A proteolytic cascade ensures that OMA1 cleaves and inactivates mitochondrial fusion protein OPA1 in times of stress, preventing damaged mitochondria from fusing with healthy organelles. (See also companion paper from Ehses et al. in this issue.)
Abstract
The mammalian mitochondrial inner membrane fusion protein OPA1 is controlled by complex patterns of alternative splicing and proteolysis. A subset of OPA1 isoforms is constitutively cleaved by YME1L. Other isoforms are not cleaved by YME1L, but they are cleaved when mitochondria lose membrane potential or adenosine triphosphate. In this study, we show that this inducible cleavage is mediated by a zinc metalloprotease called OMA1. We find that OMA1 small interfering RNA inhibits inducible cleavage, helps retain fusion competence, and slows the onset of apoptosis, showing that OMA1 controls OPA1 cleavage and function. We also find that OMA1 is normally cleaved from 60 to 40 kD by another as of yet unidentified protease. Loss of membrane potential causes 60-kD protein to accumulate, suggesting that OMA1 is attenuated by proteolytic degradation. We conclude that a proteolytic cascade controls OPA1. Inducible cleavage provides a mechanism for quality control because proteolytic inactivation of OPA1 promotes selective removal of defective mitochondrial fragments by preventing their fusion with the mitochondrial network.
Introduction
Mitochondria of healthy cells continually divide and fuse with each other (Hoppins et al., 2007). The balance between fission and fusion is controlled by the opposing actions of several large GTP-binding proteins, which are members of the dynamin family. DRP1 in mammals and Dnm1 in yeast mediate mitochondrial fission by wrapping around constricted parts of mitochondria, where they control a late stage of the fission process (Hoppins et al., 2007). Mitochondrial fusion is mediated by dynamin-related proteins in the outer (mitofusins in mammals and Fzo1 in yeast) and inner membrane (OPA1 in mammals and Mgm1 in yeast; Olichon et al., 2003; Wong et al., 2003). Mitochondrial fission and fusion proteins have antagonistic functions during apoptosis. Mutations in Drp1 inhibit cytochrome c release during apoptosis, suggesting that fission proteins are proapoptotic (Frank et al., 2001), whereas loss of OPA1 promotes cytochrome c release, suggesting that OPA1 is antiapoptotic (Olichon et al., 2003). OPA1 is intensely studied because of this connection with apoptosis and because mutations in OPA1 are the most prevalent cause of dominant optic atrophy (DOA), a progressive eye disease which affects retinal ganglion cells in the optic nerve.
OPA1 functions are controlled by complex patterns of alternative splicing and proteolysis. Humans have eight different isoforms, which all have a mitochondrial leader sequence that is cleaved by the mitochondrial matrix protease (mitochondrial processing peptidase) upon import into mitochondria. In yeast, roughly half of the protein is further cleaved by a rhomboid protease, but in mammals, the situation is less clear. It was reported that mammalian OPA1 is also cleaved by a rhomboid protease (Cipolat et al., 2006), but knockout cells show no differences in OPA1 processing (Griparic et al., 2007). Additionally, it was reported that OPA1 is cleaved by the mitochondrial matrix protease paraplegin, but the effects of paraplegin siRNA were, at best, modest (Ishihara et al., 2006). Some confusion may have arisen from the fact that OPA1 processing follows two distinct paths. OPA1 isoforms containing alternatively spliced exons 4b or 5b are constitutively cleaved by the intermembrane space AAA (ATPase associated with diverse cellular activities) protease YME1L, generating the short form of OPA1 (S-OPA1), whereas the remaining isoforms are normally not cleaved further, generating the long form of OPA1 (L-OPA1; Griparic et al., 2007; Song et al., 2007). S- and L-OPA1 are both required for fusion, so constitutive cleavage is needed for a functional fusion apparatus (Song et al., 2007; DeVay et al., 2009).
L-OPA1 isoforms do remain susceptible to cleavage by a different, inducible protease (Griparic et al., 2007). Inducible cleavage is triggered by loss of mitochondrial membrane potential, by ATP deficiency, and by apoptosis (Baricault et al., 2007; Griparic et al., 2007). This cleavage is rapid and complete, fully inactivating the remaining population of uncleaved OPA1 molecules. Loss of membrane potential caused by the protonophore carbonyl cyanide m-chlorophenyl hydrazone (CCCP) blocks mitochondrial fusion by inactivating OPA1. Mitochondria can reestablish a filamentous morphology after regaining membrane potential (CCCP washout), but this recovery requires de novo synthesis of OPA1 (Griparic et al., 2007). We now show that inducible proteolysis is mediated by OMA1, a protease with multiple membrane-spanning segments and a zinc-binding motif. Inducible proteolysis mediated by OMA1 represents a novel regulatory mechanism that will likely affect mitochondrial turnover, apoptosis, and the progression of neurodegenerative diseases such as DOA.
