Mechanical interactions between any two opposite-polarity motors are necessary and sufficient for bidirectional organelle transport in live cells.
Abstract
Intracellular transport is typically bidirectional, consisting of a series of back and forth movements. Kinesin-1 and cytoplasmic dynein require each other for bidirectional transport of intracellular cargo along microtubules; i.e., inhibition or depletion of kinesin-1 abolishes dynein-driven cargo transport and vice versa. Using Drosophila melanogaster S2 cells, we demonstrate that replacement of endogenous kinesin-1 or dynein with an unrelated, peroxisome-targeted motor of the same directionality activates peroxisome transport in the opposite direction. However, motility-deficient versions of motors, which retain the ability to bind microtubules and hydrolyze adenosine triphosphate, do not activate peroxisome motility. Thus, any pair of opposite-polarity motors, provided they move along microtubules, can activate one another. These results demonstrate that mechanical interactions between opposite-polarity motors are necessary and sufficient for bidirectional organelle transport in live cells.
Introduction
The continual reorganization of the cytoplasm is a dynamic process in all cell types. Precise temporal and spatial trafficking of cellular components (such as organelles, RNA complexes, and receptors) are critical to cell survival. Essential biological processes like chromosome segregation, cell division, secretion, dendrite formation, and organelle transport (Gross et al., 2002b; Hirokawa and Noda, 2008) depend on effective intracellular transport. In fact, many neurodegenerative diseases occur as a result of defective intracellular transport mechanisms (Hirokawa and Takemura, 2004). Furthermore, at the developmental level, accurate delivery of mRNAs to the posterior pole of the Drosophila melanogaster oocyte triggers germline specification (Duncan and Warrior, 2002; Steinhauer and Kalderon, 2006; Messitt et al., 2008). At the single-cell level, mitochondria transport must also be finely regulated to ensure timely delivery during axonal growth and migration, which is a period of high ATP requirement (Hollenbeck and Saxton, 2005).
Molecular motors use the energy of ATP hydrolysis to transport cargo along an extensive cytoskeleton network. For example, kinesins and cytoplasmic dynein move along microtubules, whereas myosins move along actin filaments. Radially organized microtubules are suited for long-range transport, whereas shorter actin filaments govern local transport at the cell periphery. During translocation along the cytoskeleton, the dimeric (or sometimes trimeric) head domains of motors alternate in a hand over hand mechanism, whereby the ATP/ADP status of each head determines the binding affinity to the cytoskeletal track (Yildiz et al., 2004; Yildiz and Selvin, 2005).
In most eukaryotic cells, multiple opposite-polarity motors (Kural et al., 2005; Shubeita et al., 2008) drive cargo transport in a bidirectional manner (Tuma et al., 1998). That is, in contrast with in vitro, a series of back and forth movements punctuate live cell transport. Lysosomes, melanosomes, lipid droplets, mitochondria, and even infected herpes viral particles all display bidirectional movements along microtubules in a variety of cell types (Freed and Lebowitz, 1970; Gross et al., 2002a; Welte, 2004; Cox and Spradling, 2006; Lyman and Enquist, 2009).
Evidence from movements of numerous different cargoes in several cell types indicates that disruption of one type of microtubule motor (through use of mutations, function-blocking antibodies, or RNAi) also abrogates transport mediated by the opposite-polarity motor. For example, fast axonal transport in squid axoplasm was completely abolished after treatment with function-blocking antibodies against dynactin (a dynein adaptor complex; Waterman-Storer et al., 1997). Similarly, fast axonal transport was also disrupted in both directions in dynein heavy chain (dhc), dynactin, or kinesin heavy chain (khc) Drosophila mutants (Martin et al., 1999; Pilling et al., 2006). Similarly, in Xenopus laevis melanophores, kinesin-II (a kinesin-2 family member) and dynein require the activity of each other to drive melanosome transport (Gross et al., 2003). Unc104 (a kinesin-3 family member) and dynein are also interdependent in function during axonal transport in Drosophila neurons; in unc104 mutants, synaptic vesicle transport is inhibited in both the anterograde and retrograde directions (Barkus et al., 2008). Recently, Uchida et al. (2009) showed that axonal neurofilament transport in cultured sympathetic neurons from kinesin-1A knockout mice is inhibited in both directions. Finally, as we have previously shown in cultured Drosophila S2 cells, depletion of either KHC or DHC using RNAi completely inhibits bidirectional motility of mRNA complexes and several classes of organelles (Ling et al., 2004; Kim et al., 2007). These examples, using multiple motor types in a variety of biological systems, all suggest that opposite-polarity motors function interdependently during transport.
Previous studies have suggested that a yet-unidentified component functions as a molecular switch between kinesin-1 and dynein and thus specifies directionality of cargo transport. For example, huntingtin, Halo, or LSD2 may alternately associate with dynein/dynactin and kinesin-1 to drive either minus or plus end–directed transport (Gross et al., 2003; Cohen, 2005; Caviston et al., 2007; Colin et al., 2008). Although these factors and many others likely contribute to a directional bias in transport, it is unclear whether the basic mechanism of bidirectional intracellular transport requires any specific factor other than the two oppositely directed motors themselves (see Discussion). In this study, we determined whether any plus end–directed molecular motor can functionally replace kinesin-1 and, conversely, whether any minus end–directed motor can functionally replace dynein in cargo transport.
In this study, we observed peroxisome transport in Drosophila S2 cells after systematically replacing endogenous kinesin-1 or dynein with motors normally not involved in peroxisome transport. These replacement motors were attached to peroxisomes via peroxisome-targeting signals. Any replacement motor that was capable of moving along microtubules activated its opposite-polarity counterpart. Thus, we suggest that opposite-polarity motors can mechanically activate one another to drive bidirectional cargo transport.
Results
Kinesin-1 and cytoplasmic dynein function in an interdependent manner during bidirectional peroxisome transport
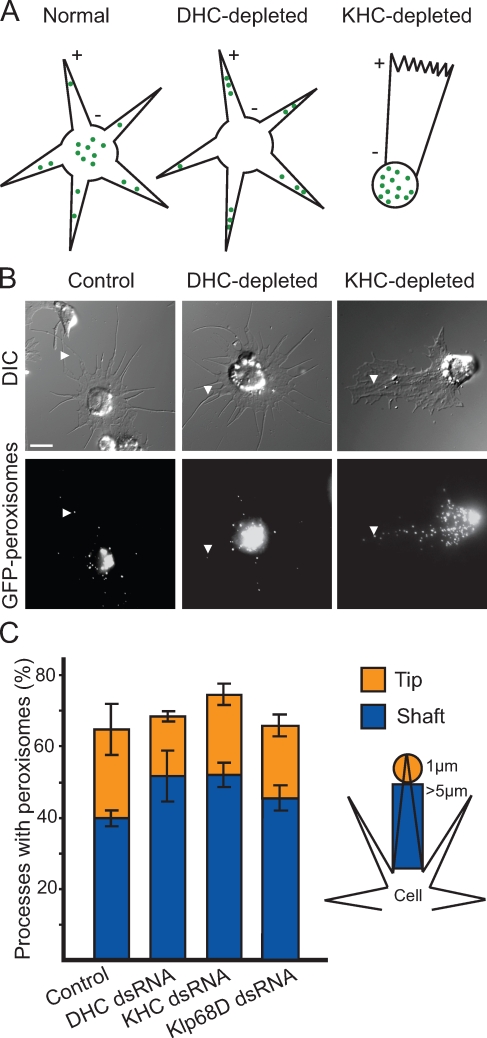
Drosophila S2 cells can be induced to form long processes filled with uniformly oriented microtubules when plated in the presence of cytochalasin D (CytoD) on a Con A–coated substrate. In these cells, at least 95% of microtubules are oriented with plus ends pointing toward the tips of processes. This system has been extensively used in our laboratory for analysis of microtubule-dependent transport (Ling et al., 2004; Kural et al., 2005; Kim et al., 2007). As mentioned in the Introduction, previous experiments in our laboratory have shown that Drosophila kinesin-1 and dynein function in an interdependent manner during cargo transport; i.e., detailed analysis of organelle movement revealed that depletion of one type of motor abrogates transport in both directions (Ling et al., 2004; Kim et al., 2007). Although we demonstrated that organelles appeared nonmotile over periods of several minutes after RNAi-based depletion of either kinesin-1 or dynein, it was unclear whether these organelles could potentially move, albeit at a much lower velocity, in the absence of one of these motors. Even a small amount of residual motility would be revealed as an altered distribution of organelles after several days of RNAi-based depletion of either motor. That is, if some kinesin-1–driven motility occurred after dynein depletion, one would expect peroxisomes to accumulate at the tips of processes (or plus ends of microtubules) over long periods of time (Fig. 1 A, middle). Conversely, one would expect some accumulation of organelles at the cell center (or minus ends of microtubules) after kinesin-1 depletion if dynein were still somewhat motile (Fig. 1 A, right).
Figure 1.
