. For immunohistochemistry, TA muscles were fixed in 4% PFA for 20 min, embedded in 2:1 optimum cutting temperature liquid/20% sucrose, and frozen with isopentane cooled in a dry ice and ethanol bath. Cross sections were cut using a Cryostat at a thickness of 8 µm. Sections were blocked for 2 h with MOM blocking reagent (Vector Laboratories) and incubated with primary antibody overnight in 5% goat serum. After washes with PBS containing 0.2% Tween 20, sections were incubated with the appropriate secondary antibody (1:1,000, Alexa Fluor fluorophores; Invitrogen), and nuclei were counterstained with DAPI (1:50,000) in PBS. Slides were coverslipped using fluorescence mounting media (Dako). For single fiber isolations from the EDL, fibers were immediately fixed after dissociation from the muscle in 4% PFA, then quenched/permeabilized with 0.2% Triton X-100 in 100 mM glycine. Nonspecific staining was blocked by incubation with 5% goat serum for 1 h. Staining was performed as indicated earlier. Pax7 antibody (Developmental Studies Hybridoma Bank) was used at a 1:10 dilution, whereas Myogenin antibody (sc578; Santa Cruz Biotechnology, Inc.) was used at a 1:100 dilution. Staining was visualized at room temperature on a microscope (Axioplan 2) using a 20× 0.75 NA Plan-Apochromat (0.17 mm coverslip) objective, a camera (AxioCam), and Axioview 3.1 software (Carl Zeiss, Inc.). Image levels were equally adjusted using Photoshop software (Adobe).
RT quantitative PCR
RNA was extracted from freshly sorted and cultured satellite cells using an RNeasy micro kit (QIAGEN), then reverse-transcribed using random hexamer primers and Superscript II enzyme (Invitrogen). RT-PCR reactions and analysis were performed as described previously (Ishibashi et al., 2005).
Online supplemental material
Fig. S1 shows reduced fiber size after regeneration in p38-γ–deficient muscle. Fig. S2 shows reduced proliferation and elevated apoptosis in p38-γ–deficient satellite cells. Fig. S3 shows mechanistic effects of p38-γ phosphorylation. Fig. S4 shows a model demonstrating the biological role of p38-γ in regulating entry into the myogenic differentiation program. Fig. S5 shows p38-γ activity and expression during differentiation. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200907037/DC1.
Acknowledgments
We thank Robert Perry for expert guidance; Daniel Mamelak of Custom Biologics; Hamid Mirzaei, Min Yuan, and Carey Sheu of Institute for Systems Biology for MS analysis; Li Dong and Zaida Ticas of University of Ottawa Pathology for tissue processing; and Iain McKinnell, Jeff Ishibashi, and F. Jeffrey Dilworth for critical reading of the manuscript.
M.A. Rudnicki holds the Canada Research Chair in Molecular Genetics and is an International Research Scholar of the Howard Hughes Medical Institute. This work was supported by grants to M.A. Rudnicki from the Canadian Institutes of Health Research (MOP12080) and the Canada Research Chair Program.
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin IP
- CTX
- cardiotoxin
- EDL
- extensor digitorum longus
- HEB
- HeLa E box binding factor
- IP
- immunoprecipitation
- LC-MS/MS
- liquid chromatography tandem mass spectrometry
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- MAPK
- mitogen-activated protein kinase
- MyHC
- myosin heavy chain
- TA
- tibialis anterior
References
- Aebersold R., Goodlett D.R. 2001. Mass spectrometry in proteomics. Chem. Rev. 101:269–295 10.1021/cr990076h [DOI] [PubMed] [Google Scholar]
- Bergstrom D.A., Penn B.H., Strand A., Perry R.L., Rudnicki M.A., Tapscott S.J. 2002. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol. Cell. 9:587–600 10.1016/S1097-2765(02)00481-1 [DOI] [PubMed] [Google Scholar]
- Berkes C.A., Bergstrom D.A., Penn B.H., Seaver K.J., Knoepfler P.S., Tapscott S.J. 2004. Pbx marks genes for activation by MyoD indicating a role for a homeodomain protein in establishing myogenic potential. Mol. Cell. 14:465–477 10.1016/S1097-2765(04)00260-6 [DOI] [PubMed] [Google Scholar]
