The protein mutated in Huntington's disease is phosphorylated by the inflammatory kinase IKK, which promotes other post-translational modifications, and protein degradation.
Abstract
Expansion of the polyglutamine repeat within the protein Huntingtin (Htt) causes Huntington's disease, a neurodegenerative disease associated with aging and the accumulation of mutant Htt in diseased neurons. Understanding the mechanisms that influence Htt cellular degradation may target treatments designed to activate mutant Htt clearance pathways. We find that Htt is phosphorylated by the inflammatory kinase IKK, enhancing its normal clearance by the proteasome and lysosome. Phosphorylation of Htt regulates additional post-translational modifications, including Htt ubiquitination, SUMOylation, and acetylation, and increases Htt nuclear localization, cleavage, and clearance mediated by lysosomal-associated membrane protein 2A and Hsc70. We propose that IKK activates mutant Htt clearance until an age-related loss of proteasome/lysosome function promotes accumulation of toxic post-translationally modified mutant Htt. Thus, IKK activation may modulate mutant Htt neurotoxicity depending on the cell's ability to degrade the modified species.
Introduction
Abnormal accumulation of misfolded and aggregated protein in affected neurons is a hallmark of many neurodegenerative diseases associated with aging. The major pathways of protein clearance in the cell are performed by the proteasome and the lysosome, which both become compromised with age (Cuervo et al., 2005; Martinez-Vicente and Cuervo, 2007; Chondrogianni and Gonos, 2008; Tonoki et al., 2009). Parallel with reduced turnover, proteins mutated in familial neurodegenerative diseases accumulate and cause dysfunction and death, and accompanying symptoms. Examples include the polyglutamine (polyQ) disease protein Huntingtin (Htt) in Huntington's disease (HD), tau in frontotemporal dementias (FTD), α-synuclein in Parkinson's disease (PD), ataxin-1 in spinocerebellar ataxia 1 (SCA1), and SOD1 in amyotrophic lateral sclerosis (ALS).
Post-translational modification of target proteins can regulate their clearance from cells. Phosphorylation regulates protein degradation, alters subcellular localization, and/or creates phosphodegrons/binding motifs for interactors that regulate secondary modifications such as ubiquitination, SUMOylation, and acetylation. For instance, phosphorylation of HSF1, MEF2, and GATA-1 activates their SUMOylation (Hietakangas et al., 2006), phosphorylation of p53 and RelA activates their acetylation (D'Orazi et al., 2002; Hofmann et al., 2002; Chen et al., 2005), and phosphorylation of IκBα and FOXO3a activates their ubiquitination (Karin and Ben-Neriah, 2000; Karin et al., 2002; Hu et al., 2004). In turn, these modifications may ultimately target the protein for degradation (Hernandez-Hernandez et al., 2006; Hietakangas et al., 2006; Hunter, 2007; Wu et al., 2007; Zuccato et al., 2007; Jeong et al., 2009). As protein clearance mechanisms become impaired upon aging, modified proteins normally targeted for degradation by post-translational modification may accumulate and disease-causing proteins take on toxic functions (Orr and Zoghbi, 2007; Shao and Diamond, 2007).
HD is a member of a family of polyQ repeat expansion diseases characterized by the accumulation and aggregation of mutant Htt protein in diseased neurons (Orr and Zoghbi, 2007). In HD, when the repeat expands above 40, disease will manifest, typically striking in mid-life (Walker, 2007). Above ∼65 repeats, a juvenile form of the disease occurs. The polyQ expansion exists within the context of a large 350-kD protein; however, expressing just the N-terminal fragment of Htt encoded by exon 1 (Httex1p), which contains a highly expanded polyQ repeat, can precipitate an aggressive HD-like disease in transgenic mice and flies (Mangiarini et al., 1996; Steffan et al., 2001). The first 17 amino acids of Htt can mediate aggregation, subcellular localization and membrane association, stability, and cellular toxicity, each of which are implicated in HD pathogenesis (Steffan et al., 2004; Luo et al., 2005; Warby et al., 2005, 2009; Anne et al., 2007; Rockabrand et al., 2007; Atwal and Truant, 2008). The potential for Htt post-translational modification to have a disease-modifying role has recently emerged as a consistent theme, with regulatory functions implicated for other sites within the full-length protein as well, including phosphorylation at S421 by Akt and S434, S1181, and S1201 by Cdk5 (Humbert et al., 2002; Luo et al., 2005; Warby et al., 2005; Anne et al., 2007), SUMOylation and ubiquitination at K6, K9, and K15 (Steffan et al., 2004), palmitoylation at C214 (Yanai et al., 2006), and acetylation at K444 (Jeong et al., 2009). The regulatory properties of post-translational modifications extend to other polyQ repeat diseases, most notably phosphorylation of S776 in expanded ataxin-1, the mutant protein in SCA1 (Orr and Zoghbi, 2007).
We evaluated the effect of phosphorylation within the first 17 amino acids of Htt on its subcellular localization, downstream post-translational modifications, and protein clearance. This domain contains two serines at positions 13 and 16, which are adjacent to the lysines found to be modified by SUMO and ubiquitin (Steffan et al., 2004). We demonstrate that the IKK complex, previously shown to directly interact with Htt (Khoshnan et al., 2004), phosphorylates Htt S13 and may activate phosphorylation of S16. Phosphorylation of these residues promotes modification of the adjacent lysine residues and activates Htt clearance in a manner requiring both the proteasome and lysosome. We find that expansion of the Htt polyQ repeat may reduce the efficiency of this phosphorylation, potentially contributing to the accumulation of mutant Htt.
Results
The IKK complex directly phosphorylates Htt
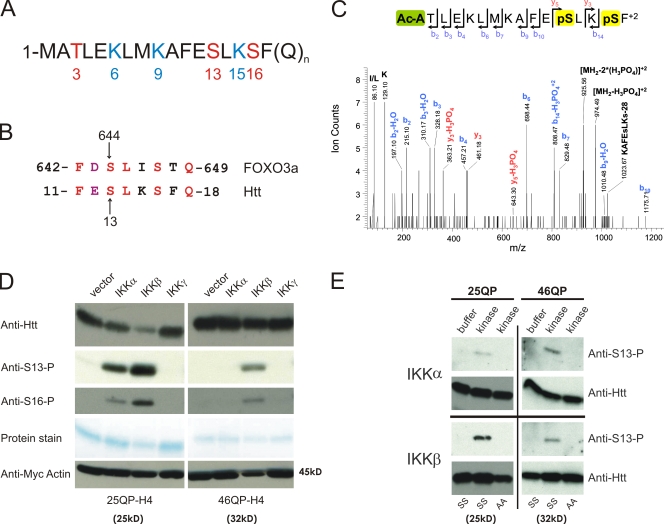
The N-terminal 17 amino acids of Htt contain a number of potentially modifiable residues (Fig. 1 A). In addition to the lysines at 6, 9, and 15, which can be SUMO modified and ubiquitinated (Steffan et al., 2004), three residues are possible phosphorylation targets (T3, S13, and S16).
Figure 1.
IKK directly phosphorylates Htt. (A) The first 17 amino acids of the Htt protein contain three residues that may be phosphorylated (red) and three modifiable lysine residues (blue). (B) Htt S13 is within a domain similar to FOXO3a S644. (C) Mass spectrometry analysis shows that Htt serines 13 and 16 can be phosphorylated. 25QP-HBH was purified under denaturing conditions from St14A cells cotransfected with IKK-αβγ. ESI-MS/MS spectra were obtained after chymotryptic digestion and collision-induced dissociation (CID) for N-terminally acetylated and diphosphorylated peptide on S13 and S16 Ac-ATLEKLMKAFEpSLKpSF, with the parent ion, [MH2]+2, at m/z 1023.03+2 (M = 2044.06 D). (D) Htt phosphorylation of S13 and S16, detected with phospho-specific antibodies, is activated with coexpression of IKK-α or IKK-β. Httex1p was purified from St14A cells transiently transfected with 25QP-H4 or 46QP-H4 with vector control or plasmids encoding subunits of IKK. Total Htt was detected with CAG53b antibody, and myc-actin transfection control detected with anti-myc antibody. (E) IKK-α and IKK-β directly phosphorylate Htt S13 in vitro. An in vitro kinase assay was performed with 75 ng recombinant IKK-α or IKK-β protein, and wt (SS) or S13,16A (AA) mutant 25Q or 46Q purified Httex1p-H4. Htt was detected with CAG53b or anti-S13-P.
Sequence evaluation for conserved motifs revealed that Htt residues 11–18 share sequence similarity with residues 642–649 of FOXO3a (Fig. 1 B), a substrate of IKK at S644 analgous to Htt S13 (Hu et al., 2004). Because expanded polyQ Httex1p activates the IKK complex in cell culture and transgenic mice, and interacts with IKK-γ in vitro (Khoshnan et al., 2004), IKK emerged as a candidate kinase that might target Htt S13. IκB kinase (IKK) is composed of three subunits: IKK-α and IKK-β are homologous catalytic subunits, and IKK-γ is a regulatory subunit. As a first step to evaluate phosphorylation of this domain and a potential role for IKK, mass spectrometry was used. 25QP-HBH, a His-tagged unexpanded form of Httex1p was transiently cotransfected with IKK-αβγ into ST14A cells, purified by nickel enrichment under denaturing conditions, digested with chymotrypsin, and analyzed by reverse-phase liquid chromatography coupled to tandem mass spectrometry. Phosphorylation of both S13 and S16 was observed in the presence of IKK (Fig. 1 C).
Using Htt peptides phosphorylated at either S13 or S16 as antigens, affinity-purified rabbit polyclonal antisera, designated anti–S13-P and anti–S16-P, were generated (Fig. S1). Overexpression of IKK-α or IKK-β but not IKK-γ increased phosphorylation of both unexpanded (25QP) and expanded (46QP) forms of Httex1p with a C-terminal His-HA-HA-His (H4) tag in ST14A cells cotransfected with Httex1p-H4 and IKK subunits (Fig. 1 D). Phosphorylation of 46QP-H4 appears less efficient than 25QP-H4 and phosphorylation of 25QP-H4 is associated with its reduced abundance. To first determine whether IKK can directly phosphorylate Htt and determine the specific residue involved, recombinant IKK phosphorylation of purified Htt was tested in vitro. S13 of both purified unexpanded (25QP) and expanded (46QP) Httex1p was phosphorylated by both IKK-α and IKK-β (Fig. 1 E), whereas phosphorylation of S16 was not observed (not depicted). These results suggest that S13 is a direct target of IKK and that the phosphorylation of S16 observed with IKK overexpression in cell culture may be primed by phosphorylation of S13 by IKK. Alternatively, the sensitivity of anti–S16-P antisera may be inadequate to detect the in vitro modification.
IKK-activated phosphorylation of Httex1p regulates its post-translational modification and subcellular localization
As described above, phosphorylation can regulate post-translational modification of adjacent lysine residues (Hunter, 2007). The role of Htt S13 and S16 phosphorylation on Htt ubiquitination, SUMOylation, and acetylation of Httex1p was tested. We previously found that 97QP Httex1p is polyubiquitinated (Steffan et al., 2004). Making a mutant that cannot be phosphorylated on S13 (S13A) reduces this polyubiquitination, whereas mimicking phosphorylation of S13 (S13D) retains its polyubiquitination (Fig. 2 A). If the dual phosphorylation we see activated by IKK is mimicked (S13,16D), a reduction in Htt polyubiquitination is again observed (Fig. 2 A). Consistent with this, overexpression of IKK reduces polyubiquitination of 97QP Httex1p (Fig. S2 A). The S13A and S13,16D mutants also demonstrate reduced mono-SUMOylation of 97QP Httex1p, whereas S13D retains its SUMOylation (Fig. 2 B). Overexpression of IKK leads to a reduction in 97QP mono-SUMOylation and an increase in its poly-SUMOylation (Fig. S2 B). Therefore, IKK may modulate ubiquitin and SUMO addition to Httex1p, two modifications globally tied to protein clearance mechanisms.
Figure 2.