Results and discussion
OMA1 is a mitochondrial protein affected by membrane potential
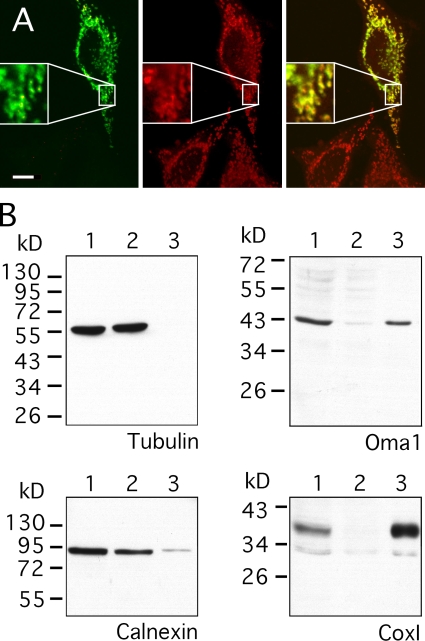
Human OMA1 is similar in sequence to the yeast mitochondrial inner membrane protease Oma1 (Kaser et al., 2003). However, human OMA1 has been localized to the ER (Bao et al., 2003). This discrepancy prompted us to reinvestigate the localization of human OMA1 using an expression construct with full-length OMA1 and two C-terminal HA tags (subsequent experiments show that this fusion protein is functional). Transfection of this construct into HeLa cells showed colocalization with MitoTracker (Fig. 1 A), whereas differential centrifugation showed cofractionation with mitochondrial markers but not with ER or cytosolic markers (Fig. 1 B). We conclude that human OMA1 is a mitochondrial protein.
Figure 1.
Localization of OMA1 to mitochondria in human cells. (A) HeLa cells were transfected with OMA1-HA and stained with HA antibody (left; green) and MitoTracker (middle; red). A merged image is shown on the right. Only one transfected cell is shown here, but many other transfected cells were observed, invariably showing colocalization of OMA1-HA and MitoTracker. The insets show enlargements to display the colocalization of HA antibody and MitoTracker staining more clearly. (B) Subcellular fractionation of OMA1-HA–transfected HeLa cells showing volume equivalents of postnuclear supernatant (lane 1), medium speed supernatant (lane 2), and medium speed pellet (lane 3) blotted and probed with HA, mitochondrial (COXI), and ER (calnexin) antibodies. Bar, 10 µm.
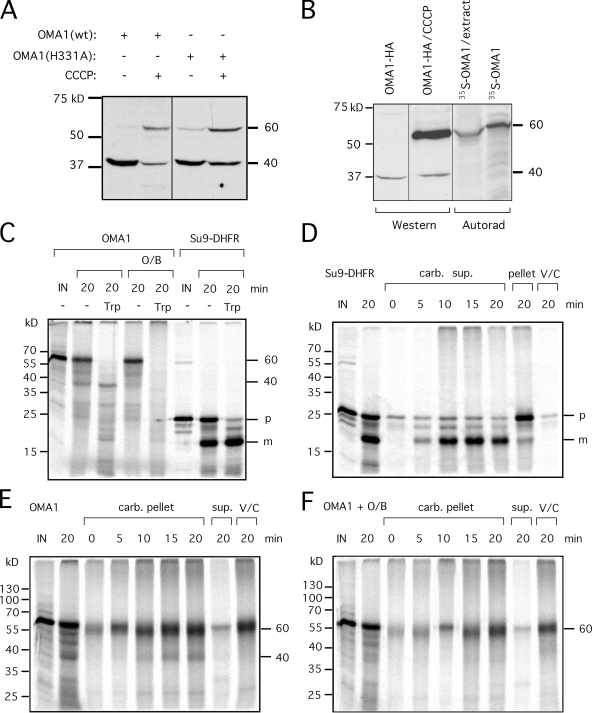
Transfections with OMA1 constructs consistently yielded a band of ∼40 kD (Fig. 1 B), whereas the predicted size of OMA1 is 62 kD. Even if OMA1 had a cleaved targeting sequence, we would have expected a protein of at least 55 kD. Instead, OMA1 is cleaved further into the N-terminal half of the protein (the product was detected with C-terminal tags). Transfections with YME1L, AFG3L2, and paraplegin siRNA had no effect on OMA1 cleavage, but there may still have been residual proteases in these experiments (unpublished data). At this point, it is still not known which protease cleaves OMA1. However, we did notice differences in OMA1 banding patterns when membrane potential was dissipated with CCCP. Transfections with two different constructs (one wild type and one with a point mutation in the zinc-binding motif) showed that the amount of 40-kD protein decreased and a 60-kD protein appeared when cells were treated with CCCP (Fig. 2 A). Rapid appearance of a 60-kD protein suggests that OMA1 continues to be synthesized and imported into mitochondria, which is similar to the CCCP-resistant import of several other mitochondrial proteins such as cytochrome b2 (Geissler et al., 2000). The newly formed 60-kD band co-migrated with in vitro–synthesized OMA1 protein (Fig. 2 B), suggesting that OMA1 does not have a cleaved mitochondrial leader sequence. High levels of 40-kD protein in untreated cells and its replacement by uncleaved protein after treatment with CCCP suggest that full-length OMA1 is cleaved at a high rate and that this cleavage is inhibited by treatment with CCCP.
Figure 2.