- Blais A., Tsikitis M., Acosta-Alvear D., Sharan R., Kluger Y., Dynlacht B.D. 2005. An initial blueprint for myogenic differentiation. Genes Dev. 19:553–569 10.1101/gad.1281105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boppart M.D., Asp S., Wojtaszewski J.F., Fielding R.A., Mohr T., Goodyear L.J. 2000. Marathon running transiently increases c-Jun NH2-terminal kinase and p38 activities in human skeletal muscle. J. Physiol. 526:663–669 10.1111/j.1469-7793.2000.00663.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briata P., Forcales S.V., Ponassi M., Corte G., Chen C.Y., Karin M., Puri P.L., Gherzi R. 2005. p38-dependent phosphorylation of the mRNA decay-promoting factor KSRP controls the stability of select myogenic transcripts. Mol. Cell. 20:891–903 10.1016/j.molcel.2005.10.021 [DOI] [PubMed] [Google Scholar]
- Cam H.P., Chen E.S., Grewal S.I. 2009. Transcriptional scaffolds for heterochromatin assembly. Cell. 136:610–614 10.1016/j.cell.2009.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y., Kumar R.M., Penn B.H., Berkes C.A., Kooperberg C., Boyer L.A., Young R.A., Tapscott S.J. 2006. Global and gene-specific analyses show distinct roles for Myod and Myog at a common set of promoters. EMBO J. 25:502–511 10.1038/sj.emboj.7600958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caretti G., Schiltz R.L., Dilworth F.J., Di Padova M., Zhao P., Ogryzko V., Fuller-Pace F.V., Hoffman E.P., Tapscott S.J., Sartorelli V. 2006. The RNA helicases p68/p72 and the noncoding RNA SRA are coregulators of MyoD and skeletal muscle differentiation. Dev. Cell. 11:547–560 10.1016/j.devcel.2006.08.003 [DOI] [PubMed] [Google Scholar]
- Cheng T.C., Wallace M.C., Merlie J.P., Olson E.N. 1993. Separable regulatory elements governing myogenin transcription in mouse embryogenesis. Science. 261:215–218 10.1126/science.8392225 [DOI] [PubMed] [Google Scholar]
- Cuenda A., Cohen P. 1999. Stress-activated protein kinase-2/p38 and a rapamycin-sensitive pathway are required for C2C12 myogenesis. J. Biol. Chem. 274:4341–4346 10.1074/jbc.274.7.4341 [DOI] [PubMed] [Google Scholar]
- Cuenda A., Cohen P., Buée-Scherrer V., Goedert M. 1997. Activation of stress-activated protein kinase-3 (SAPK3) by cytokines and cellular stresses is mediated via SAPKK3 (MKK6); comparison of the specificities of SAPK3 and SAPK2 (RK/p38). EMBO J. 16:295–305 10.1093/emboj/16.2.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R.L., Weintraub H., Lassar A.B. 1987. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 51:987–1000 10.1016/0092-8674(87)90585-X [DOI] [PubMed] [Google Scholar]
- de la Serna I.L., Carlson K.A., Imbalzano A.N. 2001. Mammalian SWI/SNF complexes promote MyoD-mediated muscle differentiation. Nat. Genet. 27:187–190 10.1038/84826 [DOI] [PubMed] [Google Scholar]
- de la Serna I.L., Ohkawa Y., Berkes C.A., Bergstrom D.A., Dacwag C.S., Tapscott S.J., Imbalzano A.N. 2005. MyoD targets chromatin remodeling complexes to the myogenin locus prior to forming a stable DNA-bound complex. Mol. Cell. Biol. 25:3997–4009 10.1128/MCB.25.10.3997-4009.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato M.D., Tjian R. 2007. Switching of the core transcription machinery during myogenesis. Genes Dev. 21:2137–2149 10.1101/gad.1583407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato M.D., Marr M.T., Sottero T., Inouye C., Hu P., Tjian R. 2008. MyoD targets TAF3/TRF3 to activate myogenin transcription. Mol. Cell. 32:96–105 10.1016/j.molcel.2008.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dignam J.D., Lebovitz R.M., Roeder R.G. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11:1475–1489 10.1093/nar/11.5.1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth F.J., Seaver K.J., Fishburn A.L., Htet S.L., Tapscott S.J. 2004. In vitro transcription system delineates the distinct roles of the coactivators pCAF and p300 during MyoD/E47-dependent transactivation. Proc. Natl. Acad. Sci. USA. 101:11593–11598 10.1073/pnas.0404192101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerber A.N., Klesert T.R., Bergstrom D.A., Tapscott S.J. 1997. Two domains of MyoD mediate transcriptional activation of genes in repressive chromatin: a mechanism for lineage determination in myogenesis. Genes Dev. 11:436–450 10.1101/gad.11.4.436 [DOI] [PubMed] [Google Scholar]
