Abstract
Heterochromatin proteins are thought to play key roles in chromatin structure and gene regulation, yet very few genes have been identified that are regulated by these proteins. We performed large-scale mapping and analysis of in vivo target loci of the proteins HP1, HP1c, and Su(var)3-9 in Drosophila Kc cells, which are of embryonic origin. For each protein, we identified ∼100–200 target genes among >6000 probed loci. We found that HP1 and Su(var)3-9 bind together to transposable elements and genes that are predominantly pericentric. In addition, Su(var)3-9 binds without HP1 to a distinct set of nonpericentric genes. On chromosome 4, HP1 binds to many genes, mostly independent of Su(var)3-9. The binding pattern of HP1c is largely different from those of HP1 and Su(var)3-9. Target genes of HP1 and Su(var)3-9 show lower expression levels in Kc cells than do nontarget genes, but not if they are located in pericentric regions. Strikingly, in pericentric regions, target genes of Su(var)3-9 and HP1 are predominantly embryo-specific genes, whereas on the chromosome arms Su(var)3-9 is preferentially associated with a set of male-specific genes. These results demonstrate that, depending on chromosomal location, the HP1 and Su(var)3-9 proteins form different complexes that associate with specific sets of developmentally coexpressed genes.
Keywords: Heterochromatin, Drosophila, target genes, gene expression, genome organization
Heterochromatin, originally defined as regions of chromatin that remain condensed throughout the cell cycle (Heitz 1928), is thought to play a major role in epigenetic regulation of gene expression. Heterochromatin is localized predominantly at pericentric chromosomal regions and at telomeres, but has also been observed in small patches within euchromatic regions, so-called intercalary heterochromatin (Zhimulev et al. 1982). Heterochromatin has certain characteristic features, such as reduced chromatin accessibility, late replication in S-phase, low gene density, low or absent recombination, and unique histone modifications (Henikoff 2000; Jenuwein 2001; Richards and Elgin 2002), although not all of these features necessarily occur simultaneously. Extensive studies of chromosome rearrangements and transgene insertions in various organisms have demonstrated repressive effects of heterochromatin on gene expression (Eissenberg and Elgin 2000; Henikoff 2000; Jenuwein 2001; Richards and Elgin 2002).
Drosophila heterochromatin contains several characteristic nonhistone chromatin proteins. Two of these, HP1 and Su(var)3-9, are strongly concentrated in pericentric heterochromatin (James et al. 1989; Schotta et al. 2002) and are required for silencing of reporter genes by heterochromatin (for review, see Weiler and Wakimoto 1995). Homologs of both proteins exist in various other organisms, including fission yeast, mice, and humans. Su(var)3-9 and its orthologs display methyltransferase activity with a high specificity for Lys 9 of histone H3 (H3-K9; Rea et al. 2000; Czermin et al. 2001; Nakayama et al. 2001; Schotta et al. 2002). Methylated H3-K9 in turn is recognized by HP1 proteins through a conserved protein domain termed the chromodomain (Bannister et al. 2001; Lachner et al. 2001; Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002). In addition, HP1 proteins and Su(var)3-9 homologs can interact directly (Aagaard et al. 1999; Schotta et al. 2002). These observations suggest that a heterochromatin complex is formed due to the triangle of interactions between HP1, Su(var)3-9, and (methylated) H3-K9. Consistent with this model, HP1 recruitment to pericentric heterochromatin is dependent on Su(var)3-9 (Ekwall et al. 1996; Bannister et al. 2001; Lachner et al. 2001; Schotta et al. 2002), and vice versa (Schotta et al. 2002). However, this model does not address the mechanism underlying the locus specificity of heterochromatin assembly. Additional mechanisms must exist that direct heterochromatin to certain parts of the genome and not to others. Investigation of such mechanisms has been difficult thus far, because a comprehensive list of genomic targets of HP1 and Su(var)3-9 is not available.
In most studies of heterochromatin function, reporter genes were used that are not naturally located within heterochromatin. Location of such euchromatic reporter genes within or near heterochromatic regions typically leads to repression of these genes. Paradoxically, two genes that are naturally embedded in pericentric heterochromatin were found not to be repressed; instead, they appear to require a heterochromatic environment for proper expression (Clegg et al. 1998; Weiler and Wakimoto 1998; Lu et al. 2000). These observations indicated that the effects of heterochromatin on gene expression are gene-specific, and that the use of reporter genes may have provided a biased view of the roles of heterochromatin in gene regulation.
To obtain insight into the global roles of HP1 and Su(var)3-9 proteins in gene regulation, we performed large-scale mapping of their target genes in Drosophila cells. The results not only provide a comprehensive overview of the sets of genes that are associated with these proteins, but also reveal that HP1 and Su(var)3-9 can bind together or independently of one another. The target specificity of these different complexes is strongly correlated with chromosomal position. Moreover, we demonstrate that distinct complexes associate with specific classes of developmentally coregulated genes, and that these classes of target genes are nonrandomly distributed along the chromosomes.
Results
Highly sensitive mapping of chromatin protein target loci
We used the previously described DamID chromatin profiling technique (van Steensel and Henikoff 2000; van Steensel et al. 2001) to map in vivo target genes of HP1 and Su(var)3-9 in cultured Drosophila melanogaster Kc167 cells. In brief, this technique involves in vivo expression of a trace amount of a chromatin protein of interest fused to Escherichia coli DNA adenine methyltransferase (Dam). As a result, DNA in the target loci of the chromatin protein is preferentially methylated by the tethered Dam. Subsequently, methylated DNA fragments are isolated, labeled with a fluorescent dye, and hybridized to a microarray. To correct for nonspecific binding of Dam and local differences in DNA accessibility, methylated DNA fragments of control cells transfected with Dam alone are labeled with a different fluorescent dye and cohybridized. The obtained ratio of fluorescent dyes reflects the extent of protein binding to the probed gene (van Steensel et al. 2001).
We refined the previously reported protocol by replacing the purification of methylated fragments with a PCR-based selective amplification of methylated fragments (Supplementary Fig. S1). Control experiments confirmed that the relative abundance of methylated sequences was conserved in the PCR amplification step (Supplementary Fig. S1; data not shown). This new protocol is much more efficient and requires considerably smaller amounts (>20-fold reduction) of genomic DNA compared with the original protocol.
Previously, we successfully used the DamID technology and a microarray of ∼300 cDNA fragments to identify a small number of target loci of HP1 (van Steensel et al. 2001). Here, we used a more comprehensive cDNA array containing over 6200 cDNA fragments, representing ∼40% of the coding Drosophila genome. The cDNA fragments were derived from seven libraries from five different tissues and developmental stages (see Materials and Methods), thus minimizing the possibility of bias in our screen. Because methylation by tethered Dam spreads ∼2 kb from the binding site of its fusion partner (van Steensel and Henikoff 2000; van Steensel et al. 2003), this cDNA array enabled us to detect protein binding to the genes themselves, as well as to most promoters and other regulatory elements in the vicinity of these genes.
Chromosomal distribution of HP1
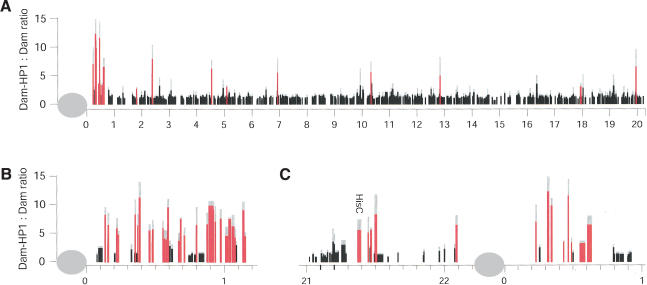
First, we generated detailed chromosomal maps of HP1 binding. An example is shown in Figure 1A, depicting the observed Dam-HP1:Dam methylation ratios for 1212 genes on the right arm of chromosome 2. Most of the genes analyzed have a methylation ratio of ∼1, indicating that they are not bound by HP1. However, a small percentage of the probed genes show a clearly elevated ratio, indicating HP1-targeted methylation. Importantly, this HP1 binding pattern is strikingly different from that of four unrelated proteins [GAGA factor (van Steensel et al. 2003), and dMyc, dMnt, and dMax (Orian et al. 2003); data not shown], indicating that our mapping technique is highly specific [see also Su(var)3-9 and HP1c data below].