Changes in the cleavage of OMA1 upon treatment with CCCP and oligomycin. (A) HeLa cells were transfected with wild-type (wt) OMA1 or OMA1(H331A) mutant constructs. Treatment for 30 min with CCCP decreases the intensity of the 40-kD band, and a previously undetectable 60-kD protein appears. (B) Comparison between in vitro–synthesized OMA1 and the 60-kD band in OMA1-transfected and CCCP-treated HeLa cells. Unlabeled cell extract was added to radiolabeled OMA1 to load similar amounts of protein in different lanes. (A and B) Black lines indicate that intervening lanes have been spliced out. (C–F) Import of in vitro–synthesized OMA1 into isolated mitochondria. (C) Mitochondria were incubated with OMA1 or Su9-DHFR for 20 min. (D) Carbonate supernatants are shown for a Su9-DHFR time course with a 20-min carbonate (carb.) pellet as control. (E and F) Carbonate pellets are shown for an OMA1 time course with a 20-min supernatant as control. (F) Oligomycin and bongkrekic acid were added to the reactions to reduce mitochondrial ATP. IN, input lanes showing 10% of OMA1 in import reactions; m, mature protein; p, Su9-DHFR precursor; O/B, oligomycin and bongkrekic acid; Trp, trypsin; V/C, valinomycin and CCCP.
To further investigate OMA1 processing, we conducted in vitro import experiments with OMA1 protein and isolated mitochondria. Su9–dihydrofolate reductase (DHFR) fusion protein, which is targeted to the mitochondrial matrix, served as a control. Su9-DHFR precursor protein was observed in the mitochondrial pellet, but this protein was removed with trypsin digestion, whereas the mature Su9-DHFR, as detected by cleavage of the mitochondrial leader sequences, was trypsin insensitive (Fig. 2 C). When mitochondria were incubated with OMA1 protein, they contained 40- and 60-kD proteins similar to those observed in vivo. The 40-kD protein was trypsin insensitive, showing that this protein is imported, whereas the 60-kD band was degraded, similar to the SU9-DHFR precursor, suggesting that the 60-kD protein remains accessible to trypsin (Fig. 2 C).
We used carbonate extractions to distinguish proteins that are genuinely inserted into membrane from peripherally bound or soluble proteins. As expected, mature Su9-DHFR was released from the mitochondrial fraction (Fig. 2 D), but both 40- and 60-kD OMA1 proteins were pelleted, showing that these are integral membrane proteins (Fig. 2 E). Dissipation of membrane potential with valinomycin and CCCP blocked import of Su9-DHFR (Fig. 2 D), and it blocked the formation of the 40-kD OMA1 protein, but it did not prevent insertion of the 60-kD OMA1 protein into mitochondrial membranes (Fig. 2 E). Oligomycin (an inhibitor of mitochondrial ATP synthase) and bongkrekic acid (an inhibitor of adenine nucleotide transporters) were used to deplete mitochondrial ATP. Like valinomycin and CCCP, these inhibitors blocked formation of the 40-kD OMA1 product but not insertion of 60-kD protein into mitochondrial membranes (Fig. 2 F). The amount of 40-kD protein in import reactions is low, perhaps because of high turnover or an abundance of unproductive translocation intermediates in the mitochondrial outer membrane. However, the formation of a trypsin-insensitive 40-kD protein, which is inhibited by valinomycin/CCCP and by oligomycin/bongkrekic acid, showed that OMA1 protein is imported and correctly processed in isolated mitochondria, faithfully recapitulating the results obtained with transfected cells. Based on our in vivo and in vitro data, we propose that the OMA1 protease is itself controlled by proteolysis.
Effects of OMA1 siRNA on inducible cleavage of OPA1
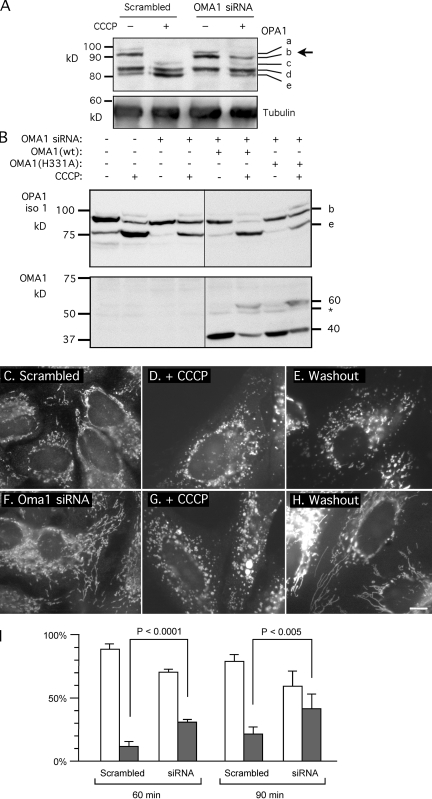
OPA1 antibodies normally detect five bands on Western blots (labeled a–e; Griparic et al., 2007). Bands b and d are the most prominent. Band b contains isoforms that lack exons 4b or 5b and are therefore not constitutively cleaved. Band d consists of isoforms that are constitutively cleaved by YME1L in exons 4b or 5b (Griparic et al., 2007). Loss of membrane potential, low ATP, and apoptosis induce cleavage of isoforms that were not cleaved by YME1L, converting band b to band e. We tested whether OMA1 is responsible for this inducible cleavage by transfection with OMA1 siRNA followed by treatment with CCCP. Western blots show that the conversion of band b to band e was inhibited by 56% in these experiments (mean of five independent experiments [SD = 12%]; as determined by densitometry of Western blots; Fig. 3 A). Similar results were obtained with three pairs of siRNA oligonucleotides targeting different regions of the OMA1 mRNA. Inhibition of inducible cleavage was never observed with siRNA for other proteases (Griparic et al., 2007). There still is some inducible cleavage, even with 90% reduction of OMA1 mRNA, so OMA1 may be partially redundant with other proteases or small amounts of residual OMA1 are enough for some cleavage of OPA1. Nevertheless, we can conclude that OMA1 siRNA inhibits inducible cleavage of OPA1.
