A new technique illuminates differential H3S10 phosphorylation dynamics in normal and cancer cells; spatial and temporal regulation of this process by aurora B kinase is required for accurate chromosome segregation.
Abstract
Posttranslational histone modifications regulate both gene expression and genome integrity. Despite the dynamic nature of these modifications, appropriate real-time monitoring systems are lacking. In this study, we developed a method to visualize histone modifications in living somatic cells and preimplantation embryos by loading fluorescently labeled specific Fab antibody fragments. The technique was used to study histone H3 Ser10 (H3S10) phosphorylation, which occurs during chromosome condensation in mitosis mediated by the aurora B kinase. In aneuploid cancer cells that frequently missegregate chromosomes, H3S10 is phosphorylated just before the chromosomes condense, whereas aurora B already accumulates in nuclei during S phase. In contrast, in nontransformed cells, phosphorylated H3S10 foci appear for a few hours during interphase, and transient exposure to an aurora B–selective inhibitor during this period induces chromosome missegregation. These results suggest that, during interphase, moderate aurora B activity or H3S10 phosphorylation is required for accurate chromosome segregation. Visualizing histone modifications in living cells will facilitate future epigenetic and cell regulation studies.
Introduction
DNA in eukaryotes is wrapped around a histone octamer containing H2A, H2B, H3, and H4, forming a nucleosome, which is the fundamental unit of chromatin. Posttranslational modifications of these histones play critical roles in genome function, including the regulation of transcription and maintenance of genome integrity (Jenuwein and Allis, 2001; Kouzarides, 2007). However, little is known about how these modifications change with time in single cells, largely because we lack the appropriate monitoring systems. Although resonance energy transfer between fluorescently tagged proteins has been used for this purpose (Kanno et al., 2004; Lin and Ting, 2004), this approach usually monitors the activity of modifying enzymes rather than the modification of endogenous proteins, and extensive probe improvements are required to obtain higher signal to noise ratios.
In this study, we detect endogenous modifications in living cells by introducing specific antibodies (Fab) directed against phosphorylated histone H3. In all organisms investigated so far (Hendzel et al., 1997; Wei et al., 1999; Johansen and Johansen, 2006), H3 is extensively phosphorylated at Ser10 (H3S10) during chromosome condensation and segregation by evolutionarily conserved aurora family kinases. In higher eukaryotes, aurora B is responsible for mitotic H3S10 phosphorylation and is essential for chromosome segregation (Ruchaud et al., 2007; Vader and Lens, 2008).
Results and discussion
Visualizing histone H3S10 phosphorylation in living HeLa cells
We first generated mouse hybridomas producing mAbs directed against phosphorylated H3S10 (H3S10ph). mAb CMA311 and CMA313 preferentially reacted with H3S10ph adjacent to un-, mono-, and dimethylated Lys9 (H3K9me0–2) and di- and trimethylated Lys9 (H3K9me2–3), respectively (Fig. 1 A). Immunofluorescence using fixed cells revealed that CMA311 and CMA313 preferentially stained mitotic chromosome arms and pericentromeric regions, respectively (Fig. 1 B and Fig. S1), which is consistent with an enrichment of H3K9me3 in pericentromeric heterochromatin (Peters et al., 2003). Some interphase nuclei were also stained weakly (Fig. S1 A), as reported previously using other H3S10ph-specific antibodies (Hendzel et al., 1997; Monier et al., 2007).
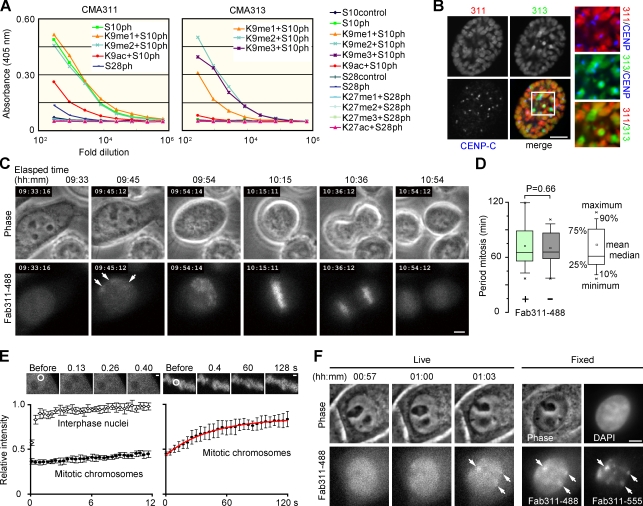
Figure 1.