Figure 1.
Chromosomal distribution of HP1 binding. Each vertical line represents the average Dam-HP1:Dam methylation ratio of a probed gene at its chromosomal position (in megabases). Red lines indicate statistically significant HP1 target genes (see Materials and Methods). Gray lines depict standard deviations. (A) Right arm of chromosome 2 (total 1212 probed genes). (B) Chromosome 4. (C) Close-up of the pericentric region of chromosome 2. Gray oval depicts the centromere. Width of the vertical lines in B and C indicates the size of the probed regions, with the exception of the repetitive histone gene cluster (HisC), which has not been fully sequenced.
In total, we identified 218 loci that are significantly associated with HP1 (see Materials and Methods). Of these target loci, 152 are unique genes (Fig. 2); in addition, 56 targets contain repetitive sequences, 46 of which correspond to 27 unique transposable elements (TEs). Our previous small-scale mapping study also revealed abundant binding of HP1 to TEs (van Steensel et al.2001). For the remaining 10 positive probes, we were unable to obtain reliable annotations.
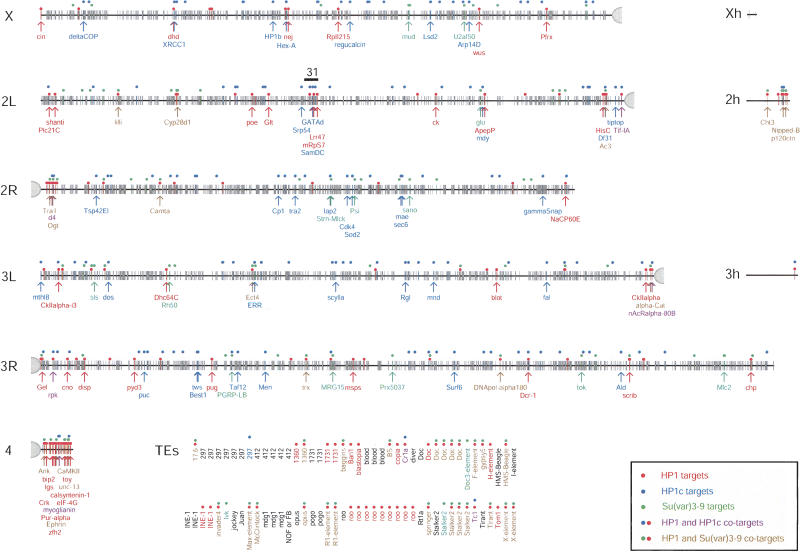
Figure 2.
Genomic distribution of HP1, HP1c, and Su(var)3-9 targets. Each tick marks a probed locus (6260 annotated loci in total). Colored dots mark significant binding sites of each protein (see Materials and Methods). See legend box for color code. Genes with a known function are marked with an arrow and the gene name. Annotations are according to BDGP release 3.1. Xh, 2h, and 3h represent heterochromatic parts of the genome that have not yet been precisely mapped. (TEs) Transposable elements. Because most TEs occur at multiple positions in the genome, they are not shown on the chromosomal maps but are listed separately, in alphabetical order. TEs listed multiple times were represented by more than one probe on the microarray. Region 31 on chromosome 2L is indicated by a black bar.
Consistent with immunofluorescent labeling of HP1 on polytene chromosomes (James et al. 1989), we observed a strong enrichment of HP1 targets in pericentric regions and on the predominantly heterochromatic chromosome 4 (Figs. 1, 2). We found significant binding of HP1 to 61 genes in pericentric regions (operationally defined as genes located <1 Mb from the proximal ends of the sequenced parts of each of the chromosomes X, 2, and 3) and on chromosome 4. The genes light and rolled, two extensively studied pericentric heterochromatic genes (Wakimoto and Hearn 1990; Hearn et al. 1991; Lu et al. 2000), were not represented on our microarray. We therefore probed these genes separately and confirmed that both are indeed bound by HP1 (Supplementary Fig. S4).
In addition to the pericentric binding, we observed association of HP1 with 91 genes dispersed on the distal chromosomal regions. The previously reported abundant association of HP1 with most of region 31 on the left arm of chromosome 2 in salivary gland chromosomes, as observed by immunofluorescence microscopy (James et al. 1989), was not detectable in our experiments. Out of 50 genes tested in region 31, we found only three genes to be associated with HP1 (Fig. 2). This discrepancy might be due to tissue-specific differences in the binding pattern of HP1 (see Discussion). We did observe binding of HP1 to subtelomeric genes (see Supplementary Table S1), consistent with immunostaining of HP1 at the tips of polytene chromosomes (Fanti et al. 1998).
HP1-containing heterochromatin is thought to be able to “spread” in cis along the chromatin fiber (Henikoff 2000; Dhillon and Kamakaka 2002; Grewal and Elgin 2002; Richards and Elgin 2002), but whether spreading over long distances occurs in a continuous or discontinous fashion has been a matter of debate (Talbert and Henikoff 2000). Closer inspection of the HP1 pattern in the pericentric regions and on chromosome 4 revealed that HP1 is not continuously distributed in these regions (Fig. 1B,C). Rather, pericentric HP1 target and nontarget genes are interspersed, in agreement with a banded HP1 staining pattern in these regions of polytene chromosomes (James et al. 1989; Sun et al. 2000; B. van Steensel, unpubl.).
Partial overlap of HP1 and Su(var)3-9 binding patterns
The reported interactions between HP1 proteins and Su(var)3-9 orthologs (Ekwall et al. 1996; Aagaard et al. 1999; Melcher et al. 2000; Bannister et al. 2001; Lachner et al. 2001), in particular between Drosophila HP1 and Su(var)3-9 (Schotta et al. 2002), predicted that the binding patterns of these two proteins match closely. To test this, we performed mapping of Su(var)3-9 binding using the same approach and conditions as for HP1. In total, we identified 127 target loci for Su(var)3-9. Of these, 86 probes corresponded to unique genes (Fig. 2); 32 targets contained repetitive sequences, 27 of which corresponded to 19 distinct TEs. Thus, like HP1, Su(var)3-9 shows a strong preference for TEs, and also binds to a large set of single-copy genes.
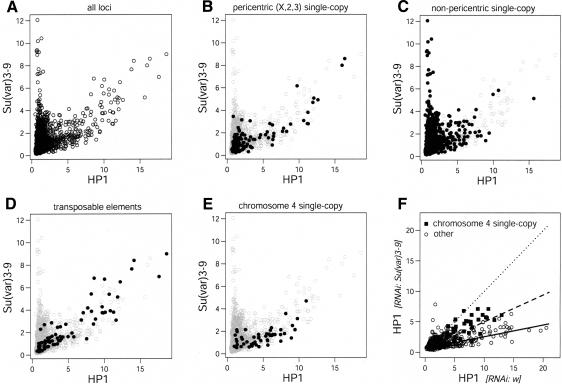
By means of a bivariate scatter plot (Fig. 3A) we compared the binding patterns of HP1 and Su(var)3-9. Surprisingly, we found clear evidence for two distinct sets of Su(var)3-9 target loci. One set of loci binds both HP1 and Su(var)3-9 and displays approximately the same methylation ratios for both proteins, suggestive of a roughly constant stoichiometry of the two proteins within this set of loci. A second set of loci displays strong binding of Su(var)3-9, but no detectable HP1 binding (see vertical “tail” in Fig. 3A). These results are in general agreement with the view that HP1 binding is dependent on Su(var)3-9, but they also demonstrate that the presence of Su(var)3-9 is not sufficient for localization of HP1. We also identified loci with significant levels of HP1 binding but no significant Su(var)3-9 levels (Fig. 2). However, these loci do not form a clearly separate group in the scatterplot (Fig. 3A).
Figure 3.