Phosphorylation of Httex1p regulates its ubiquitination, SUMOylation, acetylation, and nuclear localization. (A and B) Phosphorylation of serines 13 and 16 regulates mutant Httex1p ubiquitination and SUMOylation. St14A (A) or HeLa (B) cells were transiently transfected with vector or HIS-ubiquitin (A) or HIS-SUMO-1 (B) and control and mutant 97QP VL* Httex1p. Conjugated proteins were purified under denaturing conditions by Ni-NTA magnetic nickel beads and Htt detected with anti-Htt CAG53b by Western analysis. (C) Mass spectrometry analysis shows that Htt S13 phosphorylation can occur with K9 acetylation. 25QP-HBH was purified from St14A cells cotransfected with IKK-β and CBP and treated for 2 h with histone deacetylase inhibitors 200 mM Trichostatin A/5 mM Nicotinamide. ESI-MS/MS spectra were obtained after chymotryptic digestion and collision-induced dissociation (CID) for a peptide acetylated at K9 and phosphorylated at S13 MAcKAFEpSLKSF, [MH2]+2 at m/z 655.30+2 (M = 1308.60 D). (D) IKK-β overexpression increases phosphorylation of Htt S13 and acetylation of K9. St14A cells were transiently transfected with Httex1p-H4 with 25 or 46Qs, +/− IKK-β or with 46QP QEE-H4. Htt was purified and subjected to Western analysis with anti-K9-Ac, anti-S13-P, and CAG53b. (E) Mimicking phosphorylation significantly increases nuclear localization in primary neurons. Primary cortical neurons were cotransfected with pcDNA3.1-mRFP and 97QP-GFP or 97QP-DD-GFP plasmids. The subcellular distribution of these polypeptides was examined by measuring the fluorescence intensity of GFP, to which they are fused, in regions of the nucleus and cytoplasm for each cell. The extent to which these polypeptides localized preferentially to the nucleus or the cytoplasm was determined by calculating the ratio of nuclear/cytoplasmic GFP fluorescence intensity and comparing the distribution of the two polypeptides by t test. Error bars indicate SEM in arbitrary units.
In addition to the detection of phosphorylation at both S13 and 16, acetylation of lysine K9 was detected together with modification of S13 by mass spectrometry upon exogenous IKK overexpression (Fig. 2 C). To evaluate this acetylation further and the possible influence of phosphorylation, affinity-purified polyclonal antiserum was generated against a K9-acetyl, S13-phospho, S16-phospho Htt peptide (anti–K9-Ac; Fig. S1). The antibody recognizes Htt 25 or 46QP in the presence of exogenous IKK-β; however, it shows little to no immunoreactivity without IKK coexpression (Fig. 2 D). Acetylation can be mimicked by a lysine (K) to glutamine (Q) substitution and phosphorylation mimicked by either a serine (S) to aspartic acid (D) or to glutamic acid (E) substitution. Using a K9Q, S13,16E (QEE) mimic, immunoreactivity was observed in the absence of IKK, confirming the specificity of the antibody for this Htt species (Fig. 2 D). Collectively, these data suggest that IKK-mediated phosphorylation may regulate ubiquitination, SUMOylation, and acetylation within the first 17 amino acids of Htt.
Because the first 17 amino acids of Htt can function as a cytoplasmic retention signal and regulate the association of Htt with mitochondria, Golgi, endoplasmic reticulum, late endosomes, and autophagic vesicles (Steffan et al., 2004; Atwal et al., 2007; Rockabrand et al., 2007), and because overexpression of IKK-γ was previously demonstrated to promote expanded polyQ Httex1p nuclear localization (Khoshnan et al., 2004), we tested whether phosphorylation of this domain could influence its cellular localization. Either phosphomimetic (S13,16D/S13,16E or DD/EE) or phosphoresistant (S13,16A, or AA) forms of Httex1p were used to assess the consequence of phosphorylation on soluble cellular localization. Fluorescence of mutant Httex1p with 97Qs fused to GFP was first assessed in NIH-3T3 cells (Fig. S3 A), demonstrating largely cytoplasmic localization for both 97QP-GFP and 97QP-AA-GFP. In contrast, phosphomimetic 97QP-DD-GFP and 97QP-EE-GFP displayed increased nuclear localization. Similarly, a statistically significant increase in nuclear localization of expanded 97QP-DD-GFP over control 97QP-GFP was observed upon transfection into primary cortical neurons (Fig. 2 E). Nuclear localization was also enhanced using phophomimetics of unexpanded Httex1p (25QP-EE-GFP) compared with wild type (wt) or AA in 3T3 cells, suggesting this process may extend to normal Htt or Htt fragments (Fig. S3 B).
Phosphorylation of Htt by IKK activates Httex1p and 586aa Htt fragment clearance
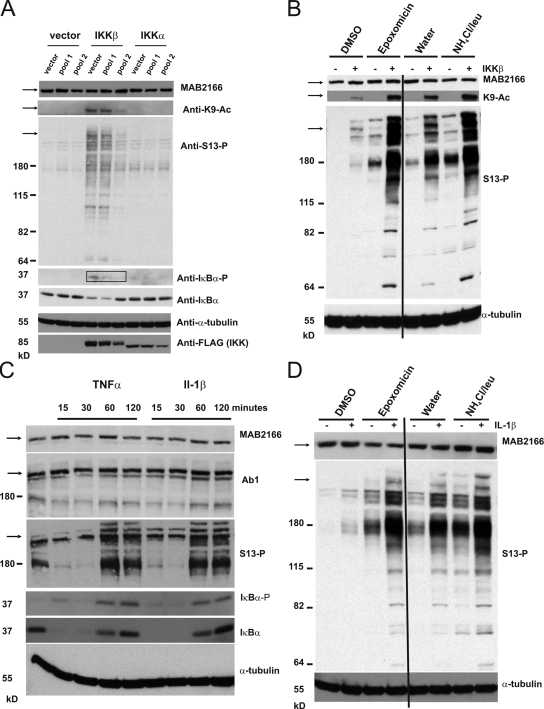
We then asked whether phosphorylation by IKK might also regulate Htt stability. Unexpanded (25QP) and expanded (46QP) polyQ Httex1p were cotransfected into ST14A cells with IKK-β, as this subunit of IKK had the greatest Htt phosphorylation activity in cell culture (Fig. 1 D), and levels of Htt evaluated. Unexpanded Htt showed a dependence on IKK-β for enhanced clearance relative to a myc-actin transfection control, and this effect persisted in the presence of either the specific proteasome inhibitor epoxomicin or the lysosome inhibitors ammonium chloride/leupeptin (Fig. 3, A and B). Proteasome inhibitors were able to block basal and IKK-β–induced degradation of 25QP; however, they were not able to abolish the differences between both types of degradation, suggesting that the reduction in cellular levels of unexpanded 25QP Httex1p mediated by IKKβ involves proteasome-dependent and -independent degradation of the protein. Inhibition of the lysosome reduced the effect of IKK-β on 25QP clearance, implicating lysosomal involvement. Interestingly, the IKK-β–induced degradation of Httex1p was largely reduced for the expanded form of the protein, although the basal degradation of the protein was unperturbed and still dependent on the proteasome system. Phosphorylation of unexpanded polyQ Httex1p may therefore target it for degradation by both the proteasome and lysosome. Expansion of the polyQ repeat to 46Qs inhibited this IKK-β–mediated reduction in Httex1p abundance (Fig. 3 A), which may at least partially be the result of its less efficient phosphorylation by IKK-β (Fig. 1 D).
Figure 3.
Phosphorylation of unexpanded polyQ Httex1p and 586aa Htt is associated with its reduced abundance in cell culture. (A) Levels of unexpanded polyQ Httex1p are reduced with overexpression of IKK-β; this effect is inhibited with expansion of the polyQ repeat. 25QP-H4 or 46QP-H4 was cotransfected with myc-actin and with vector or IKK-β into St14A cells. Cells were treated for 16 h with DMSO, 100 nM epoxomicin in DMSO, or 20 mM ammonium chloride/100 µM leupeptin in DMSO. Lysates were subjected to filter-retardation assay and Western analysis using anti-myc to detect myc-actin, and anti-HA to detect Httex1p. (B) IKK-β overexpression reduces levels of unexpanded polyQ Httex1p in the presence of proteasome or lysosome inhibition. Scion software was used to quantitate triplicate levels of 25QP-H4 from the experiment represented in A, normalized to levels of myc-actin transfection control, within each treatment group: control, epoxomicin, or ammonium chloride/leupeptin. (C) Mimicking phosphorylation of unexpanded polyQ Httex1p reduces its abundance in cell culture; this effect is reduced with expansion of the polyQ repeat. 25QP-H4 or 46QP-H4, wt control or QEE, EE, AA, or 3R were cotransfected with myc-actin into St14A cells. Cells and lysates were treated as in A. (D) Levels of phosphorylated unexpanded polyQ 586aa Htt accumulate with inhibition of the proteasome or the lysosome; phosphorylation is reduced with expansion of the polyQ repeat. 15Q or 128Q 586aa Htt constructs were cotransfected into St14A cells with myc-actin and with vector or IKK-β. Cells were treated for 4 h with DMSO or 100 nM epoxomicin in DMSO (to eliminate any possible effect on the lysosome by epoxomicin), or for 16 h with water or 20 mM ammonium chloride/100 µM leupeptin in water. Lysates were subjected to filter-retardation assay and Western analysis using anti-myc, anti–S13-P, and anti-Htt 3B5H10.
The abundance of Httex1p AA, EE, and QEE mutants were next compared with control Httex1p or to Httex1p in which all the lysines were mutated to arginine (K6,9,15R or 3R), also in the presence of epoxomicin or ammonium chloride/leupeptin. Consistent with the potential destabilization of unexpanded polyQ by phosphorylation, unexpanded (25QP) EE and QEE Httex1p levels were lower than control and AA (Fig. 3 C). In contrast, elimination of modifiable lysines (3R 25QP) had no effect on soluble Htt levels, but did increase levels of insoluble Htt. This result is in contrast to what we previously reported for a highly expanded 3R mutant (3R97QP; Steffan et al., 2004); however, we did not use filter retardation assays together with Western analysis to examine levels of insoluble Htt in those studies, and now understand that this 97QP 3R mutant is detectable, however mostly in the insoluble fraction. Inhibition of the proteasome or the lysosome increased levels of the unexpanded phosphomimetics (25QP EE and QEE), suggesting that the proteasome and lysosome may both be involved in clearance of phosphorylated and acetylated forms of unexpanded Htt fragments.
In contrast to results with unexpanded Htt, expanded control and mutant 46QP proteins accumulated in both soluble and insoluble fractions (Fig. 3 C, bottom); this accumulation was also influenced by proteasomal and lysosomal inhibition. The 46QP EE phosphomimetic consistently ran on SDS-PAGE as a doublet, suggesting the presence of a phosphorylated intermediate that is not well cleared in the presence of the expanded repeat. The doublet was not observed with the acetylation mimetic QEE, implicating lysine 9 as a critical residue in this clearance mechanism and possibly suggesting an alternative lysine 9 post-translational modification other than acetylation in doublet formation. We conclude that expansion of the polyQ repeat in Httex1p reduces the efficiency of phosphorylation-activated IKK-mediated clearance.
To further characterize IKK-mediated Htt phosphorylation and clearance, a larger Htt fragment comprised of 586 amino acids (aa) with and without IKK-β (Fig. 3 D) was expressed in ST14A cells. Co-expression of IKK-β significantly reduced levels of unexpanded Htt (586 aas with 15Qs). Although there was a reduction in levels of total unexpanded 586 (15Q) with IKK-β overexpression, as determined using anti-Htt 3B5H10 (Fig. 3 D) or anti-Htt EM48 (Fig. S4 A), a significant increase in an immunoreactive species was observed when the anti–S13-P antibody was used for detection, particularly upon longer-term lysosomal inhibition. Expanded repeat 128Q 586aa fragment levels were not reduced with overexpression of IKK-β, and S13-phosphorylated, expanded 128Q fragment was not detectible above background levels, again suggesting a reduced ability of the mutant Htt protein to be phosphorylated and cleared. Collectively, these data show that IKK-β can increase phosphorylation and reduce levels of unexpanded polyQ Htt fragments in a manner dependent on both the proteasome and the lysosome, and that expansion of the polyQ repeat inhibits this effect.
Endogenous wt full-length Htt is phosphorylated by IKK-β
The above results show that Htt fragments can be phosphorylated by IKK in cells and in vitro. We next examined whether exogenous expression of IKK subunits could phosphorylate full-length, endogenous wt Htt in ST14A cells to determine whether this type of regulation may be involved in an endogenous clearance mechanism. IKK-β overexpression increased the levels of acetylated and S13-phosphorylated full-length Htt and Htt fragments in ST14A cells (Fig. 4 A). To ensure that this effect is specific to IKK, the ability of IKK-β to enhance immunoreactivity to both antibodies was eliminated in the presence of an shRNA pool against IKK-β (Fig. 4 A). As a control, levels of phosphorylated IκBα, a defined IKK substrate targeted for degradation by its phosphorylation (Karin and Ben-Neriah, 2000), were also evaluated in this experiment, and levels (Fig. 4 A, boxed) paralleled that of phosphorylated and acetylated Htt.