Kinesin-1 and cytoplasmic dynein function in an interdependent manner during peroxisome transport in S2 cells. (A) Potential outcomes of RNAi-based motor depletion if motors work independently of each other. Schematic depicts S2 cells plated in the presence of 1 µM CytoD to induce formation of microtubule-filled processes. Microtubule polarity is indicated with + and − signs. Green dots represent GFP-labeled peroxisomes. (left) GFP-labeled peroxisomes are distributed in the cell body and along the length of processes. (middle) Plus end clusters are shown. DHC depletion allows kinesin-1 to transport GFP-labeled peroxisomes toward the tips of processes. (right) Minus end clusters are shown. KHC depletion allows dynein to transport GFP-labeled peroxisomes toward the cell center. Note that the altered morphology of KHC-depleted cells is discussed in Results. (B) Depletion of molecular motors does not alter peroxisome distribution along processes. Representative still images of S2 cells plated in the presence of CytoD on Con A–coated coverslips. (top) Differential interference contrast (DIC) images are shown. (bottom) Fluorescent images depicting GFP-labeled peroxisomes, corresponding to Videos 1–3. Arrowheads highlight the location of a single peroxisome within a process. Bar, 5 µm. (C) Bar graph representing the percentage of processes containing peroxisomes after RNAi-based depletion of motors (treatment with KHC, DHC, and Klp68D dsRNAs). Klp68D depletion served as a control. The total height of each column (blue and orange) represents the percentage of processes that contain peroxisomes anywhere along their length. The blue subcolumn represents the percentage of processes in which peroxisomes are limited to the shaft. The orange subcolumn represents the percentage of processes that contain at least one peroxisome at the tip. The schematic on the right identifies the parameters used to define shaft and tip (see Materials and methods). Data represent mean values ± SD from 120 cells per condition from three separate dsRNA treatments.
To address this possibility, we examined whether there were quantitative changes in the distribution of GFP-labeled peroxisomes after RNAi-based depletion of either KHC or DHC relative to control cells. We classified cell processes according to peroxisome distribution as (a) empty of peroxisomes, (b) containing at least one peroxisome in the shaft, or (c) containing at least one peroxisome in the tip (described in Materials and methods; Fig. 1 C, right). Depletion of either DHC or KHC did not significantly alter the distribution of peroxisomes in the shaft and tip regions of S2 processes (P > 0.05, comparing motor-depleted with control cells). This suggested that depletion of either KHC or DHC rendered its opposite-polarity motor nonmotile (Fig. 1, B and C; and Videos 1–3). We believe that these immotile peroxisomes are carried into processes by “piggy backing” on microtubules, which extend with their plus ends outward as processes are formed.
Interestingly, upon CytoD treatment, KHC-depleted cells were morphologically distinct from other motor-depleted cells. These cells form only a single large process rather than multiple processes emanating from the cell center. However, like other motor-depleted cells, microtubules within this process are still oriented with their plus ends pointing to the cell periphery (unpublished data). We are currently investigating the contribution of kinesin-1 to the formation of radial processes in cells plated in the presence of CytoD.
Unc104 replaces kinesin-1 in bidirectional peroxisome transport
Having established that depletion or mutation of kinesin-1 completely abolishes transport by dynein and vice versa, we next identified the minimal components required to restore peroxisome motility. Specifically, we investigated whether the presence of two oppositely directed motors is sufficient for bidirectional motility of organelles. We used a previously published protein replacement strategy (Hoogenraad et al., 2003) to swap either endogenous KHC or DHC with motors that normally do not transport peroxisomes but have been engineered to contain a peroxisome-targeting sequence. If the presence of an opposite-polarity motor is sufficient for bidirectional peroxisome motility, then any motor that can transport cargo along microtubules will activate the motor of the opposite polarity.
We selected the plus end–directed kinesin-3 family member Unc104 as our KHC replacement motor. Importantly, Unc104 is not involved in peroxisome transport in S2 cells: endogenous Unc104 cannot transport peroxisomes in a KHC-depleted background (unpublished data). Unc104 transports synaptic vesicle precursors (Pack-Chung et al., 2007) required for morphogenesis of axonal growth cones into synaptic boutons in Drosophila. Although normally present as a monomer, Unc104 dimerizes when present at high concentrations on lipid vesicles (Klopfenstein et al., 2002). Tomishige et al. (2002) have previously shown that Caenorhabditis elegans Unc104, truncated after the head domain (amino acids 1–389), can dimerize through use of a C-terminal leucine zipper. This dimeric motor exhibits in vitro motility rates that are similar to those obtained from full-length Unc104 in Drosophila axons (Tomishige et al., 2002; Barkus et al., 2008). Furthermore, motility rates of dimeric Unc104 and dimeric kinesin-1 are similar (Tomishige et al., 2002).
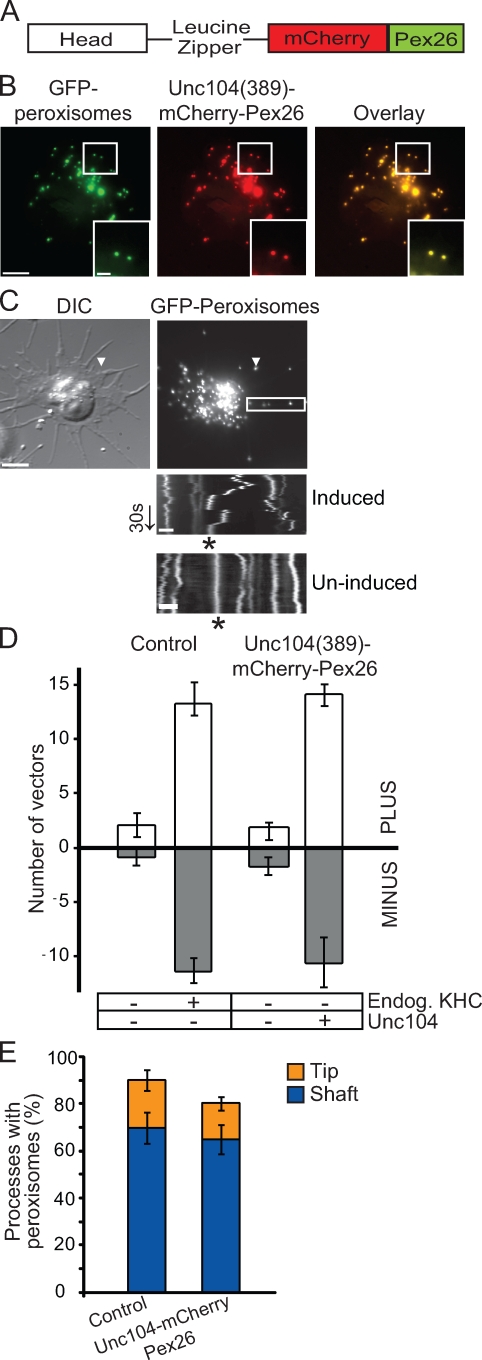
To use Unc104 as a replacement motor for KHC, we modified a previously developed strategy that targets motors onto peroxisomes (Hoogenraad et al., 2003). Peroxin (Pex) family proteins have been successfully used in HeLa cells as a method of targeting motor-related proteins onto peroxisomes (Hoogenraad et al., 2003). We used human Pex26 as an artificial N-terminal motor tag (a Drosophila homologue does not exist) to orient motor proteins onto peroxisomes. Residues 245–305 of human Pex26 are sufficient to target proteins onto peroxisomes in mammalian cells (Halbach et al., 2006). After expression of an mCherry-Pex26(245–305) fusion protein in S2 cells, ∼90% of GFP-labeled peroxisomes displayed the mCherry-Pex26(245–305) fusion protein (Fig. S2 A, top). We used this mCherry-Pex26 fusion protein to target truncated and dimeric C. elegans Unc104 (Tomishige et al., 2002) to peroxisomes in S2 cells; herein, this motor is referred to as Unc104(389)–mCherry-Pex26 (Fig. 2, A and B). We selected a cell line stably cotransfected with this construct under the control of a metallothionein (copper sulfate inducible) promoter and GFP-SKL (a peroxisome-targeting vector).
Figure 2.
Unc104 replaces kinesin-1 in bidirectional peroxisome transport. (A) Schematic of Unc104(389)–mCherry-Pex26 construct. Dimeric human Unc104 construct includes the head and the neck-linker domains (amino acids 1–389) followed by a leucine zipper for dimerization. Human Pex26 is fused to mCherry to generate a peroxisome-targeting vector. (B) Representative images showing colocalization of Unc104(389)–mCherry-Pex26 and GFP-labeled peroxisomes in stably transfected S2 cells. Boxed areas are shown at higher magnifications in the insets. (C) Unc104(389)–mCherry-Pex26 restores bidirectional peroxisome motility in KHC-depleted cells. Unc104(389)–mCherry-Pex26 expression induced with 5 mM copper sulfate. Arrowheads highlight location of a single peroxisome within a process. Images corresponds to Video 4. Boxed area delineates the region selected for kymograph analysis (bottom). Top kymograph is from a cell in which Unc104(389)–mCherry-Pex26 expression has been induced. Bottom kymograph is from a cell in which Unc104(389)–mCherry-Pex26 expression has not been induced. Endogenous KHC has been depleted in both cases. Asterisks indicate the track of a peroxisome. DIC, differential interference contrast. Arrow delineates time (30 s). (D) Graph showing the number of peroxisome vectors >0.2 µm in S2 cells expressing Unc104(389)–mCherry-Pex26 in an endogenous KHC-depleted background. Data represent mean values ± SD from 30 cells per condition (from three separate dsRNA treatments). PLUS refers to those peroxisomes moving toward the tips of processes, whereas MINUS refers to those moving toward the cell center. (E) Replacement of endogenous KHC with Unc104(389)–mCherry-Pex26 does not affect peroxisome distribution along processes. Graph representing the percentage of processes containing peroxisomes after induction of Unc104(389)–mCherry-Pex26 in a KHC-depleted background. Data are presented as mean values ± SD from 120 cells (from three separate dsRNA treatments). Bars: (B and C [top]) 5 µm; (C [bottom]) 1 µm.