Position-dependent relationships between HP1 and Su(var)3-9 binding. (A–E) Comparison of HP1 and Su(var)3-9 binding. Data are average ratios from three independent experiments. (A) Bivariate scatterplot of Dam-fusion:Dam ratios of HP1 and Su(var)3-9 of all probed loci. (B–E) Same as in A, with solid dots marking pericentric single-copy genes on chromosomes X, 2, and 3 (B); nonpericentric single-copy genes (C); transposable elements (D); and genes on chromosome 4 (E). (F) Loss of HP1 binding after RNAi-mediated knockdown of Su(var)3-9. HP1 binding (expressed as Dam-HP1:Dam ratios) was mapped in cells treated with either Su(var)3-9 dsRNA or with white dsRNA. Solid squares represent genes on chromosome 4; open circles mark genes on other chromosomes and TEs. Dotted line marks the diagonal (slope = 1); other lines show linear regression fitting for the data from genes on chromosome 4 (broken line, slope = 0.47) and from genes on other chromosomes and TEs (solid line, slope = 0.22). Data are average ratios of two independent experiments.
When we compared the chromosomal distribution of HP1 and Su(var)3-9 targets, we found that colocalization of the two proteins occurs primarily at pericentric genes of the three large chromosomes (Fig. 3B), whereas genes that bind exclusively Su(var)3-9 are enriched in nonpericentric regions (Fig. 3C). Repetitive elements, most of which are concentrated in pericentric regions (Bartolomé et al. 2002), also display cobinding of HP1 and Su(var)3-9, and rarely bind Su(var)3-9 alone (Fig. 3D). These results demonstrate that the interaction between HP1 and Su(var)3-9 is not universal, but occurs predominantly in pericentric regions.
Many genes on chromosome 4 show very low levels of Su(var)3-9 binding (Fig. 3E), suggesting that on this chromosome the binding of HP1 may often occur independently of Su(var)3-9. Earlier microscopy analysis of green fluorescent protein (GFP)-tagged HP1 localization in salivary gland nuclei (Schotta et al. 2002) demonstrated that HP1 binding to the chromocenter is strongly reduced in Su(var)3-9-deficient larvae compared to wild-type larvae. In contrast, HP1 binding to chromosome 4 was mostly retained in a Su(var)3-9 mutant background (Schotta et al. 2002). We therefore tested whether the binding of HP1 to genes on chromosome 4 was independent of the presence of Su(var)3-9 in Kc cells.
We performed DamID mapping of HP1 binding after RNA interference (RNAi) of Su(var)3-9. As a control, we treated cells with dsRNA of the white gene, which is not functional in Kc cells. Immunofluorescence microscopy and Western blotting confirmed that Su(var)3-9 was largely depleted in Su(var)3-9 RNAi cells compared to white RNAi cells (Supplementary Fig. S2; data not shown). Moreover, in nuclei of Su(var)3-9 dsRNA-treated cells, we observed reduced HP1 staining of the chromocenter accompanied by a somewhat increased staining of the euchromatic regions of the nucleus (data not shown). After DamID mapping of HP1 binding in these cells, we observed that, in agreement with the findings of Schotta and colleagues (2002), the general binding of HP1 to its target genes is strongly reduced (Fig. 3F). On average, the Dam-HP1:Dam ratios in Su(var)3-9 dsRNA-treated cells were reduced by a factor of 3–4 compared with white dsRNA-treated cells. Importantly, HP1 targets on the fourth chromosome were clearly less affected than those situated on other chromosomes. Taken together with the absence of Su(var)3-9 from many HP1 target genes on chromosome 4 under normal conditions (Fig. 3E), these results indicate that binding of HP1 to genes on chromosome 4 is largely independent of Su(var)3-9.
In contrast to the genes on chromosome 4, many genes on chromosomes X, 2, and 3 that were scored as targets of HP1 but not of Su(var)3-9 (i.e., genes marked by red but not by green dots in Fig. 2) displayed a loss of HP1 binding upon depletion of Su(var)3-9 (Fig. 3F; data not shown). This suggests that these genes were incorrectly scored as Su(var)3-9-negative, possibly due to somewhat higher noise levels in the Su(var)3-9 binding data. Indeed, genes that are located on the large chromosomes and scored as HP1+/Su(var)3-9- show on average a somewhat elevated level of Su(var)3-9 binding (average ratio 1.6 ± 0.7, n = 68). It is possible that the interaction of Su(var)3-9 with these genes is relatively weak or transient, and therefore difficult to detect.
Multiple targeting mechanisms for HP1c
Several studies have indicated that binding of HP1 proteins to pericentric heterochromatin by Su(var)3-9 orthologs is at least partially established through methylation of H3-K9, which in turn is specifically recognized by the chromodomain of HP1 proteins. We speculated that another chromodomain protein might accompany Su(var)3-9 when HP1 is absent. We considered two other Drosophila HP1 family members, HP1b and HP1c (Smothers and Henikoff 2001). Microscopy studies have shown that both proteins decorate at least partially the arms of polytene chromosomes (Smothers and Henikoff 2001), and protein structure analysis has shown that most of the amino acid residues that contact H3meK9 in HP1 and in mouse HP1β (Jacobs and Khorasanizadeh 2002; Nielsen et al. 2002) are conserved in HP1b and HP1c (see Supplementary Fig. S3).
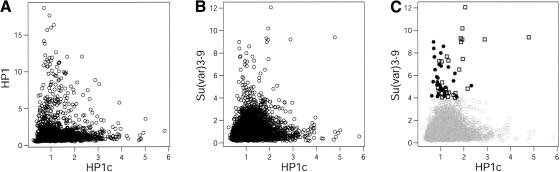
Our attempts to obtain evidence for locus-specific binding of HP1b failed, because all probes on the microarray showed Dam-HP1b:Dam methylation ratios close to unity. However, we did detect significant binding of HP1c to 165 loci, the vast majority of which are dispersed along the chromosome arms (Fig. 2). Relatively few target loci were found among pericentric genes (16 targets) or repetitive sequences (seven targets, three of which are TEs), in agreement with the previously reported absence of HP1c from the chromocenter (Smothers and Henikoff 2001). Bivariate scatter plots showed poor correlations between the binding patterns of HP1c and Su(var)3-9 and between the patterns of HP1c and HP1 (Fig. 4A,B). This lack of correlation was confirmed by randomization of the binding data, which yielded similar scatter plots (data not shown). Thus, most loci that bind HP1c strongly do not bind either of the two other proteins. Interestingly, we noticed that in loci with strong Su(var)3-9 binding, absence of HP1 correlates with a moderate but significant level of HP1c binding (Fig. 4C; P = 2.3 × 10-5, Mann-Whitney U-test). This indicates that HP1c is recruited at moderate levels to strong Su(var)3-9 target loci that lack HP1. It must be emphasized, however, that HP1c binds predominantly to loci that lack Su(var)3-9.
Figure 4.
Comparison of HP1c binding with HP1 and Su(var)3-9 binding. (A–C) Bivariate scatterplot of Dam-fusion:Dam ratios of HP1c and HP1 (A), HP1c and Su(var)3-9 (B), and HP1c and Su(var)3-9 (C). Highlighted symbols in C mark loci with strong Su(var)3-9 binding (ratio > 4). (Solid circles) HP1 targets; (open squares) HP1 nontargets. Data are average ratios from three independent experiments.
Target loci of HP1 and HP1c are enriched in distinct sequence motifs
Because the binding patterns of HP1, HP1c, and Su(var)3-9 are only partially overlapping, their targeting mechanisms must be independent to some extent. To gain insight into the nature of these specific targeting mechanisms, we tested whether specific sequence motifs present in the target genes might play a role. We applied the REDUCE algorithm (Bussemaker et al. 2001), which allows for a completely unbiased search for motifs whose occurrence correlates with the binding profiles of each of the three proteins (Orian et al. 2003; van Steensel et al. 2003). To avoid bias by repetitive elements, we performed this analysis on single-copy genes only. Table 1 shows a summary of sequence motifs that correlate with binding of each of the proteins. We found that HP1 binding correlates with the occurrence of AT-rich motifs that predominantly consist of stretches of adenosines and stretches of thymidines. These “AT stretches” were also found for HP1c. In addition, the binding of HP1c correlates with the occurrence of motifs that share the consensus (AT)ACATA(TGT). This motif was not found for HP1. Interestingly, this consensus motif contains the 8-mer palindrome ACATATGT, suggestive of a sequence-specific DNA-binding factor that associates with the motif as a dimer. No motifs were found to correlate significantly with Su(var)3-9 binding, possibly due to a somewhat lower signal/noise ratio of the Su(var)3-9 binding profiles.
Table 1.
