Summary
Transcription in human mitochondria is carried out by a single-subunit, T7-like RNA polymerase assisted by several auxiliary factors. We demonstrate that an essential initiation factor, TFB2, forms a network of interactions with DNA near the transcription start site and facilitates promoter melting but may not be essential for promoter recognition. Unexpectedly, catalytic autolabeling reveals that TFB2 interacts with the priming substrate, suggesting that TFB2 acts as a transient component of the catalytic site of the initiation complex. Mapping of TFB2 identifies a region of its N-terminal domain that is involved in simultaneous interactions with the priming substrate and the templating (+1) DNA base. Our data indicate that the transcriptional machinery in human mitochondria has evolved into a system that combines features inherited from self-sufficient, T7-like RNA polymerase and those typically found in systems comprising cellular multi-subunit polymerases, and provide insights into the molecular mechanisms of transcription regulation in mitochondria.
Introduction
All eukaryotic organisms employ two major types of DNA-dependent RNA polymerases (RNAPs). The first type includes large multi-subunit RNAPs (pol I, pol II, and pol III) and is required for transcription of chromosomal DNA. The second, structurally unrelated family of RNAPs includes a single-subunit, T7-like mitochondrial RNAP (mitoRNAP). MitoRNAP is responsible for transcription of the mitochondrial genome, which encodes several subunits of the key enzymes involved in oxidative phosphorylation. Since oxidative phosphorylation is the major source of energy production during respiration, an understanding of the molecular mechanisms underlying regulation of transcription in mitochondria is critical.
In humans bidirectional transcription of the relatively small (16,569 bps) mitochondrial genome is driven by a nuclear-encoded mitoRNAP that initiates at two major promoters located in both “light” (light strand promoter, LSP) and “heavy” (heavy strand promoter, HSP1) strands of genomic DNA resulting in synthesis of near-genome-size nascent RNA that undergoes further processing (Bonawitz et al., 2006; Falkenberg et al., 2007). Transcription from LSP results in synthesis of eight tRNAs and mRNA for subunit 6 of NADH dehydrogenase, while transcription from HSP1 generates 14 tRNAs, two ribosomal RNAs, and mRNA for 12 subunits of proteins involved in oxidative phosphorylation. MitoRNAP is also responsible for priming of DNA replication at two origins of replication, one of which is located immediately downstream of LSP (Wanrooij et al., 2008).
Although human mitoRNAP and T7 RNAP share significant sequence homology, this is limited to the C-terminal domain that is required for catalytic activity (45% identity for residues 647 - 1230). Unlike T7 RNAP, which requires no additional factors for initiation, human mitoRNAP needs two transcription factors (TFA and TFB2) for efficient initiation (Gaspari et al., 2004). The mechanisms of action of these factors are currently unknown. TFA is a high mobility group (HMG) protein that is involved in formation of nucleoid-like structures on mitochondrial DNA and binds to the promoters 15 to 35 bp upstream of the transcription start site (Dairaghi et al., 1995a). HMG group proteins are known to cause local distortion of the DNA, suggesting the possibility that TFA may assist in the initial melting of a portion of the transcription bubble during initiation (Falkenberg et al., 2007). Consistent with this hypothesis, the distance between the TFA-binding region and the promoter start site appears to be crucial for efficient initiation (Dairaghi et al., 1995b); however, the high abundance of this factor in human mitochondria argues against a specific mechanism for its action and its involvement in transcription initiation in vivo is still being debated (Alam et al., 2003; Cotney et al., 2007). On the other hand, genetic and biochemical data suggest that the C-terminus of TFA interacts with the other transcription factors that are essential for initiation, TFB2 and TFB1, possibly recruiting the initiation complex (IC) to the promoter (Falkenberg et al., 2002; McCulloch et al., 2002). These transcription factors belong to a large family of methyltransferases, as revealed by presence of the conserved catalytic residues and their ability to dimethylate bacterial 16S rRNA, which is modified in a manner similar to human mitochondrial 12S rRNA (McCulloch et al., 2002). TFB2 has a significantly weaker methyltransferase activity but stimulates transcription 10- to 100-fold more efficiently than its paralogue TFB1 (Cotney and Shadel, 2006; Falkenberg et al., 2002; Metodiev et al., 2009).
While the mechanism of stimulation of human mitochondrial transcription initiation by TFB transcription factors has not yet been identified, the yeast mitochondrial analog of these proteins Mtf1 has been implicated in melting of promoter DNA and stabilization of the promoter open complex (Matsunaga and Jaehning, 2004). It remains unclear whether TFBs, which are bifunctional proteins, are able to bind nascent transcripts and thus modulate initiation of transcription. Importantly, experiments in both human and yeast transcription systems suggested that neither TFB2 nor Mtf1 is responsible for promoter recognition, because this function is likely preserved in mitoRNAP itself (Gaspari et al., 2004; Matsunaga and Jaehning, 2004). It is clear, therefore, that the transcriptional machinery in human mitochondria has evolved into a system that combines a number of features inherited from self-sufficient, single-subunit, T7-like RNAPs and those typically found in systems comprising cellular multi-subunit polymerases. In this work we demonstrate that TFB2 makes direct contacts with promoter DNA at the transcription start site and assists mitoRNAP in promoter melting and binding of a priming substrate. Our findings provide insights into the molecular organization of the human mitochondrial IC and its relation to other transcription systems.
Results
TFB2 is involved in promoter melting
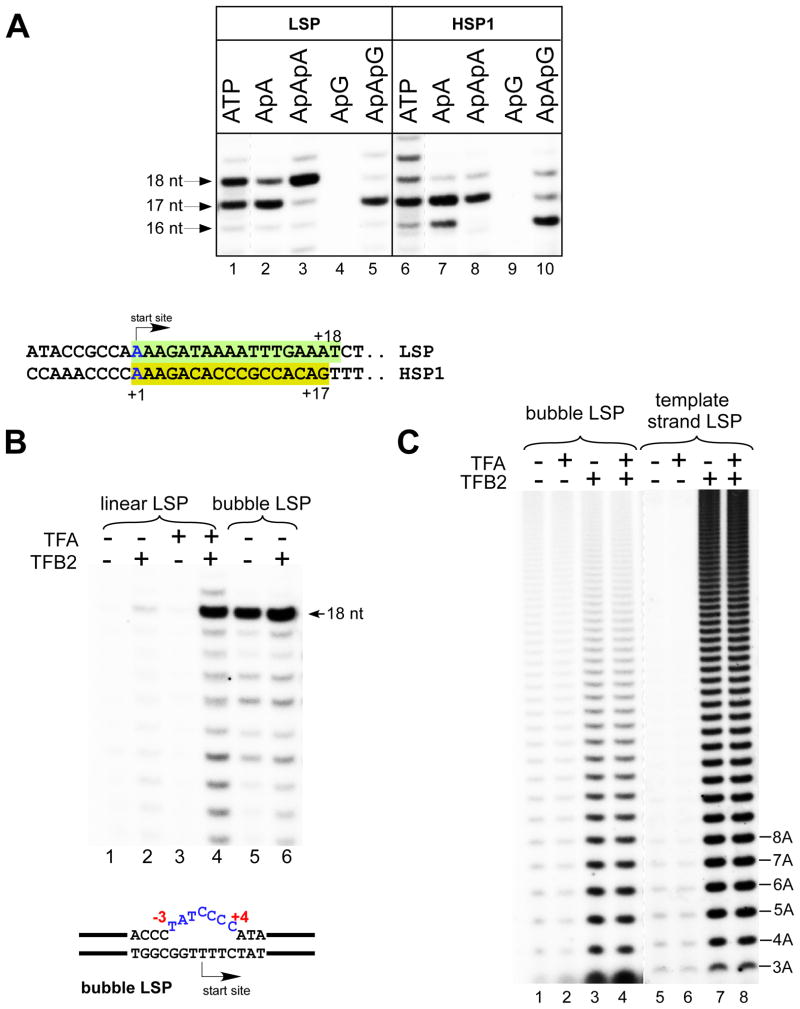
Previous studies demonstrated that human mitochondrial in vitro transcription requires the presence of two transcription factors, TFA and TFB2 (Gaspari et al., 2004). Transcription of a template containing the LSP promoter in the presence of mitoRNAP, TFA, TFB2, and a limited set of substrate NTPs (ATP, GTP, and UTP) (Figure 1A) results in the accumulation of 17- and 18-nt transcripts, consistent with previous observations that the predominant in vivo start site is located in the middle of the four-thymidine stretch in the template strand (Chang and Clayton, 1984). Transcription of a template containing the HSP1 promoter resulted in accumulation of predominately a 17-nt RNA product (Figure 1A); this is also consistent with the in vivo data (Chang and Clayton, 1984). Similar to other RNAPs, human mitoRNAP can initiate transcription from short two- to three-nt RNA primers (Figure 1A). Thus, we observed robust transcription with ApApA and ApApG primers resulting in the accumulation of 17- and 18-nt RNA products and 16- and 17-nt RNA products on LSP and HSP1, respectively. As expected, no transcription was observed when ApG primer was used (Figure 1A, lanes 4 and 9).