Treatment of wild-type cells with double-stranded RNA (dsRNA) directed against the 5′ untranslated region of KHC mRNA depleted endogenous kinesin-1 and completely stopped peroxisome movement (compare Video 1 with Video 2). However, expression of exogenous full-length KHC in these cells fully restores peroxisome transport (see Fig. 5 C, top left). Peroxisomes in cells stably transfected with Unc104(389)–mCherry-Pex26 but not induced with copper sulfate were also immotile (Fig. 2, C [bottom] and D). However, upon induction of Unc104(389)–mCherry-Pex26 in an endogenous KHC-depleted background (Fig. S1 A, lanes 5–8), bidirectional transport of peroxisomes was restored (Fig. 2, C and D; and Video 4). We computed the relative number of vectors >0.2 µm as a measure of peroxisome motility. We have previously used the same method of vector analysis to demonstrate that peroxisomes do not move in the absence of either KHC or DHC (Kim et al., 2007). A single vector was defined as the distance a single peroxisome moved in 1 s (see Materials and methods). In fact, the number of vectors was similar in both Unc104(389)–mCherry-Pex26–expressing and control cells (P = 0.568; Fig. 2 D). This suggested that Unc104(389)–mCherry-Pex26, an equally fast and processive motor, replaced kinesin-1 in bidirectional peroxisome transport. Furthermore, in KHC-depleted and Unc104(389)–mCherry-Pex26–expressing cells, peroxisome distributions in the shafts and tips of processes were the similar to that of control; i.e., in both cases, ∼70% of processes contained peroxisomes in the shafts of processes (P = 0.529; Fig. 2 E). Thus, replacement of endogenous KHC with Unc104 restores peroxisome motility but does not bias transport in either direction.
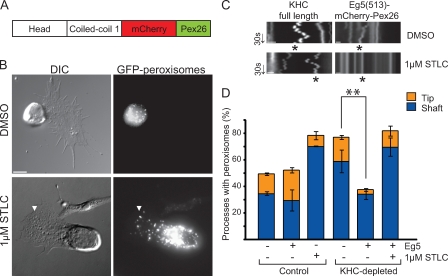
Figure 5.
Eg5 restores dynein-driven transport. (A) Schematic of Eg5(513)–mCherry-Pex26. Dimeric Xenopus Eg5 encodes the head, neck-linker, and first coiled-coil domains (amino acids 1–513). Dimeric Eg5 was cloned into the mCherry-Pex26 peroxisome-targeting vector. (B) Eg5(513)–mCherry-Pex26 expression results in peroxisome clustering at the cell center. S2 cells plated in the presence of CytoD and stably transfected with Eg5(513)–mCherry-Pex26 in an endogenous KHC-depleted background. (bottom) Plated in the presence of 1 µM STLC for 2 h. Arrowheads highlight location of a single peroxisome within a process. DIC, differential interference contrast. (C) Kymographs of GFP-labeled peroxisomes present along the length of a single process. Kymographs were derived from S2 cells expressing either full-length KHC (left) or Eg5(513)–mCherry-Pex26 (right) in an endogenous KHC-depleted background. Cells were plated in the presence of either DMSO (top) or 1 µM STLC (bottom). Asterisk indicates the track of a single peroxisome over the course of 1 min. Arrows delineate time (30 s). (D) Quantitative representation of results showing changes in the distribution of peroxisomes along processes before and after KHC depletion and expression of Eg5(513)–mCherry-Pex26. Data represent mean values ± SD from 120 cells per condition (from three separate dsRNA treatments). Asterisks indicate statistical significance using the Student’s t test. Bars: (B) 5 µm; (C) 1 µm.
Because Unc104(389)–mCherry-Pex26 is only targeted to peroxisomes, other organelles served as an internal control. Using MitoTracker green, we fluorescently labeled mitochondria in Unc104(389)–mCherry-Pex26–expressing cells in both control and KHC-depleted backgrounds. Mitochondria in control cells moved robustly along processes (Fig. S3, A and B). However, unlike peroxisomes, mitochondria did not display bidirectional movements upon Unc104(389)–mCherry-Pex26 induction in a KHC-depleted background (Fig. S3, A and B). This confirms that Unc104(389)–mCherry-Pex26 specifically drives the bidirectional motility of peroxisomes only.
Plus end–directed movement activates dynein-driven transport
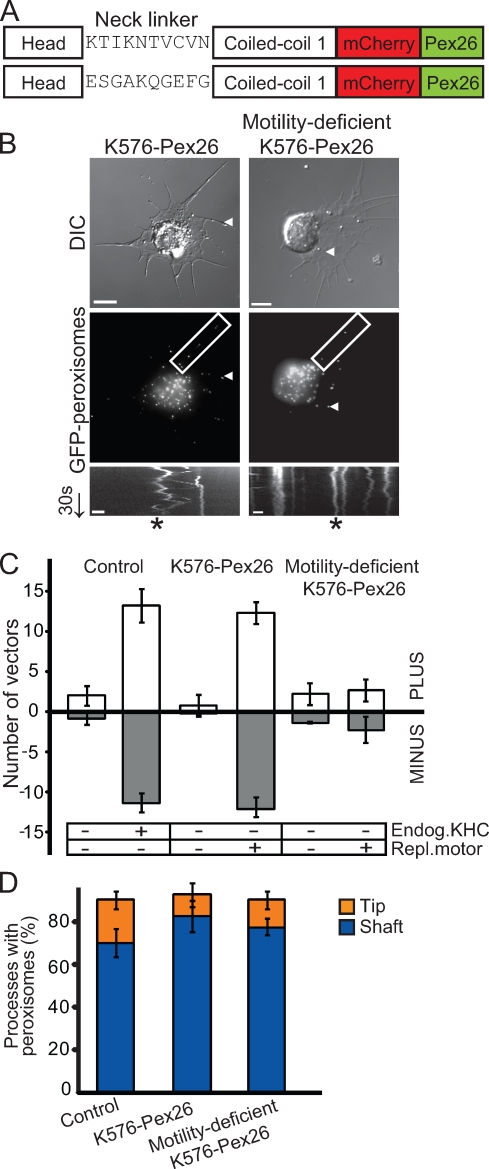
Thus far, our findings indicated that an exogenous, peroxisome-targeted, plus end–directed motor was sufficient to rescue bidirectional peroxisome motility in a KHC-depleted background.However, these results did not address whether motor motility was also essential for restoration of peroxisome transport. Perhaps microtubule-bound KHC (or any replacement motor) provided an extra link between cargo and microtubules and thus allowed processive runs by dynein. To address this possibility, we compared peroxisome transport in cells expressing peroxisome-targeted versions of either wild-type or motility-deficient KHC. The first kinesin-1 construct encoded amino acids 1–576 of Drosophila KHC (corresponding to the head, neck-linker, and first coiled-coil domains) cloned into our mCherry-Pex26–targeting vector, herein referred to as K576–mCherry-Pex26 (Fig. 3 A; and Fig. S2 B, top). The second kinesin construct was mutated such that the 10 amino residues in the neck linker were replaced with ESGAKQGEKG. This replacement, designed to form a random coil, rendered this motor largely immotile: it moves at a velocity 400-fold slower than wild type (Case et al., 2000). However, it has relatively normal catalytic properties and microtubule affinity (Case et al., 2000), indicating that it is comparable with wild-type kinesin in its ability to bind and release from microtubules in an ATP-dependent manner. This construct is herein referred to as motility-deficient K576ran10–mCherry-Pex26 (Fig. 3 A).
Figure 3.
Plus end–directed movement activates dynein-driven transport. (A) Schematic of K576–mCherry-Pex26 constructs. Dimeric Drosophila KHC includes amino acids 1–576 (head and first coiled-coil domains) cloned into the mCherry-Pex26 peroxisome–targeting vector. (top) K576–mCherry-Pex26 with wild-type neck linker. (bottom) Motility-deficient K576ran10–mCherry-Pex26 with mutated neck linker (Case et al., 2000). (B) K576–mCherry-Pex26 recues bidirectional peroxisome motility in KHC-depleted cells, but a motility-deficient version does not. (top) Representative micrographs of S2 cells plated in the presence of CytoD and stably transfected with either K576–mCherry-Pex26 (left) or motility-deficient K576ran10–mCherry-Pex26 (right) in endogenous KHC-depleted backgrounds. Boxed areas delineate regions selected for kymograph analysis (bottom). Arrowheads highlight location of a single peroxisome within a process. Arrow delineates time (30 s). DIC, differential interference contrast. Bars, 5 µm. (bottom) Kymographs showing peroxisome tracks over 1 min. Asterisks indicate the track of a single peroxisome. Bars, 1 µm. Video 5 corresponds to peroxisome motility in K576–mCherry-Pex26–expressing cells. (C) Graph showing the relative number of peroxisome vectors >0.2 µm in S2 cells expressing K576–mCherry-Pex26 in an endogenous KHC-depleted background. Data represent mean values ± SD from 30 cells per condition (from three separate dsRNA treatments). PLUS refers to those peroxisomes moving toward the tips of processes, whereas MINUS refers to those moving toward the cell center. (D) Motility-deficient K576ran10–mCherry-Pex26 does not rescue dynein-driven peroxisome motility toward the minus ends of microtubules. Graph represents the percentage of processes containing peroxisomes after induction of replacement motors in KHC-depleted backgrounds. Data represent mean values ± SD from 120 cells (from three separate dsRNA treatments).
In the absence of endogenous KHC (Fig. S1 B, lanes 1–4), K576–mCherry-Pex26 rescued bidirectional transport of peroxisomes (Fig. 3, B and C; and Video 5). In fact, the observed peroxisome motility was similar to control cells, with roughly the same relative number of vectors >0.2 µm (P = 0.582, comparing control with K576–mCherry-Pex26–expressing cells in endogenous KHC-depleted backgrounds; Fig. 3 C). Like Unc104(389)–mCherry-Pex26, this rescue of motility was indeed limited to peroxisomes in KHC-depleted cells, as mitochondria did not display the same bidirectional movements upon induction of K576–mCherry-Pex26 (Fig. S3, A and B).