Representative sequence motifs enriched in HP1 and HP1c binding sites
| HP1
|
HP1c
|
|||
|---|---|---|---|---|
| Motif | P-value | Motif | P-value | |
| AAAAAT | <10-12 | AAATAAA | <10-12 | |
| AAAAATT | <10-12 | TTTTTT | <10-12 | |
| AAAATT | <10-12 | ATTTTT | <10-12 | |
| AATTTTT | <10-12 | TATTTTT | <10-12 | |
| AAAAT | <10-12 | TTTTTA | <10-12 | |
| ATTTTT | <10-12 | — | ||
| AAATTT | <10-12 | TACATA | <10-12 | |
| AAAATTT | <10-12 | ATACATA | <10-12 | |
| TTTTTA | <10-12 | ATATGTA | 2 × 10-12 | |
| TAAAA | <10-12 | TACATAT | 7 × 10-10 | |
| TAAAAA | <10-12 | TATGTA | 1 × 10-9 | |
| AATTTT | <10-12 | ACATATG | 6 × 10-9 | |
| — | — | |||
| AT-stretches | AT-stretches + (at)ACATA(tgt) | |||
No motifs were positively correlated with Su(var)3-9 binding. Motifs were obtained by applying the REDUCE algorithm (Bussemaker et al. 2001; see Materials and Methods). See Supplemental Material for a complete list of motifs and associated P-values.
HP1 and Su(var) 3-9 binding is inversely correlated with gene expression levels in Kc cells
HP1 and Su(var)3-9 have been implicated in gene silencing (Eissenberg and Elgin 2000; Henikoff 2000; Jenuwein 2001; Richards and Elgin 2002), whereas the function of HP1c is unknown. Most studies of gene regulation by HP1 proteins and Su(var)3-9 orthologs were based on artificial reporter constructs or chromosomal rearrangements, or were focused on a small set of genes. We analyzed the genome-wide correlations between the binding pattern of each protein and the pattern of gene expression. Because absolute expression levels of individual genes in Kc cells were not known, we isolated total RNA samples from Kc cells, labeled and hybridized these to microarrays, and used the spot fluorescence intensities as a rough approximation of mRNA abundances.
We found that target genes of Su(var)3-9 and HP1 showed on average significantly lower expression levels than genes that do not bind the respective protein (Fig. 5A,B; P = 2.5 × 10-8 and P = 8.5 × 10-4, respectively, Mann-Whitney U-test). This is consistent with roles for these proteins in repression of transcription. Interestingly, this inverse correlation is strongest for Su(var)3-9, which displays a strong preference for genes with extremely low expression levels or that are not expressed (Fig. 5A). Genes that bind HP1 and Su(var)3-9 together are rarely expressed at very low levels, in contrast to genes that bind Su(var)3-9 alone (Fig. 5C). Thus, chromatin marked by Su(var)3-9 alone appears to be more repressive than chromatin containing both HP1 and Su(var)3-9. Importantly, HP1 and Su(var)3-9 bind only to a subset (<10%, data not shown) of all genes with low expression levels, ruling out the possibility that these proteins are general markers of inactive genes. It should be stressed that a considerable number of target genes of Su(var)3-9 and HP1 display moderate or even high expression levels, indicating that recruitment of these proteins is not sufficient to repress transcription completely. We did not find any evidence for reduced expression of genes that bind HP1c (Fig. 5D, P = 0.38), suggesting that this protein does not play a major role in repression of transcription.
Figure 5.
Expression levels of target genes bound by Su(var)3-9, HP1, and HP1c. Histograms are shown of relative gene expression levels of single-copy genes. Expression levels are calculated as normalized log2-transformed spot intensities after hybridization of labeled mRNA from untreated Kc cells to microarrays (average of three experiments; see Materials and Methods). (A) Su(var)3-9 target genes (gray) compared with an equal number of nontarget genes (black). (B) HP1 target genes (gray) compared with an equal number of nontarget genes (black). (C) Genes that are targets of both Su(var)3-9 and HP1 (dark gray) compared with genes that bind only Su(var)3-9 (light gray). (D) HP1c target genes (gray) compared with an equal number of nontarget genes (black). (E) Pericentric (white) compared with nonpericentric (gray) Su(var)3-9 target genes. Pericentric genes include all genes on chromosome 4. (F) Pericentric (white) compared with nonpericentric (gray) HP1 target genes.
Lack of repression of pericentric genes in Kc cells
A few genes that are naturally located in pericentric heterochromatin have been reported to be expressed (Weiler and Wakimoto 1998; Lu et al. 2000); mutation of HP1 or translocation of these genes to a euchromatic environment reduces their expression (Clegg et al. 1998; Weiler and Wakimoto 1998; Lu et al. 2000), arguing that heterochromatin facilitates rather than represses the expression of these genes. We investigated whether the lack of repression by heterochromatin might be a general property of pericentric genes, by comparing the overall expression levels of Su(var)3-9 and HP1 targets within pericentric regions to those of targets elsewhere. Strikingly, virtually all pericentric genes bound by Su(var)3-9 (Fig. 5E) and HP1 (Fig. 5F) are expressed at medium to high levels. In comparison, nonpericentric genes bound by either protein are much more frequently expressed at very low levels. This suggests that, as a general rule, pericentric genes in Kc cells are not subject to the repressive effect of HP1- and Su(var)3-9-containing chromatin, whereas binding of HP1 or Su(var)3-9 to nonpericentric genes is overall associated with reduced expression levels.
Su(var)3-9 and HP1 associate with genes of specific developmental programs depending on chromosomal location
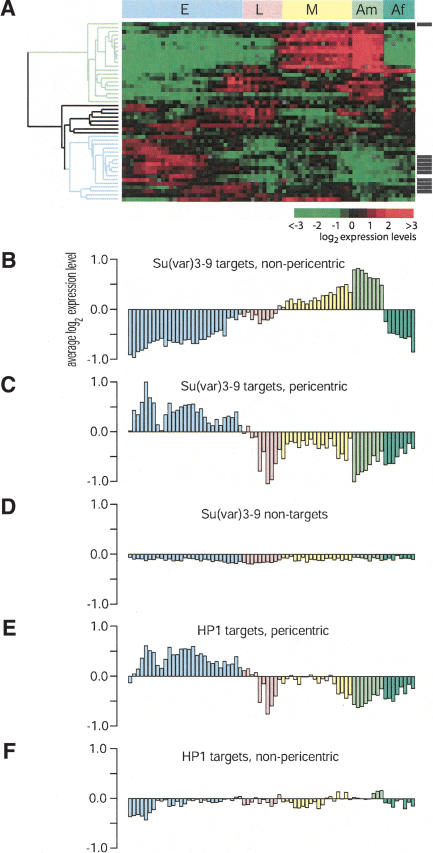
We wondered whether the identified target genes of each of the investigated proteins have similar expression patterns. To test this, we took advantage of the recently reported database of genome-wide expression profiles throughout development (Arbeitman et al. 2002). For each protein, we compared the developmental expression patterns of its target genes by cluster analysis. Of the 85 single-copy Su(var)3-9 targets, complete developmental expression profiles were available for 46 genes. Strikingly, we found that these targets fall into a small number of distinct categories based on their developmental expression patterns (Fig. 6A).
Figure 6.
Genes bound by Su(var)3-9 and HP1 belong to specific developmental programs. (A) Cluster analysis of the developmental expression time course of genes that are Su(var)3-9 targets in Kc cells. All gene expression data were taken from Arbeitman et al. (2002). (E) Embryos; (L) larvae; (M) metamorphosis; (Am) adult male; (Af) adult female. Note that pericentric target genes (indicated by black bars to the right of the cluster-gram) are generally genes that are predominantly expressed during embryogenesis (blue tree branch). The male-specific gene cluster is marked by the green tree branch. (B–F) Average expression of Su(var)3-9 (B,C) and HP1 (E,F) target genes and Su(var)3-9 nontarget genes (D) during fly development, divided into pericentric (C,E) and nonpericentric (B,F). Color coding of the developmental stages is as in A.