Figure 1. TFB2 is required for promoter melting.
A. Top panel - Human mitochondrial in vitro transcription system. Transcription was performed with the nucleotide sets lacking CTP (LSP) or UTP (HSP1) for 30 min at 35°C. RNA transcripts in reactions using ATP (lanes 1 and 6) were treated with phosphatase to match their mobility to the mobility of the transcripts generated by incorporation of the RNA primers. Transcripts obtained using ApApG primers (lanes 5 and 10) served as the size markers as they could only anneal to the complementary sequence in the promoter DNA and thus generated 17 and 16 nt RNA products on LSP and HSP1, respectively. Lanes 1 and 6 are overexposed to compensate for the more efficient label incorporation in reactions containing RNA primers. Bottom panel - Sequences of the LSP and the HSP1 promoters aligned at their transcription start sites. Shaded boxes indicate transcribed regions in the presence of the limited nucleotide sets as described above.
B. MitoRNAP can initiate transcription in the absence of TFA and TFB2 on bubble promoter templates. Transcription assays were performed on linear (lanes 1–4) and bubble (lanes 5,6) templates containing the LSP promoter in the presence of ApApA primer, GTP, ATP and UTP for 30 min.
C. TFB2 stimulates A-ladder production on bubble and single-stranded templates. The bubble LSP promoter template (lanes 1–4) and the template strand of the LSP promoter (lanes 5–8) were transcribed in the presence of ATP (0.3 mM) and [α-32P]ATP for 30 min.
Yeast mitoRNAP (Rpo41), which is related to the human mitoRNAP, can initiate transcription in the absence of an initiation transcription factor (Mtf1) on a double-stranded DNA template in which the promoter region is “premelted” by virtue of the presence of a noncomplementary region in the nontemplate (NT) strand near the transcription start site (a “bubble” template) (Matsunaga and Jaehning, 2004). We employed similar template constructs with human mitochondrial promoters in in vitro assays involving human mitoRNAP. We observed efficient sequence-specific transcription when the LSP bubble template contained a noncomplementary region involving nucleotide positions −3 to +4 (Figure 1B). This observation suggests that (i) human mitoRNAP is primarily responsible for promoter recognition and (ii) TFB2 (along with TFA) is likely involved in promoter melting. Interestingly, in contrast to observations made in the yeast mitochondrial transcription system (Matsunaga and Jaehning, 2004), we did not observe inhibition of transcription initiation on bubble templates in the presence of TFB2 (Figure 1B).
While the experiments described above demonstrate that TFB2 is not required for efficient runoff transcription on premelted bubble templates, we observed a stimulatory effect of TFB2 during early initiation steps (Figure 1C). When ATP was the only NTP present in the reaction mixture human mitoRNAP, similar to T7 RNAP (Martin et al., 1988), became engaged in transcript slippage, resulting in synthesis of homopolymeric transcripts and producing an A-ladder. On bubble templates, as well as on single-stranded DNA templates, A-ladder production was greatly stimulated in the presence of TFB2; addition of TFA had no significant effect, suggesting a role for TFB2 during the early steps of initiation (Figure 1C).
TFB2 bridges upstream and downstream promoter contacts of the IC
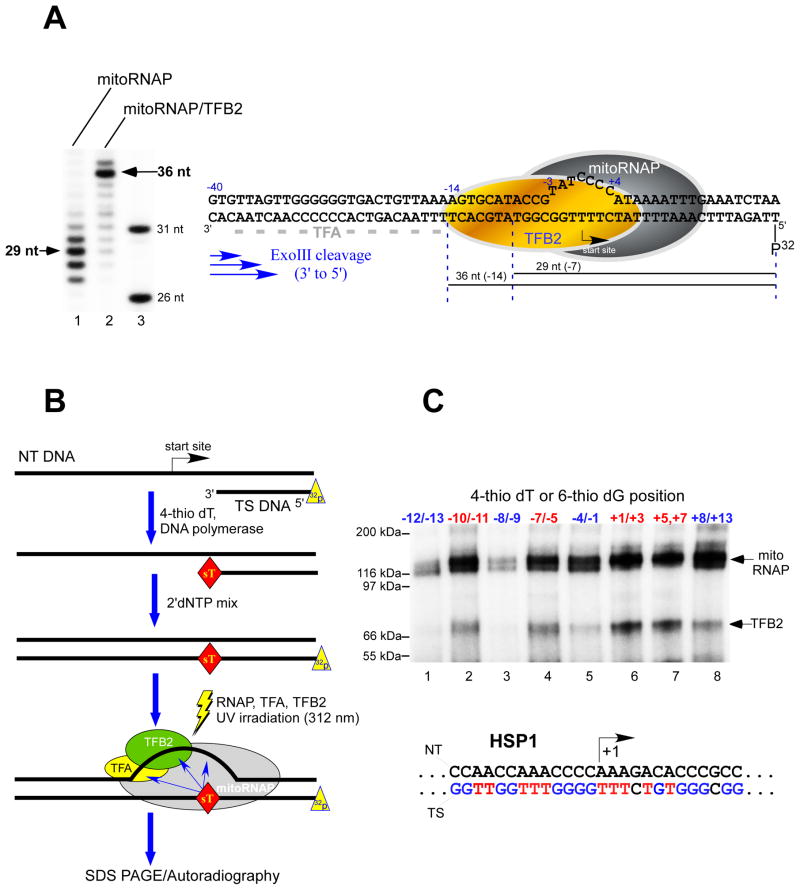
DNase I footprinting assays that employed linear promoter DNA templates indicated that while TFA protects the region including nucleotides −35 to −15 on promoter DNA, mitoRNAP/TFB2 appears to occupy the relatively small region that includes nucleotides −4 to +10, leaving a gap of 11 nt between these components of the IC (Gaspari et al., 2004). We used the bubble promoter template described above to determine the position of the “trailing” edge of the mitoRNAP/TFB2 complex in an exonuclease footprinting assay (Figure 2A). This approach allowed us to omit TFA from the IC assembly since it would otherwise complicate interpretation of the exonuclease cleavage pattern. The exonuclease footprint of mitoRNAP alone (7 – 11 bp downstream of the start site) was somewhat diffuse; this most likely reflects the enzyme s lateral mobility. However, the mitoRNAP/TFB2 footprint revealed a distinct band 14-nt upstream of the promoter start site, indicating an extended region of interaction with the promoter DNA. Similar results were obtained using an HSP1 bubble template, where the mitoRNAP/TFB2 complex protected a 15-bp region of the upstream promoter (data not shown). Thus, during transcription initiation TFA and TFB2 are likely to be located in close proximity to each other on the promoter DNA, consistent with the finding that these proteins interact directly (McCulloch and Shadel, 2003).
Figure 2. TFB2 makes direct interactions with the promoter DNA.
A. Exo III footprints of the TFB2/mitoRNAP complex
Left panel. The bubble LSP template was prepared by annealing of 32P-labeled template strand with the NT DNA and preincubated with the mitoRNAP (lane 1) or with the TFB2/mitoRNAP complex (lane 2) for 10 min at room temperature prior to the treatment with Exo III as described in Experimental Procedures. Products of the reaction were resolved using 15% PAGE containing 6M urea. DNA size markers (lane 3) were obtained by 5′ end labeling of 26 nt and 31 nt long oligonucleotides having sequence identical to the 5′ end of the template DNA strand. Right panel. Schematics of the mitochondrial IC. Major DNA products (36 nt and 29 nt) generated by Exo III cleavage are indicated with the black lines below the DNA sequence. Grey dash line indicates TFA footprint (−38 to −15) as determined in (Gaspari et al., 2004).