In contrast, motility-deficient K576ran10–mCherry-Pex26 did not rescue any movement of peroxisomes in an endogenous KHC-depleted background in comparison with control cells (P = 0.001; Fig. 3, B and C). Bidirectional motility was not observed in these cells, and the distribution of peroxisomes in the shafts versus tips of processes in cells expressing K576ran10–mCherry-Pex26 was similar to control and KHC-depleted cells (P = 0.461; Fig. 3 D; and Fig. S1 B, lanes 5–8). Thus, motility-deficient K576ran10–mCherry-Pex26 was unable to activate dynein-driven transport. This suggested that the ability of kinesin-1 to bind microtubules and hydrolyze ATP was not sufficient to activate dynein-driven motility. Therefore, replacement motors did not function simply as an extra link on microtubules. Rather, an actively moving replacement motor was required to rescue organelle motility.
Ncd rescues kinesin-1–driven peroxisome transport
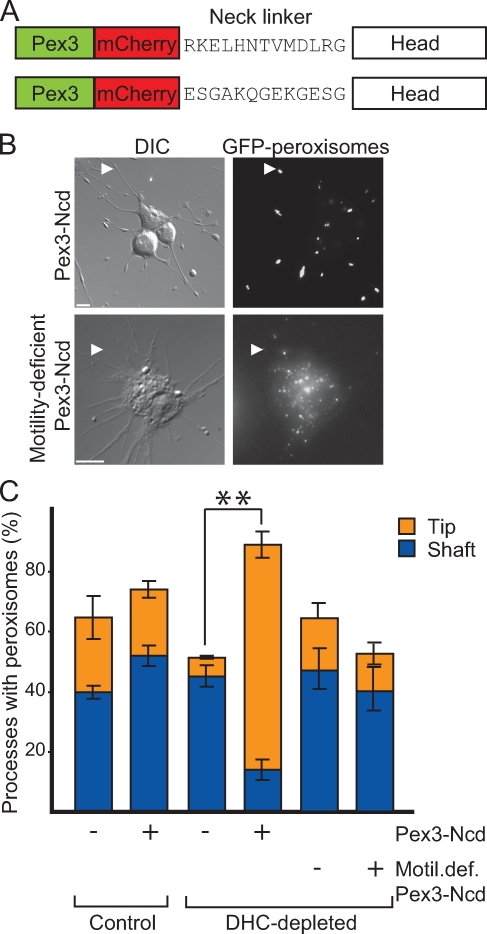
Replacing endogenous kinesin-1 with Unc104 clearly demonstrated that a different plus end motor, which moved along microtubules, was sufficient to restore peroxisome transport in KHC-depleted cells. We next performed the converse experiment: replacing DHC with a different minus end motor also targeted to peroxisomes. In this way, we determined whether another minus end motor can activate its opposite-polarity counterpart, i.e., kinesin-1. We selected kinesin-14 member Ncd as our replacement motor. Ncd is responsible for microtubule sliding during mitosis (Sharp et al., 2000; Hallen et al., 2008). Like Unc104, Ncd depletion does not affect peroxisome transport in S2 cells (unpublished data). However, unlike the Unc104/KHC replacement, its motile properties are somewhat different from those of dynein. In comparison with in vitro gliding assays, purified Ncd moved at a rate 10-fold less than that of dynein (Crevel et al., 1997; Sablin et al., 1998; Foster and Gilbert, 2000; Shima et al., 2006). We used a similar peroxisome-targeting strategy to artificially attach a dimeric Drosophila Ncd construct onto peroxisomes. In this instance, we used a different targeting signal (amino acids 1–40 of Drosophila Pex3; Fig. S2 A, bottom) to properly orient the C-terminal motor domain of Ncd onto peroxisomes (Fig. S2 B, middle; Hoogenraad et al., 2003). Peroxisome-targeted Ncd is herein referred to as Pex3-mCherry–Ncd(700) (Fig. 4 A).
Figure 4.
Ncd restores kinesin-1–driven transport. (A) Schematic of Pex3-mCherry–Ncd(700) constructs. Drosophila Pex3 is fused to mCherry and human Ncd encoded by amino acids 201–700. (top) Pex3-mCherry–Ncd(700) with wild-type neck linker. (bottom) Motility-deficient Pex3-mCherry–Ncd(700)ran12 with mutated neck linker (Sablin et al.,1998). (B) Pex3-mCherry–Ncd(700) expression results in accumulation of peroxisomes at the tips of processes. S2 cells stably transfected and plated in the presence of CytoD in an endogenous DHC-depleted background. DIC, differential interference contrast. Arrowheads highlight peroxisomes in processes. Bars, 5 µm. (C) Quantitative representation of results showing changes in the distribution of peroxisomes along processes before and after DHC depletion and expression of Pex3-mCherry–Ncd. Data represent mean values ± SD from 120 cells per condition from three separate dsRNA treatments. Asterisks indicate statistical significance using the Student’s t test.
In a DHC-depleted background (Fig. S1 C, lanes 1–4), Pex3-mCherry–Ncd(700) caused dramatic accumulation of peroxisomes at the tips of ∼80% of processes, whereas the cell body was almost devoid of peroxisomes (P = 0.02, comparing tip localization in induced vs. uninduced cells in the absence of DHC; Fig. 4, B and C). Very often, cells expressing Pex3-mCherry–Ncd(700) in a DHC-depleted background displayed swellings at the tips of processes. Upon further examination using fluorescence microscopy, it was clear that tightly packed peroxisomes induced formation of these swellings (Fig. 4 B, top). Only peroxisomes clustered in this manner at the tips of processes, as mitochondria did not accumulate similarly upon Pex3-mCherry–Ncd(700) induction in a DHC-depleted background (Fig. S3, A–C). These data suggested that Pex3-mCherry–Ncd(700) was sufficient to activate kinesin-1–driven peroxisome transport toward the tips of processes. Most likely, Ncd also moved peroxisomes, albeit briefly, toward the minus ends of microtubules. However, Ncd was unable to compete with kinesin-1, and thus, plus end–directed transport was dominant.
We then tested whether motile activity of the replacement Ncd motor was required to rescue peroxisome transport. We replaced dynein with a motility-deficient Pex3-mCherry–Ncdran12(700). Sablin et al. (1998) have shown that replacement of the neck-linker domain of Ncd with a 12–amino acid sequence (ESGAKQGEKGESG) renders the motor very nearly immotile but does not significantly affect microtubule binding or ATP hydrolysis. We stably expressed this construct in S2 cells (Fig. 4 A). In a DHC-depleted background, expression of motility-deficient Pex3-mCherry–Ncdran12(700) does not result in accumulation of peroxisomes at process tips (Fig. 4, B [bottom] and C). Rather, peroxisomes were immotile, and their distribution in the tips and shafts of processes was similar to control, KHC-, and DHC-depleted cells (Fig. 4 C). This suggests that kinesin-1 could not transport peroxisomes toward the tips of processes in the presence of motility-deficient Pex3-mCherry–Ncdran12(700). Clearly, opposite-polarity motors must be able to move along microtubules to support any kind of peroxisome transport. In addition, the accumulation of peroxisomes at the tips of processes is specifically caused by actively moving Pex3-mCherry–Ncd(700).
Eg5 rescues dynein-driven peroxisome transport
Thus far, we have shown that replacement motors Unc104 and Ncd activate their opposite-polarity counterparts (DHC and KHC, respectively) and restore peroxisome transport. This reciprocal activation appears to be contingent upon both motors retaining the ability to move along microtubules. We demonstrated this using largely immotile KHC and Ncd mutants in which the catalytic and microtubule-binding properties of both motors are mostly intact. However, would catalytic inhibition of a replacement motor also prevent motility driven by motor of the opposite polarity? That is, would a chemically inhibited motor have the same effect as expression of motility-deficient KHC? In this study, the plus end motor would also be targeted to peroxisomes, except inhibition of this motor would disrupt ATP hydrolysis. For these experiments, we used Eg5 (a plus end kinesin-5 family member) as a KHC replacement motor because it can be chemically inhibited in vitro (Brier et al., 2006) and in vivo (Skoufias et al., 2006), and the inhibited motor does not form a rigor complex with microtubules (Crevel et al., 2004).
Eg5 is a homotetrameric mitotic motor involved in microtubule sliding during cell division (Kashina et al., 1996; Kapitein et al., 2005). A dimeric version of Xenopus Eg5 (amino acids 1–513) displays the same motor properties in vitro as the full-length tetramer (Krzysiak et al., 2006; Kwok et al., 2006; Valentine et al., 2006; Korneev et al., 2007). However, its motile speed is 10-fold less than those of kinesin-1 or Unc104 (Hirose et al., 2000; Tomishige et al., 2002; Block et al., 2003; Crevel et al., 2004; Valentine et al., 2006; Valentine and Gilbert, 2007).
We created a peroxisome-targeted version of dimeric Eg5 (Fig. S2 B, bottom), herein referred to as Eg5(513)–mCherry-Pex26 (Fig. 5 A). Remarkably, Eg5(513)–mCherry-Pex26 induction resulted in a phenotype opposite to that seen in Pex3-mCherry–Ncd(700)-expressing cells. In an endogenous KHC-depleted background (Fig. S1 A, lanes 1–4), Eg5(513)–mCherry-Pex26 induction caused peroxisome clustering at the cell center (Fig. 5 B). In these cells, <30% of processes contained peroxisomes in the shaft (Fig. 5 D). This phenotype of peroxisome clustering is in stark contrast to cells that we did not induce with Eg5(513)–mCherry-Pex26 expression. In those cells, ∼70% of processes contained peroxisomes in the shaft (P = 0.03, comparing induced vs. uninduced cells in the absence of endogenous KHC; Fig. 5 D). Thus, induction of this slower plus end–directed motor activated dynein-dependent transport, resulting in peroxisome clusters at the cell center. This phenotype was limited to peroxisomes, as mitochondria did not cluster at the cell center upon Eg5(513)–mCherry-Pex26 induction (compare Fig. 5 B [top] with Fig. S3 A; and compare Fig. 5 D with Fig. S3 C).