Figure 4.
Phosphorylated and acetylated endogenous wt Htt accumulates with inhibition of the proteasome and lysosome. (A) Overexpression of IKK-β increases phosphorylation of endogenous Htt and IκBα in cell culture. St14A cells were transiently transfected with vector, IKK-β, or IKK-α, together with vector or two different pools of anti–IKK-αβ shRNAs; a control pool that did not silence IKK well (pool 1) and one pool that was effective in silencing IKK (pool 2), as assessed by levels of FLAG-IKK-α and -β by Western. Lysates were subjected to Western analysis with anti-Htt MAB2166, anti–S13-P, anti–K9-Ac, anti-phosphoserine 32 IκBα (IκBα-P), anti–IκBα, anti–α-tubulin, and anti-FLAG to detect FLAG-tagged IKK-β and IKK-α. Bands the size of full-length endogenous Htt (350 kD) are shown by the arrow on the left. Boxed bands show the reduction in phosphorylated IκBα with IKK shRNA. (B) Phosphorylated and acetylated Htt accumulate with inhibition of the proteasome or the lysosome. St14A cells were transiently transfected with IKK-β or vector, and were treated as in Fig. 3 D. Lysates were subjected to Western analysis with anti–α-tubulin, and anti-Htt antibodies MAB2166, anti–S13-P or anti–K9-Ac. (C) Pharmacological activation of the IKK complex modulates levels of phosphorylated Htt and IκBα. St14A cells were incubated with 20 ng/ml TNF-α or IL-1β over a time course. Lysates were subjected to Western analysis as in A with additional detection by anti-Htt Ab1. (D) IL-1β–induced Htt S13-phosphorylated species accumulate with inhibition of the proteasome or lysosome. St14A cells were treated for 2 h with 20 ng/ml Il-1β before lysis. The proteasome and lysosome were inhibited, and Western analysis performed as in B.
Full-length acetylated and phosphorylated Htt, as well as phosphorylated Htt fragments, were again increased by inhibition of either the proteasome or the lysosome (Fig. 4 B), suggesting that they may serve as intermediates in the wt Htt degradation process. High molecular weight full-length S13-phosphorylated Htt species were observed upon IKK-β overexpression (Fig. 4 B), which may reflect full-length Htt post-translational modification by SUMO or ubiquitin as we observed for Httex1p (Fig. 2, A and B). Using a well-described anti-Htt antibody, MAB2166 (Millipore), which is uniquely sensitive enough to detect endogenous rat Htt in ST14A whole-cell lysate, we did not observe either a loss of endogenous Htt in cells overexpressing IKK-β or an accumulation with inhibition of the proteasome or lysosome, possibly suggesting that the levels of modified Htt that are modulated represent a small percentage of the total. However, MAB2166 may not interact well with the phosphorylated Htt species. Within the epitope recognized by MAB2166, Htt residues 414–503, S421 (Humbert et al., 2002), and S434 (Luo et al., 2005) are phosphorylated by Akt and Cdk5, respectively, and K444 is acetylated (Jeong et al., 2009). This acetylation at K444, involved in Htt lysosomal clearance, destroys the MAB2166 epitope (Krainc, D., personal communication), supporting the possibility that modifications eliminate reactivity with MAB2166. Although we have not determined whether these modifications occur at the same time as phosphorylation of S13 by IKK, Akt acts upstream of IKK activation (Dan et al., 2008) and it is possible that modification of this epitope concurrent with S13 phosphorylation may reduce the ability of MAB2166 to recognize the Htt species being cleared. We find that MAB2166 does not strongly recognize immunoprecipitated S13/S16-phosphorylated or K9-acetylated Htt species in cells overexpressing IKK-β, whereas Ab1 and MAB5490/1H6, antibodies that recognize the N-terminal domain of Htt, do recognize these modified forms (Fig. S4 B). In addition, Htt antibodies EM48 (recognizing the first 256 aa of human Htt with a deletion of the polyQ stretch [Gutekunst et al., 1999]) and 3B5H10 (raised against GST-human Htt 171aa-66Q [Peters-Libeu et al., 2005]) detect reduced abundance of unexpanded human 15Q 586aa fragment with exogenous expression of IKK-β, whereas MAB2166 does not, supporting the idea that the cleared Htt species may not be not recognized well by this antibody (Fig. S4 A).
In addition to genetic modulation of IKK, pharmacological activation of IKK with IL-1β or TNF-α was tested for activation of Htt S13 phosphorylation. ST14A cells were treated with these standard IKK-activating cytokines for 15, 30, 60, and 120 min (Fig. 4 C). Both Il-1β and TNF-α increased levels of phosphorylated full-length and fragmented Htt at 60 and 120 min, similar to phosphorylated IκBα. S13-phosphorylated Htt and phosphorylated IκBα were both reduced at 15 and 30 min, paralleling the increased clearance of total IκBα at these time points. Total full-length Htt levels were not reduced as assessed by MAB2166 on Western analysis, but an ∼180-kD-sized set of N-terminal fragments recognized by anti-Htt Ab1 showed a possible decrease in abundance at 15 and 30 min and increased abundance at 60 and 120 min, similar in trend to robust effects on total IκBα. S13-phosphorylated Htt and Htt fragments accumulated in cells treated for 60 and 120 min with Il-1β. These phosphorylated Htt species accumulated even further at 120 min of IL-1β treatment with either epoxomicin or ammonium chloride/leupeptin, suggesting both proteasomal and lysosomal involvement (Fig. 4 D).
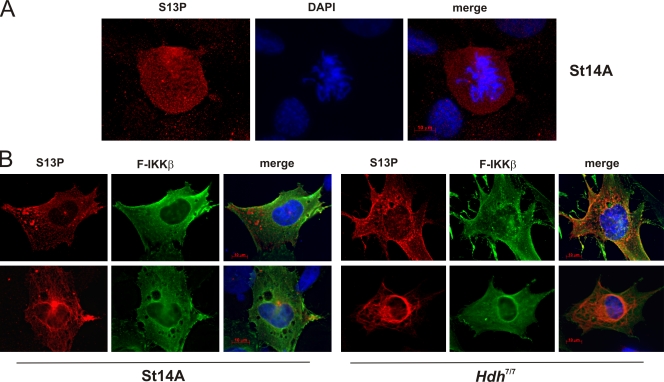
Endogenous S13-phosphorylated Htt was next analyzed by immunofluorescence. Without exogenous IKK-β, only mitotic cells stained with anti–S13-P (Fig. 5 A), representing a small fraction of the cell population. In cells that were transiently transfected with FLAG–IKK-β, the phosphorylated species of Htt was detected in cells with exogenous IKK-β expression in a variety of localization patterns (Fig. 5 B and Fig. S3 C). This staining was specific for phosphorylated S13, as it could only be competed away with a 1–17aa peptide phosphorylated on serine 13, but not with the corresponding unmodified peptide (not depicted). S13-phosphorylated Htt and K9-acetyl Htt immunoreactivity was also detected in FLAG–IKK-β nucleofected mouse striatal progenitor cells, Hdh7/7 (Fig. 5 B; unpublished data).
Figure 5.
Phosphorylated Htt can be detected by immunofluorescence in cell culture. (A) S13-phosphorylated Htt is present in untransfected mitotic rat S14A cells. (B) S13-phosphorylated Htt colocalizes with FLAG immunoreactivity in rat St14A cells lipofected with FLAG–IKK-β, and S13-phosphorylated Htt colocalizes with FLAG immunoreactivity in mouse Hdh7/7 cells nucleofected with FLAG–IKK-β.
Proteins involved in lysosomal and proteasomal clearance mechanisms modify levels of phosphorylated Htt
The pronounced effect of the lysosomal inhibitors on the intracellular levels of phosphorylated S13 Htt compared with the unmodified protein (Fig. 3 D and Fig. 4, B and D), and the distinctive punctuate pattern observed in the immunofluorescence studies with the anti–S13-P antibody compatible with lysosomal association of the modified protein (Fig. 5 and Fig. S3 C), led us to further characterize the mechanism mediating the lysosomal degradation of S13-phophorylated Htt. Because lysosomal inhibition increases levels of the S13 phospho-species, the expectation is that proteins involved in regulating lysosomal activity could also influence levels of phosphorylated Htt. The lysosomal-associated membrane protein 2A (LAMP-2A) mediates selective autophagy of proteins that contain KFERQ-like Hsc70 binding sequences in mammalian cells through a process known as chaperone-mediated autophagy (CMA; Massey et al., 2006b). Similarly, Atg7 is essential for autophagy and a loss of Atg7 function in mouse brain causes neurodegeneration (Mizushima et al., 1998; Komatsu et al., 2006). We therefore examined whether LAMP-2A or Atg7 might be involved in the clearance of phosphorylated Htt by the lysosome. When either endogenous LAMP-2A or Atg7 is knocked down by shRNA, accumulation of endogenous S13-phosphorylated Htt and Htt fragments is observed (Fig. 6 A), suggesting that both proteins may be involved in lysosomal clearance of phosphorylated Htt. Likewise, shRNA against rat LAMP-2A increased Httex1p levels and aggregation, whereas overexpression of human LAMP-2A had the opposite effect (Fig. 6 B). These combined data are consistent with a role for LAMP-2A in the lysosomal clearance of Htt.
Figure 6.
LAMP-2A, Atg7, and Hsc70 may modulate Htt clearance and toxicity. (A) Reducing levels of LAMP-2A or Atg7 in cell culture increases abundance of S13-phosphorylated Htt. St14A cells were transiently transfected with shRNA for rat LAMP-2A or Atg7, or pSUPER vector control; antibodies used for Western analysis are shown to the left of the Western panels. (B) LAMP-2A levels modulate Httex1p abundance. St14A cells were transiently cotransfected with LAMP-2A shRNA, human HA-tagged LAMP-2A or vector control, and myc-actin transfection control. Lysates were subjected to Western analysis and filter-retardation assay, detected with anti-Htt CAG53b, anti–rat/mouse LAMP-2A, anti-HA to detect HA-hLAMP-2A, anti-myc, and MemCode protein stain. (C) Hsc70 interacts with phosphomimetic unexpanded polyQ Httex1p in vitro. Purified 25QP-H4 or 46QP-H4 wt and mutant proteins radiolabeled with 35S were incubated with isolated GST-Hsc70 or GST protein bound to glutathione-agarose beads. Where indicated, 5 mM ATP or ADP was added to the reaction. Bound proteins were washed, subjected to SDS-PAGE, and detected by phosphoimager autoradiography. (D) Hsc70 reduces Htt-mediated toxicity in Hdh111/111 cells. Hdh7/7 or Hdh111/111 cells were nucleofected with vector, Hsp70, or Hsc70 together with GFP. 47 h later, XTT cell viability assays were performed, and the percentage of relative survival was calculated correcting for transfection efficiency of the GFP control ± SEM from triplicates.
Although Httex1p does not contain a bonafide “KFERQ”-like Hsc70-binding motif in its sequence, phosphorylation of Htt serine 16 (14-LKpSFQ-18) could provide the negative charge required to convert this sequence to an Hsc70-binding motif (mimic 14-LKEFQ-18, where the phosphorylated serine at residue 16 resembles a glutamic acid [E]). We therefore tested the ability of phosphomimetic Httex1p to interact with GST-Hsc70 in vitro and found that mimicking phosphorylation of Htt serines 13 and 16 on unexpanded polyQ Httex1p (25QP-EE) increased the in vitro binding of Httex1p to Gst-Hsc70 by a specific ADP-dependent mechanism (Fig. 6 C, left), whereas expansion of the polyQ repeat to 46Qs reduced this interaction (Fig. 6 C, right).