B. Schematics of the synthesis of 32P-labeled templates containing photo crosslinking reagents. The 5′ end 32P-labeled DNA primer annealed to the NT DNA strand was extended by DNAP I in the presence of the photo reactive probe (4-thio dTMP). Upon the probe incorporation, the DNAP I was allowed to complete the primer extension by providing dNTPs mixture. The labeled DNA templates were then incubated with mitoRNAP and transcription factors to form the IC, followed by UV irradiation to activate the probe.
C. DNA crosslinking of the IC. ICs (50 nM) were UV-irradiated for 10 min at 30°C and the products of the reaction resolved in 4–12 % Bis-Tris MES-SDS gel. Positions of the photoreactive probes, 4-thio dTMP (red letters) and 6-thio dGMP (blue letters) in the template strand of the HSP1 promoter are indicated.
TFB2 interacts with promoter DNA at the transcription start site
To monitor interactions of the mitochondrial transcription IC with the promoter DNA we incorporated the photocrosslinking nucleoside analogs 4-thiothymidine (sT) and 6-thioguanidine (sG) into specific positions within the promoter (Figure 2B). Both probes differ from the corresponding natural bases in that they have an oxygen-to-sulfur atom substitution and are known to photoreact with the target protein within the distance of a chemical bond. The crosslinking was highly specific since no radioactive species were observed when either mitoRNAP, TFA, or TFB2 were omitted from the reaction (Figure S2). TFB2 crosslinking to a number of bases in promoter DNA was observed, with the strongest interaction with the region including nucleotides +1 to +3 at the promoter start site (Figure 2C). In a separate experiment we determined that TFB2 efficiently crosslinked to DNA having a photoreactive probe exclusively at the +1 position (data not shown). Crosslinking to TFB2 was reduced when the probe was incorporated at positions +5 to +7 downstream of the promoter start site and was weak at positions −10 to −11 and −7 to −5 upstream of the promoter start site. Crosslinking to mitoRNAP was registered along a significant portion of promoter DNA with particularly strong interactions observed near the transcription start site (Figure 2C), consistent with the footprinting data discussed above.
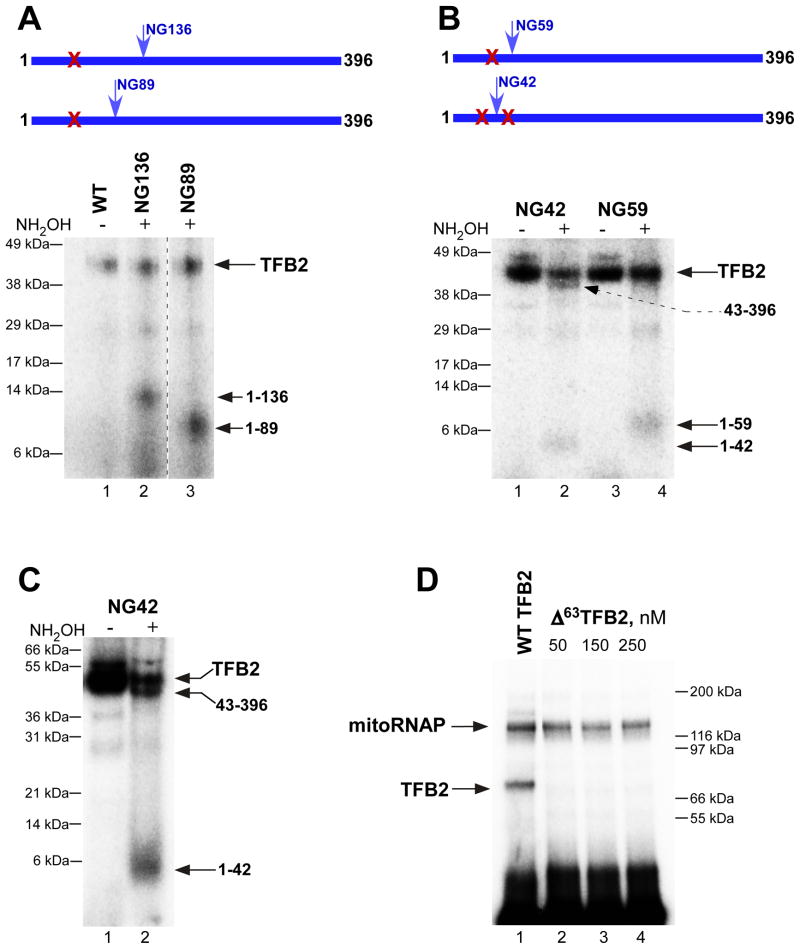
Mapping of TFB2-DNA interactions at the transcription start site
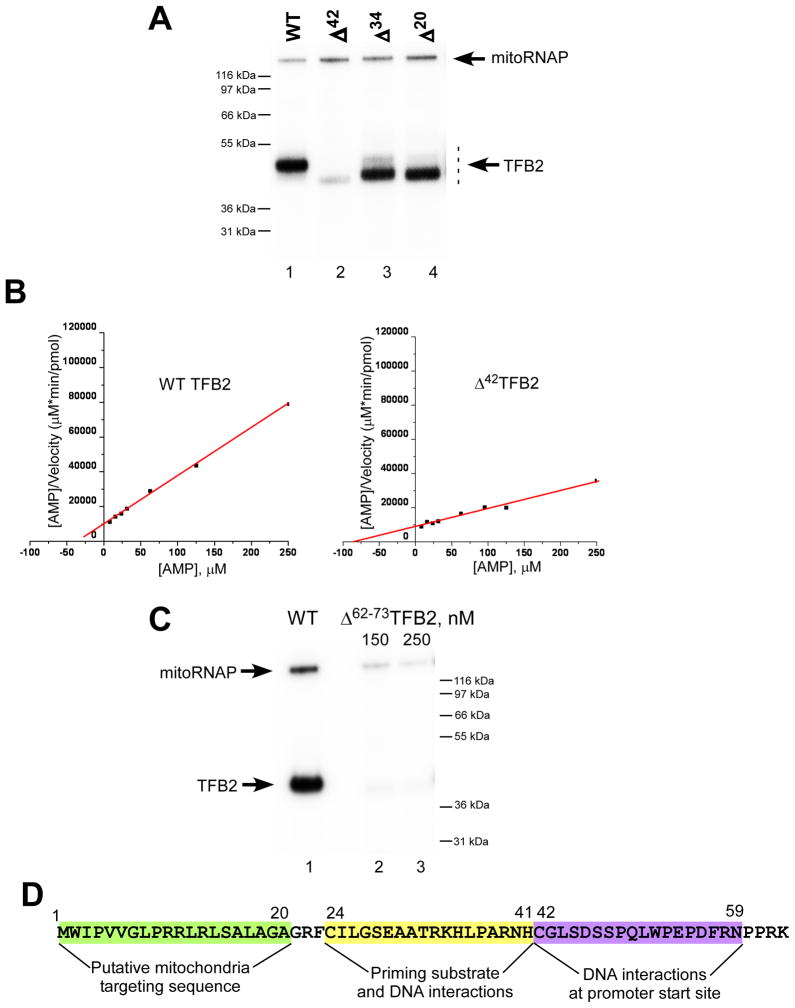
Because the strongest TFB2-DNA crosslinking was observed when the photoreactive probes were incorporated at the transcription start site, we performed mapping of this region of TFB2 using chemical cleavage of the peptide bond between asparagine (N) and glycine (G) residues by hydroxylamine. Since wild-type (WT) TFB2 does not contain such NG pairs, we created mutants containing a single NG pair at positions 136, 89, 59, or 42 (Figure 3). These mutants were indistinguishable from the WT TFB2 in all transcription assays (data not shown). To simplify interpretation of the hydroxylamine cleavage pattern we also constructed a mutant mitoRNAP in which all four naturally occurring NG pairs were eliminated. This enzyme, while exhibiting a WT phenotype, crosslinked to the DNA efficiently but was not cleaved when treated with hydroxylamine (Figure 3). At the same time, we observed generation of small cleavage products corresponding to the segments containing amino acid residues 1 to 136 and 1 to 89 in mutants NG136 and NG89 TFB2, respectively (Figure 3A). A small fragment (residues 1-59) was also generated by cleavage of mutant NG59, indicating that the crosslink is to the N-terminus of this residue (Figure 3B). However, the hydroxylamine cleavage pattern of mutant NG42 revealed the presence of two labeled fragments corresponding to the segments containing amino acid residues 1 to 42 (35%) and 43 to 396 (65%) (Figure 3B and C). We thus conclude that two adjacent regions in TFB2 interact with the promoter DNA at the transcription start site - one at the extreme N-terminus between residues 1 and 42 and another between residues 43 and 59.