- Halevy O., Novitch B.G., Spicer D.B., Skapek S.X., Rhee J., Hannon G.J., Beach D., Lassar A.B. 1995. Correlation of terminal cell cycle arrest of skeletal muscle with induction of p21 by MyoD. Science. 267:1018–1021 10.1126/science.7863327 [DOI] [PubMed] [Google Scholar]
- Han J., Lee J.D., Bibbs L., Ulevitch R.J. 1994. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 265:808–811 10.1126/science.7914033 [DOI] [PubMed] [Google Scholar]
- Hasegawa M., Cuenda A., Spillantini M.G., Thomas G.M., Buée-Scherrer V., Cohen P., Goedert M. 1999. Stress-activated protein kinase-3 interacts with the PDZ domain of alpha1-syntrophin. A mechanism for specific substrate recognition. J. Biol. Chem. 274:12626–12631 10.1074/jbc.274.18.12626 [DOI] [PubMed] [Google Scholar]
- Hasty P., Bradley A., Morris J.H., Edmondson D.G., Venuti J.M., Olson E.N., Klein W.H. 1993. Muscle deficiency and neonatal death in mice with a targeted mutation in the myogenin gene. Nature. 364:501–506 10.1038/364501a0 [DOI] [PubMed] [Google Scholar]
- Hollenberg S.M., Cheng P.F., Weintraub H. 1993. Use of a conditional MyoD transcription factor in studies of MyoD trans-activation and muscle determination. Proc. Natl. Acad. Sci. USA. 90:8028–8032 10.1073/pnas.90.17.8028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holterman C.E., Le Grand F., Kuang S., Seale P., Rudnicki M.A. 2007. Megf10 regulates the progression of the satellite cell myogenic program. J. Cell Biol. 179:911–922 10.1083/jcb.200709083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishibashi J., Perry R.L., Asakura A., Rudnicki M.A. 2005. MyoD induces myogenic differentiation through cooperation of its NH2- and COOH-terminal regions. J. Cell Biol. 171:471–482 10.1083/jcb.200502101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones N.C., Tyner K.J., Nibarger L., Stanley H.M., Cornelison D.D., Fedorov Y.V., Olwin B.B. 2005. The p38α/β MAPK functions as a molecular switch to activate the quiescent satellite cell. J. Cell Biol. 169:105–116 10.1083/jcb.200408066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J.S., Bae G.U., Yi M.J., Yang Y.J., Oh J.E., Takaesu G., Zhou Y.T., Low B.C., Krauss R.S. 2008. A Cdo-Bnip-2-Cdc42 signaling pathway regulates p38α/β MAPK activity and myogenic differentiation. J. Cell Biol. 182:497–507 10.1083/jcb.200801119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Nesvizhskii A.I., Kolker E., Aebersold R. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- Kitzmann M., Vandromme M., Schaeffer V., Carnac G., Labbé J.C., Lamb N., Fernandez A. 1999. cdk1- and cdk2-mediated phosphorylation of MyoD Ser200 in growing C2 myoblasts: role in modulating MyoD half-life and myogenic activity. Mol. Cell. Biol. 19:3167–3176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapp J.R., Davie J.K., Myer A., Meadows E., Olson E.N., Klein W.H. 2006. Loss of myogenin in postnatal life leads to normal skeletal muscle but reduced body size. Development. 133:601–610 10.1242/dev.02249 [DOI] [PubMed] [Google Scholar]