But would chemically inhibited Eg5 still activate dynein-driven peroxisome transport? To answer this question, we used the small molecule S-trityl-l-cysteine (STLC) to inhibit Eg5(513)–mCherry-Pex26. STLC is a monastrol-like inhibitor that specifically inhibits ATP hydrolysis by Eg5 in vitro and in vivo (Brier et al., 2006; Skoufias et al., 2006). Importantly, STLC addition did not affect KHC, as peroxisomes moved robustly along processes in cells expressing full-length KHC (Fig. 5 C, left). Furthermore, the distribution of peroxisomes in the shafts and tips of processes was similar to control cells (unpublished data).
We depleted endogenous KHC and induced Eg5(513)–mCherry-Pex26 expression cells in the presence of STLC. Peroxisome motility in these cells was completely inhibited; we did not observe any movements in either direction (Fig. 5 C, bottom right). Significantly, peroxisomes did not cluster at the cell center. Instead, peroxisomes were distributed along the shafts of processes (Fig. 5 B, bottom). We confirmed this result through quantification of peroxisome distribution along processes in the presence and absence of STLC (Fig. 5 D). The distribution of peroxisomes in STLC-treated cells was similar to that of control cells but in sharp contrast to cells expressing actively moving Eg5(513)–mCherry-Pex26 (P = 0.03, comparing Eg5-expressing cells plated in the presence or absence of STLC). This result suggested that dynein could not transport peroxisomes toward the cell center in the presence of chemically inhibited Eg5(513)–mCherry-Pex26. Thus, peroxisome clustering was specifically caused by the expression of an actively moving Eg5(513)–mCherry-Pex26 in the presence of endogenous dynein. Collectively, these data suggested that actively moving Eg5(513)–mCherry-Pex26 was sufficient for restoration of dynein-driven motility toward the minus end. Only a few, brief plus end runs were detected (Fig. 5 C, top right), suggesting that minus end–directed motility was dominant.
Discussion
Many types of cargo in numerous cell types move in a bidirectional manner along cytoplasmic microtubules, with plus and minus end motors present on the surface of cargo at the same time. Furthermore, typically, neither kinesin nor dynein can transport cargo in the absence of an opposite-polarity motor (Martin et al., 1999; Gross et al., 2002b; Ling et al., 2004; Pilling et al., 2006; Kim et al., 2007; Barkus et al., 2008; Uchida et al., 2009). Our results indicate that opposite-polarity motors activate one another, thereby driving bidirectional peroxisome transport in live cells. Thus, replacement of endogenous Drosophila KHC with plus end motors Eg5 or Unc104 restores dynein-driven peroxisome transport. In the converse experiment, replacement of endogenous DHC with the minus end motor Ncd restores kinesin-1–driven peroxisome transport. These results show that opposite-polarity motors are both necessary and sufficient for activation of peroxisome transport provided that they (a) are cargo bound and (b) move along microtubules.
Interestingly, although all motile replacement motors activated the opposite-polarity motor, the peroxisome distributions we observed were dependent on the replacement motor used. For example, Unc104, which has motile properties similar to those of kinesin-1, rescued bidirectional peroxisome transport. In addition, the overall peroxisome distribution in the shafts and tips of processes was similar to that observed in wild-type cells. In contrast, replacement of DHC or KHC with either Ncd or Eg5 resulted in accumulation of peroxisomes at either the tips of processes or cell body, respectively. These differences in peroxisome localization are unlikely the result of variable protein expression, as all of the replacement motors in this study are kinesins targeted to peroxisomes in a similar manner and expressed at equivalent levels. Similarly, morphological differences in cells after KHC depletion and/or expression of replacement motors did not likely contribute to the aforementioned difference in peroxisome distribution. For example, despite having altered morphology, organelle distribution in KHC-depleted cells was similar to that of wild-type cells. Furthermore, chemical inhibition of Eg5 induced a shift in peroxisome distribution in cells that were otherwise identical.
In this study, the velocity of the replacement motors may in fact be a key parameter in determining the distribution of motile peroxisomes. Although Unc104 and kinesin-1 move at similar speeds in vivo and in vitro, Eg5 and Ncd are approximately 10-fold slower (Crevel et al., 1997; Hirose et al., 2000; Tomishige et al., 2002; Block et al., 2003; Valentine et al., 2006; Barkus et al., 2008; Furuta and Toyoshima, 2008). In our experiments, the presence of equally fast opposite-polarity motors (Unc104 and DHC) produces numerous and long plus and minus end–directed runs. In contrast, motors with a greater velocity differential (like Ncd and KHC or Eg5 and DHC) result in peroxisome clusters at either the tips of processes or the cell center. In this study, we show that Eg5 and Ncd activate but cannot compete with their opposite-polarity counterparts (DHC and kinesin-1, respectively). Thus, peroxisome transport appears to be restored only in the direction of the faster motor.
Several possible explanations may account for the reciprocal activation of motors described in this study. Perhaps a plus end motor pulls against a minus end motor (and vice versa), and that pulling force is necessary for activation of the opposite-polarity motor. Interestingly, two motors of the same polarity cannot support peroxisome transport altogether (unpublished data), suggesting that opposing forces are required for activation. Recently, using in vitro–reconstituted endosome motility and bead assays, Soppina et al. (2009) have shown that dissimilar teams of dynein and kinesin-1 motors are engaged in a tug of war to drive net cargo transport. These data support our hypothesis that motors may be cooperating mechanically to activate one another and thus promote bidirectional transport.
Several in silico models of bidirectional cargo transport support the idea that opposite-polarity motors are coordinated through their mechanical interactions (Müller et al., 2008; Howard, 2009). Müller et al. (2008) developed an in silico model in which multiple motors rapidly engage and disengage from cargo, predominantly using their motor properties, and thus, independent of any additional regulatory components. Our experiments presented in this study also suggest that opposite-polarity motors activate one another, and thus, can transport cargo seemingly independent of additional components.
However, we do not believe that regulatory pathways and the reciprocal activation of motors described in this study are mutually exclusive during bidirectional cargo transport. Most likely, these higher order pathways fine tune transport and integrate motor–motor activation with overall cellular responses. For example, upon stimulation in Xenopus melanophores, PKA (cyclic AMP–dependent protein kinase A) triggers motor-mediated melanosome aggregation at the cell center or dispersion toward the cell periphery (Tuma et al., 1998). Other upstream regulators also have a potentially critical role in dictating the final destination of cargo within a cell. JNK interacts with several JNK-interacting proteins to regulate KHC-mediated transport of mitochondria in axons (Verhey et al., 2001; Horiuchi et al., 2007). A plethora of other signaling pathways linked to developmental processes also affect cargo transport (Cohen, 2003; Gross et al., 2003; Kelkar et al., 2005; Cox and Spradling, 2006; Colin et al., 2008; Kovacs et al., 2008; Caviston and Holzbaur, 2009; Montagnac et al., 2009; Wang and Schwarz, 2009). Such signaling pathways are critical in determining net distributions of cargo but may not directly affect the reciprocal activation of opposite-polarity motors described in this study.
Many experiments have explored the biological significance of bidirectional transport (Welte, 2004). Although it may appear inefficient in comparison with unidirectional movement, bidirectional transport is far better suited for maneuvering through a crowded cytoplasm. In contrast with in vitro conditions, during intracellular cargo transport, motors must navigate through a tightly packed cytoplasmic milieu where they may encounter multiple “roadblocks” en route to their final destination (Telley et al., 2009). Such roadblocks could include organelles, microtubule-associated proteins, and other cytoskeletal elements. If a cargo-bound motor encounters such an obstruction, the opposite-polarity motor can take back steps to move around the obstruction. This leads to numerous back and forth movements, which is characteristic of bidirectional transport.
Furthermore, bidirectional transport could serve as a “proofreading mechanism,” whereby a cargo-bound motor can rapidly search the cytoplasm for the correct destination of its cargo. Such mechanisms are in fact more efficient because unidirectional transport does not allow error correction if a cargo is delivered to the wrong location. This model is similar to the classical search and capture mechanism used by dynamic microtubules during mitosis (Mitchison and Kirschner, 1984). Like a microtubule end searching for a suitable location before capture, motor-bound cargo can also explore numerous destinations before selective stabilization in the correct position.