Figure 3. Mapping of TFB2-DNA interactions at the promoter start site.
A. The crosslinking region is located in the N-terminal part of TFB2. Crosslinking was performed as described in Experimental Procedures using NG-less mitoRNAP, TBF2 mutants indicated and the HSP1 template containing 4-thio dTMP at positions from +1 to +3 and internal [α-32P] dCMP label. After UV irradiation, the complexes were first treated with DNAse I and then with hydroxylamine for 6h at 43°C, and resolved using 4–12% Bis-Tris MES-SDS gel.
B, C. Fine mapping of TFB2 crosslink with hydroxylamine. Crosslinking was performed as above using NG42 and NG59 TFB2 mutants. The products of the hydroxylamine reaction (5h at 43°C) were resolved using 4–12% Bis-Tris MES-SDS (B) or 10% Bis-Tris MES-SDS gel (C).
D. Cross-linking efficiency of Δ 63TFB2 mutant. Crosslinking reaction was performed as described above with the WT (lane 1) or Δ 63 TFB2 mutant (lane 2–4). The products of the reaction were treated with DNAse I prior to their resolution in 4–12% Bis-Tris MES-SDS gel.
We next constructed several N-terminal TFB2 deletion mutants:Δ20TFB2 (eliminates presumptive mitochondrial targeting sequence located at residues 1-20); Δ34TFB2, Δ42TFB2, and Δ63TBF2. All mutants exhibited the WT phenotype in run-off transcription assays (Figure S3). Analysis of these mutants in DNA crosslinking experiments revealed only minor (15% to 20%) reduction of the crosslinking in the case of Δ42TFB2 mutant (data not shown). When DNA crosslinking was carried out with Δ63TBF2 no crosslinking species were detected, confirming our mapping data (Figure 3D).
TFB2 interacts with the priming nucleotide
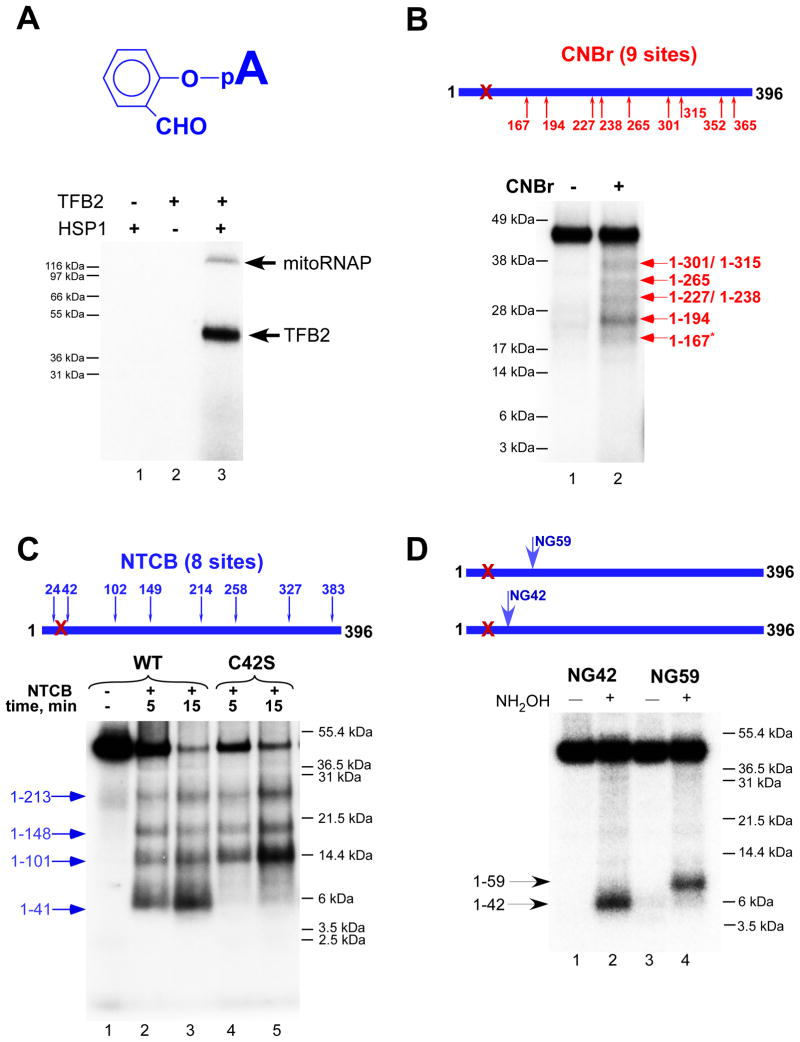
Since the crosslinking experiments described above suggest that TFB2 interacts with the templating (+1) DNA base, we asked whether this transcription factor might also be involved in interactions with substrate NTP. We took advantage of a procedure known as a catalytic autolabeling (Grachev et al., 1987). In these experiments a reactive derivative of the priming substrate (+1 nucleotide) is first crosslinked to the IC. Labeling of the derivative (and the protein components to which it is crosslinked) is subsequently performed by allowing formation of the first phosphodiester bond with the next incoming 32P-labeled NTP (+2 NTP). Thus, labeling depends strictly upon the catalytic activity of RNAP and functional activity of the IC. We employed a 2-hydroxybenzaldehyde derivative of AMP as a crosslinking agent (Figure 4); this reagent was originally used in analyses of T7 RNAP, E.coli RNAP, yeast RNAP II, human RNAPII, and, more recently, RNAPs from the N4 virion to identify residues in the vicinity of a catalytic site involved in substrate binding (Grachev et al., 1987; Kazmierczak et al., 2002; Maksimova et al., 1989; Riva et al., 1990).
Figure 4. TFB2 interacts with the priming substrate in the IC.
A. Catalytic autolabeling with 2-hydroxybenzaldehyde AMP depends on the functional activity of the IC. Autolabeling with 2-hydroxybenzaldehyde AMP (top panel) was performed with the reaction mixtures that lacked TFB2 (lane 1), DNA template (lane 2) or with the IC (lane 3) after the crosslinking (bottom panel). Reaction products were resolved using 4–12% Bis-Tris MES-SDS gel.
B. Mapping of TFB2 crosslink with CNBr. Crosslinking reactions containing WT TFB2 were treated with CNBr for 10 min at 37°C and the products resolved as indicated above. The pattern of labeled bands observed corresponds to a nested set of N-terminal fragments that extend to the CNBr cleavage sites indicated. The asterisk indicates resistant to CNBr cleavage methionine-serine pair.
C. Mapping of TFB2 crosslink with NTCB. Crosslinking reactions containing WT (lanes 1–3) or mutant C42S TFB2 (lanes 4,5) were treated with NTCB for the time indicated at 37°C and resolved using SDS PAGE as above.
D. Mapping of TFB2 crosslink with hydroxylamine. Crosslinking reactions containing NG59 (lanes 1,2) and NG42 TFB2 mutants (lanes 3,4) were treated with 0.2 M hydroxylamine for 5 h at 43°C. Reaction products were resolved using 10% Bis-Tris MES-SDS gel.