- Kuang S., Kuroda K., Le Grand F., Rudnicki M.A. 2007. Asymmetric self-renewal and commitment of satellite stem cells in muscle. Cell. 129:999–1010 10.1016/j.cell.2007.03.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner C., Zahalka M.A., Giot J.F., Møller N.P., Ullrich A. 1996. ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation. Proc. Natl. Acad. Sci. USA. 93:4355–4359 10.1073/pnas.93.9.4355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluís F., Ballestar E., Suelves M., Esteller M., Muñoz-Cánoves P. 2005. E47 phosphorylation by p38 MAPK promotes MyoD/E47 association and muscle-specific gene transcription. EMBO J. 24:974–984 10.1038/sj.emboj.7600528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A.K. 2006. Histone methyltransferase Suv39h1 represses MyoD-stimulated myogenic differentiation. EMBO J. 25:3323–3334 10.1038/sj.emboj.7601229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A., Harter M.L. 2003. MyoD is functionally linked to the silencing of a muscle-specific regulatory gene prior to skeletal myogenesis. Proc. Natl. Acad. Sci. USA. 100:1735–1739 10.1073/pnas.0437843100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mal A., Sturniolo M., Schiltz R.L., Ghosh M.K., Harter M.L. 2001. A role for histone deacetylase HDAC1 in modulating the transcriptional activity of MyoD: inhibition of the myogenic program. EMBO J. 20:1739–1753 10.1093/emboj/20.7.1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnell I.W., Ishibashi J., Le Grand F., Punch V.G., Addicks G.C., Greenblatt J.F., Dilworth F.J., Rudnicki M.A. 2008. Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex. Nat. Cell Biol. 10:77–84 10.1038/ncb1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows E., Cho J.H., Flynn J.M., Klein W.H. 2008. Myogenin regulates a distinct genetic program in adult muscle stem cells. Dev. Biol. 322:406–414 10.1016/j.ydbio.2008.07.024 [DOI] [PubMed] [Google Scholar]
- Megeney L.A., Kablar B., Garrett K., Anderson J.E., Rudnicki M.A. 1996. MyoD is required for myogenic stem cell function in adult skeletal muscle. Genes Dev. 10:1173–1183 10.1101/gad.10.10.1173 [DOI] [PubMed] [Google Scholar]
- Mertens S., Craxton M., Goedert M. 1996. SAP kinase-3, a new member of the family of mammalian stress-activated protein kinases. FEBS Lett. 383:273–276 10.1016/0014-5793(96)00255-4 [DOI] [PubMed] [Google Scholar]
- Molkentin J.D., Black B.L., Martin J.F., Olson E.N. 1995. Cooperative activation of muscle gene expression by MEF2 and myogenic bHLH proteins. Cell. 83:1125–1136 10.1016/0092-8674(95)90139-6 [DOI] [PubMed] [Google Scholar]
- Nabeshima Y., Hanaoka K., Hayasaka M., Esumi E., Li S., Nonaka I., Nabeshima Y. 1993. Myogenin gene disruption results in perinatal lethality because of severe muscle defect. Nature. 364:532–535 10.1038/364532a0 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii A.I., Keller A., Kolker E., Aebersold R. 2003. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75:4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- Olguin H.C., Yang Z., Tapscott S.J., Olwin B.B. 2007. Reciprocal inhibition between Pax7 and muscle regulatory factors modulates myogenic cell fate determination. J. Cell Biol. 177:769–779 10.1083/jcb.200608122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.H., Perry R.L., Fauteux M.C., Berkes C.A., Rudnicki M.A. 2006. MyoD synergizes with the E-protein HEB beta to induce myogenic differentiation. Mol. Cell. Biol. 26:5771–5783 10.1128/MCB.02404-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn B.H., Bergstrom D.A., Dilworth F.J., Bengal E., Tapscott S.J. 2004. A MyoD-generated feed-forward circuit temporally patterns gene expression during skeletal muscle differentiation. Genes Dev. 18:2348–2353 10.1101/gad.1234304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdiguero E., Ruiz-Bonilla V., Gresh L., Hui L., Ballestar E., Sousa-Victor P., Baeza-Raja B., Jardí M., Bosch-Comas A., Esteller M., et al. 2007. Genetic analysis of p38 MAP kinases in myogenesis: fundamental role of p38alpha in abrogating myoblast proliferation. EMBO J. 26:1245–1256 10.1038/sj.emboj.7601587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry R.L., Parker M.H., Rudnicki M.A. 2001. Activated MEK1 binds the nuclear MyoD transcriptional complex to repress transactivation. Mol. Cell. 8:291–301 10.1016/S1097-2765(01)00302-1 [DOI] [PubMed] [Google Scholar]