Approximately one-third of all Su(var)3-9 target genes display low activity during embryogenesis and in female adults, but show increased activity in pupae and male adults. This pattern is typical of male-specific genes (Arbeitman et al. 2002). In total, we found 16 Su(var)3-9 targets (34.8%) that display significantly higher expression levels in males compared to females (using a paired t-test for each gene; P < 0.001). According to the same criteria, only 16.6% (430 of 2788) of the genes that were present in both the Arbeitman et al. data set and our data set are male-specific, indicating a significant enrichment in the Su(var)3-9 target set (twofold, P = 6.3 × 10-4, hypergeometric test). In contrast, female-specific genes are underrepresented (8.7% vs. 25.6%). The expression of about half (9/16) of the male-specific targets is reduced more than twofold in tudor mutant male flies, which lack testes (Arbeitman et al. 2002; Supplementary Table S2). Thus, some of the male-specific Su(var)3-9 targets may be preferentially expressed in germline tissue, whereas others are likely to be somatically expressed.
Another third of the Su(var)3-9 target genes displays elevated expression specifically during embryogenesis. The remaining target genes show heterogeneous expression patterns but are expressed mostly during the larval stage. We looked for a dependence of these expression patterns on chromosomal location of Su(var)3-9 target genes. Surprisingly, we found that 9/17 (52.9%) of the embryo-specific Su(var)3-9 target genes were pericentric, whereas only one of the remaining 29 genes (3.4%) was pericentric (Fig. 6A). Direct comparison of the developmental expression patterns confirmed that pericentric Su(var)3-9 target genes are predominantly expressed during embryogenesis, whereas nonpericentic Su(var)3-9 target genes display mostly the male-specific expression pattern (Fig. 6B–D). In agreement with the observation that cobinding of HP1 and Su(var)3-9 is largely restricted to pericentric regions, we found that pericentric HP1 target genes are also predominantly expressed during embryogenesis (Fig. 6E). However, nonpericentric HP1 target genes do not show a clear male-specific expression pattern (Fig. 6F). We also did not find any preference of HP1c for specific sets of coordinately expressed genes (data not shown). In summary, in pericentric regions, both HP1 and Su(var)3-9 associate with genes that are preferentially expressed during embryogenesis, whereas in nonpericentric regions only Su(var)3-9 binds to a large set of genes that are preferentially expressed in male adults.
Discussion
Mapping of in vivo protein targets using DamID
We used the DamID technique to map target loci of three chromatin proteins: HP1, HP1c, and Su(var)3-9. Previously, this technique was successfully used to identify genomic binding sites of several transcription factors and chromatin-associated proteins in Drosophila, including Myc, Max, Mad/Mnt (Orian et al. 2003), GAGA factor (van Steensel et al. 2001, 2003), and Sir2 (van Steensel et al. 2001). An excellent agreement was found between chromatin immunoprecipitation and DamID data for GAGA factor for 12 tested loci (Sun et al. 2003). The binding maps reported here are distinct from the maps of GAGA factor and the Max/Mad/Mnt proteins (data not shown), further supporting the specificity of DamID. The specific effects of Su(var)3-9 depletion on HP1 binding patterns, which are in perfect agreement with microscopy observations (Schotta et al. 2002), also argue that the binding patterns reported here are specific and reliable. Thus, DamID is a reliable method for large-scale mapping of genomic targets of the HP1 and Su(var)3-9 proteins.
The use of cDNA arrays in these experiments limited our genomic view of protein binding to transcribed regions and ∼2 kb of flanking sequences (van Steensel and Henikoff 2000; van Steensel et al. 2003). Thus, our maps provide little insight into the interaction of the studied proteins with the large noncoding fraction of the fly genome. We recently employed genomic “tiling path” arrays for more detailed studies of protein binding in a contiguous 2.9-Mb segment of chromosome 2L (Sun et al. 2003). Interestingly, these studies revealed that HP1 binds to the transcribed region but not to the promoter of the gene ck. Likewise, HP1 was reported to bind to the transcribed region of the activated hsp70 gene (Piacentini et al. 2003). Binding of HP1 [and possibly of Su(var)3-9 and HP1c] to transcribed regions may occur at some or many of the other loci that we identified, and this may explain the large number of target loci detected using cDNA arrays.
HP1 and Su(var)3-9 proteins occur in several distinct complexes
In agreement with the prevalent model of the interactions between HP1, Su(var)3-9, and H3-K9 methylation, we found that HP1 and Su(var)3-9 bind together to many loci. We confirmed that in most of these loci, HP1 binding is dependent on the presence of Su(var)3-9. These HP1/Su(var)3-9 cotargets consist predominantly of pericentric genes and TEs, which are also concentrated in pericentric heterochromatin (Bartolomé et al. 2002). This explains the pericentric colocalization of HP1 and Su(var)3-9 observed by fluorescence microscopy in polytene and diploid nuclei (James et al. 1989; van Steensel and Henikoff 2000; Schotta et al. 2002).
Strikingly, we found that HP1 and Su(var)3-9 can also bind independently of each other at distinct sets of genes. Several genes located on the chromosome arms display strong Su(var)3-9 binding but no detectable HP1 binding. Possibly, at these loci Su(var)3-9 fails to methylate H3-K9, either because H3-K9 is acetylated or because the methyltransferase activity of Su(var)3-9 is inhibited by another (unknown) mechanism. Alternatively, access of HP1 to methylated H3-K9 may be blocked at these loci. This may be either due to a chromatin conformation that renders the methylated H3-K9 inaccessible to HP1, or due to the presence of another methyl-H3-K9-binding protein that competes with HP1 for binding at these loci. HP1c may contribute to this competition, because we observed a weak but significant enrichment of HP1c at this group of loci. In agreement with this, the amino acid residues required for HP1 binding to methylated H3-K9 are conserved in the chromodomain of HP1c.
In addition, we find that many genes on chromosome 4 are bound by HP1 in a manner that is relatively independent of Su(var)3-9. In agreement with earlier microscopic observations in polytene chromosomes (Schotta et al. 2002), we found that HP1 is retained at genes on chromosome 4 but not on other chromosomes after loss of Su(var)3-9 (Fig. 3F). Consistently, most of the HP1-binding genes on chromosome 4 show only weak or undetectable binding of Su(var)3-9. Because immunofluorescence microscopy experiments have demonstrated that H3-K9 on chromosome 4 is overall strongly methylated in a Su(var)3-9-independent fashion (Schotta et al. 2002), it is likely that a different, yet unidentified H3-K9 methyltransferase is involved in the binding of HP1 to the chromosome 4 target genes that we identified. The physiological relevance of the different mechanism of HP1 binding to chromosome 4 is unclear.
In addition to methylation of H3-K9, other molecular signals may confer target specificity to the HP1 proteins. We found that HP1 binding correlates with the occurrence of certain AT-rich motifs, whereas HP1c binding correlates both with AT-rich motifs and the ACATATGT motif. These motifs suggest the presence of sequence-specific factors that may facilitate binding of the HP1 proteins. A factor that could bind to the ACATATGT motif is as yet unidentified, but the palindromic nature of the motif suggests that the factor binds to DNA as a dimer. Additionally, AT stretches may facilitate binding of HP1- or HP1c-containing complexes by affecting the DNA or nucleosome structure (Iyer and Struhl 1995).
Roles of HP1 and Su(var)3-9 in gene regulation
Based on the well documented roles of HP1 and Su(var)3-9 in silencing of reporter genes, we expected that the genes identified as targets would be mostly inactive. Indeed, we found that Su(var)3-9 and HP1 preferentially associate with genes of low expression levels. This preference is more prominent for Su(var)3-9 than for HP1; in fact, genes that are bound by Su(var)3-9 without HP1 are more often inactive than genes that are bound by both proteins. Formally, binding of Su(var)3-9 may be either the cause or the consequence of gene silencing. In the latter case, Su(var)3-9 complexes would mark genes that are already inactive due to other silencing mechanisms. However, if Su(var)3-9 is indeed actively involved in silencing of its target genes, then Su(var)3-9 complexes may be more potent silencers if they lack HP1.