Because mitoRNAP shares significant homology with T7 RNAP in the region of its catalytic core we expected that the autolabeling procedure would reveal strong crosslinking to mitoRNAP. Moreover, 2-hydroxybenzaldehyde AMP specifically interacts with lysine and arginine residues and, in T7 RNAP, crosslinks to residue K631 of the O-helix (Maksimova et al., 1989), which is highly conserved in all single-subunit RNAPs and is also present in the vicinity of the putative active site of human mitoRNAP. Surprisingly, when we carried out the crosslinking procedure using mitochondrial ICs, we found that the crosslink was made almost exclusively and very efficiently to TFB2 and not to mitoRNAP (Figure 4A). Derivatives of priming ATP of broader crosslinking specificity produced efficient crosslinks to both RNAP and TFB2 (Figure S4). Since the reactive aldehyde group of 2-hydroxybenzaldehyde AMP had the shortest radius of action (approximately 4 Å, counting from the phosphorus atom) we used this compound in the subsequent experiments.
Mapping of TFB2 interactions with the priming substrate
To map the region of TFB2 that interacts with the priming substrate we employed chemical reagents that cleave proteins at specific residues. We first used CNBr digestion under single-hit conditions to cleave the crosslinked protein at methionine residues. Using this approach we found that the region of interaction is located in the N-terminal part of the protein, between residues M1 and M167 (Figure 4B). For fine-mapping of this region we used 2-nitro-5-thiocyano-benzoic acid (NTCB), which cleaves the protein before cysteine residues (Figure 4C). The single-hit cleavage pattern suggested that the crosslinked region was located between residues 24 and 41 because the fragment containing residues 1 to 41 (4.6 kDa), but not that containing residues 1 to 23 (2.3 kDa), was observed. To confirm these findings we constructed an additional TFB2 mutant in which cysteine 42 was substituted with serine (C42S) and used it in NTCB cleavage experiments (Figure 4C, lanes 4 and 5). In the absence of C42, the smallest fragment generated by NTCB corresponded to the region containing residues 1 to 101, confirming our interpretation of the cysteine cleavage pattern for WT TFB2.
To further confirm the location of the crosslinked region we used hydroxylamine cleavage and TFB2 mutants containing single NG pairs at positions 136, 116, 89, 59, or 42. The cleavage pattern revealed 32P-labeled N-terminal fragments progressively decreasing in size, indicating that the crosslinked region is located to the amino terminal side of residue N42 (Figures 4D and S5). These data are in agreement with NTCB and CNBr cleavage experiments and confirm that the crosslinked region is between residues 24 and 42 in WT TFB2.
We next analyzed the N-terminal TFB2 deletion mutants described above (Δ20TFB2, Δ34 TFB2, and Δ42 TFB2) in catalytic autolabeling experiments (Figure 5A). WT TBF2 and mutant Δ20TFB2 demonstrated similar efficiencies of crosslinking (Figure 5A, lanes 1 and 4). At the same time, the mutant Δ34 TFB2 showed 1.5- to 2-fold lower crosslinking efficiency, while crosslinking of Δ42TFB2 with the AMP derivative was not detected, consistent with the mapping data (Figure 5A). Finally, NTCB cleavage of the crosslink formed with Δ20TFB2 resulted in generation of a small peptide (residues 24-41) confirming interpretation of the mapping data (Figure S5B).
Figure 5. The ICs formed with Δ42TFB2 mutant have low affinity to the priming nucleotide.
A. Catalytic autolabeling of TFB2 deletion mutants. Reactions containing WT (lane 1), Δ42 (lane 2), Δ34 (lane 3) and Δ20 (lane 4) TFB2 were resolved using 4–12% Bis-Tris MES-SDS.
B. Steady state kinetic experiments. Transcription reactions were performed using AGU-LSP promoter in the presence of 7–1000 μM AMP (priming substrate) and 50 μM GTP to produce pApG transcripts. The data are presented using Hanes-Woolf plots for WT (left panel) and Δ42 (right panel) TFB2. Kmapp was calculated using the data obtained in four independent experiments.
C. Catalytic autolabeling using Δ62–73TFB2 mutant. Reactions containing WT (lane 1) and Δ62–73 (lanes 2 and 3) TFB2 were resolved using 4–12% Bis-Tris MES-SDS.
D. Schematics of TFB2 interactions due to its unique N-terminal region. The amino acid sequence of the N-terminal part (residues 1–63) of H.s. TFB2 is indicated.
Catalytic functions are shared between TBF2 and core RNAP during initiation
As mentioned above, deletion of the region of TFB2 that interacts with the priming nucleotide (residues 1 - 42) did not result in any significant changes in standard run-off transcription assays performed at a relatively high NTP concentration. Considering that mutations in the regions of RNAP implicated as potential priming substrate binding sites by catalytic autolabeling result in a decreased affinity to the substrate (Kazmierczak et al., 2002; Woody et al., 1998) we performed steady-state kinetics experiments using WT, Δ20, Δ42, and Δ63 TBF2 (Figure 5B).
Since both natural human mitochondrial promoters contain several T residues at their start sites, we used a mutant LSP promoter ( AGU” LSP; transcribed sequence is AGU…; Figure S1) and AMP as the priming NTP to monitor kinetics of formation of the first phosphodiester bond. No difference was observed in Km values for the WT and Δ20 TFB2. At the same time we found that Kmapp for AMP on the AGU LSP promoter was 2.5- to 3-fold higher for the IC containing Δ42TFB2 (or Δ63TFB2, data not shown) mutant than for the IC containing WT TFB2 (105 ± 67 μM and 42 ± 27 μM, respectively) (Figure 5B). We also detected no significant difference in Km values for the +2 substrate NTP (data not shown). We conclude that the observed transcription defects are related to the reduced ability of Δ42TFB2 to interact with the priming nucleotide.
To assess how the orientation of the N-terminal domain of TFB2 relative to its core affects interactions with the priming substrate, we constructed mutant Δ62–73 TFB2 (Figure 5C and S6). When analyzed in autolabeling experiments this mutant, similar to Δ42 TFB2, did not produce crosslinked species (Figure 5C). At the same time, we found that the activity of Δ62–73 TFB2 in run-off transcription assays was four to five times lower than that of WT TFB2 (Figure S6, A). While the exact correlation between efficiency of the cross-linking and activity of the IC is not known, it is possible that the observed phenotype of Δ62–73 TFB2 mutant was due to changes in orientation/folding of its N-terminal domain.” The Kmapp for AMP on the LSP promoter was about four–fold higher for the IC containing Δ62–73TFB2 than for that containing the WT TFB2 (157.2 ± 10.5 μM and 39.1 ± 5.6 μM, respectively), which also indicates defects in interactions with the priming nucleotide. However, this mutant also had a Kcatapp 1.5-fold lower than that of WT TFB2 and demonstrated defects during A-ladder synthesis (Figure S6, B and C), suggesting that the region containing residues 62 to 72 may alone contribute to the TFB2 functions.
To summarize our biochemical and genetic data, we determined that the first 20 amino acids of TFB2 are dispensable for its function in vitro and may constitute a mitochondrial targeting signal consistent with MitoProt (ExPASy Tools Server) prediction (Figure 5D). The region of TFB2 located between residues 24 and 42 is primarily involved in interactions with the priming NTP but also constitutes a minor site of interaction with the template DNA at the transcription start site. Finally, the region of TFB2 consisting of residues 43 to 59 is involved in major interactions with the +1,+2, and +3 bases in the template strand of the promoter (Figure 5D). While the N-terminal region of TFB2 is conserved among mammals, it shares no sequence homology with the other related mitochondrial transcription factors such as human TFB1 or yeast Mtf1, nor does it contain the residues important for methyltransferase activity found in all mitochondrial initiation factors of this family (McCulloch et al., 2002).