- Puri P.L., Avantaggiati M.L., Balsano C., Sang N., Graessmann A., Giordano A., Levrero M. 1997a. p300 is required for MyoD-dependent cell cycle arrest and muscle-specific gene transcription. EMBO J. 16:369–383 10.1093/emboj/16.2.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P.L., Sartorelli V., Yang X.J., Hamamori Y., Ogryzko V.V., Howard B.H., Kedes L., Wang J.Y., Graessmann A., Nakatani Y., Levrero M. 1997b. Differential roles of p300 and PCAF acetyltransferases in muscle differentiation. Mol. Cell. 1:35–45 10.1016/S1097-2765(00)80005-2 [DOI] [PubMed] [Google Scholar]
- Puri P.L., Iezzi S., Stiegler P., Chen T.T., Schiltz R.L., Muscat G.E., Giordano A., Kedes L., Wang J.Y., Sartorelli V. 2001. Class I histone deacetylases sequentially interact with MyoD and pRb during skeletal myogenesis. Mol. Cell. 8:885–897 10.1016/S1097-2765(01)00373-2 [DOI] [PubMed] [Google Scholar]
- Raingeaud J., Whitmarsh A.J., Barrett T., Dérijard B., Davis R.J. 1996. MKK3- and MKK6-regulated gene expression is mediated by the p38 mitogen-activated protein kinase signal transduction pathway. Mol. Cell. Biol. 16:1247–1255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampalli S., Li L., Mak E., Ge K., Brand M., Tapscott S.J., Dilworth F.J. 2007. p38 MAPK signaling regulates recruitment of Ash2L-containing methyltransferase complexes to specific genes during differentiation. Nat. Struct. Mol. Biol. 14:1150–1156 10.1038/nsmb1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber F., O'Carroll D., Strahl B.D., Sun Z.W., Schmid M., Opravil S., Mechtler K., Ponting C.P., Allis C.D., Jenuwein T. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 406:593–599 10.1038/35020506 [DOI] [PubMed] [Google Scholar]
- Rosenblatt J.D., Lunt A.I., Parry D.J., Partridge T.A. 1995. Culturing satellite cells from living single muscle fiber explants. In Vitro Cell. Dev. Biol. Anim. 31:773–779 10.1007/BF02634119 [DOI] [PubMed] [Google Scholar]
- Rudnicki M.A., Braun T., Hinuma S., Jaenisch R. 1992. Inactivation of MyoD in mice leads to up-regulation of the myogenic HLH gene Myf-5 and results in apparently normal muscle development. Cell. 71:383–390 10.1016/0092-8674(92)90508-A [DOI] [PubMed] [Google Scholar]
- Ruiz-Bonilla V., Perdiguero E., Gresh L., Serrano A.L., Zamora M., Sousa-Victor P., Jardí M., Wagner E.F., Muñoz-Cánoves P. 2008. Efficient adult skeletal muscle regeneration in mice deficient in p38beta, p38gamma and p38delta MAP kinases. Cell Cycle. 7:2208–2214 [DOI] [PubMed] [Google Scholar]
- Sabio G., Arthur J.S., Kuma Y., Peggie M., Carr J., Murray-Tait V., Centeno F., Goedert M., Morrice N.A., Cuenda A. 2005. p38gamma regulates the localisation of SAP97 in the cytoskeleton by modulating its interaction with GKAP. EMBO J. 24:1134–1145 10.1038/sj.emboj.7600578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabourin L.A., Girgis-Gabardo A., Seale P., Asakura A., Rudnicki M.A. 1999. Reduced differentiation potential of primary MyoD−/− myogenic cells derived from adult skeletal muscle. J. Cell Biol. 144:631–643 10.1083/jcb.144.4.631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartorelli V., Puri P.L., Hamamori Y., Ogryzko V., Chung G., Nakatani Y., Wang J.Y., Kedes L. 1999. Acetylation of MyoD directed by PCAF is necessary for the execution of the muscle program. Mol. Cell. 4:725–734 10.1016/S1097-2765(00)80383-4 [DOI] [PubMed] [Google Scholar]
- Seale P., Sabourin L.A., Girgis-Gabardo A., Mansouri A., Gruss P., Rudnicki M.A. 2000. Pax7 is required for the specification of myogenic satellite cells. Cell. 102:777–786 10.1016/S0092-8674(00)00066-0 [DOI] [PubMed] [Google Scholar]