Materials and methods
Molecular cloning
Full-length Drosophila KHC (amino acids 1–975) was cloned KpnI–NotI into pMT-A (Invitrogen) to generate pMT KHC. Peroxisome-targeting vectors were cloned as follows: mCherry-Pex26 includes mCherry and amino acids 245–305 of Pex26 (cDNA clone 391035; Thermo Fisher Scientific). Both sequences were amplified by PCR and annealed together by overlap PCR. mCherry-Pex26 was cloned into pMT-A (Invitrogen) using NotI–XbaI cloning sites. Pex3-mCherry includes amino acids 1–40 of Pex3 (amplified from pSMART6 Pex3 cDNA; Drosophila Genomics Resource Center, Indiana University, Bloomington, IN) and mCherry. Both sequences were amplified via PCR and annealed together by overlap PCR. Pex3-mCherry was cloned into pMT-A (Invitrogen) using KpnI–NotI. Restriction sites were included in primer design. Amino acids 1–576 of KHC (from pMT KHC) were amplified by PCR and cloned EcoRI–NotI into pMT mCherry-Pex26 to generate K576–mCherry-Pex26. The neck linker in K576–mCherry-Pex26 was replaced with the sequence ESGAKQGEKG using site-directed mutagenesis (Agilent Technologies) to generate motility-deficient K576ran10–mCherry-Pex26 (Drosophila equivalent of the human kinesin-1 neck-linker mutation in Case et al., 2000). Amino acids 1–513 of Xenopus Eg5 were amplified from pBS KS+ Eg5 FL (provided by C. Walczak, Indiana University) and cloned Kpn–NotI into pMT mCherry-Pex26 to generate Eg5(513)–mCherry-Pex26. Amino acids 1–389 of C. elegans Unc104 followed by a leucine zipper (Tomishige et al., 2002) were amplified from pET17 U389-LZ (provided by R.Vale, University of California, San Francisco, San Francisco, CA) and cloned KpnI–NotI into pMT mCherry-Pex26 to generate Unc104(389)–mCherry-Pex26. Amino acids 209–700 of Drosophila Ncd were amplified by PCR from pET17b Ncd (provided by R. Vale) and cloned NotI–Xba into the pMT Pex3-mCherry vector to generate Pex3-mCherry–Ncd(700). Amino acids 209–700 of motility-deficient Ncd were amplified from pET17b Ncdran12 (provided by R. Vale) and cloned NotI–XbaI into the pMT Pex3-mCherry vector to generate motility-deficient Pex3-mCherry–Ncdran12(700).
Cell culture
Conditions for S2 cell culture have been described previously (Ling et al., 2004). In brief, S2 cells were maintained in a humidified RT incubator in S2 media: Schneider’s media (Sigma-Aldrich) supplemented with 10% FBS (Atlanta Biologicals). Selection of stable cell lines was performed as described previously (Kim et al., 2007). In brief, motor-mCherry-Pex fusions were cotransfected into S2 cells in 20-fold excess over pGG101 (encoding GFP-SKL; provided by G. Goshima, University of California, San Francisco) and pCoHygro (hygromycin selection plasmid obtained from Invitrogen). 10 µl of the transfection reagent Cellfectin (Invitrogen) was used for each stable cell line.
RNAi
The procedures used for RNAi-based depletion and primer sequences have been described previously (Clemens et al., 2000; Goshima and Vale, 2003) with the following changes. Drosophila KHC cDNA was purchased from the Drosophila Genomics Resource Center. Primers for amplification of in vitro transcription templates are as follows: T7-5′ untranslated region KHC, 5′-TAATACGACTCACTATAGGGTGCTTACAGGGCGGAGATAG-3′ and 5′-TAATACGACTCACTATAGGGCTCCAGCGATATCGCCATTT-3′; T7-Klp68D, 5′-TAATACGACTCACTATAGGGCATGATCAAAATCGAGATGTGC-3′ and 5′-TAATACGACTCACTATAGGGAAGTTGACCCTCCAATTCTGC-3′; T7-DHC, 5′-TAATACGACTCACTATAGGGAAACTCAACAGAATTAACGCCC-3′ and 5′-TAATACGACTCACTATAGGGTTGGTACTTGTCACACCACTCC-3′. dsRNA was generated using an in vitro transcription reaction. 1 µg of T7-appended PCR product was mixed with 12 mM rNTPS (Sigma-Aldrich), transcription buffer (80 mM Hepes, pH 7.5, 12 mM MgCl2, 2 mM spermidine, and 40 mM DTT), 1 µl RNAsin (1 µg/ml; Sigma-Aldrich), 0.1 U/ml pyrophosphatase (Sigma-Aldrich), and 5 µl purified T7 enzyme (100 µg/ml). The transcription mixture was incubated in a 37°C water bath for 2 h. dsRNA was precipitated using lithium chloride and 70% ethanol, and the resulting pellet was resuspended in DEPC-treated water. 105 cells/well were plated in a 12-well plate. Cells were brought up to a volume of 1 ml in S2 media. To deplete a protein of interest, 12 µg dsRNA was added per well for 2 d at RT. Cells were split 1:2 on the third day and brought up to 1 ml with fresh S2 media. An additional 12 µg dsRNA was added at this time, and cells were incubated at RT for an additional day. 4 d after cells were first plated, cells were induced (if needed) using copper sulfate (final concentration of 5 mM) for at least 12 h.
Antibodies
DHC hybridoma cell line was provided by J. Scholey (University of California, Davis, Davis, CA). Hybridoma cell line SUK4 (against KHC) was obtained from the Developmental Studies Hybridoma Bank. Both antibodies were purified at the Northwestern University Monclonal Antibody Facility (Feinberg School of Medicine, Chicago, IL). The mouse anti–rabbit Living Colors dsRed antibody (Takara Bio Inc.) was used at 1:1,000 dilution to detect expression of mCherry-Pex fusions. The anti-Klp68D antibody was provided by L.S.B. Goldstein (University of California, San Diego, La Jolla, CA).
Cell lysate preparation
For whole cell lysates, ∼105 cells were collected for analysis of protein expression by Western blotting. Cells were harvested and pelleted by centrifugation at 1,000 g for 2 min at RT. The cell pellet was rinsed and pelleted twice in sterile PBS. Finally, the cell pellet was resuspended in 200 µl 4× SDS-PAGE loading buffer. 10 µl was loaded onto an SDS-PAGE gel for Western blotting.
Preparation of cells for imaging
Cells were treated with 0.4 mM copper sulfate (final concentration) for 12 h to induce expression of mCherry-Pex fusions. Induced cells were plated on 25-mm coverslips precoated with 500 µg/ml Con A (Sigma-Aldrich) for 1 h. Cells were plated in the presence of 1 µm CytoD (Sigma-Aldrich) for a minimum of 2 h to depolymerize actin. Where required, cells were plated in the presence of 1 µM STLC (Sigma-Aldrich) in addition to CytoD. Cells used for analyses of peroxisome motility expressed both mCherry fusions and a plasmid encoding GFP-SKL at the time of imaging. Cells used for analyses of mitochondria motility expressed only mCherry-Pex fusions. Mitochondria were labeled using 100 nM MitoTracker green (Invitrogen) for 15 min at RT. Cells were washed at least three times in S2 media without MitoTracker before imaging in fresh S2 media with CytoD.
Cell fixation
Before fixation, cells were rinsed twice in HL3 buffer (70 mM NaCl, 5 mM KCl, 1.5 mM NaCl, 20 mM MgCl2, 10 mM NaHCO3, 5 mM trehalose, 115 mM sucrose, and 5 mM Hepes, pH 7.2). Cells were fixed in 3.7% formaldehyde (Sigma-Aldrich) in HL3 buffer for 15 min. Coverslips were transferred to clean, 35-mm dishes and rinsed three times in HL3 buffer. Rinsed coverslips were mounted onto glass slides for imaging.
Microscopy
Images of live and fixed cells were acquired using an inverted microscope (Eclipse U2000 Perfect Focus; Nikon) with a 100× 1.49 NA objective. Either a 100-W mercury bulb or a light-emitting diode light source (precisExcite) was used for fluorescence excitation. Images of motile GFP-labeled peroxisomes were captured at RT every 1 s for 1 min using an EM charge-coupled device camera (Cascade II; Roper Industries) run by MetaMorph software (MDS Analytical Technologies). MitoTracker green–labeled mitochondria were imaged using the same set up and conditions as for peroxisomes, except images were captured every 1 s over a period of 3 or 5 min.
Analysis of live cell imaging
Peroxisome movements were analyzed using DiaTrak software (version 3.01; Semasopht). DiaTrak discarded particles that were deemed as noise (from either the camera or illumination). A single vector was defined as the distance moved by a single peroxisome in 1 s. Vectors were separated into two categories: PLUS (toward the tip of a given process) or MINUS (toward the cell center). The number of vectors >0.2 µm for either direction was divided by the mean number of peroxisomes. This value was known as the number of vectors > 0.2 µm. Three independent experiments were conducted, and 10 cells were analyzed per condition per experiment. The number of vectors for each condition (± SD) was calculated using a spreadsheet (Excel; Microsoft). All kymographs were made using ImageJ software (National Institutes of Health). The Student’s two-tailed t test for independent variables using a 95% confidence interval was used to determine statistical significance.
Mitochondria movements were analyzed using the MTtrack plug-in in ImageJ. Three independent experiments (three separate dsRNA treatments) were conducted, and five cells were analyzed per condition (total of 11 different conditions) per experiment. In each cell, 10 mitochondria were selected at random for analysis provided that they were (a) within processes ≥5 µm in length and (b) remained visible for at least 15 frames. Each mitochondrion was tracked manually by selecting the same tip of a given mitochondrion for a maximum of 60 frames. The distance moved by a single mitochondrion in 1 s (defined as a single vector) was tabulated. The number of vectors for each condition was calculated using a spreadsheet (Excel). The number of vectors >0.2 µm was determined (±SD) and plotted as shown in Fig. S3. The Student’s two-tailed t test for independent variables using a 95% confidence interval was used to determine statistical significance.