Figure 3.
Effects of OMA1 on CCCP-induced cleavage of OPA1 and recovery of fusion competence after CCCP washout. (A) HeLa cells were transfected with OMA1 siRNA or scrambled oligonucleotides followed by 30 min with 10 µM CCCP (OMA1 RNA was reduced by 90%). CCCP-induced cleavage normally converts band b to band e, but this cleavage is inhibited by OMA1 siRNA (arrow). (B) Effects of Oma1 siRNA on CCCP-induced cleavage are suppressed by cotransfected wild-type OMA1 but not by the OMA1(H331A) mutant. Cotransfected OPA1 isoform 1 was detected with an myc tag (Griparic et al., 2007). Isoform 1 is not cleaved by YME1L, but it is cleaved by OMA1, showing a CCCP-induced shift from band b to band e. OMA1 expression and the shift from 40- to 60-kD proteins are shown on the bottom. Black lines indicate that intervening lanes have been spliced out. (C–I) Effects of OMA1 siRNA on recovery from CCCP-induced mitochondrial fragmentation. HeLa cells were transfected with scrambled (C–E) or OMA1 siRNA (F–H). Mitochondria are normally filamentous (C and F) but fragment after 30 min with CCCP (D and G). At 90 min after CCCP washout, the filamentous morphology of mitochondria returns more quickly in OMA1 siRNA cells (H) than in control cells (E). (I) Percentages of cells with fully fragmented mitochondria (open bars) or with partial or full recovery of tubular mitochondria (closed bars) are shown at 60 or 90 min after washout. Means and SD were determined with six plates in two independent experiments (400 cells per plate). P-values were determined with an unpaired Student's t test. OMA1 expression was reduced by 70% (means with real-time PCR). Bar, 10 µm.
Fig. 3 A shows reduced intensity of band e in cells transfected with OMA1 siRNA even without CCCP treatment, suggesting that a small fraction of protein in band e might result from constitutive cleavage by OMA1. However, the differences were not significant when the relative intensities of band e were compared in four different experiments (Fig. S1 A). It would appear that inducible proteolysis of OPA1 is exquisitely sensitive to slight disruptions in the overall conditions of cells, particularly at the time of harvest. OMA1 siRNA also had no obvious effect on steady-state levels of mitochondrial connectivity, but there were some localized swellings, which could be caused by another as of yet unknown function of OMA1 (Fig. S2, D and D′).
We further tested the role of OMA1 in inducible cleavage with a rescue experiment in which OMA1 siRNA–transfected cells were cotransfected with wild-type or mutant (H331A) OMA1 constructs. Transfection with either construct did not change mitochondrial morphology in wild-type cells (Fig. S2 A) nor did it cause more cleavage of OPA1 (Fig. 3 B), perhaps because OMA1 is rapidly degraded without CCCP. Nevertheless, wild-type OMA1 was able to compensate for OMA1 siRNA by restoring CCCP-induced cleavage of OPA1, whereas OMA1(H331A) did not restore cleavage (Fig. 3 B). The differences were statistically significant, as shown by densitometry of Western blots from three independent experiments (mean of 23% in band b [SD = 10.5; n = 3] with OMA1 siRNA, wild-type OMA1, and CCCP; mean of 55% in band b [SD = 6.1; n = 3] with OMA1 siRNA, OMA1(H331A), and CCCP; P = 0.010 in an unpaired Student's t test). Together, these experiment show that OMA1 mediates inducible cleavage of OPA1 and that this cleavage depends on intact protease sequences. In addition, we observed changes in OMA1 bands upon treatment with CCCP, confirming that these changes correspond with changes in OMA1 function.
To determine which cleavage sites are affected by OMA1, we transfected HeLa cells with expression constructs encoding three representative OPA1 isoforms with different cleavage sites (isoforms 1, 5, and 7; Griparic et al., 2007). These constructs were cotransfected with OMA1 or YME1L siRNA followed by CCCP treatment to compare constitutive with induced cleavage. Isoform 1 has only one cleavage site (S1 in exon 5), which is not cleaved by YME1L but is cleaved by the inducible protease (Griparic et al., 2007). Our results show that isoform 1 cleavage was induced by CCCP but not in cells transfected with OMA1 siRNA (Fig. 3 B). Isoforms 5 and 7 are constitutively cleaved by YME1L in exons 4b and 5b, respectively (Griparic et al., 2007). This was confirmed in this study with YME1L siRNA (Fig. S1 B). These isoforms became susceptible to CCCP-induced cleavage upon transfection with YME1L siRNA (Fig. 3 and Fig. S1 B). We conclude that OMA1 cleaves OPA1 in the S1 site (exon 5), which is present in all three isoforms, but isoforms 5 and 7 are only cleaved by OMA1 when constitutive cleavage in exons 4b and 5b is prevented with YME1L siRNA (Fig. S1 B).