Because an interaction with Hsc70 could regulate clearance of phosphorylated mutant Htt, the ability of Hsc70 to reduce Htt-mediated toxicity (Fig. 6 D) was tested. Overexpression of Hsc70 increased the survival of HdhQ111/Q111-expressing cells more than overexpression of Hsp70, the latter being 85% identical to Hsc70. Exogenous expression of Hsp70 or Hsc70 showed that Hsc70, but not Hsp70, increased levels of Htt acetylation and S13 phosphorylation, suggesting that Hsc70 may specifically activate an IKK-regulated Htt degradation process (Fig. S5). Minor differences in heat-shock proteins have previously been demonstrated to define their function. For instance, in yeast, the homologues of Hsp70/Hsc70, Ssa1p, and Ssa2p are 97% identical and yet only Ssa2p is required to target protein substrates to the yeast vacuole, the functional equivalent of the mammalian lysosome (Brown et al., 2000). We propose that Hsc70 may increase Htt clearance by the proteasome and the lysosome by activating S13 phosphorylation and K9 acetylation.
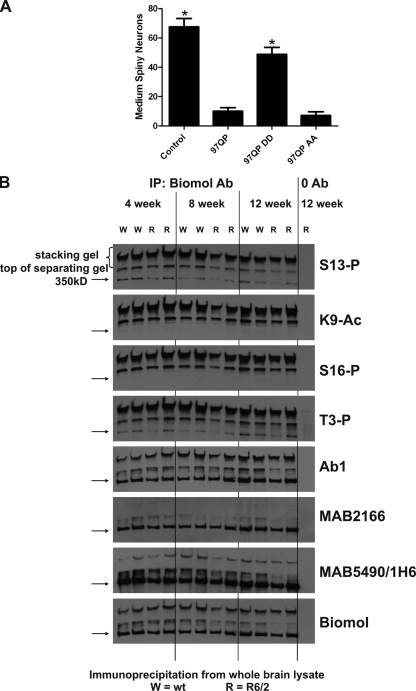
Mimicking phosphorylation of mutant Htt in rat brain slice cultures reduces its toxicity
The data presented show that IKK-β can enhance the level of a phosphorylated form of Htt that appears to be more readily cleared. To test whether this phosphorylation is functionally significant, toxicity of Htt phosphomimetics was compared with control expanded repeat Htt in an acutely transfected rat cortico-striatal slice culture model where toxicity is dependent on expansion of the polyQ repeat (Khoshnan et al., 2004). Phosphomimetic (DD) or phosphoresistant (AA) forms of expanded polyQ Httex1p (97QP) were tested for their effects on Htt-mediated toxicity in this model. The phosphomimetic 97QP-DD displayed significantly reduced toxicity compared with either control or 97QP-AA (Fig. 7 A), suggesting that phosphorylation of Httex1p may reduce neurotoxicity. Because we showed that the phosphomimetic 97QP-DD Httex1p is more nuclear localized (Fig. 2 E), but is less toxic than 97QP in the slice cultures, the data present a potential contradiction to the extensive studies showing that nuclear accumulation of mutant Htt significantly enhances neurodegeneration (Saudou et al., 1998; Schilling et al., 1999; Cornett et al., 2005). These results suggest that while nuclear accumulation of mutant Htt is toxic, nuclear localization facilitated by phosphorylation could be part of a normal process of protein degradation that becomes impaired upon expansion of the polyQ repeat, thus promoting the accumulation of nuclear, toxic Htt that is implicated in HD. Confirming a potential in vivo role for this modification, mutant Htt-mediated neurotoxicity was significantly reduced when the phosphomimetic was expressed compared with control or phosphoresistant mutant Htt in BACHD mice, suggesting that Htt phosphorylation may slow the progression of HD in vivo and represents a valid therapeutic target (Gu et al., 2009).
Figure 7.
Phosphorylation may reduce toxicity but increase insolubility of Htt. (A) Mimicking 97QP Httex1p phosphorylation reduces its toxicity in acutely transfected rat cortico-striatal slice explants. Plasmids encoding YFP and 97QP-CFP, 97QP DD-CFP, 97QP AA-CFP, or CFP control were biolistically cotransfected into rat cortico-striatal brain slices. The number of healthy medium spiny neurons in the striatal region of each slice was scored 5 d after transfection. n = 9 for each condition. Error bars represent SEM. Asterisk = difference from 97QP at P < 0.0001. (B) Immunoprecipitated phosphorylated/acetylated wt Htt purified from mouse brain runs in an insoluble fraction in the SDS-PAGE stacking gel. 12 independent wt control (W) or R6/2 (R) mouse brains were collected at 4, 8, and 12 wk of age, snap frozen, and lysed. 500 µg of lysate was subjected to immunoprecipitation with PW0595 anti-Htt antibody or zero antibody control (lysate from lanes 12 and 13 are identical). Western analysis was performed with a series of antibodies listed to the right of the panel. Immunoprecipitated Htt is present as an insoluble species in the stacking gel and at the top of the separating gel, and as a soluble form at the standard 350-kD size, marked with an arrow. MAB2166 does not detect the insoluble species well.
Phosphorylated and acetylated Htt can be detected in mouse brain
We next examined whether modified Htt species could be detected in an extensively studied HD mouse model, R6/2, which contains two wt copies of the mouse HD gene and is transgenic for exon 1 of the human huntingtin gene originally carrying ∼150 CAG repeats (Mangiarini et al., 1996). R6/2 mice have a rapid phenotypic progression (are severely impaired by 14 wk) and intranuclear inclusions throughout the brain. Htt was immunoprecipitated from whole brain tissue from 4-, 8-, and 12-wk R6/2 versus wt control mice (Fig. 7 B; antibody PW0595, Enzo Life Sciences, Inc.). S13- and S16-phosphorylated and K9-acetylated endogenous mouse Htt showed reactivity to relatively insoluble Htt species in both R6/2 and wt control brain. A major percentage of modified Htt remained in the stacking gel and at the top of the separating gel with less migrating to the size of the standard 350-kD full-length Htt band. No fragments originating from the transgene were visualized using this method with the modification-specific antibodies, suggesting that the transgene was not efficiently phosphorylated and acetylated, although it could be detected after immunoprecipititation with the Enzo Life Sciences, Inc. antibody (unpublished data). Wt Htt antibodies Ab1, MAB5490/1H6, and the Enzo Life Sciences, Inc. antibody, each raised against unique Htt species, recognized wt-soluble and the insoluble Htt species in the stacking gel, whereas MAB2166 did not recognize the insoluble Htt, consistent with our data above suggesting that MAB2166 does not substantially detect the phosphorylated/acetylated Htt species (Fig. S4) or that the insoluble fraction consists primarily of truncated N-terminal Htt species, which do not contain the MAB2166 epitope. We also find that Htt phosphorylated on threonine 3 (T3) is present in the insoluble fraction, consistent with our recent observation that mimicking phosphorylation of T3 increases Htt aggregation (Aiken et al., 2009). Overall, our data suggest that the S13/S16-phosphorylated and K9-acetylated forms of wt endogenous Htt are detectable in both wt and R6/2 mice, and that these modified forms represent relatively insoluble species.
Discussion
Delineation of Htt clearance mechanisms is of great significance because an accumulation of mutant Htt is implicated in HD pathogenesis. We demonstrate that the IKK complex phosphorylates Htt at S13 and may activate its degradation, similar to IKK-mediated degradation of IκBα and FOXO3a (Karin and Ben-Neriah, 2000; Karin et al., 2002; Hu et al., 2004). The proposed selective degradation of phosphorylated wt Htt, which involves both the proteasome and lysosome, may include a transient nuclear localization mediated by phosphorylation of Htt, where it is subsequently acetylated, ubiquitinated, and SUMOylated in an order that remains to be established. Proteins involved in lysosomal degradation pathways, Hsc70, LAMP-2A, and Atg7, appear to modulate the levels of these modified forms of Htt in mammalian cells. Our data also suggest that IKK-β–mediated Htt S13 phosphorylation is more efficient for wt than for expanded polyQ truncated Htt polypeptides (Fig. 1 D and Fig. 3 C), which may result in reduced clearance of mutant Htt by inhibiting this phosphorylation-driven mechanism, and ultimately contribute to disease. Finally, modified species recognized by phospho- and acetyl-specific antibodies are present in mouse brain.
Mimicking phosphorylation of Htt serines 13 and 16 increases soluble Httex1p-GFP nuclear localization (Fig. 2 E). In previous studies, we showed that Htt interacts with the acetyltransferase CBP (Steffan et al., 2000, 2001); therefore CBP, a nuclear protein, is a candidate acetyltransferase for Htt lysine 9, as was demonstrated for lysine 444 (Jeong et al., 2009). CBP/p300 contains ubiquitin ligase activity which regulates protein degradation (Grossman et al., 2003); therefore, this E3 ligase activity of CBP could also be involved in regulating Htt ubiquitination. A futher connection between the IKK-mediated phosphorylation of Htt and CBP activites may exist, as CBP interacts directly with IKK-γ in the nucleus (Verma et al., 2004) and is a substrate for IKK-α (Huang et al., 2007), suggesting that CBP and the IKK complex could function together to modulate Htt stability.
Relevant to the potential role for S13 and S16 phosphorylation in nuclear localization, nuclear caspase-6 cleavage of mutant Htt has been implicated in its pathogenic potential (Graham et al., 2006; Warby et al., 2008). We find that IKK activates phosphorylation of Htt fragments, one of which is consistent with the predicted size of a wt Htt caspase-6 cleavage product (Fig. 4 B) and the fragment recently shown to be generated in neurons with activation of IKK-β (Khoshnan et al., 2009). It is therefore an intriguing possibility that phosphorylation by IKK ultimately promotes Htt nuclear localization, poly-SUMOylation, acetylation by CBP, and subsequent caspase-6 cleavage, which all facilitate a regulated form of clearance.
Phosphorylation of Htt by IKK appears to activate its degradation at least in part by the lysosome, dependent on LAMP-2A levels (Fig. 6), the integral membrane receptor protein that can directly import proteins across the lysosomal membrane for CMA (Massey et al., 2006b). The CMA chaperone Hsc70 preferentially interacts with phosphomimetic wt Httex1p and reduces Htt-mediated toxicity (Fig. 6, C and D). Combined, this data may suggest that phosphorylated Htt is degraded in a LAMP-2A–dependent mechanism through CMA. Because phosphorylation can trigger nuclear localization and acetylation of specific Htt species (Fig. 2, C–E) and overexpression of Hsc70 increases levels of acetylated and phosphorylated endogenous full-length and fragmented Htt (Fig. S5), the findings implicate Hsc70 in either mediating an interaction of Htt with the IKK complex, or alternatively activating the IKK complex, as has been demonstrated for the ubiquitin ligase parkin, which is mutated in Parkinson's disease (Henn et al., 2007).
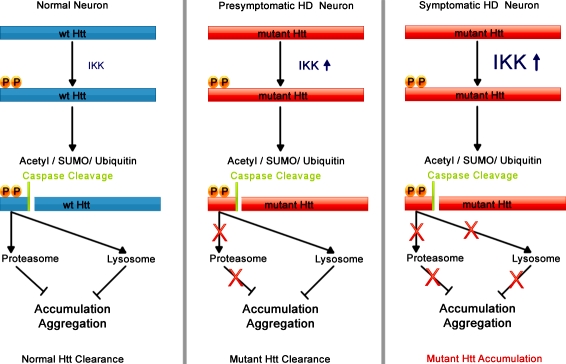
CMA activity declines with age due to a gradual decrease of LAMP-2A levels in lysosomes (Cuervo and Dice, 2000), whereas artificially maintaining LAMP-2A levels in aging rat liver similar to those in young animals can restore CMA activity to youthful levels and improve organ function (Zhang and Cuervo, 2008). Because HD is a neurodegenerative disease associated with aging and we have found clearance of phosphorylated Htt dependent on LAMP-2A, a reduction in LAMP-2A levels over time may be tied to HD pathogenesis. We propose a hypothetical model for the progression of HD at the molecular level (Fig. 8), where IKK phosphorylates Htt and activates a cascade of Htt post-translational modifications and caspase cleavage (Khoshnan et al., 2009) associated with rapid Htt degradation by the proteasome and the lysosome in unaffected neurons. In a presymptomatic HD neuron, IKK could be induced by the presence of the mutant protein (Khoshnan et al., 2004), thus stimulating an IKK-mediated mechanism of Htt clearance, consistent with the innate immune activation that occurs in premanifest patients well before symptom onset (Björkqvist et al., 2008). As long as LAMP-2A levels remain high, patients can degrade mutant Htt before it can cause toxicity, despite progressively inhibited proteasome activity and reduced efficiency of mutant Htt phosphorylation. As aging and mutant Htt together progressively impair proteasome and overall lysosomal activity, and as LAMP-2A levels decline with age, modified Htt may accumulate, enhancing HD pathogenesis.