Quantification of peroxisome and mitochondria distribution
Quantification of the number of processes containing peroxisomes or mitochondria was conducted as follows. The “tip” of the process was defined as a circular region of 1-µm diameter at the very extreme end of each process (Fig. 1 C, right). A 1-µm diameter circle was drawn in ImageJ. In each case, the outer circumference of the circle was kept in alignment with the extreme tip of each process. The “shaft” was defined the region between the cell center and the tip. Only processes with a shaft length ≥5 µm were used in analyses. Processes were scored as containing (a) no peroxisomes/mitochondria, (b) at least one peroxisome/mitochondrion in the shaft, (c) at least one peroxisome/mitochondrion in the tip, or (d) at least one peroxisome/mitochondrion in both the shaft and tip. 40 cells were imaged per condition and repeated in triplicate (i.e., a sum total of 120 cells per condition). The mean ± SD were calculated from all 120 cells using a spreadsheet. The Student’s two-tailed t test for independent variables using a 95% confidence interval was used to determine statistical significance.
Online supplemental material
Fig. S1 shows that replacement motors can be expressed in S2 cells in the absence of endogenous motors. Western blots confirming RNAi-based motor depletion and expression of replacement motors are depicted. Fig. S2 shows that truncated motors can be targeted to peroxisomes using Pex family proteins. Differential interference contrast and fluorescent micrographs of S2 cells demonstrating colocalization between GFP-labeled peroxisomes and motor-mCherry-Pex fusions are shown. Fig. S3 shows that expression of peroxisome-targeted motors does not affect mitochondria motility and distribution. Detailed analyses of mitochondria motility (vector analysis) and distribution (DIC and fluorescent micrographs and quantification) in S2 cells expressing replacement motors are shown. Video 1 shows peroxisome motility in control S2 cells. Video 2 shows inhibition of bidirectional peroxisome motility in a KHC-depleted cell. Video 3 shows inhibition of bidirectional peroxisome motility in a DHC-depleted cell. Video 4 shows that expression of Unc104(389)–mCherry-Pex26 in a KHC-depleted background restores bidirectional peroxisome transport. Video 5 shows that expression of K576–mCherry-Pex26 in a KHC-depleted background restores bidirectional peroxisome transport. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908075/DC1.
Acknowledgments
We thank Claire Walczak and Ron Vale for constructs and Steven Gould for advice on peroxisome targeting.
This work was supported by the National Institutes of Health (grants GM52111 to V.I. Gelfand and GM72656 to S.E. Rice).
Footnotes
Abbreviations used in this paper:
- CytoD
- cytochalasin D
- DHC
- dynein heavy chain
- dsRNA
- double-stranded RNA
- KHC
- kinesin heavy chain
- Pex
- peroxin
- STLC
- S-trityl-l-cysteine
References
- Barkus R.V., Klyachko O., Horiuchi D., Dickson B.J., Saxton W.M. 2008. Identification of an axonal kinesin-3 motor for fast anterograde vesicle transport that facilitates retrograde transport of neuropeptides. Mol. Biol. Cell. 19:274–283 10.1091/mbc.E07-03-0261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block S.M., Asbury C.L., Shaevitz J.W., Lang M.J. 2003. Probing the kinesin reaction cycle with a 2D optical force clamp. Proc. Natl. Acad. Sci. USA. 100:2351–2356 10.1073/pnas.0436709100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brier S., Lemaire D., DeBonis S., Forest E., Kozielski F. 2006. Molecular dissection of the inhibitor binding pocket of mitotic kinesin Eg5 reveals mutants that confer resistance to antimitotic agents. J. Mol. Biol. 360:360–376 10.1016/j.jmb.2006.04.062 [DOI] [PubMed] [Google Scholar]
- Case R.B., Rice S., Hart C.L., Ly B., Vale R.D. 2000. Role of the kinesin neck linker and catalytic core in microtubule-based motility. Curr. Biol. 10:157–160 10.1016/S0960-9822(00)00316-X [DOI] [PubMed] [Google Scholar]
- Caviston J.P., Holzbaur E.L. 2009. Huntingtin as an essential integrator of intracellular vesicular trafficking. Trends Cell Biol. 19:147–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviston J.P., Ross J.L., Antony S.M., Tokito M., Holzbaur E.L. 2007. Huntingtin facilitates dynein/dynactin-mediated vesicle transport. Proc. Natl. Acad. Sci. USA. 104:10045–10050 10.1073/pnas.0610628104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens J.C., Worby C.A., Simonson-Leff N., Muda M., Maehama T., Hemmings B.A., Dixon J.E. 2000. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc. Natl. Acad. Sci. USA. 97:6499–6503 10.1073/pnas.110149597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen R.S. 2003. Halo: a guiding light for transport. Curr. Biol. 13:R869–R870 10.1016/j.cub.2003.10.046 [DOI] [PubMed] [Google Scholar]
- Cohen R.S. 2005. Microtubule motors: LSD2 trips the toggle. Curr. Biol. 15:R651–R653 10.1016/j.cub.2005.08.027 [DOI] [PubMed] [Google Scholar]
- Colin E., Zala D., Liot G., Rangone H., Borrell-Pagès M., Li X.J., Saudou F., Humbert S. 2008. Huntingtin phosphorylation acts as a molecular switch for anterograde/retrograde transport in neurons. EMBO J. 27:2124–2134 10.1038/emboj.2008.133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R.T., Spradling A.C. 2006. Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development. 133:3371–3377 10.1242/dev.02514 [DOI] [PubMed] [Google Scholar]
- Crevel I.M., Lockhart A., Cross R.A. 1997. Kinetic evidence for low chemical processivity in ncd and Eg5. J. Mol. Biol. 273:160–170 10.1006/jmbi.1997.1319 [DOI] [PubMed] [Google Scholar]
- Crevel I.M., Alonso M.C., Cross R.A. 2004. Monastrol stabilises an attached low-friction mode of Eg5. Curr. Biol. 14:R411–R412 10.1016/j.cub.2004.05.030 [DOI] [PubMed] [Google Scholar]
- Duncan J.E., Warrior R. 2002. The cytoplasmic dynein and kinesin motors have interdependent roles in patterning the Drosophila oocyte. Curr. Biol. 12:1982–1991 10.1016/S0960-9822(02)01303-9 [DOI] [PubMed] [Google Scholar]
- Foster K.A., Gilbert S.P. 2000. Kinetic studies of dimeric Ncd: evidence that Ncd is not processive. Biochemistry. 39:1784–1791 10.1021/bi991500b [DOI] [PubMed] [Google Scholar]
- Freed J.J., Lebowitz M.M. 1970. The association of a class of saltatory movements with microtubules in cultured cells. J. Cell Biol. 45:334–354 10.1083/jcb.45.2.334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuta K., Toyoshima Y.Y. 2008. Minus-end-directed motor Ncd exhibits processive movement that is enhanced by microtubule bundling in vitro. Curr. Biol. 18:152–157 10.1016/j.cub.2007.12.056 [DOI] [PubMed] [Google Scholar]
- Goshima G., Vale R.D. 2003. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J. Cell Biol. 162:1003–1016 10.1083/jcb.200303022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.P., Tuma M.C., Deacon S.W., Serpinskaya A.S., Reilein A.R., Gelfand V.I. 2002a. Interactions and regulation of molecular motors in Xenopus melanophores. J. Cell Biol. 156:855–865 10.1083/jcb.200105055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.P., Welte M.A., Block S.M., Wieschaus E.F. 2002b. Coordination of opposite-polarity microtubule motors. J. Cell Biol. 156:715–724 10.1083/jcb.200109047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross S.P., Guo Y., Martinez J.E., Welte M.A. 2003. A determinant for directionality of organelle transport in Drosophila embryos. Curr. Biol. 13:1660–1668 10.1016/j.cub.2003.08.032 [DOI] [PubMed] [Google Scholar]
- Halbach A., Landgraf C., Lorenzen S., Rosenkranz K., Volkmer-Engert R., Erdmann R., Rottensteiner H. 2006. Targeting of the tail-anchored peroxisomal membrane proteins PEX26 and PEX15 occurs through C-terminal PEX19-binding sites. J. Cell Sci. 119:2508–2517 [DOI] [PubMed] [Google Scholar]
- Hallen M.A., Liang Z.Y., Endow S.A. 2008. Ncd motor binding and transport in the spindle. J. Cell Sci. 121:3834–3841 10.1242/jcs.038497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirokawa N., Noda Y. 2008. Intracellular transport and kinesin superfamily proteins, KIFs: structure, function, and dynamics. Physiol. Rev. 88:1089–1118 10.1152/physrev.00023.2007 [DOI] [PubMed] [Google Scholar]
- Hirokawa N., Takemura R. 2004. Molecular motors in neuronal development, intracellular transport and diseases. Curr. Opin. Neurobiol. 14:564–573 10.1016/j.conb.2004.08.011 [DOI] [PubMed] [Google Scholar]
- Hirose K., Henningsen U., Schliwa M., Toyoshima C., Shimizu T., Alonso M., Cross R.A., Amos L.A. 2000. Structural comparison of dimeric Eg5, Neurospora kinesin (Nkin) and Ncd head-Nkin neck chimera with conventional kinesin. EMBO J. 19:5308–5314 10.1093/emboj/19.20.5308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck P.J., Saxton W.M. 2005. The axonal transport of mitochondria. J. Cell Sci. 118:5411–5419 10.1242/jcs.02745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogenraad C.C., Wulf P., Schiefermeier N., Stepanova T., Galjart N., Small J.V., Grosveld F., de Zeeuw C.I., Akhmanova A. 2003. Bicaudal D induces selective dynein-mediated microtubule minus end-directed transport. EMBO J. 22:6004–6015 10.1093/emboj/cdg592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi D., Collins C.A., Bhat P., Barkus R.V., Diantonio A., Saxton W.M. 2007. Control of a kinesin-cargo linkage mechanism by JNK pathway kinases. Curr. Biol. 17:1313–1317 10.1016/j.cub.2007.06.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. 2009. Mechanical signaling in networks of motor and cytoskeletal proteins. Annu Rev Biophys. 38:217–234 10.1146/annurev.biophys.050708.133732 [DOI] [PubMed] [Google Scholar]