Discussion
Molecular organization of the human mitochondrial transcription IC
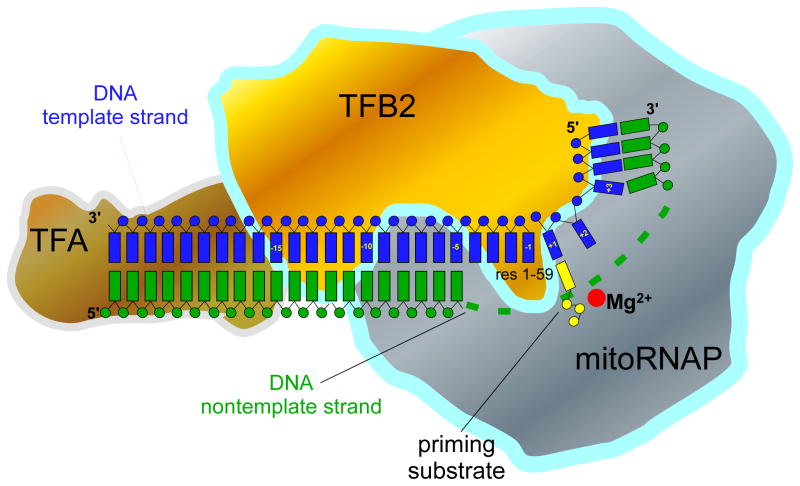
The data obtained in the course of this study outline the general organization of the IC and suggest multiple roles of TFB2 during transcription initiation. We demonstrated that the TFB2/mitoRNAP complex interacts with the promoter DNA in close proximity to the TFA recognition sequence (positions −35 to −15), starting at least 15 to14 bp upstream of the promoter start site and extends its interactions to the downstream portion of the promoter (Figure 6). MitoRNAP, in turn, has a slightly less extended footprint, which most likely starts 10 to 11 bp upstream of the promoter start site and extends to the +10 position downstream, as determined earlier (Gaspari et al., 2004). Therefore, the entire promoter region from −35 to +10 bp is covered by the transcription IC, suggesting that other DNA-binding transcription factors (e.g., MTERFs), unless they bind directly to the components of IC, can only interact with the DNA outside that region (Figure 6). The organization of ICs appears to be similar for both LSP and HSP1 since DNA crosslinking, Exo III footprinting, and experiments with bubble templates produced very similar or identical data. Thus, melting of 3 bps upstream and 4 bps downstream of the transcription start site is sufficient for transcription initiation using both promoters, as further melting does not increase efficiency of initiation in the absence of TFB2 (data not shown).
Figure 6.
Schematics of the molecular organization of human mitochondrial transcription IC
TFB2 contacts promoter DNA near the transcription start site and interacts with TFA, mitoRNAP and the priming nucleotide. The N-terminal region of TFB2 implicated in interactions with the first base pair of the RNA-DNA hybrid is shown in the vicinity of the mitoRNAP active site. We are thankful to Drs William T McAllister and Vadim G Nikiforov for fruitful discussion and critical reading of the manuscript. This work was supported in part by UMDNJ Foundation grant (to D.T.) and by National Institutes of Health Grants GM30317 (to A.M.) and GM38147 (to W.T.M).
Multiple roles of TFB2 in transcription initiation
Since TFB2 interacts with both TFA and mitoRNAP (McCulloch and Shadel, 2003) and makes a number of contacts with the promoter DNA, one of its roles is likely to “cement” the IC by bridging the upstream and downstream promoter contacts, and stabilizing it during initiation (Figure 6). However, by itself TFB2 is unable to bind promoter DNA and leaves no footprints on promoter template when used in Exo III footprinting experiments (data not shown). In the related T7 RNAP promoter melting is facilitated by insertion of a so-called intercalating hairpin (located in the N-terminal domain of the polymerase) in the minor groove of DNA upstream of the TATA box (Cheetham et al., 1999). Due to the lack of sequence homology in the N-terminal regions of single-subunit RNAPs, it is difficult to predict whether mitoRNAP has a similar element involved in promoter melting. However, the fact that mitoRNAP can transcribe single-stranded and bubble templates, but not double-stranded promoters, indicates that the promoter melting function depends on TFB2. The extent of DNA melting sufficient for mitoRNAP activity (7 bp) correlates well with the TFB2-DNA crosslinking data and suggests that TFB2 may be involved in maintenance of the upstream edge of the transcription bubble during initiation and in the positioning of the templating (+1) base in the active site of the polymerase that is required for an efficient catalysis.
One of the unexpected findings of this work is the interactions of TFB2 with the priming substrate. In most prokaryotic and eukaryotic transcription complexes studied thus far, binding of the priming substrate is an exclusive function of the core RNAP. Thus, mapping of the crosslink formed with the AMP derivative used in the current work, resulted in identification of residue K631 in T7 RNAP and residue K1065 in the β subunit of E.coli RNAP (Maksimova et al., 1989; Mustaev et al., 1991). Similarly, residue K670 (K631 in T7 RNAP) in a related N4 RNAP was also shown to affect binding of the priming NTP during initiation (Kazmierczak et al., 2002). In contrast, our experiments revealed that during initiation it is TFB2, not mitoRNAP, that becomes crosslinked to the priming NTP. We propose that NTP-TFB2 interactions are essential during initiation since this factor also assists in promoter melting and presentation of the templating base (+1) in the DNA strand for base-pairing with the priming substrate, as suggested by DNA photocrosslinking data. These functions require that the same protein (TFB2) is responsible for promoter melting and NTP binding, which would assure correct positioning of the priming nucleotide (Figure 6). Thus, it is possible that the concurrent interactions of TFB2 with the priming substrate and DNA templating base (+1) serve to stabilize the open promoter complex and prevent competing re-annealing of the NT strand. During elongation, when the 3′ end of the nascent RNA occupies the binding site for the priming substrate (and thus is stabilized by complementary interactions with the template DNA), TFB2 is no longer required and likely dissociates from the complex as suggested for yeast Mtf1 and Rpo41 (Mangus et al., 1994). Similarly, during transcription of the bubble templates, lack of the complementary region in the NT DNA strand results in the absence of the force required to displace the priming nucleotide. Consistent with this, addition of TFB2 does not increase the efficiency of the run-off transcription on bubble templates. A different situation is observed when mitoRNAP is engaged in an A-ladder production. It is possible, that due to the extremely low stability of the short RNA-DNA hybrids formed during slippage (Martin et al., 1988), TFB2 participation in substrate (or small transcript) binding becomes critical and results in significant stimulation of a A-ladder production (Figure 1C). However, transcription slippage is a complex process, and therefore other mechanisms of A-ladder stimulation by TFB2 (e.g. RNA release) cannot be excluded.
The location of TFB2 on promoter DNA and its interactions with TFA and a priming substrate (and thus, with the 5′ end of the nascent RNA) suggests that TFB2 displacement can be a major event triggering the transition from the initiation stage of transcription to the elongation stage. In that respect, promoter clearance by the mitoRNAP IC should resemble (mechanistically) release of a sigma subunit during initiation on bacterial promoters (Murakami and Darst, 2003). Interestingly, E.coli sigma region 3.2 was also shown to interact with an ATP derivative, though the crosslinking reagent had a substantially longer reactive “arm” than 2-hydroxybenzaldehyde AMP, which was used in the present study (Kulbachinskiy and Mustaev, 2006).
Molecular mechanisms of regulation of transcription in human mitochondria
The anticipated regulatory pathways of transcription in human mitochondria involve regulation of transcription by changing ATP concentration (Gaines et al., 1987), negative regulation by the MTERF family of transcription factors (Park et al., 2007), and differential efficiency of transcription of complexes containing TFB1 or TFB2 (Gaspari et al., 2004). Recently, another model of transcription regulation was proposed, which postulates that gene expression in yeast mitochondria may be linked to the energetic needs of cells (Amiott and Jaehning, 2006a), The authors noted that since yeast mitochondrial promoters, which require relatively high levels of ATP for initiation, have different binding affinities for ATP, the consumption of ATP may selectively decrease the production of mRNA from low-affinity promoters (Amiott and Jaehning, 2006a; Amiott and Jaehning, 2006b), a mechanism related to the molecular sensing of changing initiation NTP concentrations by multisubunit RNAPs (Murray et al., 2003). Much earlier studies revealed that transcription in human mitochondria was also sensitive to ATP concentration (Gaines et al., 1987). Considering that the LSP promoter encodes only one mRNA, it is unclear whether the proposed mechanism (Amiott and Jaehning, 2006a) would be operational in human mitochondria as well. While additional studies are required to determine whether differential initiation on human promoters is related to the energetic needs of cells, we speculate that, if such regulation exists, it could be based on the attenuation of the ability of TFB2 to interact with the priming nucleotide rather than on differences in promoter affinities for ATP. In this respect, it is worth noting that a mutation has been identified in the yeast mitochondrial initiation factor Mtf1 that affects ATP binding (10-fold increase in Km), suggesting that, similar to TFB2, this factor could be involved in interactions with the substrate (Amiott and Jaehning, 2006a). Although the region of TFB2 implicated in ATP binding in this study bears no sequence homology to any region of Mtf1, it is remarkable that both essential initiation factors in different mitochondrial systems may serve as molecular “sensors” for changing levels of ATP (or AMP) during respiration, given that RNA synthesis on all mitochondrial promoters starts with ATP.