Figure 8.
Proposed molecular mechanism for the development of HD. Normal neuronal function: IKK phosphorylates wt Htt activating its post-translational modification, caspase cleavage, and clearance by the proteasome and lysosome. Presymptomatic HD neuronal function: With chronic expression of mutant Htt, proteasome activity is reduced, and lysosomal degradation of mutant Htt becomes essential. Mutant Htt triggers activation of the IKK complex; however, it is less efficiently phosphorylated than wt Htt. With the clearance mechanism activated, mutant and wt Htt in the presymptomatic cell are degraded by the lysosome. Symptomatic HD neuronal function: Lysosomal degradation of Htt is impaired through reduction of LAMP-2A levels or other loss of lysosomal function caused by aging and by mutant Htt expression. Uncleared mutant Htt and Htt fragments accumulate and take on toxic functions, enhancing HD pathogenesis.
From this model, it follows that increasing the efficiency of Htt clearance by the lysosome, or increasing levels or mobility (Kaushik et al., 2006) of functional LAMP-2A in the lysosomal membrane early in the disease process, could delay HD onset and serve as a therapeutic strategy. Treatment choice may vary depending on the stage of HD. In mammals, early treatment to increase Htt phosphorylation and acetylation may be useful when levels of LAMP-2A are adequate, as suggested by the reduced toxicity in acutely transfected rat slice cultures (Fig. 7 A) and complete lack of neurotoxicity in BACHD mice when Htt is mutated to mimic the phosphorylated form (Gu et al., 2009). However, when LAMP-2A levels are low or its function is impaired, drugs that activate the formation of the post-translationally modified Htt species normally targeted for degradation by the lysosome might actually increase HD pathogenesis through increased nuclear accumulation and aggregation of the mutant protein. At this stage of disease, serine-modified Htt would accumulate, leading to increased pathology as a result of the intrinisic toxicity of the modified Htt. Consistent with this, we find that mimicking Httex1p phosphorylation increases its toxicity and aggregation in Drosophila photoreceptor neurons where components of the mammalian machinery to degrade phosphorylated Htt, specifically LAMP-2A, are not present (unpublished data). Thus, during late-stage HD, it may be harmful to increase pathways involved in IKK activation, SUMOylation, and acetylation, whereas in presymptomatic stages, these pathways may be protective.
The Htt protein itself may play an integral role in autophagic clearance of proteins. Conditional knockout of Htt in the mouse central nervous system results in an accumulation of neuropil protein aggregates containing ubiquitin and p62/SQSTM1 (unpublished data). Htt has been shown to associate with autophagosomes (Atwal and Truant, 2008) and lysosomes (unpublished data) and may therefore play a functional and regulatory role in a selective protein clearance mechanism ultimately involved in its own processing. Extensive investigation will be necessary to test this possibility and elucidate the precise mechanisms involved.
Materials and methods
Plasmid constructs
pcDNA3.1-based plasmids (Invitrogen) containing the Htt exon 1 DNA between the HindIII and BamHI sites were used as described previously (Steffan et al., 2004). These plasmids contained alternating CAG/CAA repeats, coding for either a normal range (25) or expanded (46 or 97) polyglutamine tracts followed by the proline-rich region of Htt ending with the amino acids HRP to create Httex1p. The plasmids were opened at BamHI and XbaI and DNA encoding various tags was inserted in frame with Httex1p to create C-terminal tagged Httex1p. The following tags were used: GS*, VL*, HBH (a gift from Peter Kaiser and Christian Tagwerker, University of California, Irvine, Irvine, CA [Tagwerker et al., 2006]), EGFP, and H4 (HIS-HA-HA-HIS: GSHHHHHHMGYPYDVPDYAEFYPYDVPDYAVHHHHHH*) where a stop codon is denoted by the asterisk. The H4 tag was created by a two-step double-stranded oligonucleotide ligation.
Mutations in Httex1p were created using double-stranded oligonucleotides containing HindIII-compatible ends encoding the first 17 amino acids of Huntingtin, which were ligated between the HindIII site of pcDNA3.1 in the polylinker and the HindIII site in exon I, immediately 5′ to the CAG repeat. K6R K9R K15R (3R) was used as described previously (Steffan et al., 2004). To create S16 mutations, the HindIII site was shifted using oligonucleotides containing BspMI (BfuAI) sites, then double-stranded oligonucleotides were again used to create S13 together with S16 mutants. pHsc70 was constructed by ligating the BamHI fragment from pGST-Hsc70 encoding Hsc70 into the BamHI site of pcDNA3.1. For plasmids used in the acutely transfected striatal slice culture assay, 97QP Httex1p wt and mutants were cloned into pGWIZ (Gene Therapy Systems) in frame with CFP using the PstI site.
The following plasmids were obtained collaboratively or as gifts: pHis-SUMO-1 (A. Dejean, Institut Pasteur, Paris, France); pHis-Ubiquitin (D. Bohmann, University of Rochester, Rochester, NY); p-human-HA-LAMP-2A (pAMC1) and pGST-Hsc70 (A.M. Cuervo, Albert Einstein College of Medicine, Bronx, NY; Cuervo and Dice, 1996); pCMV-Hsp70 (P. Muchowski [University of California, San Francisco, San Francisco, CA] and H. Kampinga [University of Groningen, Groningen, Netherlands; Michels et al., 1997); pMyc-Actin (H. Rommelaere, Ghent University, Ghent, Belgium); pFLAG-IKKα, pFLAG-IKKβ, and pHA-IKKγ (A. Khoshnan and P. Patterson, California Institute of Technology, Pasadena, CA); pYFP (L. Kaltenbach and D. Lo; Duke University, Durham, NC); 586aa Htt constructs 15Q (pCINeoHtttt586-15) and 128Q (pCINeoHtt 586–128; M. Hayden, University of British Columbia, Vancouver, Canada); pcDNA3.1-CBP (A. Kazantsev and D. Housman, Massachusetts Institute of Technology, Cambridge, MA; Kazantsev et al., 1999); pcDNA3.1-mRFP (S. Finkbeiner, University of California, San Francisco, San Francisco, CA); and pcDNA3-CHIP and pcDNA-CHIPΔUbox (C. Patterson, University of North Carolina at Chapel Hill, Chapel Hill, NC; Jiang et al., 2001).
shRNA for rat Atg7 5′-GAAGTACCACTTCTACTAC-3′was cloned into pSUPER vector (Appllied Biosystems). shRNA for rat LAMP-2A, 5′-GACTGCAGTGCAGATGAAG-3′ in pSUPER was previously constructed (Massey et al., 2006a). IKK pool 1 contained the following shRNAs in pLKO.1 (Addgene): 5-CCGGGCACTGGGAAAGTATCTGAAACTCGAGTTTCAGATACTTTCCCAGTGCTTTTT-3′, 5′-CCGGCCAGCCAAGAAGAGTGAAGAACTCGAGTTCTTCACTCTTCTTGGCTGGTTTTT-3′, 5′-CCGGCTTACCTGAATCAGACAAGAACTCGAGTTCTTGTCTGATTCAGGTAAGTTTTT-3′, 5′-CCGGGCATCTAGTAGAGCGGATGATCTCGAGATCATCCGCTCTACTAGATGCTTTTT-3′, 5′-CCGGCGTTGTTAGTGAAGACTTGAACTCGAGTTCAAGTCTTCACTAACAACGTTTTT-3′. IKK pool 2 shRNAs in pLKO.1 were: 5′-CCGGGCATCATAAGGAGTTGGTGTACTCGAGTACACCAACTCCTTATGATGCTTTTT-3′, 5′-CCGGCCAGATTATGAAGAAGTTGAACTCGAGTTCAACTTCTTCATAATCTGGTTTTT-3′, 5′-CCGGCCAGCCTCTCAATGTGTTCTACTCGAGTAGAACACATTGAGAGGCTGGTTTTT-3′, 5′-CCGGGCAAATGAGGAACAGGGCAATCTCGAGATTGCCCTGTTCCTCATTTGCTTTTT-3′, 5′-CCGGGCGTGCCATTGATCTATATAACTCGAGTTATATAGATCAATGGCACGCTTTTT-3′.
Cell culture and transfections
The Hela, St12.7, ST14A, and N548mu and the wild-type STHdhQ7/HdhQ7 and homozygous mutant STHdhQ111/HdhQ111 cell lines were propagated as described previously (Steffan et al., 2004; Apostol et al., 2008). NIH-3T3 cells were grown in DMEM and 10% newborn calf serum (Hyclone). All cells were transfected with Lipofectamine 2000 according to the manufacturer's instructions (Invitrogen) except STHdhQ7/HdhQ7, which were nucleofected. rhTNF-α and rmIl-1β (R&D Systems) were used to pharmacologically activate IKK.
Primary antibodies
Three affinity-purified rabbit polyclonal antibodies were generated (New England Peptide) against post-translationally modified 1–17aa Htt peptides. Antibodies were generated against the following peptides: anti-S16-P: H2N-CMATLEKLMKAFESLK(pS)F-amide; anti-S13-P: H2N-CMATLEKLMKAFE(pS)LKSF-amide; anti-K9-Ac: Ac-CLEKLM(Ac-K)AFE(pS)LK(pS)F-amide. Two rabbits were immunized with each peptide; antisera were pooled and run over an unmodified Htt 1–17aa peptide column (affinity matrix 20401; Thermo Fisher Scientific) to remove antibodies recognizing unmodified Htt species. The flow-through from each was then run over the modified peptide column for each respective project. The elutions from these columns were used as the modification-specific antibodies for this study, and tested for specificity using a peptide dot blot (Fig. S1). These three antibodies were also tested on Westerns of lysates from Hdh7/7 and Hdh111/111 cells nucleofected with IKKβ or vector +/− siRNA for Htt (a gift from R. Friedlander, Brigham and Women's Hospital, Boston, MA); levels of antigenic species were reduced with Htt siRNA in both cell lines, demonstrating modified Htt specificity. JG1 is another rabbit polyclonal anti-Htt 1–17aa antibody generated by our laboratory.
We also used the following antibodies for this work: CAG53b (a gift from E. Wanker, Max Delbrueck Center for Molecular Medicine, Berlin, Germany); anti-Htt PW0595 (Enzo Life Sciences, Inc.); anti-Myc 9E10 (Millipore); anti-FLAG (Sigma-Aldrich), anti-HA 16B12 (Covance); anti–α-tubulin clone B-5-1-2 (Sigma-Aldrich); anti-Htt Ab1 (M. DiFiglia, Harvard University, Cambridge, MA); anti-Htt EM48 MAB5374 (Millipore); anti-Htt 1H6 (Abnova; Fig. S4); anti-Htt MAB2166 (Millipore); anti-Htt MAB5490 (Millipore; Fig. 7); anti-Htt 3B5H10 (S. Finkbeiner, Univeristy of California, San Francisco); anti–rat LAMP-2A Igp96 (Invitrogen); anti-nestin (Millipore); anti-Atg7 (Abcam); anti–SUMO-1 (PW9460; Enzo Life Sciences, Inc.), anti-ubiquitin (13–1600, Invitrogen; and sc8017, Santa Cruz Biotechnology, Inc.); anti-IκBα clone IκB-245 (Invitrogen); anti–phospho-IκBα Ser 32 14D4 (Cell Signaling Technology); and anti-phosphorylated Htt threonine 3 [anti-T3-P; Aiken et al. [2009]). Secondary antibodies used for Western analysis were goat anti–mouse HRP (The Jackson Laboratory) and goat anti–rabbit HRP (Thermo Fisher Scientific).
Immunofluorescence analysis
Cells were transfected (Lipofectamine 2000) or nucleofected (Lonza) with IKK-β (1/2 IKK-β and 1/2 pcDNA) and 24 h later fixed at room temperature with 4% PFA, permeabilized, and blocked with 5% BSA and 4% donkey serum. Primary antibodies were diluted at 1:1,000. Secondary antibodies were anti–rabbit conjugated with Cy3 (Jackson ImmunoResearch Laboratories, Inc.) and anti–mouse conjugated with FITC (Jackson ImmunoResearch Laboratories, Inc.), and they were used at 1:1,000. Slides were stained with DAPI to detect nuclei. ProLong Gold Antifade (Invitrogen) imaging medium was used. Images were collected at room temperature with an inverted microscope (Observer.Z1; Carl Zeiss, Inc.) with a Plan-Apochromat 63X objective, NA 1.40. AxioVision AxioVs40 v 4.7.1.0 software (Carl Zeiss, Inc.) was used to generate 3D deconvoluted images. The camera used was an AxioCam MRm (Carl Zeiss, Inc.).