Effects of OMA1 siRNA on mitochondrial fusion after CCCP washout
The effects of OMA1 on mitochondrial fusion were tested by transfecting cells with OMA1 siRNA, fragmenting their mitochondria with CCCP and monitoring recovery of filamentous mitochondria through fusion after CCCP washout. Fragmentation still occurred in OMA1 siRNA cells (Fig. 3 G) because fragmentation is mediated by DRP1 (Griparic et al., 2007), but these cells recovered tubular mitochondrial morphologies more quickly than cells transfected with scrambled siRNA (Fig. 3, E and H). The differences were most pronounced at 60 and 90 min after CCCP washout (Fig. 3 I). After 120 min, control cells also regained tubular morphologies through de novo synthesis of OPA1 protein, as shown previously with cycloheximide (Griparic et al., 2007). We conclude that mitochondria of OMA1 siRNA cells fuse under conditions that would normally prevent fusion because of proteolytic inactivation of OPA1.
Effects of OMA1 siRNA on apoptosis
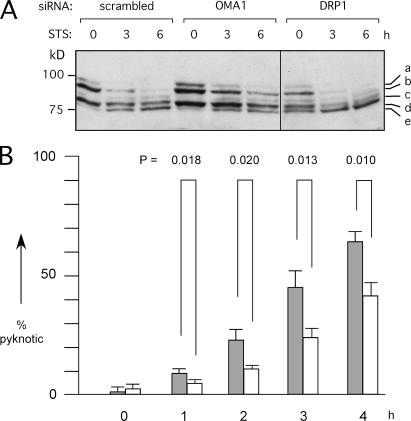
It was shown previously that OPA1 is cleaved concurrent with cytochrome c release during apoptosis (Duvezin-Caubet et al., 2006; Baricault et al., 2007; Griparic et al., 2007). In this study, we show that OMA1 siRNA slowed the onset of staurosporine-induced cleavage of OPA1 (Fig. 4 A), indicating that apoptosis and CCCP induce cleavage through the same mechanism. Loss of DRP1 also inhibits apoptosis (Frank et al., 2001), but it did not inhibit induced cleavage of OPA1 (Fig. 4 A). The effects of DRP1 and OPA1 on apoptosis may be independent; DRP1 contributes to mitochondrial outer membrane permeabilization (Cassidy-Stone et al., 2008), whereas OPA1 may affect cytochrome c release from within cristae (Duvezin-Caubet et al., 2006).
Figure 4.
Effects of OMA1 siRNA on apoptosis. (A) HeLa cells were transfected with siRNA oligonucleotides followed by 0, 3, and 6 h with staurosporine (STS). Extracts were blotted and probed with OPA1 antibody. Staurosporine converts band b to band e, similar to CCCP-induced cleavage. OMA1 siRNA slows this process, whereas DRP1 siRNA does not, even though DRP1 siRNA inhibits apoptosis (detected with PARP and caspase cleavage; not depicted). The black line indicates that intervening lanes have been spliced out. (B) The effects of OMA1 siRNA on staurosporine-induced apoptosis were determined by counting the numbers of pyknotic nuclei in 300 cells for each time point. Mean values for three independent experiments are given with SD. The significance of differences between scrambled (closed bars) and OMA1 siRNA (open bars) was determined with an unpaired Student's t test.
We could now also test whether OPA1 cleavage actively promotes apoptosis. To this end, HeLa cells were transfected with OMA1 siRNA, followed by treatment with staurosporine and counting the numbers of cells with pyknotic nuclei. The percentages of pyknotic nuclei were significantly lower in cells transfected with OMA1 siRNA than in control cells up to 3 h after treatment with staurosporine (Fig. 4 B). At longer time points, all cells had pyknotic nuclei, showing that the inhibition of apoptosis is not absolute (unpublished data). Similar effects were observed with cytochrome c release induced by actinomycin D (Fig. S3). We conclude that OMA1 siRNA inhibits apoptotic release of cytochrome c and further progression of apoptosis similar to the inhibitory effects of Drp1 and Fis1 siRNA, but these effects are not complete, perhaps because knockdown of OMA1 was incomplete or because apoptosis does not absolutely require loss of OPA1 function.
Conclusions
Our results show that OMA1 is responsible for CCCP-induced cleavage of OPA1 in mammalian cells. Evidence for this role is provided by the specific inhibition of inducible proteolysis with OMA1 siRNA and by the rescue of inducible proteolysis with cotransfected wild-type OMA1 but not with mutant OMA1. Inducible proteolysis was never inhibited by knockdown of other mitochondrial proteases (Griparic et al., 2007), nor did OMA1 siRNA affect constitutive proteolysis of OPA1. Functional evidence for the inhibitory effects on proteolytic inactivation of OPA1 is provided by CCCP washout experiments in which we showed that cells transfected with OMA1 siRNA recovered fused mitochondria more quickly than mock-transfected cells and with apoptosis experiments in which we showed that OMA1 siRNA inhibited apoptosis. Cleavage of OPA1 by OMA1, as described in this study, and constitutive proteolysis by YME1L, as described previously (Griparic et al., 2007; Song et al., 2007; Guillery et al., 2008), account for the banding patterns of OPA1 on Western blots.