Comparison of human mitochondrial transcription with other transcription systems
Although the two large families of DNA-dependent RNAPs, i.e., single- and multi-subunit RNAPs, share no structural homology, they have evolved surprisingly similar features that are engaged at different stages of transcription. These include the conserved configuration of the nucleic acid scaffold of the ternary complexes, closing of the catalytic site by movement of a particular functional element (the O-helix in phage-like RNAPs and the “trigger loop” in multisubunit RNAPs), termination on hairpin terminators, etc (Temiakov et al., 2004; Wang et al., 2006). The dramatic difference between these two classes of RNAPs, however, is apparent during transcription initiation and regulation. Multisubunit RNAPs display an array of mechanisms and a variety of interactions with auxiliary factors, whereas T7 RNAP, the most studied of the single-subunit RNAPs, performs all essential steps by itself. Indeed, in all transcription systems that have been analyzed, the core RNAP is responsible for the binding of the priming nucleotide while in mitoRNAP this function appears to be shared with TFB2. This is particularly surprising considering that all residues involved in catalytic functions seems to be highly conserved (if not identical) between mitoRNAPs and T7 RNAP. At the same time, mitoRNAP has lost its ability to melt promoter DNA on its own and, in that respect, resembles structurally unrelated multisubunit RNAPs that rely on auxiliary factors for both promoter melting and recognition. Yet, promoter recognition in human mitochondria is performed by mitoRNAP, possibly in a self-sufficient fashion similar to T7 RNAP (Cheetham et al., 1999). Finally, unlike T7 RNAP, promoter clearance in human mitochondria would likely require dissociation of TFB2 similar to the release of the sigma subunit in prokaryotes or TFBII in eukaryotes. It is apparent that human mitochondrial transcription shares a number of similar features with phage, prokaryotic, and eukaryotic transcription and represents a unique solution adapted by eukaryotic cells to the highly specific task of mitochondrial gene expression. Future studies of initiation and elongation complexes will shed light on the complex nature of human mitochondrial transcription and its mechanisms of regulation.
Experimental Procedures
Cloning, expression and purification of the components of human mitochondrial transcription
The coding sequences of human mitoRNAP (without leader peptide, residues 44-1203), human mitochondrial transcription factors A (without leader peptide, residues 43-246) and B2 (WT) were amplified by PCR from human heart cells cDNA (Panomics). An amplified gene encoding mitoRNAP was cloned into the expression vector pProEx(Hb) (Invitrogen), that allowed expression of N-terminally His6-tagged version of a protein. PCR fragments containing TFA and TFB2 genes were cloned into pTYB11 expression vector (NEB) and expressed as intein-fusion proteins (See Supplemental material).
Construction of TFB2 and mitoRNAP mutants
TBF2 mutants were obtained by site-directed mutagenesis (Quick-change, Stratagene) starting with the plasmid containing intein-WT TBF2 fusion (see above). Two groups of mutants were constructed. The first group of mutants contained single amino acid substitutions designed to construct a single hydroxylamine cleavage site (NG pair) in TFB2 by substituting Cys42 to Asp (NG42 mutant), Pro60 to Gly (NG59), Leu89 to Asn (NG89), Ala116 to Asn (NG116) and Leu136 to Asn (NG136). This group of mutants also includes mutant C42S TFB2 used in NTCB mapping experiments. The second group of TFB2 mutants contained N-terminal TFB2 deletions: Δ20TFB2, Δ34TFB2, Δ42TFB2, Δ63TFB2 and Δ62–73TFB2. All mutants were purified similarly to the WT TFB2. Mutant mitoRNAP that lacks four natural hydroxylamine cleavage sites was constructed by introducing G711K, N926S, N1103S, and N1117S substitutions using Quick-Change Multi mutagenesis kit (Stratagene).
Transcription assays
Standard transcription reactions were carried out using synthetic DNA templates (50 nM) containing human mitochondrial promoters and mitoRNAP (150nM), TFA (50nM), TFB2 (150nM), substrate NTPs (0.3 mM) in a transcription buffer containing 20 mM Tris (pH=7.9), 10 mM MgCl2, 10 mM DTT and 0.05% Tween 20. HSP1 templates contained 82 bps of WT human mitochondrial DNA sequence (with G to C substitution 11 bps downstream of the promoter start site to improve the yield during chemical synthesis), LSP – 62 bps of DNA (see Figure S1). Reactions were carried out at 35°C for the time indicated in the figure legends and stopped by addition of an equal volume of 95% formamide/0.05M EDTA. The products were resolved using 20% PAGE containing 6 M urea and visualized by Phosphoimager (GE Health).
Exonuclease footprinting
Template DNA strand containing LSP promoter was 32P-labeled by using N4 kinase (NEB) prior to annealing with the NT DNA strand to form a bubble template (Figure S1). IC was formed by incubation of mitoRNAP (150nM), TFB2 (150nM) and template DNA (50 nM) in transcription buffer for 15 min at RT. Exonuclease III (NEB) was added to 0.1 U/μl and the complexes were incubated for 5 min at 35°C. Products of the reaction were resolved and visualized as described above.
Protein-DNA photo crosslinking
To generate templates for protein-DNA photo crosslinking 5′ 32P-radiolabeled DNA primers (Figure S1) were annealed to the NT strand of the HSP1 promoter and extended by DNA polymerase I (NEB) in the presence of the 50 μM photo-reactive nucleotide analogs 4-thio-dTTP or 6-thio-dGTP (Trilink Biotech) for 5 min at room temperature. Synthesis of the modified template strand was completed by the addition of dNTPs (1 mM). A modification of this procedure was used to prepare a template for mapping of TFB2-DNA crosslinks. Thus, H11 primer (see Figure S1) was annealed to the NT strand of HSP1 promoter and extended with 4-thio-dTTP (50 μM), dCTP (5 μM) and [α-32P] dCTP (1.25 μM, 3000 Ci/mmol) for 5 min at RT. Synthesis of the modified template strand was completed by addition of dNTP mix (1 mM). The ICs were formed as described above and UV irradiated (312 nm) for 15 min at 30°C. Crosslinking products were resolved using a 4–12% Bis-Tris NuPAGE gel (Invitrogen).
Crosslinking of the priming nucleotide by catalytic autolabeling
Crosslinking using 2-hydroxybenzaldehyde AMP was performed essentially as described previously (Mustaev et al., 1991). Specifically, IC was formed by incubating template DNA (HSP1, 50nM) with mitoRNAP (150mM), TBF2 (150nM), TFA (50 nM) and 2-hydroxybenzaldehyde AMP (0.1 mM) in transcription buffer for 10 min at RT. The crosslinking was activated by addition of 0.1 mM of NaBH4 for 15 min at RT. Auto labeling was carried out by addition of 1 μL of [α-32P]ATP (800 Ci/mmol) followed by incubation of the reaction for 45 min at 35°C. The reaction was stopped by addition of SDS loading buffer and the products were resolved using SDS-PAGE as described above.
Mapping of the crosslinking sites in TFB2
Mapping of the region in TFB2 that interacts with the priming substrate with CNBr and NTCB (2-nitro-5-thiocyano-benzoic acid) was performed as described previously (Temiakov et al., 2000). For hydroxylamine (NH2OH) cleavage, 2 μl of the crosslinked material was lyophilized, dissolved in 30 μl of the solution containing 2M hydroxylamine and 7M Guanidine (pH 9.0 adjusted with 4.5 M LiOH), and incubated for 2–6 h at 43°C. The reaction was stopped by addition of 70 μl of H2O and 400 μl of acetone, the products precipitated at −70°C for 20 min, re-suspended in SDS gel loading buffer and resolved using SDS-PAGE. In mapping experiments involving TFB2-DNA photo crosslinking, the crosslinked species were treated with DNAse I (0.1 U, 1 h at 30°C) prior to hydroxylamine cleavage to trim the DNA and improve resolution of the protein fragments in SDS-PAGE.