Fluorescently labeled Fab binds to phosphorylated histone H3S10 during mitosis in HeLa cells. (A) Specificity of mAbs evaluated by ELISA using the indicated peptides. The peptides that reacted with individual mAbs are indicated in the graphs. (B) Immunofluorescence. Fixed cells were stained with Fab311-488 (Alexa Fluor 488–labeled Fab from CMA311; red), Fab313-555 (Alexa Fluor 555–labeled Fab from CMA313; green), and anti–CENP-C (blue). Images of a prophase cell are shown with magnified views of boxed area. See Fig. S1 for other cells at different phases of the cell cycle. (C) Time-lapse images of a cell loaded with Fab311-488. Arrows indicate concentrations of Fab311-488. See Video 1. (D) Period of mitosis in Fab311-488–loaded and control cells. By collecting three z-stack images every 3 min, the period from prophase to anaphase was measured (n = 27) using phase-contrast images. No significant difference was seen; P = 0.66 (Student’s t test). (E) The mobility of Fab311-488 by FRAP. After bleaching a 2-µm spot (white circle), images were collected for 12 s every 0.13 s (left) or for 120 s every 0.4 s (right). Means ± SD are shown (n = 12). The red line shows the fitted curve using single exponential association kinetics. Residence time (k−1; k = association coefficient) of the transiently bound fraction is calculated as 44 s. (F) H3S10ph detected in live and fixed samples. Cells loaded with Fab311-488 were imaged every 3 min. When foci were detected in two consecutive frames (01:00 and 01:03), cells were fixed and immunolabeled with Fab311-555. Foci observed during live imaging (arrows) were also detected after fixation and immunolabeling. Bars: (B, C, and F) 5 µm; (E) 1 µm.
To visualize H3S10 phosphorylation in living cells, Fab from CMA311 was tagged with Alexa Fluor 488 (Fab311-488) and loaded into HeLa cells (Fig. 1 C and Video 1). They passed through nuclear pores to become distributed throughout the interphase nucleus (09:33; the elapsed time from the start of acquisition). They then became concentrated in a few nuclear foci (Fig. 1 C, 09:45, arrows) and on condensed chromosomes during prophase (09:54) to metaphase (10:15) and eventually dispersed when daughter nuclei formed (10:54). Imaging cells loaded with both Fab311 and Fab313 revealed that the latter, which can react with H3S10ph next to H3K9me3, became concentrated in more discrete heterochromatin foci, which were surrounded by the former (Video 2). This confirms previous results using fixed cells (Fig. S1; Hendzel et al., 1997; Monier et al., 2007) and shows that our mAbs target H3S10ph in living cells. Imaged cells went through at least two to three cell divisions (Video 3) without affecting the duration of mitosis (Fig. 1 D). These results suggest that Fab loaded at the concentrations used in this study, which are sufficient to bind to, at most, only a few percent of the total histone H3 (Table S1), has little effect on cell cycle progression.
To examine the mobility of Fab311-488 in living cells, we performed FRAP (Kimura and Cook, 2001) by bleaching a small spot in loaded cells. Homogenously distributed Fab311 in interphase nuclei redistributed rapidly within seconds after bleaching, which is consistent with it being free to diffuse (Fig. 1 E, left). However, Fab311 on mitotic chromosomes, where H3S10 is phosphorylated, redistributed slowly (over seconds) with a residence time of ∼44 s (Fig. 1 E, right). As most H3 remains stably integrated into nucleosomes for hours (Kimura and Cook, 2001), these kinetics probably reflect the association of Fab with its target phosphorylated epitope. Substantial fluorescence recoveries were also observed when interphase foci were bleached in the presence of a protein phosphatase inhibitor, tautomycetin (TMC), which inhibits dephosphorylation of H3S10 (see Fig. 4 and not depicted), indicating that Fab311 could dissociate from H3S10ph spontaneously rather than as a result of phosphorylation turnover. Thus, Fab311 does not seem to permanently block H3S10ph from accessing cellular proteins, including phosphatases.