Protein purification and Western analysis
His-tagged proteins were purified under denaturing conditions using magnetic Ni-NTA nickel beads (QIAGEN) as described previously (Steffan et al., 2004) for Fig. 1 D, Fig. 2, A and B, and Fig. S2, A and B. Western blots were processed with SuperSignal West Pico and Dura reagents (Thermo Fisher Scientific). Quantitative densiometric analyses (Fig. 6 A and Fig. S4) were performed on digitalized images of immunoblots using Scion Image 4.0 software (Scion Corporation) and SEM calculated from densiometric levels of Western signal from triplicate preparations of protein extracts. Densitometric levels of phosphorylated or acetylated Htt protein were normalized to levels of α-tubulin loading control. Filter retardation assays were performed as follows: Cell debris pellets were taken after centrifugation for 10 min at 16,000 g. Pellets were resuspended in 100 µl of Tris buffer (20 mM Tris and 15 mM MgCl2 at pH 8.0) and 100 µl of 4% SDS-100 mM DTT in PBS was added. These samples were boiled for 5 min and then filtered through nitrocellulose membrane via a dot-blot apparatus. The membranes were then dried at room temperature for 30 min, stained with MemCode reversible protein stain (Thermo Fisher Scientific), blocked, and primary antibodies were added for Western analysis. Native buffer A, used for lysis of cells in Figs. 3, 4, 6, S3, S4, and S5: 10 mM Tris-HCl, pH 7.5, 10% glycerol, 400 mM NaCl, 1 mM EDTA, 1 mM PMSF, 0.5% NP-40, 20 mM N-ethylmaleimide, 1 mM PMSF, phosphatase inhibitors 1 and 2 (Sigma-Aldrich), complete mini protease inhibitor pellet (Roche), 10 ng/ml aprotenin, 10 ng/ml leupeptin, 5 mM nicotinamide and 5 mM butyrate, pH 7.5. Native buffer B, used for purification in Fig. 2 D: 50 mM NaH2PO4, pH 8.0, 150 mM NaCl, 0.1% Tween 20, 1 mM DTT, 5 mM ADP, 10 mM Imidazole, 1 mM PMSF, 10 ng/ml aprotenin, 10 ng/ml leupeptin, complete mini protease inhibitor pellet (Roche), and phosphatase inhibitors 1 and 2 (Sigma-Aldrich). All St14A cells used in Westerns showing phosphorylated Htt were treated with phosphatase inhibitor Calyculin A (Enzo Life Sciences, Inc.) 10–30 min (20 nM) before lysis.
In vitro kinase assay
The in vitro kinase assay was performed as described previously (Liu et al., 2007) using 75 ng recombinant IKK-α or IKK-β protein (Millipore) and 25QP-H4 or 46QP-H4 purified from St14A cells using magnetic Ni-NTA nickel beads (QIAGEN) under native conditions with the following buffers. Lysis buffer: 50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 10 mM imidazole, 0.05% Tween 20, 1 mM PMSF, 10 ng/ml aprotenin, 10 ng/ml leupeptin, and phosphatase inhibitors 1 and 2 (Sigma-Aldrich). Wash buffer: 50 mM NaH2PO4, pH 8.0, 300 mM NaCl, 20 mM imidazole, 0.05% Tween 20, and phosphatase inhibitors 1 and 2 (Sigma-Aldrich). Recombinant IKK was diluted in enzyme dilution buffer: 20 mM MOPS/NaOH, pH 7.0, 1 mM EDTA, 5% glycerol, 0.01% Brij35, 0.1% β-mercaptoethanol, and 1 mg/ml BSA. The kinase assay was performed in 5X kinase buffer: 40 mM MOPS/NaOH, pH 7.0, and 1 mM EDTA. Before the assay, purified Htt bound to Ni-NTA beads was incubated at 95°C for 3 min, and then placed on ice for 5 min. The assay was performed at 30°C with light agitation for 10 min under the following conditions: 2.5 µl recombinant IKK subunit, 5 µl 5X kinase buffer, 2.5 µl 1 mM ATP, 2.5 µl 0.1 M MgAc, and 12.5 µl purified Htt in water. The assay was stopped with addition of Western sample loading buffer, boiled 10 min, run on 12% SDS-PAGE, and Western analysis was performed.
Mass spectrometry analysis
ST14A cells were transiently transfected with Htt25QP-HBH + IKK-αβγ (Fig. 1 C) or Htt25QP-HBH + CBP + IKK-β (Fig. 2 C); 48 h after transfection, the cells reached confluency. Cells were then treated with fresh media containing 10 nM Calyculin A (EMD) for 30 min, or 20 nM Calyculin A (Enzo Life Sciences, Inc.) for 10 min. Cells were washed with cold 1x PBS, then harvested and lysed in 1 ml lysis buffer each (50 mM Tris-HCl, pH 8.0, 8 M urea, 500 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, 0.5% Triton X-100, and complete mini protease inhibitor [Roche]). The DNA was sheared and the cells further lysed by passing through a 20-guage needle 20 times, and cellular debris was removed by centrifugation. Clarified lysates were then incubated with 25 µl Ni-Sepharose 6 Fast Flow (GE Healthcare) or Ni-NTA magnetic nickel (QIAGEN) bead slurry for 3 h or overnight at room temperature. The beads were then washed twice in the lysis buffer, and four times with wash buffer (50 mM Tris-HCl, pH 6.3, 8 M urea, 500 mM NaCl, 50 mM NaH2PO4, 20 mM imidazole, and 0.5% Triton X-100). The beads of the same condition were pooled and the urea buffer was replaced with 50 mM NH4CO3 before digestion.
Chymotryptic digestion (2% by weight) of 25QP-HBH was performed on the Ni beads used to purify the protein to maximize peptide recovery. The digestion occurred overnight at 37°C. Resulting peptides were extracted from the beads with 25% acetonitrile, 0.1% formic acid three times. The extracts were pooled, concentrated using a SpeedVac, and acidified by 0.1% formic acid before mass spectrometric analysis. Resultant peptides were then separated and analyzed by reverse-phase liquid chromatography coupled to tandem mass spectrometry (LC MS/MS) on a quadropole-orthogonoal-time-of-flight tandem (Guerrero et al., 2008) (QSTAR XL; Applied Biosystems/PE Sciex) or an ultra-high performance Thermo Electron linear trap quadropole (LTQ)-Orbitrap hybrid (Fang et al., 2008) (Thermo Fisher Scientific) mass spectrometer.
Extraction of the monoisotopc masses (m/z) of parent ions, their charge states, and their corresponding fragment ions was performed automatically using Analyst software (Applied Biosystems) for QSTAR data or using extract MSn (Matrix Science) for LTQ-Orbitrap data. These data were then submitted for automated database searching for protein identification using the Protein Prospector (University of California, San Francisco) search engine. Post-translational modifications were confirmed by manual inspection of the MS/MS spectra.
GST pull-down assay
For GST pull-down assays, 35S-labeled His-tagged Httex1p-H4 proteins were synthesized in a TNT coupled reticulocyte lysate system (Promega), purified, and eluted under native conditions using magnetic Ni-NTA nickel beads (QIAGEN) in the buffers suggested by the manufacturer. The eluted proteins were dialyzed using SlideALyzer 3.5K Dialysis Casettes (Thermo Fisher Scientific) against 20 mM MOPS, pH 7.3/0.25 M sucrose buffer. GST-Hsc70 was purified from Escherichia coli using glutathione-agarose beads, and incubated with purified radioactive Httex1p-H4 proteins in 20 mM Tris-HCl, pH 7.2, 150 mM NaCl, 0.1% Tween 20, and 1 mM DTT supplemented with ATP or ADP corresponding to the incubation conditions, washed five times, and subjected to SDS-PAGE and autoradiography.
Toxicity assays
For XTT cell viability assays, STHdhQ7 and STHdhQ111 and cell lines were plated in 24-well plates (0.75 × 105 cells per well) in complete media as described previously (Apostol et al., 2008). The next day, cells were shifted to nonpermissive conditions (i.e., 39°C and low serum media) for ∼48 h for STHdhQ111 and STHdhQ7 lines followed by incubation for 4 h with XTT and phenazine methosulfate (0.2 mg/ml and 0.2 µg/ml, respectively; Sigma-Aldrich) for 4 h, and plates were read at 450 nM.
Rat cortico-striatal brain slice neurodegeneration assay
All animal experiments were performed in accordance with the Institutional Animal Care and Use Committee and Duke University Medical Center Animal Guidelines. Brain slice preparation and biolistic transfection were performed as described previously (Lo et al., 1994; Southwell et al., 2008) with some modifications. In brief, brain tissue was dissected from postnatal day 10 CD Sprague Dawley rats (Charles River Laboratory) and placed in ice-cold Neurobasal A culture medium containing 10% heat-inactivated pig serum, 5% heat-inactivated rat serum (Lampire), 10 mM KCl, 10 mM Hepes (Sigma-Aldrich), 1 mM sodium pyruvate (Sigma-Aldrich), 100 U/ml penicillin/streptomycin, and 1 mMl-glutamine. All media reagents were obtained from Invitrogen unless otherwise noted. Brain tissue was cut into 250-µm-thick coronal slices using a Vibratome and incubated for 30 min at 32°C/5% CO2 before biolistic transfection. Gold particles (1.6 µm; Bio-Rad Laboratories) were coated with the indicated plasmids, loaded into Tefzel tubing (McMaster-Carr), and transfected with the Helios Gene Gun (Bio-Rad Laboratories) at 95 psi. Brain slices were incubated at 32°C/5%CO2 until analysis at 5 d post-transfection. Medium spiny neurons were visualized by fluorescence microscopy of YFP and scored by neuron morphology. Medium spiny neurons were considered healthy if they were of uniform size and shape and contained visible dendrites. Data were analyzed with Prism software (GraphPad Software, Inc.) and significance was determined by unpaired Student's t test.
Calculation of the nucleus/cytoplasm ratio for 97QP-GFP versus 97QP-DD-GFP
Cell culture and transfection.
Primary cultures of rat cortical neurons were prepared from embryonic rats (E 19–20) and transfected using calcium phosphate with plasmids at 5 d in vitro, as described previously (Xia et al., 1996; Finkbeiner et al., 1997). Specifically, neurons were cotransfected with pcDNA3.1-mRFP (Arrasate et al., 2004) and 97QP-GFP or 97QP-PP-GFP in a 1:1 molar ratio using a total of 3 µg of DNA in each well of a 24-well plate. After transfection, neurons were maintained in serum-free medium with Forskolin (10 µM; Sigma-Aldrich) and IBMX (100 µM; Sigma-Aldrich). Neurons were fixed with 4% paraformaldehyde in PBS (15 min) 20 h after transfection, permeabilized with 0.1% Triton X-100 in PBS (30 min), and incubated with 1 M glycine in PBS (20 min). Finally, neurons were washed twice for 5 min each with 2.5 µg/ml of Hoechst 33258 in PBS dye to stain the nuclei.
Robotic microscope imaging system.
The microscope imaging system has been described previously (Arrasate et al., 2004; Arrasate and Finkbeiner, 2005). Basically, the system is based on an inverted microscope (TE300 Quantum; Nikon). Xenon lamp (175 W) illumination was supplied by a liquid light guide. The Nikon working distance objective 20X (NA 0.45) was used. Fluorescence excitation and emission filters were moved into or out of the optical path by two ten position filter wheels (Sutter Instrument Co.) under computer control. Images were collected with a 12/14 bit digital cooled CCD camera (Orca II; Hamamatsu Photonics) and digitized with MetaMorph software (Universal Imaging). Before image acquisition, care was taken to adjust the gain and offset of the camera and the analogue-to-digital converter to ensure that the intensities of all the pixels of each image were within the detection range of the instrument, and that the settings were the same across the samples that were examined. The whole system is mounted on a vibration isolation table.
Image analysis.
Measurements of htt expression were extracted from images generated with the microscope imaging system described above. The validity of estimating htt expression levels in live cells from images of the fluorescence of the GFP fusion tag to which it is fused has been demonstrated previously by directly comparing this approach to other methods that are highly quantitative but unsuitable for live-cell imaging (Arrasate et al., 2004). The expression of 97QP-GFP or 97QP-DD-GFP was estimated by measuring GFP fluorescence intensity within a region of interest from the image that corresponded to a portion of the cytoplasm or the nucleus in neurons that had not developed inclusion bodies. Hoechst staining was used to localize the nucleus. Pixel intensities within a similar sized region from an adjacent acellular portion of the image were collected as a measurement of background signal. Pixel values from these background measurements were subtracted from the corresponding pixel intensity measurements made from the nucleus and the cytoplasm. These calculations produced background-corrected measurements of htt 97QP-GFP or 97P-DD-GFP from the cytoplasm and nucleus.