Our results also suggest a mechanism for the control of OMA1 activity. Transfections with OMA1 expression constructs show a dominant 40-kD cleavage product of OMA1 in untreated cells. The amount of 40-kD protein is reduced in cells treated with CCCP, suggesting that this protein is subjected to further proteolytic degradation. However, CCCP also leads to the accumulation of uncleaved OMA1. Because this accumulation coincides with OMA1 cleavage of OPA1, we propose that the 60-kD band is the active species and the 40-kD band is a breakdown product. These observations lead to a model in which OMA1 activity is normally attenuated by high rates of turnover. This turnover is slowed when mitochondria lose membrane potential or matrix ATP, making it possible to cleave the OPA1 protein (Fig. 5).
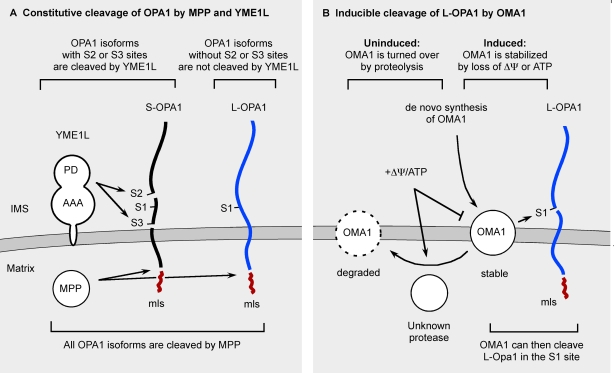
Figure 5.
Pathways for proteolytic processing of OPA1. (A) The constitutive cleavage pathway. All isoforms of OPA1 have a mitochondrial leader sequence, which is constitutively cleaved by the matrix protease mitochondrial processing peptidase (MPP). Some isoforms contain exon 4b, which has a cleavage site that we name S3, and some isoforms have exon 5b, which has a cleavage site named S2 (Ishihara et al., 2006). Isoforms with exon 4b or 5b are constitutively cleaved by YME1L to produce S-OPA1. Isoforms lacking exons 4b and 5b are not cleaved by YME1L, thus producing L-OPA1. Mitochondrial fusion requires a mixture of S- and L-OPA1. (B) The pathway for inducible cleavage. L-OPA1, which is not cleaved by YME1L, remains susceptible to inducible cleavage in the S1 site of exon 5 by OMA1. This inducible cleavage occurs when mitochondria lose membrane potential or ATP. Loss of membrane potential also leads to an accumulation of 60-kD OMA1 and reduced amounts of 40-kD OMA1, suggesting that OMA1 activity is regulated by OMA1 turnover. Another, as of yet unknown, protease may attenuate OMA1 activity by proteolytic degradation. This attenuation is slowed when mitochondria lose membrane potential or ATP, thus allowing for cleavage of OPA1. Alternatively, membrane potential or ATP levels may also directly affect OMA1 activity. IMS, intermembrane space; PD, protease domain.
Rapid proteolytic inactivation of OPA1 may have coevolved with other inducible processes such as cytochrome c release from mitochondria during apoptosis. Alternatively, rapid proteolytic inactivation of OPA1 might help eliminate defective fragments of mitochondria because those fragments lose membrane potential and will thus lose their ability to refuse with other mitochondria (Twig et al., 2008). Both of these scenarios suggest that OMA1-mediated cleavage of OPA1 is beneficial for growth and development. However, inducible proteolysis could also have pathogenic effects in neurodegenerative diseases such as Parkinson's (Yang et al., 2008) or DOA. Identification of OMA1 as the protease responsible for inducible cleavage of OPA1 may lead to selective inhibitors, which could help reduce the debilitating effects of these neurodegenerative diseases.
Materials and methods
Cell culture, protein analysis, and microscopy
For the experiments in Fig. 1, HeLa cells were stained with MitoTracker red CMXRos (Invitrogen), fixed with formaldehyde, and processed for antibody staining and detection with FITC (Griparic et al., 2007). For the experiments in Fig. 3 and Fig. S2, mitochondria were detected by transfection with mitochondrial DsRed2 (Takara Bio Inc.). Fluorescence microscopy was performed with a microscope (Axiovert 200M; Carl Zeiss, Inc.) using a Plan-NeoFluar 40× NA 1.3 oil objective (Fig. S2) or an α Plan-Fluar 100× NA 1.45 oil objective (all other figures). Images were acquired at room temperature (24°C) with a charge-coupled device camera (ORCA ER; Hamamatsu Photonics) controlled by Axiovision software (Carl Zeiss, Inc.). Image files were processed with Photoshop software (Adobe). Membrane potential was disrupted with 10 µM CCCP (Sigma-Aldrich). In washout experiments, cells were rinsed twice with PBS and incubated with fresh medium. Controls were treated with solvent (DMSO). Apoptosis was induced with 2 µM staurosporine (Sigma-Aldrich) or 20 µM actinomycin D (EMD) with 200 µM zVAD-fmk (MP Biomedicals) added 30 min before actinomycin D. Pyknotic nuclei were counted by staining with Hoechst (Gandre-Babbe and van der Bliek, 2008). Cells were lysed in Laemmli sample buffer. Blots were probed with OPA1 and calnexin (BD), HA (Roche), CoxI (MitoSciences), tubulin (Sigma-Aldrich), and c-myc (AbD Serotec) antibodies. Blots were developed with horseradish peroxidase secondary antibody and chemiluminescence (GE Healthcare). Subcellular fractions were isolated by differential centrifugation (Gandre-Babbe and van der Bliek, 2008). For densitometry, film was scanned with a phosphorimager (Typhoon 9410; GE Healthcare) and analyzed with ImageQuant software (GE Healthcare).