- Seale P., Ishibashi J., Scimè A., Rudnicki M.A. 2004. Pax7 is necessary and sufficient for the myogenic specification of CD45+:Sca1+ stem cells from injured muscle. PLoS Biol. 2:E130 10.1371/journal.pbio.0020130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra C., Palacios D., Mozzetta C., Forcales S.V., Morantte I., Ripani M., Jones D.R., Du K., Jhala U.S., Simone C., Puri P.L. 2007. Functional interdependence at the chromatin level between the MKK6/p38 and IGF1/PI3K/AKT pathways during muscle differentiation. Mol. Cell. 28:200–213 10.1016/j.molcel.2007.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simone C., Forcales S.V., Hill D.A., Imbalzano A.N., Latella L., Puri P.L. 2004. p38 pathway targets SWI-SNF chromatin-remodeling complex to muscle-specific loci. Nat. Genet. 36:738–743 10.1038/ng1378 [DOI] [PubMed] [Google Scholar]
- Sims R.J., III, Mandal S.S., Reinberg D. 2004. Recent highlights of RNA-polymerase-II-mediated transcription. Curr. Opin. Cell Biol. 16:263–271 10.1016/j.ceb.2004.04.004 [DOI] [PubMed] [Google Scholar]
- Song A., Wang Q., Goebl M.G., Harrington M.A. 1998. Phosphorylation of nuclear MyoD is required for its rapid degradation. Mol. Cell. Biol. 18:4994–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu G., Kang J.S., Bae G.U., Yi M.J., Lee C.M., Reddy E.P., Krauss R.S. 2006. Activation of p38α/β MAPK in myogenesis via binding of the scaffold protein JLP to the cell surface protein Cdo. J. Cell Biol. 175:383–388 10.1083/jcb.200608031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapscott S.J. 2005. The circuitry of a master switch: Myod and the regulation of skeletal muscle gene transcription. Development. 132:2685–2695 10.1242/dev.01874 [DOI] [PubMed] [Google Scholar]
- Tapscott S.J., Davis R.L., Thayer M.J., Cheng P.F., Weintraub H., Lassar A.B. 1988. MyoD1: a nuclear phosphoprotein requiring a Myc homology region to convert fibroblasts to myoblasts. Science. 242:405–411 10.1126/science.3175662 [DOI] [PubMed] [Google Scholar]
- Tintignac L.A., Sirri V., Leibovitch M.P., Lécluse Y., Castedo M., Metivier D., Kroemer G., Leibovitch S.A. 2004. Mutant MyoD lacking Cdc2 phosphorylation sites delays M-phase entry. Mol. Cell. Biol. 24:1809–1821 10.1128/MCB.24.4.1809-1821.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorella L.L., Lin C.B., Pilch P.F. 2003. ERK6 is expressed in a developmentally regulated manner in rodent skeletal muscle. Biochem. Biophys. Res. Commun. 306:163–168 10.1016/S0006-291X(03)00936-7 [DOI] [PubMed] [Google Scholar]
- Wang H., Xu Q., Xiao F., Jiang Y., Wu Z. 2008. Involvement of the p38 mitogen-activated protein kinase alpha, beta, and gamma isoforms in myogenic differentiation. Mol. Biol. Cell. 19:1519–1528 10.1091/mbc.E07-08-0817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weintraub H., Genetta T., Kadesch T. 1994. Tissue-specific gene activation by MyoD: determination of specificity by cis-acting repression elements. Genes Dev. 8:2203–2211 10.1101/gad.8.18.2203 [DOI] [PubMed] [Google Scholar]
- Wright W.E., Sassoon D.A., Lin V.K. 1989. Myogenin, a factor regulating myogenesis, has a domain homologous to MyoD. Cell. 56:607–617 10.1016/0092-8674(89)90583-7 [DOI] [PubMed] [Google Scholar]
- Wu Z., Woodring P.J., Bhakta K.S., Tamura K., Wen F., Feramisco J.R., Karin M., Wang J.Y., Puri P.L. 2000. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 20:3951–3964 10.1128/MCB.20.11.3951-3964.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetser A., Gredinger E., Bengal E. 1999. p38 mitogen-activated protein kinase pathway promotes skeletal muscle differentiation. Participation of the Mef2c transcription factor. J. Biol. Chem. 274:5193–5200 10.1074/jbc.274.8.5193 [DOI] [PubMed] [Google Scholar]
- Zhang C.L., McKinsey T.A., Olson E.N. 2002. Association of class II histone deacetylases with heterochromatin protein 1: potential role for histone methylation in control of muscle differentiation. Mol. Cell. Biol. 22:7302–7312 10.1128/MCB.22.20.7302-7312.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M., New L., Kravchenko V.V., Kato Y., Gram H., di Padova F., Olson E.N., Ulevitch R.J., Han J. 1999. Regulation of the MEF2 family of transcription factors by p38. Mol. Cell. Biol. 19:21–30 [DOI] [PMC free article] [PubMed] [Google Scholar]