- Kapitein L.C., Peterman E.J., Kwok B.H., Kim J.H., Kapoor T.M., Schmidt C.F. 2005. The bipolar mitotic kinesin Eg5 moves on both microtubules that it crosslinks. Nature. 435:114–118 10.1038/nature03503 [DOI] [PubMed] [Google Scholar]
- Kashina A.S., Baskin R.J., Cole D.G., Wedaman K.P., Saxton W.M., Scholey J.M. 1996. A bipolar kinesin. Nature. 379:270–272 10.1038/379270a0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelkar N., Standen C.L., Davis R.J. 2005. Role of the JIP4 scaffold protein in the regulation of mitogen-activated protein kinase signaling pathways. Mol. Cell. Biol. 25:2733–2743 10.1128/MCB.25.7.2733-2743.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Ling S.C., Rogers G.C., Kural C., Selvin P.R., Rogers S.L., Gelfand V.I. 2007. Microtubule binding by dynactin is required for microtubule organization but not cargo transport. J. Cell Biol. 176:641–651 10.1083/jcb.200608128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klopfenstein D.R., Tomishige M., Stuurman N., Vale R.D. 2002. Role of phosphatidylinositol(4,5)bisphosphate organization in membrane transport by the Unc104 kinesin motor. Cell. 109:347–358 10.1016/S0092-8674(02)00708-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneev M.J., Lakämper S., Schmidt C.F. 2007. Load-dependent release limits the processive stepping of the tetrameric Eg5 motor. Eur. Biophys. J. 36:675–681 10.1007/s00249-007-0134-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovacs J.J., Whalen E.J., Liu R., Xiao K., Kim J., Chen M., Wang J., Chen W., Lefkowitz R.J. 2008. Beta-arrestin-mediated localization of smoothened to the primary cilium. Science. 320:1777–1781 10.1126/science.1157983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzysiak T.C., Wendt T., Sproul L.R., Tittmann P., Gross H., Gilbert S.P., Hoenger A. 2006. A structural model for monastrol inhibition of dimeric kinesin Eg5. EMBO J. 25:2263–2273 10.1038/sj.emboj.7601108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kural C., Kim H., Syed S., Goshima G., Gelfand V.I., Selvin P.R. 2005. Kinesin and dynein move a peroxisome in vivo: a tug-of-war or coordinated movement? Science. 308:1469–1472 10.1126/science.1108408 [DOI] [PubMed] [Google Scholar]
- Kwok B.H., Kapitein L.C., Kim J.H., Peterman E.J., Schmidt C.F., Kapoor T.M. 2006. Allosteric inhibition of kinesin-5 modulates its processive directional motility. Nat. Chem. Biol. 2:480–485 10.1038/nchembio812 [DOI] [PubMed] [Google Scholar]
- Ling S.C., Fahrner P.S., Greenough W.T., Gelfand V.I. 2004. Transport of Drosophila fragile X mental retardation protein-containing ribonucleoprotein granules by kinesin-1 and cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 101:17428–17433 10.1073/pnas.0408114101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman M.G., Enquist L.W. 2009. Herpesvirus interactions with the host cytoskeleton. J. Virol. 83:2058–2066 10.1128/JVI.01718-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M., Iyadurai S.J., Gassman A., Gindhart J.G., Jr, Hays T.S., Saxton W.M. 1999. Cytoplasmic dynein, the dynactin complex, and kinesin are interdependent and essential for fast axonal transport. Mol. Biol. Cell. 10:3717–3728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messitt T.J., Gagnon J.A., Kreiling J.A., Pratt C.A., Yoon Y.J., Mowry K.L. 2008. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev. Cell. 15:426–436 10.1016/j.devcel.2008.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Kirschner M. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Montagnac G., Sibarita J.B., Loubéry S., Daviet L., Romao M., Raposo G., Chavrier P. 2009. ARF6 Interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 19:184–195 10.1016/j.cub.2008.12.043 [DOI] [PubMed] [Google Scholar]
- Müller M.J., Klumpp S., Lipowsky R. 2008. Tug-of-war as a cooperative mechanism for bidirectional cargo transport by molecular motors. Proc. Natl. Acad. Sci. USA. 105:4609–4614 10.1073/pnas.0706825105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pack-Chung E., Kurshan P.T., Dickman D.K., Schwarz T.L. 2007. A Drosophila kinesin required for synaptic bouton formation and synaptic vesicle transport. Nat. Neurosci. 10:980–989 10.1038/nn1936 [DOI] [PubMed] [Google Scholar]
- Pilling A.D., Horiuchi D., Lively C.M., Saxton W.M. 2006. Kinesin-1 and Dynein are the primary motors for fast transport of mitochondria in Drosophila motor axons. Mol. Biol. Cell. 17:2057–2068 10.1091/mbc.E05-06-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin E.P., Case R.B., Dai S.C., Hart C.L., Ruby A., Vale R.D., Fletterick R.J. 1998. Direction determination in the minus-end-directed kinesin motor ncd. Nature. 395:813–816 10.1038/27463 [DOI] [PubMed] [Google Scholar]
- Sharp D.J., Rogers G.C., Scholey J.M. 2000. Microtubule motors in mitosis. Nature. 407:41–47 10.1038/35024000 [DOI] [PubMed] [Google Scholar]
- Shima T., Kon T., Imamula K., Ohkura R., Sutoh K. 2006. Two modes of microtubule sliding driven by cytoplasmic dynein. Proc. Natl. Acad. Sci. USA. 103:17736–17740 10.1073/pnas.0606794103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shubeita G.T., Tran S.L., Xu J., Vershinin M., Cermelli S., Cotton S.L., Welte M.A., Gross S.P. 2008. Consequences of motor copy number on the intracellular transport of kinesin-1-driven lipid droplets. Cell. 135:1098–1107 10.1016/j.cell.2008.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., DeBonis S., Saoudi Y., Lebeau L., Crevel I., Cross R., Wade R.H., Hackney D., Kozielski F. 2006. S-trityl-L-cysteine is a reversible, tight binding inhibitor of the human kinesin Eg5 that specifically blocks mitotic progression. J. Biol. Chem. 281:17559–17569 10.1074/jbc.M511735200 [DOI] [PubMed] [Google Scholar]
- Soppina V., Rai A.K., Ramaiya A.J., Barak P., Mallik R. 2009. Tug-of-war between dissimilar teams of microtubule motors regulates transport and fission of endosomes. Proc. Natl. Acad. Sci. USA. 106:19381–19386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhauer J., Kalderon D. 2006. Microtubule polarity and axis formation in the Drosophila oocyte. Dev. Dyn. 235:1455–1468 10.1002/dvdy.20770 [DOI] [PubMed] [Google Scholar]
- Telley I.A., Bieling P., Surrey T. 2009. Obstacles on the microtubule reduce the processivity of Kinesin-1 in a minimal in vitro system and in cell extract. Biophys. J. 96:3341–3353 10.1016/j.bpj.2009.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomishige M., Klopfenstein D.R., Vale R.D. 2002. Conversion of Unc104/KIF1A kinesin into a processive motor after dimerization. Science. 297:2263–2267 10.1126/science.1073386 [DOI] [PubMed] [Google Scholar]
- Tuma M.C., Zill A., Le Bot N., Vernos I., Gelfand V. 1998. Heterotrimeric kinesin II is the microtubule motor protein responsible for pigment dispersion in Xenopus melanophores. J. Cell Biol. 143:1547–1558 10.1083/jcb.143.6.1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida A., Alami N.H., Brown A. 2009. Tight functional coupling of kinesin-1A and dynein motors in the bidirectional transport of neurofilaments. Mol. Biol. Cell. 20:4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine M.T., Gilbert S.P. 2007. To step or not to step? How biochemistry and mechanics influence processivity in Kinesin and Eg5. Curr. Opin. Cell Biol. 19:75–81 10.1016/j.ceb.2006.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valentine M.T., Fordyce P.M., Krzysiak T.C., Gilbert S.P., Block S.M. 2006. Individual dimers of the mitotic kinesin motor Eg5 step processively and support substantial loads in vitro. Nat. Cell Biol. 8:470–476 10.1038/ncb1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhey K.J., Meyer D., Deehan R., Blenis J., Schnapp B.J., Rapoport T.A., Margolis B. 2001. Cargo of kinesin identified as JIP scaffolding proteins and associated signaling molecules. J. Cell Biol. 152:959–970 10.1083/jcb.152.5.959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Schwarz T.L. 2009. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 136:163–174 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C.M., Karki S.B., Kuznetsov S.A., Tabb J.S., Weiss D.G., Langford G.M., Holzbaur E.L. 1997. The interaction between cytoplasmic dynein and dynactin is required for fast axonal transport. Proc. Natl. Acad. Sci. USA. 94:12180–12185 10.1073/pnas.94.22.12180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte M.A. 2004. Bidirectional transport along microtubules. Curr. Biol. 14:R525–R537 10.1016/j.cub.2004.06.045 [DOI] [PubMed] [Google Scholar]
- Yildiz A., Selvin P.R. 2005. Kinesin: walking, crawling or sliding along? Trends Cell Biol. 15:112–120 10.1016/j.tcb.2004.12.007 [DOI] [PubMed] [Google Scholar]
- Yildiz A., Tomishige M., Vale R.D., Selvin P.R. 2004. Kinesin walks hand-over-hand. Science. 303:676–678 10.1126/science.1093753 [DOI] [PubMed] [Google Scholar]