Although HP1 and Su(var)3-9 generally display a preference for genes of low activity, a considerable fraction of their target genes are expressed, sometimes even at high levels. Many of these active target genes are located in pericentric regions and on chromosome 4. Earlier findings already demonstrated that the pericentric genes lt and rl are active (Lu et al. 2000), and we confirmed that these genes are also bound by HP1 and Su(var)3-9. Association of HP1 was reported recently with ecdysone- and heat shock-induced puffs on polytene chromosomes (Piacentini et al. 2003). The expression of lt, rl, and hsp70 genes is reduced in HP1-deficient larvae (Lu et al. 2000; Piacentini et al. 2003), suggesting that HP1 may facilitate rather than suppress transcription of certain genes. We attempted to extend these observations in Kc cells by microarray mRNA expression profiling after RNA interference of HP1 and Su(var)3-9 (Supplementary Fig. S5). We were not able to detect any changes in expression of the HP1 or Su(var)3-9 target genes after knockdown of either of the two proteins. According to these results, HP1 and Su(var)3-9 may have only redundant roles in gene regulation in Kc cells. However, it should be noted that the dsRNA-induced reduction of HP1 and Su(var)3-9, although substantial, may have been insufficient or not long enough to cause detectable alterations in gene regulation. In addition, the previously reported changes in expression of the lt and rl genes in HP1-deficient larvae were only ∼2.5-fold (Lu et al. 2000). Such modest changes in gene expression may have been missed in our microarray-based assay. Finally, HP1 and Su(var)3-9 complexes may not be essential for gene regulation in Kc cells. Heterochromatin-mediated silencing of a reporter gene is not fully developed until late embryogenesis (Lu et al. 1998). Kc cells appear to be embryonic (Supplementary Fig. S6), so it is possible that the regulatory functions of HP1 and Su(var)3-9 initiate only later in development. Furthermore, the lack of a visible phenotype of Su(var)3-9 null mutants (Tschiersch et al. 1994) suggests that a role of Su(var)3-9 in gene regulation may be redundant.
Specific gene programs
Among the target loci of HP1 and Su(var)3-9, we identified two conspicuous groups of developmentally coregulated genes. First, many of the nonpericentric genes that are exclusively bound by Su(var)3-9 in Kc cells are highly expressed in adult males, but much less in females, embryos, larvae, and Kc cells. Extrapolating our data to the entire genome [our comparison of Su(var)3-9 binding and developmental expression patterns was only possible for ∼2700 genes], we anticipate that 50–100 male-specific genes are bound by Su(var)3-9 in Kc cells. This could be an underestimate, because testis-specific genes may be underrepresented in the cDNA libraries present on our microarray (see Materials and Methods).
Su(var)3-9 may contribute to repression of these male-specific genes in early stages of development and in adult females. Kc cells are female, as judged from the absence of a Y chromosome and expression of the female-specific but not the male-specific splicing variant of doublesex (R. Lascaris and B. van Steensel, unpubl.). Therefore binding of Su(var)3-9 to these genes may reflect either the female or the embryonic origin of the Kc cells. Alternatively, aberrant expression of these genes in embryos, larvae, and female adults may not lead to a detectable phenotype under laboratory conditions.
The second group of developmentally coregulated target genes is formed by a set of embryo-specific genes. Strikingly, these embryo-specific target genes are strongly concentrated in pericentric regions, and are typically bound by both HP1 and Su(var)3-9. This suggests a specialized role for pericentric HP1 and Su(var)3-9 in the embryonic gene expression program. In Kc cells, these pericentric target genes are generally not repressed, consistent with the embryonic origin of these cells. Both HP1 and Su(var)3-9 are present in embryos (Tschiersch et al. 1994; Kellum and Alberts 1995), suggesting that the lack of repression of pericentric target genes cannot be attributed to the absence of either HP1 or Su(var)3-9 during this developmental stage. Rather, these proteins may facilitate gene expression in the embryo, or perhaps mark the embryonic pericentric genes for silencing later in development. The clustering of these genes in the pericentric region may play a role in their coordinated regulation.
It is likely that the genomic binding pattern of the proteins studied here depends at least in part on the cell type or developmental stage. Evidence that the target specificity of heterochromatin proteins is dynamic comes from recent observations that HP1 binds to induced but not to uninduced heat-shock and ecdysone-responsive genes (Piacentini et al. 2003). Our HP1 binding map obtained in Kc cells showed only limited overlap with the banding pattern of HP1 staining on polytene chromosome arms in salivary glands (Fanti et al. 2003). Of 91 nonpericentric target loci that we identified, only nine coincided with HP1 bands in polytene chromosomes (Supplementary Table S3). Although this comparison should be interpreted with caution because of the different methodology and a >10-fold difference in mapping resolution (the median size of the stained polytene chromosome regions listed by Fanti and colleagues (2003) is 74 kb, whereas the median size of the genomic regions probed by our microarray is 3.7 kb), it suggests that many target loci may be cell type-specific. An example of this is region 31, a broad (∼0.5 Mb) region on chromosome 2 that is bound by HP1 in polytene chromosomes in salivary gland tissue. In Kc cells we found that three out of 50 probed loci in this region are associated with HP1, which is unlikely to account for the extensive HP1 staining of region 31 in polytene chromosomes. This suggests that much of the binding of HP1 to this region in salivary gland cells is cell type-specific.
The difficulties of defining heterochromatin
The heterogeneity of the HP1 and Su(var)3-9 complexes, in terms of both protein composition and target gene expression status, further complicates the matter of defining heterochromatin (Henikoff 2000). By morphological criteria, heterochromatin in Drosophila chromosomes is concentrated in pericentric regions. However, most pericentric genes, although bound by HP1 and Su(var)3-9, are transcriptionally active, contrary to the repressive role that is generally attributed to heterochromatin. On the “euchromatic” chromosome arms, genes bound by Su(var)3-9 are often repressed, yet these genes typically lack the classical heterochromatin marker protein HP1. Over the long term, it may be more useful to define different types of chromatin according to their protein composition, including posttranslational modifications and histone variants. This will require a much more sophisticated nomenclature than “euchromatin” and “heterochromatin”. The transcriptional status of a gene may be expected to be controlled by the combinatorial action of the proteins that are associated with it. Global approaches to study chromatin composition on a gene-by-gene basis, such as described here, will be essential to catalog the different chromatin types and to understand their role in gene regulation.
Materials and methods
Chromatin profiling
Full-length Su(var)3-9 and HP1c were fused to the N terminus of Dam through a myc epitope-tag linker by cloning of the open reading frames (ORFs) into pCMycDam (van Steensel and Henikoff 2000). The Dam-HP1 construct was described earlier (van Steensel and Henikoff 2000). Immunofluorescent labeling of Kc cells expressing the fusion proteins (van Steensel and Henikoff 2000) showed that Dam-HP1 and Su(var)3-9-Dam were concentrated in the chromocenter, whereas HP1c-Dam was located in the nucleoplasm but mostly excluded from the chromocenter (data not shown), in agreement with previous localization studies of the respective proteins (Smothers and Henikoff 2001; Schotta et al. 2002). Chromatin profiling was performed as described (van Steensel et al. 2001), except that methylated genomic DNA fragments were not purified by sucrose gradient centrifugation, but instead specifically amplified using a methylation-specific PCR protocol (Supplementary Fig. S1).
Microarrays
For hybridizations we used spotted microarrays containing 5930 unique cDNA clones from release 1 of the Drosophila Gene Collection (DGCr1), and ∼370 additional cDNA and genomic fragments contributed by members of the Northwest Flychip Consortium. DGCr1 consists of cDNA fragments, mostly cDNAs containing the full ORF, from adult head, ovary, embryo, larvae and pupae, and Schneider cells (Rubin et al. 2000).
dsRNA production
As a template for Su(var)3-9 dsRNA production, a 606-bp fragment containing bp 1093–1602 of the ORF of Su(var)3-9 flanked by a T7 promoter at both the 5′ and 3′ end was amplified using the primer pair 5′-GAATTAATACGACTCACTATAGG GAGAAATTCACCCTTACGCAAGGCAGTTG-3′ and 5′-GA ATTAATACGACTCACTATAGGGAGAAATTGGAGAACG AGCCCTGAAAAGC-3′. Because the gene coding for Su(var)3-9 also codes for the transcription elongation factor eLF-2γ, with two exons being shared by both transcripts, the fragment used as a template in dsRNA production was chosen from an exon unique for Su(var)3-9. For RNAi of HP1 and white, the complete ORF of HP1 was excised from pDam-HP1 by digestion with NotI and NaeI, and a 630-bp fragment of the white+mc gene was excised from pCaSpeR-hs by digestion with DpnI. Each fragment was cloned in both orientations into the EcoRV site of vector pcDNA3 (Invitrogen), downstream of the T7 promoter. The Su(var)3-9 PCR product, as well as the linearized and combined plasmids with forward or reverse sequences of either white or HP1, were purified with the QIAquick PCR purification kit (QIAGEN) and used as a template in a transcription reaction to produce dsRNA with the RiboMAX large-scale RNA production system T7 (Promega). The dsRNAs were purified by phenol/chloroform extraction, dissolved in DEPC-treated water and stored at -70°C. An aliquot of each dsRNA was analyzed on a formaldehyde-containing agarose gel in order to check the size and integrity of the RNA.