Plasmids, siRNA oligonucleotides, and transfection techniques
OMA1 cDNA was modified using PCR to add two C-terminal HA tags in pCMV-SPORT6 (Invitrogen). OMA1(H331A) was made by site-directed mutagenesis. OPA1 isoforms were cloned in pCDNA3.1 with a C-terminal myc tag (Griparic et al., 2007). Su9-DHFR plasmid was a gift from C. Koehler (University of California, Los Angeles, Los Angeles, CA). Oligonucleotides for siRNA were synthesized by Sigma-Aldrich. OMA1 sequences were 5′-GGCUGGCAUGGUUCAUUUGdTdT-3′ (first), 5′-CCGAUAUUCAUCAACUUUCdTdT-3′ (second), and 5′-GAAGUGCUUUGUCAUCUAAdTdT-3′ (third). Most experiments were performed with the third set, but effects on OPA1 cleavage were confirmed with the first two pairs as well. Scrambled controls had inverted sequences. HeLa cells were transfected with 60 pmol siRNA and Fugene6 HD (Roche). Cells were split on day 2 and transfected again on day 3. On day 4, cells were stained for microscopy or harvested for RNA and protein analysis. RNA was quantified in triplicate with real-time PCR (Griparic et al., 2007). For CCCP recovery experiments and OMA1 overexpression, cells were cotransfected with MitoDsRed (Takara Bio Inc.).
Import of proteins into isolated mitochondria
OMA1 and Su9-DHFR proteins were synthesized with [35S]methionine (PerkinElmer) and TNT SP6-coupled reticulocyte lysates (Promega). Mitochondria were isolated from mouse liver (provided by C. Koehler) by homogenization and differential centrifugation. Mitochondria were resuspended at 2 mg protein/ml in 250 mM sucrose, 5 mM MgAc, 100 mM KAc, 10 mM Na-succinate, and 20 mM Hepes/KOH, pH 7.4, adding 2.5 mM ATP, 0.1 mM ADP, 1 mM DTT, and 10 mM methionine before starting reactions with in vitro–synthesized protein. Reactions were stopped with 1 µM valinomycin and 20 µM CCCP. Where indicated, 5 µM oligomycin and 50 µM bongkrekic acid were added before the reaction. Where indicated, samples were incubated on ice with 100 µg/ml trypsin (Sigma-Aldrich) for 20 min, followed with 200 µg/ml trypsin inhibitor (Sigma-Aldrich). 2 vol of 0.6 M sorbitol, 50 mM Hepes/KOH, pH 7.4, and 80 mM KCl were added, and mitochondria were pelleted by centrifugation. Untreated and trypsinized samples were dissolved in Laemmli sample buffer. Remaining samples were subjected to alkaline extraction with 30 min on ice in 0.2 M Na2CO3/NaOH, pH 12. Membranes were pelleted for 30 min at 21,000 g. Proteins in supernatants were precipitated with trichloroacetic acid. Carbonate and trichloroacetic acid pellets were resuspended in Laemmli sample buffer and size fractionated by SDS-PAGE. Gels were dried and analyzed with a Typhoon 9410 phosphorimager.
Online supplemental material
Fig. S1 shows that OMA1 does not affect constitutive proteolysis of OPA1. Fig. S2 shows the effects of OMA1 expression and siRNA on mitochondrial morphology. Fig. S3 shows the effects of OMA1 siRNA on actinomycin D–induced apoptosis. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906083/DC1.
Acknowledgments
We are grateful to Dr. Carla Koehler for expert advice on mitochondrial protein import.
This project was supported by grants from the National Institutes of Health (GM051866), the Stein Oppenheimer Endowment, and the Jonsson Comprehensive Cancer Research Center.