Statistical analysis.
The ratio of GFP intensity in the nucleus over the cytoplasm was calculated for individual neurons by dividing the background-corrected pixel intensities from the region of interest within the nucleus by the background-corrected pixel intensities from the region of interest within the cytoplasm of the same neuron. Differences in the mean of these ratio measurements were compared by t test with commercially available software (Prism 3.0; GraphPad Software, Inc.).
Htt immunoprecipitation from mouse brain
Age-matched R6/2 and wt control brains were collected and snap frozen. Whole brain tissue was dounced 20 times on ice in T-Per lysis buffer (Thermo Fisher Scientific) containing an EDTA-free mini protease inhibitor pellet (Roche), a Phos-Stop pellet (Roche), 5 mM sodium fluoride, and 1 mM sodium orthovanadate. Lysates were microfuged at 16,000 g at 4°C for 15 min, and the supernatant saved. Htt was immunoprecipitated from the 500 µg of supernatant using Protein G-Plus Agarose (Santa Cruz Biotechnology, Inc.) with 1 μl PW0595 antibody (Enzo Life Sciences, Inc.) or zero antibody control, and run on 8% SDS-PAGE and blotted to nitrocellulose for standard Western analysis.
Online supplemental material
Fig. S1 shows the specificity of the modification-specific antibodies. Fig. S2 shows the role of IKK in the regulation of mutant Httex1p ubiquitination and SUMOylation. Fig. S3 shows cellular localization of phosphorylated Htt. Fig. S4 shows loss of an epitope for popular anti-Htt antibody MAB2166 with post-translational modification of Htt. Fig. S5 shows the role of Hsc70 and the ubiquitin ligase CHIP in the regulation of levels of phosphorylated and acetylated Htt. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200909067/DC1.
Acknowledgments
This paper is dedicated to the memory of Dr. Charles H. Sawyer, whose example was its constant inspiration.
We thank Drs. David Housman, Matthew Blurton-Jones, Masashi Kitazawa, Peter Kaiser, Alex Osmand, Christian Landes, Bin Liu, and Daniel Keys for insightful discussion; Denise Dunn and Emily Mitchell for technical assistance; and Anne Dejean, Dirk Bohmann, Paul Muchowski, Harm Kampinga, Peter Kaiser, Christian Tagwerker, Cam Patterson, Heidi Rommelaere, Alex Kazantsev, Robert Friedlander, Erich Wanker, and Marian DiFiglia for their generous gifts of reagents for these experiments.
This work was supported by the Hereditary Disease Foundation (J.S. Steffan, L.M. Thompson, J.L. Marsh, D.C. Lo, and P.H. Patterson); the Fox Family Foundation (J.S. Steffan and L.M. Thompson); the High Q Foundation (J.S. Steffan, L.M. Thompson, J.L. Marsh, and D.C. Lo); the Huntington's Disease Society of America Coalition for the Cure (L.M. Thompson); the Taube-Koret Center for Huntington's Disease Research (S. Finkbeiner); and National Institutes of Health awards NS52789 (L.M. Thompson and J.L. Marsh), HD36081 (J.L. Marsh), NS045283 (J.L. Marsh and L.M. Thompson), NS043466 (S.O. Zeitlin), GM74830 (L. Huang), 2R01NS039074 (S. Finkbeiner), 2R01NS045191 (S. Finkbeiner), 2P01AG022074 (S. Finkbeiner), P01AG031782 (A.M. Cuervo) and NS055298 (P.H. Patterson), and T32GM0731130 (C.T. Aiken).
Footnotes
Abbreviations used in this paper:
- CMA
- chaperone-mediated autophagy
- HD
- Huntington's disease
- Htt
- Huntingtin
- LAMP-2A
- lysosomal-associated membrane protein 2A
- polyQ
- polyglutamine
- wt
- wild type
References
- Aiken C.T., Steffan J.S., Guerrero C.M., Khashwji H., Lukacsovich T., Simmons D., Purcell J.M., Menhaji K., Zhu Y.Z., Green K., et al. 2009. Phosphorylation of threonine 3: implications for Huntingtin aggregation and neurotoxicity. J. Biol. Chem. 284:29427–29436 10.1074/jbc.M109.013193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne S.L., Saudou F., Humbert S. 2007. Phosphorylation of huntingtin by cyclin-dependent kinase 5 is induced by DNA damage and regulates wild-type and mutant huntingtin toxicity in neurons. J. Neurosci. 27:7318–7328 10.1523/JNEUROSCI.1831-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostol B.L., Simmons D.A., Zuccato C., Illes K., Pallos J., Casale M., Conforti P., Ramos C., Roarke M., Kathuria S., et al. 2008. CEP-1347 reduces mutant huntingtin-associated neurotoxicity and restores BDNF levels in R6/2 mice. Mol. Cell. Neurosci. 39:8–20 10.1016/j.mcn.2008.04.007 [DOI] [PubMed] [Google Scholar]
- Arrasate M., Finkbeiner S. 2005. Automated microscope system for determining factors that predict neuronal fate. Proc. Natl. Acad. Sci. USA. 102:3840–3845 10.1073/pnas.0409777102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrasate M., Mitra S., Schweitzer E.S., Segal M.R., Finkbeiner S. 2004. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 431:805–810 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- Atwal R.S., Truant R. 2008. A stress sensitive ER membrane-association domain in Huntingtin protein defines a potential role for Huntingtin in the regulation of autophagy. Autophagy. 4:91–93 [DOI] [PubMed] [Google Scholar]
- Atwal R.S., Xia J., Pinchev D., Taylor J., Epand R.M., Truant R. 2007. Huntingtin has a membrane association signal that can modulate huntingtin aggregation, nuclear entry and toxicity. Hum. Mol. Genet. 16:2600–2615 10.1093/hmg/ddm217 [DOI] [PubMed] [Google Scholar]
- Björkqvist M., Wild E.J., Thiele J., Silvestroni A., Andre R., Lahiri N., Raibon E., Lee R.V., Benn C.L., Soulet D., et al. 2008. A novel pathogenic pathway of immune activation detectable before clinical onset in Huntington's disease. J. Exp. Med. 205:1869–1877 10.1084/jem.20080178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown C.R., McCann J.A., Chiang H.L. 2000. The heat shock protein Ssa2p is required for import of fructose-1, 6-bisphosphatase into Vid vesicles. J. Cell Biol. 150:65–76 10.1083/jcb.150.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L.F., Williams S.A., Mu Y., Nakano H., Duerr J.M., Buckbinder L., Greene W.C. 2005. NF-kappaB RelA phosphorylation regulates RelA acetylation. Mol. Cell. Biol. 25:7966–7975 10.1128/MCB.25.18.7966-7975.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chondrogianni N., Gonos E.S. 2008. Proteasome activation as a novel antiaging strategy. IUBMB Life. 60:651–655 10.1002/iub.99 [DOI] [PubMed] [Google Scholar]
- Cornett J., Cao F., Wang C.E., Ross C.A., Bates G.P., Li S.H., Li X.J. 2005. Polyglutamine expansion of huntingtin impairs its nuclear export. Nat. Genet. 37:198–204 10.1038/ng1503 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., Dice J.F. 1996. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 273:501–503 10.1126/science.273.5274.501 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., Dice J.F. 2000. Age-related decline in chaperone-mediated autophagy. J. Biol. Chem. 275:31505–31513 10.1074/jbc.M002102200 [DOI] [PubMed] [Google Scholar]
- Cuervo A.M., Bergamini E., Brunk U.T., Dröge W., Ffrench M., Terman A. 2005. Autophagy and aging: the importance of maintaining “clean” cells. Autophagy. 1:131–140 10.4161/auto.1.3.2017 [DOI] [PubMed] [Google Scholar]
- D'Orazi G., Cecchinelli B., Bruno T., Manni I., Higashimoto Y., Saito S., Gostissa M., Coen S., Marchetti A., Del Sal G., et al. 2002. Homeodomain-interacting protein kinase-2 phosphorylates p53 at Ser 46 and mediates apoptosis. Nat. Cell Biol. 4:11–19 10.1038/ncb714 [DOI] [PubMed] [Google Scholar]
- Dan H.C., Cooper M.J., Cogswell P.C., Duncan J.A., Ting J.P., Baldwin A.S. 2008. Akt-dependent regulation of NF-kappaB is controlled by mTOR and Raptor in association with IKK. Genes Dev. 22:1490–1500 10.1101/gad.1662308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L., Wang X., Yamoah K., Chen P.L., Pan Z.Q., Huang L. 2008. Characterization of the human COP9 signalosome complex using affinity purification and mass spectrometry. J. Proteome Res. 7:4914–4925 10.1021/pr800574c [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner S., Tavazoie S.F., Maloratsky A., Jacobs K.M., Harris K.M., Greenberg M.E. 1997. CREB: a major mediator of neuronal neurotrophin responses. Neuron. 19:1031–1047 10.1016/S0896-6273(00)80395-5 [DOI] [PubMed] [Google Scholar]
- Graham R.K., Deng Y., Slow E.J., Haigh B., Bissada N., Lu G., Pearson J., Shehadeh J., Bertram L., Murphy Z., et al. 2006. Cleavage at the caspase-6 site is required for neuronal dysfunction and degeneration due to mutant huntingtin. Cell. 125:1179–1191 10.1016/j.cell.2006.04.026 [DOI] [PubMed] [Google Scholar]
- Grossman S.R., Deato M.E., Brignone C., Chan H.M., Kung A.L., Tagami H., Nakatani Y., Livingston D.M. 2003. Polyubiquitination of p53 by a ubiquitin ligase activity of p300. Science. 300:342–344 10.1126/science.1080386 [DOI] [PubMed] [Google Scholar]
- Gu X., Greiner E.R., Mishra R., Kodali R., Osmand A., Finkbeiner S., Steffan J.S., Thompson L.M., Wetzel R., Yang X.W. 2009. Serines 13 and 16 are critical determinants of full-length human mutant Huntingtin induced disease pathogenesis in HD mice. Neuron. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero C., Milenkovic T., Przulj N., Kaiser P., Huang L. 2008. Characterization of the proteasome interaction network using a QTAX-based tag-team strategy and protein interaction network analysis. Proc. Natl. Acad. Sci. USA. 105:13333–13338 10.1073/pnas.0801870105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutekunst C.A., Li S.H., Yi H., Mulroy J.S., Kuemmerle S., Jones R., Rye D., Ferrante R.J., Hersch S.M., Li X.J. 1999. Nuclear and neuropil aggregates in Huntington's disease: relationship to neuropathology. J. Neurosci. 19:2522–2534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn I.H., Bouman L., Schlehe J.S., Schlierf A., Schramm J.E., Wegener E., Nakaso K., Culmsee C., Berninger B., Krappmann D., et al. 2007. Parkin mediates neuroprotection through activation of IkappaB kinase/nuclear factor-kappaB signaling. J. Neurosci. 27:1868–1878 10.1523/JNEUROSCI.5537-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hernandez A., Ray P., Litos G., Ciro M., Ottolenghi S., Beug H., Boyes J. 2006. Acetylation and MAPK phosphorylation cooperate to regulate the degradation of active GATA-1. EMBO J. 25:3264–3274 10.1038/sj.emboj.7601228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietakangas V., Anckar J., Blomster H.A., Fujimoto M., Palvimo J.J., Nakai A., Sistonen L. 2006. PDSM, a motif for phosphorylation-dependent SUMO modification. Proc. Natl. Acad. Sci. USA. 103:45–50 10.1073/pnas.0503698102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann T.G., Möller A., Sirma H., Zentgraf H., Taya Y., Dröge W., Will H., Schmitz M.L. 2002. Regulation of p53 activity by its interaction with homeodomain-interacting protein kinase-2. Nat. Cell Biol. 4:1–10 10.1038/ncb715 [DOI] [PubMed] [Google Scholar]