RNAi
Drosophila Kc167 cells of a near confluent dish were seeded in HyQ SFX-Insect medium (Hyclone). For chromatin profiling experiments, 840 μL cell suspension was added to a 10-cm culture dish with 7 mL medium. For expression profiling, 120 μL cell suspension was added to each well of a six-well culture dish with 1 mL medium per well. To each 10-cm dish, 200–210 μg of either Su(var)3-9 or white dsRNA was added, and to each well of a six-well dish, 30–40 μg of dsRNA was added. The cells were grown at 23°C for 3 d. For chromatin profiling, cells were transfected with either Dam-HP1, Su(var)3-9-Dam, or Dam only. Subsequently, the protocol for chromatin profiling was followed as stated above. For expression profiling, RNA was isolated with Trizol (Invitrogen) according to the manufacturer's protocol and stored at -70°C. The RNA was labeled and hybridized to microarrays as described (Pritchard et al. 2001).The knockdown of Su(var)3-9 and HP1 was confirmed by antibody staining using a rabbit polyclonal anti-Su(var)3-9 antibody (gift from G. Reuter, Martin Luther University Halle, Halle, Germany; Schotta et al. 2002) and a rabbit anti-HP1 antibody (Platero et al. 1998), respectively.
mRNA expression profiling
Total RNA was extracted from Kc cells, reverse transcribed to Cy3- and Cy5-labeled cDNA, and hybridized to microarrays. For each probed gene, the relative expression was calculated as [spot intensity (Cy3 + Cy5)]/[summed intensity of all spots (Cy3 + Cy5)] and subsequently log2-transformed and normalized to the median.
Data analysis
For statistical analysis, all measured ratios were log2-transformed and normalized to the median value of the entire array. Data from three independent experiments were averaged, with one experiment performed with reversed dye orientation. For the Su(var)3-9 data, we noticed that a set of 2026 probes, which were all cloned into pBlueScript vectors, gave a slight positive bias of log-ratios (average bias ∼0.6) compared to the remaining probes, which were cloned into pOT2. We assume that this is due to a weak cross-hybridization of a sequence in the transfected Su(var)3-9-Dam expression vector, which coamplifies with methylated genomic fragments. We therefore normalized the probe sets in the two different vectors separately. Importantly, this does not affect any of the main conclusions of this report. This vector bias was not observed with HP1 or HP1c. To define target loci, we tested whether log-ratios were significantly greater than 0 using the CyberT algorithm (Long et al. 2001), followed by a correction for multiple testing (Benjamini and Hochberg 1995), adjusting the estimated false discovery rate to 10%.
Chromosomal maps
Sequences of DGCr1 (Stapleton et al. 2002) were obtained from the Berkeley Drosophila Genome Project (BDGP). For 5890 probes we were able to identify unique matching genomic regions <100 kb (BLAST search against release 3 whole-genome sequence using default settings). For each of these, the precise chromosomal coordinates of the 5′ and 3′ boundaries of the matching region were determined and used to generate chromosomal maps of protein binding. The sequences of 205 probes matched to more than one genomic locus; by subsequent BLAST search against the BDGP database of TE sequences (Kaminker et al. 2002) we found that 85 of these match to 40 distinct TEs (see Figs. 2, 3D). For 163 probes, no reliable sequence information was available, and therefore we were unable to accurately identify the corresponding genomic loci.
REDUCE analysis
REDUCE analysis was performed as described (van Steensel et al. 2003) using software available at http://bussemaker.bio.columbia.edu/reduce. REDUCE analysis was restricted to microarray data obtained from 5145 cDNA probes that matched to a single genomic region <10 kb in size. The sequences of the probed loci (including 2 kb of flanking genomic sequence on both sides to account for the mapping resolution; van Steensel et al. 2003) were extracted from the BDGP release 3 whole-genome sequence (Celniker et al. 2002) using dedicated Perl scripts. We tested the REDUCE analysis method on similar binding data obtained for GAGA factor. This yielded predominantly GA-repeat motifs, in perfect agreement with the in vitro sequence specificity of GAGA factor (van Steensel et al. 2003).
Acknowledgments
We thank Gunter Reuter for Su(var)3-9 plasmid and antibody, Kevin White for critically reading the manuscript, members of the NKI microarray facility for help, and Eleanor Williams (Fred Hutchinson Cancer Research Center, Seattle, WA) for sharing a Perl script for generating chromosomal maps.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Supplemental material is available at http://www.genesdev.org.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.281503.
References
- Aagaard L., Laible, G., Selenko, P., Schmid, M., Dorn, R., Schotta, G., Kuhfittig, S., Wolf, A., Lebersorger, A., Singh, P.B., et al. 1999. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 18: 1923-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbeitman M.N., Furlong, E.E., Imam, F., Johnson, E., Null, B.H., Baker, B.S., Krasnow, M.A., Scott, M.P., Davis, R.W., and White, K.P. 2002. Gene expression during the life cycle of Drosophila melanogaster. Science 297: 2270-2275. [DOI] [PubMed] [Google Scholar]
- Bannister A.J., Zegerman, P., Partridge, J.F., Miska, E.A., Thomas, J.O., Allshire, R.C., and Kouzarides, T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120-124. [DOI] [PubMed] [Google Scholar]
- Bartolomé C., Maside, X., and Charlesworth, B. 2002. On the abundance and distribution of transposable elements in the genome of Drosophila melanogaster. Mol. Biol. Evol. 19: 926-937. [DOI] [PubMed] [Google Scholar]
- Benjamini Y. and Hochberg, Y. 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Statistical Soc. Ser. B—Methodological 57: 289-300. [Google Scholar]
- Bussemaker H.J., Li, H., and Siggia, E.D. 2001. Regulatory element detection using correlation with expression. Nat. Genet. 27: 167-171. [DOI] [PubMed] [Google Scholar]
- Celniker S.E., Wheeler, D.A., Kronmiller, B., Carlson, J.W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S.P., Frise, E., et al. 2002. Finishing a whole-genome shotgun: Release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 3: RESEARCH0079. [DOI] [PMC free article] [PubMed]
- Clegg N.J., Honda, B.M., Whitehead, I.P., Grigliatti, T.A., Wakimoto, B., Brock, H.W., Lloyd, V.K., and Sinclair, D.A. 1998. Suppressors of position-effect variegation in Drosophila melanogaster affect expression of the heterochromatic gene light in the absence of a chromosome rearrangement. Genome 41: 495-503. [PubMed] [Google Scholar]
- Czermin B., Schotta, G., Hulsmann, B.B., Brehm, A., Becker, P.B., Reuter, G., and Imhof, A. 2001. Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep. 2: 915-919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N. and Kamakaka, R.T. 2002. Breaking through to the other side: Silencers and barriers. Curr. Opin. Genet. Dev. 12: 188-192. [DOI] [PubMed] [Google Scholar]
- Eissenberg J.C. and Elgin, S.C. 2000. The HP1 protein family: Getting a grip on chromatin. Curr. Opin. Genet. Dev. 10: 204-210. [DOI] [PubMed] [Google Scholar]
- Ekwall K., Nimmo, E.R., Javerzat, J.P., Borgstrom, B., Egel, R., Cranston, G., and Allshire, R. 1996. Mutations in the fission yeast silencing factors clr4+ and rik1+ disrupt the localisation of the chromo domain protein Swi6p and impair centromere function. J. Cell Sci. 109: 2637-2648. [DOI] [PubMed] [Google Scholar]
- Fanti L., Giovinazzo, G., Berloco, M., and Pimpinelli, S. 1998. The heterochromatin protein 1 prevents telomere fusions in Drosophila. Mol. Cell 2: 527-538. [DOI] [PubMed] [Google Scholar]
- Fanti L., Berloco, M., Piacentini, L., and Pimpinelli, S. 2003. Chromosomal distribution of heterochromatin protein 1 (HP1) in Drosophila: A cytological map of euchromatic HP1 binding sites. Genetica 117: 135-147. [DOI] [PubMed] [Google Scholar]