Footnotes
Abbreviations used in this paper:
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- DHFR
- dihydrofolate reductase
- DOA
- dominant optic atrophy
References
- Bao Y.C., Tsuruga H., Hirai M., Yasuda K., Yokoi N., Kitamura T., Kumagai H. 2003. Identification of a human cDNA sequence which encodes a novel membrane-associated protein containing a zinc metalloprotease motif. DNA Res. 10:123–128 10.1093/dnares/10.3.123 [DOI] [PubMed] [Google Scholar]
- Baricault L., Ségui B., Guégand L., Olichon A., Valette A., Larminat F., Lenaers G. 2007. OPA1 cleavage depends on decreased mitochondrial ATP level and bivalent metals. Exp. Cell Res. 313:3800–3808 10.1016/j.yexcr.2007.08.008 [DOI] [PubMed] [Google Scholar]
- Cassidy-Stone A., Chipuk J.E., Ingerman E., Song C., Yoo C., Kuwana T., Kurth M.J., Shaw J.T., Hinshaw J.E., Green D.R., Nunnari J. 2008. Chemical inhibition of the mitochondrial division dynamin reveals its role in Bax/Bak-dependent mitochondrial outer membrane permeabilization. Dev. Cell. 14:193–204 10.1016/j.devcel.2007.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipolat S., Rudka T., Hartmann D., Costa V., Serneels L., Craessaerts K., Metzger K., Frezza C., Annaert W., D'Adamio L., et al. 2006. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 126:163–175 10.1016/j.cell.2006.06.021 [DOI] [PubMed] [Google Scholar]
- DeVay R.M., Dominguez-Ramirez L., Lackner L.L., Hoppins S., Stahlberg H., Nunnari J. 2009. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. J. Cell Biol. 186:793–803 10.1083/jcb.200906098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvezin-Caubet S., Jagasia R., Wagener J., Hofmann S., Trifunovic A., Hansson A., Chomyn A., Bauer M.F., Attardi G., Larsson N.G., et al. 2006. Proteolytic processing of OPA1 links mitochondrial dysfunction to alterations in mitochondrial morphology. J. Biol. Chem. 281:37972–37979 10.1074/jbc.M606059200 [DOI] [PubMed] [Google Scholar]
- Frank S., Gaume B., Bergmann-Leitner E.S., Leitner W.W., Robert E.G., Catez F., Smith C.L., Youle R.J. 2001. The role of dynamin-related protein 1, a mediator of mitochondrial fission, in apoptosis. Dev. Cell. 1:515–525 10.1016/S1534-5807(01)00055-7 [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S., van der Bliek A.M. 2008. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol. Biol. Cell. 19:2402–2412 10.1091/mbc.E07-12-1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler A., Krimmer T., Bömer U., Guiard B., Rassow J., Pfanner N. 2000. Membrane potential-driven protein import into mitochondria. The sorting sequence of cytochrome b(2) modulates the deltapsi-dependence of translocation of the matrix-targeting sequence. Mol. Biol. Cell. 11:3977–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griparic L., Kanazawa T., van der Bliek A.M. 2007. Regulation of the mitochondrial dynamin-like protein Opa1 by proteolytic cleavage. J. Cell Biol. 178:757–764 10.1083/jcb.200704112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillery O., Malka F., Landes T., Guillou E., Blackstone C., Lombès A., Belenguer P., Arnoult D., Rojo M. 2008. Metalloprotease-mediated OPA1 processing is modulated by the mitochondrial membrane potential. Biol. Cell. 100:315–325 10.1042/BC20070110 [DOI] [PubMed] [Google Scholar]
- Hoppins S., Lackner L., Nunnari J. 2007. The machines that divide and fuse mitochondria. Annu. Rev. Biochem. 76:751–780 10.1146/annurev.biochem.76.071905.090048 [DOI] [PubMed] [Google Scholar]
- Ishihara N., Fujita Y., Oka T., Mihara K. 2006. Regulation of mitochondrial morphology through proteolytic cleavage of OPA1. EMBO J. 25:2966–2977 10.1038/sj.emboj.7601184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaser M., Kambacheld M., Kisters-Woike B., Langer T. 2003. Oma1, a novel membrane-bound metallopeptidase in mitochondria with activities overlapping with the m-AAA protease. J. Biol. Chem. 278:46414–46423 10.1074/jbc.M305584200 [DOI] [PubMed] [Google Scholar]
- Olichon A., Baricault L., Gas N., Guillou E., Valette A., Belenguer P., Lenaers G. 2003. Loss of OPA1 perturbates the mitochondrial inner membrane structure and integrity, leading to cytochrome c release and apoptosis. J. Biol. Chem. 278:7743–7746 10.1074/jbc.C200677200 [DOI] [PubMed] [Google Scholar]
- Song Z., Chen H., Fiket M., Alexander C., Chan D.C. 2007. OPA1 processing controls mitochondrial fusion and is regulated by mRNA splicing, membrane potential, and Yme1L. J. Cell Biol. 178:749–755 10.1083/jcb.200704110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twig G., Elorza A., Molina A.J., Mohamed H., Wikstrom J.D., Walzer G., Stiles L., Haigh S.E., Katz S., Las G., et al. 2008. Fission and selective fusion govern mitochondrial segregation and elimination by autophagy. EMBO J. 27:433–446 10.1038/sj.emboj.7601963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong E.D., Wagner J.A., Scott S.V., Okreglak V., Holewinske T.J., Cassidy-Stone A., Nunnari J. 2003. The intramitochondrial dynamin-related GTPase, Mgm1p, is a component of a protein complex that mediates mitochondrial fusion. J. Cell Biol. 160:303–311 10.1083/jcb.200209015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Ouyang Y., Yang L., Beal M.F., McQuibban A., Vogel H., Lu B. 2008. Pink1 regulates mitochondrial dynamics through interaction with the fission/fusion machinery. Proc. Natl. Acad. Sci. USA. 105:7070–7075 10.1073/pnas.0711845105 [DOI] [PMC free article] [PubMed] [Google Scholar]