- Hu M.C., Lee D.F., Xia W., Golfman L.S., Ou-Yang F., Yang J.Y., Zou Y., Bao S., Hanada N., Saso H., et al. 2004. IkappaB kinase promotes tumorigenesis through inhibition of forkhead FOXO3a. Cell. 117:225–237 10.1016/S0092-8674(04)00302-2 [DOI] [PubMed] [Google Scholar]
- Huang W.C., Ju T.K., Hung M.C., Chen C.C. 2007. Phosphorylation of CBP by IKKalpha promotes cell growth by switching the binding preference of CBP from p53 to NF-kappaB. Mol. Cell. 26:75–87 10.1016/j.molcel.2007.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humbert S., Bryson E.A., Cordelières F.P., Connors N.C., Datta S.R., Finkbeiner S., Greenberg M.E., Saudou F. 2002. The IGF-1/Akt pathway is neuroprotective in Huntington's disease and involves Huntingtin phosphorylation by Akt. Dev. Cell. 2:831–837 10.1016/S1534-5807(02)00188-0 [DOI] [PubMed] [Google Scholar]
- Hunter T. 2007. The age of crosstalk: phosphorylation, ubiquitination, and beyond. Mol. Cell. 28:730–738 10.1016/j.molcel.2007.11.019 [DOI] [PubMed] [Google Scholar]
- Jeong H., Then F., Melia T.J.J., Jr., Mazzulli J.R., Cui L., Savas J.N., Voisine C., Paganetti P., Tanese N., Hart A.C., et al. 2009. Acetylation targets mutant huntingtin to autophagosomes for degradation. Cell. 137:60–72 10.1016/j.cell.2009.03.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J., Ballinger C.A., Wu Y., Dai Q., Cyr D.M., Höhfeld J., Patterson C. 2001. CHIP is a U-box-dependent E3 ubiquitin ligase: identification of Hsc70 as a target for ubiquitylation. J. Biol. Chem. 276:42938–42944 10.1074/jbc.M101968200 [DOI] [PubMed] [Google Scholar]
- Karin M., Ben-Neriah Y. 2000. Phosphorylation meets ubiquitination: the control of NF-[kappa]B activity. Annu. Rev. Immunol. 18:621–663 10.1146/annurev.immunol.18.1.621 [DOI] [PubMed] [Google Scholar]
- Karin M., Cao Y., Greten F.R., Li Z.W. 2002. NF-kappaB in cancer: from innocent bystander to major culprit. Nat. Rev. Cancer. 2:301–310 10.1038/nrc780 [DOI] [PubMed] [Google Scholar]
- Kaushik S., Massey A.C., Cuervo A.M. 2006. Lysosome membrane lipid microdomains: novel regulators of chaperone-mediated autophagy. EMBO J. 25:3921–3933 10.1038/sj.emboj.7601283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazantsev A., Preisinger E., Dranovsky A., Goldgaber D., Housman D. 1999. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc. Natl. Acad. Sci. USA. 96:11404–11409 10.1073/pnas.96.20.11404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnan A., Ko J., Watkin E.E., Paige L.A., Reinhart P.H., Patterson P.H. 2004. Activation of the IkappaB kinase complex and nuclear factor-kappaB contributes to mutant huntingtin neurotoxicity. J. Neurosci. 24:7999–8008 10.1523/JNEUROSCI.2675-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnan A., Ko J., Tescu S., Brundin P., Patterson P.H. 2009. IKKalpha and IKKbeta regulation of DNA damage-induced cleavage of huntingtin. PLoS One. 4:e5768 10.1371/journal.pone.0005768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., Tanaka K. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Liu B., Yang Y., Chernishof V., Loo R.R., Jang H., Tahk S., Yang R., Mink S., Shultz D., Bellone C.J., et al. 2007. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell. 129:903–914 10.1016/j.cell.2007.03.056 [DOI] [PubMed] [Google Scholar]
- Lo D.C., McAllister A.K., Katz L.C. 1994. Neuronal transfection in brain slices using particle-mediated gene transfer. Neuron. 13:1263–1268 10.1016/0896-6273(94)90412-X [DOI] [PubMed] [Google Scholar]
- Luo S., Vacher C., Davies J.E., Rubinsztein D.C. 2005. Cdk5 phosphorylation of huntingtin reduces its cleavage by caspases: implications for mutant huntingtin toxicity. J. Cell Biol. 169:647–656 10.1083/jcb.200412071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangiarini L., Sathasivam K., Seller M., Cozens B., Harper A., Hetherington C., Lawton M., Trottier Y., Lehrach H., Davies S.W., Bates G.P. 1996. Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause a progressive neurological phenotype in transgenic mice. Cell. 87:493–506 10.1016/S0092-8674(00)81369-0 [DOI] [PubMed] [Google Scholar]
- Martinez-Vicente M., Cuervo A.M. 2007. Autophagy and neurodegeneration: when the cleaning crew goes on strike. Lancet Neurol. 6:352–361 10.1016/S1474-4422(07)70076-5 [DOI] [PubMed] [Google Scholar]
- Massey A.C., Kaushik S., Sovak G., Kiffin R., Cuervo A.M. 2006a. Consequences of the selective blockage of chaperone-mediated autophagy. Proc. Natl. Acad. Sci. USA. 103:5805–5810 10.1073/pnas.0507436103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A.C., Zhang C., Cuervo A.M. 2006b. Chaperone-mediated autophagy in aging and disease. Curr. Top. Dev. Biol. 73:205–235 10.1016/S0070-2153(05)73007-6 [DOI] [PubMed] [Google Scholar]
- Michels A.A., Kanon B., Konings A.W., Ohtsuka K., Bensaude O., Kampinga H.H. 1997. Hsp70 and Hsp40 chaperone activities in the cytoplasm and the nucleus of mammalian cells. J. Biol. Chem. 272:33283–33289 10.1074/jbc.272.52.33283 [DOI] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., Ohsumi Y. 1998. A protein conjugation system essential for autophagy. Nature. 395:395–398 10.1038/26506 [DOI] [PubMed] [Google Scholar]
- Orr H.T., Zoghbi H.Y. 2007. Trinucleotide repeat disorders. Annu. Rev. Neurosci. 30:575–621 10.1146/annurev.neuro.29.051605.113042 [DOI] [PubMed] [Google Scholar]
- Peters-Libeu C., Newhouse Y., Krishnan P., Cheung K., Brooks E., Weisgraber K., Finkbeiner S. 2005. Crystallization and diffraction properties of the Fab fragment of 3B5H10, an antibody specific for disease-causing polyglutamine stretches. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 61:1065–1068 10.1107/S1744309105036547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockabrand E., Slepko N., Pantalone A., Nukala V.N., Kazantsev A., Marsh J.L., Sullivan P.G., Steffan J.S., Sensi S.L., Thompson L.M. 2007. The first 17 amino acids of Huntingtin modulate its sub-cellular localization, aggregation and effects on calcium homeostasis. Hum. Mol. Genet. 16:61–77 10.1093/hmg/ddl440 [DOI] [PubMed] [Google Scholar]
- Saudou F., Finkbeiner S., Devys D., Greenberg M.E. 1998. Huntingtin acts in the nucleus to induce apoptosis but death does not correlate with the formation of intranuclear inclusions. Cell. 95:55–66 10.1016/S0092-8674(00)81782-1 [DOI] [PubMed] [Google Scholar]
- Schilling G., Becher M.W., Sharp A.H., Jinnah H.A., Duan K., Kotzuk J.A., Slunt H.H., Ratovitski T., Cooper J.K., Jenkins N.A., et al. 1999. Intranuclear inclusions and neuritic aggregates in transgenic mice expressing a mutant N-terminal fragment of huntingtin. Hum. Mol. Genet. 8:397–407 10.1093/hmg/8.3.397 [DOI] [PubMed] [Google Scholar]
- Shao J., Diamond M.I. 2007. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum. Mol. Genet. 16 Spec No. 2:R115–R123 10.1093/hmg/ddm213 [DOI] [PubMed] [Google Scholar]
- Southwell A.L., Khoshnan A., Dunn D.E., Bugg C.W., Lo D.C., Patterson P.H. 2008. Intrabodies binding the proline-rich domains of mutant huntingtin increase its turnover and reduce neurotoxicity. J. Neurosci. 28:9013–9020 10.1523/JNEUROSCI.2747-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S., Kazantsev A., Spasic-Boskovic O., Greenwald M., Zhu Y.Z., Gohler H., Wanker E.E., Bates G.P., Housman D.E., Thompson L.M. 2000. The Huntington's disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc. Natl. Acad. Sci. USA. 97:6763–6768 10.1073/pnas.100110097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffan J.S., Bodai L., Pallos J., Poelman M., McCampbell A., Apostol B.L., Kazantsev A., Schmidt E., Zhu Y.Z., Greenwald M., et al. 2001. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 413:739–743 10.1038/35099568 [DOI] [PubMed] [Google Scholar]
- Steffan J.S., Agrawal N., Pallos J., Rockabrand E., Trotman L.C., Slepko N., Illes K., Lukacsovich T., Zhu Y.Z., Cattaneo E., et al. 2004. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 304:100–104 10.1126/science.1092194 [DOI] [PubMed] [Google Scholar]
- Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. 2006. A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivo cross-linking. Mol. Cell. Proteomics. 5:737–748 [DOI] [PubMed] [Google Scholar]
- Tonoki A., Kuranaga E., Tomioka T., Hamazaki J., Murata S., Tanaka K., Miura M. 2009. Genetic evidence linking age-dependent attenuation of the 26S proteasome with the aging process. Mol. Cell. Biol. 29:1095–1106 10.1128/MCB.01227-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma U.N., Yamamoto Y., Prajapati S., Gaynor R.B. 2004. Nuclear role of I kappa B Kinase-gamma/NF-kappa B essential modulator (IKK gamma/NEMO) in NF-kappa B-dependent gene expression. J. Biol. Chem. 279:3509–3515 10.1074/jbc.M309300200 [DOI] [PubMed] [Google Scholar]
- Walker F.O. 2007. Huntington's disease. Lancet. 369:218–228 10.1016/S0140-6736(07)60111-1 [DOI] [PubMed] [Google Scholar]
- Warby S.C., Chan E.Y., Metzler M., Gan L., Singaraja R.R., Crocker S.F., Robertson H.A., Hayden M.R. 2005. Huntingtin phosphorylation on serine 421 is significantly reduced in the striatum and by polyglutamine expansion in vivo. Hum. Mol. Genet. 14:1569–1577 10.1093/hmg/ddi165 [DOI] [PubMed] [Google Scholar]
- Warby S.C., Doty C.N., Graham R.K., Carroll J.B., Yang Y.Z., Singaraja R.R., Overall C.M., Hayden M.R. 2008. Activated caspase-6 and caspase-6-cleaved fragments of huntingtin specifically colocalize in the nucleus. Hum. Mol. Genet. 17:2390–2404 10.1093/hmg/ddn139 [DOI] [PubMed] [Google Scholar]
- Warby S.C., Doty C.N., Graham R.K., Shively J., Singaraja R.R., Hayden M.R. 2009. Phosphorylation of huntingtin reduces the accumulation of its nuclear fragments. Mol. Cell. Neurosci. 40:121–127 10.1016/j.mcn.2008.09.007 [DOI] [PubMed] [Google Scholar]
- Wu R.C., Feng Q., Lonard D.M., O'Malley B.W. 2007. SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell. 129:1125–1140 10.1016/j.cell.2007.04.039 [DOI] [PubMed] [Google Scholar]
- Xia Z., Dudek H., Miranti C.K., Greenberg M.E. 1996. Calcium influx via the NMDA receptor induces immediate early gene transcription by a MAP kinase/ERK-dependent mechanism. J. Neurosci. 16:5425–5436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai A., Huang K., Kang R., Singaraja R.R., Arstikaitis P., Gan L., Orban P.C., Mullard A., Cowan C.M., Raymond L.A., et al. 2006. Palmitoylation of huntingtin by HIP14 is essential for its trafficking and function. Nat. Neurosci. 9:824–831 10.1038/nn1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C., Cuervo A.M. 2008. Restoration of chaperone-mediated autophagy in aging liver improves cellular maintenance and hepatic function. Nat. Med. 14:959–965 10.1038/nm.1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuccato E., Blott E.J., Holt O., Sigismund S., Shaw M., Bossi G., Griffiths G.M. 2007. Sorting of Fas ligand to secretory lysosomes is regulated by mono-ubiquitylation and phosphorylation. J. Cell Sci. 120:191–199 10.1242/jcs.03315 [DOI] [PubMed] [Google Scholar]