Steady-state kinetics
The experiments were carried out using templates containing the mutant AGU LSP promoter (Figure S1). Reactions contained DNA templates (50 nM), mitoRNAP (150 nM), TFA (50 nM), TFB2 (150 nM), GTP (50 μM), [α-32P] GTP (800 Ci/mmol) and increasing concentrations of AMP (0.007 –1 mM). Reactions were carried out for 30 min at 35°C, stopped by addition of 10 μl of stop solution, resolved in 25% PAGE containing 6M urea and quantified by PhosphorImager™. The apparent Km and Vmax values were determined as described previously (Osumi-Davis et al., 1992).
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alam TI, Kanki T, Muta T, Ukaji K, Abe Y, Nakayama H, Takio K, Hamasaki N, Kang D. Human mitochondrial DNA is packaged with TFAM. Nucleic Acids Res. 2003;31:1640–1645. doi: 10.1093/nar/gkg251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiott EA, Jaehning JA. Mitochondrial transcription is regulated via an ATP “sensing” mechanism that couples RNA abundance to respiration. Mol Cell. 2006a;22:329–338. doi: 10.1016/j.molcel.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Amiott EA, Jaehning JA. Sensitivity of the yeast mitochondrial RNA polymerase to +1 and +2 initiating nucleotides. J Biol Chem. 2006b;281:34982–34988. doi: 10.1074/jbc.M608638200. [DOI] [PubMed] [Google Scholar]
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Chang DD, Clayton DA. Precise identification of individual promoters for transcription of each strand of human mitochondrial DNA. Cell. 1984;36:635–643. doi: 10.1016/0092-8674(84)90343-x. [DOI] [PubMed] [Google Scholar]
- Cheetham GM, Jeruzalmi D, Steitz TA. Structural basis for initiation of transcription from an RNA polymerase-promoter complex. Nature. 1999;399:80–83. doi: 10.1038/19999. [DOI] [PubMed] [Google Scholar]
- Cotney J, Shadel GS. Evidence for an early gene duplication event in the evolution of the mitochondrial transcription factor B family and maintenance of rRNA methyltransferase activity in human mtTFB1 and mtTFB2. J Mol Evol. 2006;63:707–717. doi: 10.1007/s00239-006-0075-1. [DOI] [PubMed] [Google Scholar]
- Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dairaghi DJ, Shadel GS, Clayton DA. Addition of a 29 residue carboxyl-terminal tail converts a simple HMG box-containing protein into a transcriptional activator. J Mol Biol. 1995a;249:11–28. doi: 10.1006/jmbi.1995.9889. [DOI] [PubMed] [Google Scholar]
- Dairaghi DJ, Shadel GS, Clayton DA. Human mitochondrial transcription factor A and promoter spacing integrity are required for transcription initiation. Biochim Biophys Acta. 1995b;1271:127–134. doi: 10.1016/0925-4439(95)00019-z. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- Falkenberg M, Larsson NG, Gustafsson CM. DNA replication and transcription in mammalian mitochondria. Annu Rev Biochem. 2007;76:679–699. doi: 10.1146/annurev.biochem.76.060305.152028. [DOI] [PubMed] [Google Scholar]
- Gaines G, Rossi C, Attardi G. Markedly different ATP requirements for rRNA synthesis and mtDNA light strand transcription versus mRNA synthesis in isolated human mitochondria. J Biol Chem. 1987;262:1907–1915. [PubMed] [Google Scholar]
- Gaspari M, Falkenberg M, Larsson NG, Gustafsson CM. The mitochondrial RNA polymerase contributes critically to promoter specificity in mammalian cells. Embo J. 2004;23:4606–4614. doi: 10.1038/sj.emboj.7600465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grachev MA, Kolocheva TI, Lukhtanov EA, Mustaev AA. Studies on the functional topography of Escherichia coli RNA polymerase. Highly selective affinity labelling by analogues of initiating substrates. Eur J Biochem. 1987;163:113–121. doi: 10.1111/j.1432-1033.1987.tb10743.x. [DOI] [PubMed] [Google Scholar]
- Kazmierczak KM, Davydova EK, Mustaev AA, Rothman-Denes LB. The phage N4 virion RNA polymerase catalytic domain is related to single-subunit RNA polymerases. Embo J. 2002;21:5815–5823. doi: 10.1093/emboj/cdf584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulbachinskiy A, Mustaev A. Region 3.2 of the sigma subunit contributes to the binding of the 3′-initiating nucleotide in the RNA polymerase active center and facilitates promoter clearance during initiation. J Biol Chem. 2006;281:18273–18276. doi: 10.1074/jbc.C600060200. [DOI] [PubMed] [Google Scholar]
- Maksimova TG, Mustaev AA, Zaichikov EF, Baranova LV, Kumarev VP. Localization of a lysine residue near the site of initiating substrate binding of T7 bacteriophage RNA polymerase. Bioorg Khim. 1989;15:18–23. [PubMed] [Google Scholar]
- Mangus DA, Jang SH, Jaehning JA. Release of the yeast mitochondrial RNA polymerase specificity factor from transcription complexes. J Biol Chem. 1994;269:26568–26574. [PubMed] [Google Scholar]
- Martin CT, Muller DK, Coleman JE. Processivity in early stages of transcription by T7 RNA polymerase. Biochemistry. 1988;27:3966–3974. doi: 10.1021/bi00411a012. [DOI] [PubMed] [Google Scholar]
- Matsunaga M, Jaehning JA. Intrinsic promoter recognition by a “core” RNA polymerase. J Biol Chem. 2004;279:44239–44242. doi: 10.1074/jbc.C400384200. [DOI] [PubMed] [Google Scholar]
- McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Murakami KS, Darst SA. Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol. 2003;13:31–39. doi: 10.1016/s0959-440x(02)00005-2. [DOI] [PubMed] [Google Scholar]
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. doi: 10.1016/s1097-2765(03)00266-1. [DOI] [PubMed] [Google Scholar]
- Mustaev A, Kashlev M, Lee JY, Polyakov A, Lebedev A, Zalenskaya K, Grachev M, Goldfarb A, Nikiforov V. Mapping of the priming substrate contacts in the active center of Escherichia coli RNA polymerase. J Biol Chem. 1991;266:23927–23931. [PubMed] [Google Scholar]
- Osumi-Davis PA, de Aguilera MC, Woody RW, Woody AY. Asp537, Asp812 are essential and Lys631, His811 are catalytically significant in bacteriophage T7 RNA polymerase activity. Journal of Molecular Biology. 1992;226:37–45. doi: 10.1016/0022-2836(92)90122-z. [DOI] [PubMed] [Google Scholar]
- Park CB, Asin-Cayuela J, Camara Y, Shi Y, Pellegrini M, Gaspari M, Wibom R, Hultenby K, Erdjument-Bromage H, Tempst P, et al. MTERF3 is a negative regulator of mammalian mtDNA transcription. Cell. 2007;130:273–285. doi: 10.1016/j.cell.2007.05.046. [DOI] [PubMed] [Google Scholar]
- Riva M, Carles C, Sentenac A, Grachev MA, Mustaev AA, Zaychikov EF. Mapping the active site of yeast RNA polymerase B (II) J Biol Chem. 1990;265:16498–16503. [PubMed] [Google Scholar]
- Temiakov D, Mentesana PE, Ma K, Mustaev A, Borukhov S, McAllister WT. The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc Natl Acad Sci U S A. 2000;97:14109–14114. doi: 10.1073/pnas.250473197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Structural basis for substrate selection by T7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–954. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanrooij S, Fuste JM, Farge G, Shi Y, Gustafsson CM, Falkenberg M. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc Natl Acad Sci U S A. 2008;105:11122–11127. doi: 10.1073/pnas.0805399105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody AY, Osumi-Davis PA, Hiremath MM, Woody RW. Pre-steady-state and steady-state kinetic studies on transcription initiation catalyzed by T7 RNA polymerase and its active-site mutants K631R and Y639F. Biochemistry. 1998;37:15958–15964. doi: 10.1021/bi9805801. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.