- Grewal S.I. and Elgin, S.C. 2002. Heterochromatin: New possibilities for the inheritance of structure. Curr. Opin. Genet. Dev. 12: 178-187. [DOI] [PubMed] [Google Scholar]
- Hearn M.G., Hedrick, A., Grigliatti, T.A., and Wakimoto, B.T. 1991. The effect of modifiers of position-effect variegation on the variegation of heterochromatic genes of Drosophila melanogaster. Genetics 128: 785-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz E. 1928. Das Heterochromatin der Moose. Jahrb. Wiss. Bot. 69: 762-818. [Google Scholar]
- Henikoff S. 2000. Heterochromatin function in complex genomes. Biochim Biophys Acta 1470: 1-8. [DOI] [PubMed] [Google Scholar]
- Iyer V. and Struhl, K. 1995. Poly(dA:dT), a ubiquitous promoter element that stimulates transcription via its intrinsic DNA structure. EMBO J. 14: 2570-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs S.A. and Khorasanizadeh, S. 2002. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080-2083. [DOI] [PubMed] [Google Scholar]
- James T.C., Eissenberg, J.C., Craig, C., Dietrich, V., Hobson, A., and Elgin, S.C. 1989. Distribution patterns of HP1, a heterochromatin-associated nonhistone chromosomal protein of Drosophila. Eur. J. Cell Biol. 50: 170-180. [PubMed] [Google Scholar]
- Jenuwein T. 2001. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 11: 266-273. [DOI] [PubMed] [Google Scholar]
- Kaminker J.S., Bergman, C.M., Kronmiller, B., Carlson, J., Svirskas, R., Patel, S., Frise, E., Wheeler, D.A., Lewis, S.E., Rubin, G.M., et al. 2002. The transposable elements of the Drosophila melanogaster euchromatin: A genomics perspective. Genome Biol. 3: RESEARCH0084. [DOI] [PMC free article] [PubMed]
- Kellum R. and Alberts, B.M. 1995. Heterochromatin protein 1 is required for correct chromosome segregation in Drosophila embryos. J. Cell Sci. 108: 1419-1431. [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Carroll, D., Rea, S., Mechtler, K., and Jenuwein, T. 2001. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature 410: 116-120. [DOI] [PubMed] [Google Scholar]
- Long A.D., Mangalam, H.J., Chan, B.Y., Tolleri, L., Hatfield, G.W., and Baldi, P. 2001. Improved statistical inference from DNA microarray data using analysis of variance and a Bayesian statistical framework. Analysis of global gene expression in Escherichia coli K12. J. Biol. Chem. 276: 19937-19944. [DOI] [PubMed] [Google Scholar]
- Lu B.Y., Ma, J., and Eissenberg, J.C. 1998. Developmental regulation of heterochromatin-mediated gene silencing in Drosophila. Development 125: 2223-2234. [DOI] [PubMed] [Google Scholar]
- Lu B.Y., Emtage, P.C., Duyf, B.J., Hilliker, A.J., and Eissenberg, J.C. 2000. Heterochromatin protein 1 is required for the normal expression of two heterochromatin genes in Drosophila. Genetics 155: 699-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher M., Schmid, M., Aagaard, L., Selenko, P., Laible, G., and Jenuwein, T. 2000. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol. Cell Biol. 20: 3728-3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayama J., Rice, J.C., Strahl, B.D., Allis, C.D., and Grewal, S.I. 2001. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science 292: 110-113. [DOI] [PubMed] [Google Scholar]
- Nielsen P.R., Nietlispach, D., Mott, H.R., Callaghan, J., Bannister, A., Kouzarides, T., Murzin, A.G., Murzina, N.V., and Laue, E.D. 2002. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103-107. [DOI] [PubMed] [Google Scholar]
- Orian A., Van Steensel, B., Delrow, J., Bussemaker, H.J., Li, L., Sawado, T., Williams, E., Loo, L.W., Cowley, S.M., Yost, C., et al. 2003. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes & Dev. 17: 1101-1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piacentini L., Fanti, L., Berloco, M., Perrini, B., and Pimpinelli, S. 2003. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J. Cell Biol. 161: 707-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platero J.S., Csink, A.K., Quintanilla, A., and Henikoff, S. 1998. Changes in chromosomal localization of heterochromatin-binding proteins during the cell cycle in Drosophila. J. Cell Biol. 140: 1297-1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard C.C., Hsu, L., Delrow, J., and Nelson, P.S. 2001. Project normal: Defining normal variance in mouse gene expression. Proc. Natl. Acad. Sci. 98: 13266-13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea S., Eisenhaber, F., O'Carroll, D., Strahl, B.D., Sun, Z.W., Schmid, M., Opravil, S., Mechtler, K., Ponting, C.P., Allis, C.D., et al. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406: 593-599. [DOI] [PubMed] [Google Scholar]
- Richards E.J. and Elgin, S.C. 2002. Epigenetic codes for heterochromatin formation and silencing: Rounding up the usual suspects. Cell 108: 489-500. [DOI] [PubMed] [Google Scholar]
- Rubin G.M., Hong, L., Brokstein, P., Evans-Holm, M., Frise, E., Stapleton, M., and Harvey, D.A. 2000. A Drosophila complementary DNA resource. Science 287: 2222-2224. [DOI] [PubMed] [Google Scholar]
- Schotta G., Ebert, A., Krauss, V., Fischer, A., Hoffmann, J., Rea, S., Jenuwein, T., Dorn, R., and Reuter, G. 2002. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 21: 1121-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers J.F. and Henikoff, S. 2001. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol. Cell Biol. 21: 2555-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stapleton M., Liao, G., Brokstein, P., Hong, L., Carninci, P., Shiraki, T., Hayashizaki, Y., Champe, M., Pacleb, J., Wan, K., et al. 2002. The Drosophila gene collection: Identification of putative full-length cDNAs for 70% of D. melanogaster genes. Genome Res. 12: 1294-1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun F.L., Cuaycong, M.H., Craig, C.A., Wallrath, L.L., Locke, J., and Elgin, S.C. 2000. The fourth chromosome of Drosophila melanogaster: Interspersed euchromatic and heterochromatic domains. Proc. Natl. Acad. Sci. 97: 5340-5345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L.V., Chen, L., Greil, F., Negre, N., Li, T.-R., Cavalli, G., Zhao, H., van Steensel, B., and White, K.P. 2003. Protein–DNA interaction mapping using genomic tiling path microarrays in Drosophila. Proc. Natl. Acad. Sci. 100: 9428-9433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert P.B. and Henikoff, S. 2000. A reexamination of spreading of position-effect variegation in the white-roughest region of Drosophila melanogaster. Genetics 154: 259-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tschiersch B., Hofmann, A., Krauss, V., Dorn, R., Korge, G., and Reuter, G. 1994. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 13: 3822-3831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B. and Henikoff, S. 2000. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 18: 424-428. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow, J., and Henikoff, S. 2001. Chromatin profiling using targeted DNA adenine methyltransferase. Nat. Genet. 27: 304-308. [DOI] [PubMed] [Google Scholar]
- van Steensel B., Delrow, J., and Bussemaker, H.J. 2003. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc. Natl. Acad. Sci. 100: 2580-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakimoto B.T. and Hearn, M.G. 1990. The effects of chromosome rearrangements on the expression of heterochromatic genes in chromosome 2L of Drosophila melanogaster. Genetics 125: 141-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler K.S. and Wakimoto, B.T. 1995. Heterochromatin and gene expression in Drosophila. Annu. Rev. Genet. 29: 577-605. [DOI] [PubMed] [Google Scholar]
- ____. 1998. Chromosome rearrangements induce both variegated and reduced, uniform expression of heterochromatic genes in a development-specific manner. Genetics 149: 1451-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhimulev I.F., Semeshin, V.F., Kulichkov, V.A., and Belyaeva, E.S. 1982. Intercalary heterochromatin in Drosophila. Chromosoma 87: 197-228. [Google Scholar]