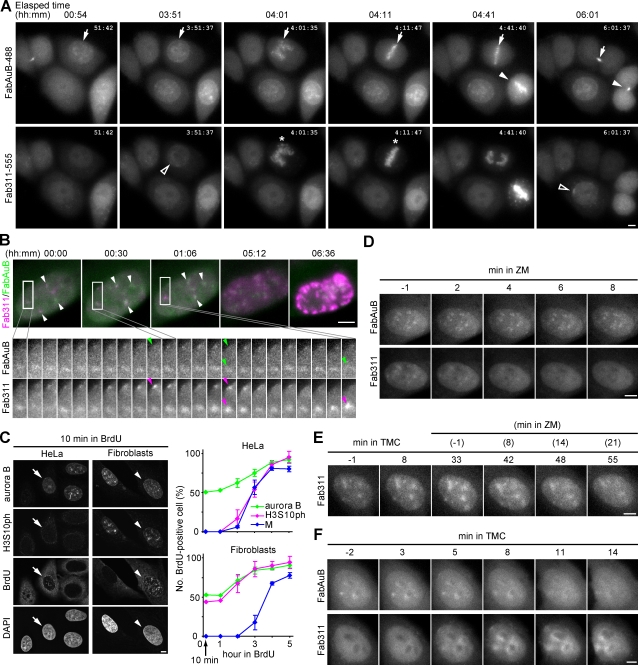
Figure 4.
Visualization of aurora B with H3S10ph in living cells and the effects of kinase and phosphatase inhibitors. (A) HeLa cells loaded with FabAuB-488 and Fab311-555. Maximum projections of three z-stack images at 1.5-µm intervals are shown. Arrows and closed arrowheads indicate FabAuB concentrations in heterochromatin, the midzone, and the midbody. Open arrowheads indicate Fab311 foci before the accumulation on condensed chromosomes (asterisks). See Video 8. (B) A fibroblast loaded with FabAuB-488 (green) and Fab311-555 (magenta). Deconvolved and maximum-projected images of five z-stack images at 0.75-µm intervals taken every 2 min are shown. Full time series images of the boxed areas are shown. (top) White arrowheads indicate FabAuB and Fab311 foci. (bottom) The positions of Fab311 foci with the highest intensities are indicated (arrowheads). See Video 9. (C) BrdU incorporation in cells exhibiting aurora B and H3S10ph. Cells were incubated in BrdU for 10 min to 5 h, fixed, and immunolabeled for aurora B, H3S10ph, and BrdU. (left) When treated with BrdU for 10 min, aurora B–positive cells often exhibit BrdU signals, which are unassociated (arrows; HeLa) or associated (arrowheads; fibroblasts) with H3S10ph. (right) BrdU-positive cells were counted among those positive for aurora B, H3S10ph, or condensed chromosomes (n > 20); cells in early mitotic phase (i.e., prophase to prometaphase; M) were counted in HeLa because of their prolonged mitosis (> 1 h vs. ∼30 min in fibroblasts). The means ± SD from three independent experiments are shown. (D) A fibroblast treated with ZM. (E) A fibroblast treated with TMC followed by ZM. (F) A HeLa cell treated with TMC. Bars, 5 µm.
We next compared images of Fab311-488 foci in living cells taken under low light dose with those by postfixation immunostaining to evaluate the sensitivity of live imaging. HeLa cells were fixed immediately after foci were detected and labeled with the same Fab conjugated with another fluorophore (Fab311-555; Fig. 1 F). Most large foci detected in the postfixed nuclei were also identifiable in the live image (Fig. 1 F, arrows), although smaller foci, clearly detected with Fab311-555, were obscured by background fluorescence from unbound Fab molecules. Although careful analysis is needed to define when and where foci are formed, the presence of faint foci was often identifiable from the background by investigating consecutive time series images (e.g., Video 2, arrowheads).
Visualizing H3S10 phosphorylation in mouse preimplantation embryos
Fab311-488 or Fab313-488 was injected with mRNA encoding histone H2B tagged with monomeric RFP (mRFP; H2B-mRFP) into mouse fertilized eggs, which were then allowed to develop to morula stage under a confocal microscope (Yamagata et al., 2009). Fab311 became concentrated in the centromere-rich perinucleolar region (Fig. 2 A, 06:29, arrow; and Video 4; Yamazaki et al., 2007) ∼1 h before chromosomes condensed at the first division (07:44). During the first mitosis, Fab311 highlighted both maternal and paternal chromosomes. At the two-cell stage, H3S10ph foci appeared distant from the division plane (Fig. 2 A, 21:15, arrow), which is consistent with heterochromatin maintaining the Rabl configuration (Marshall et al., 1997; Cowell et al., 2002). In contrast, Fab313 preferentially associated with maternal chromosomes (Fig. 2 B and Video 5), which is consistent with H3K9me3 on maternal chromatin being a heritable epigenetic mark (Cowell et al., 2002; Santos et al., 2005). As seen previously (Mayer et al., 2000), maternal and paternal chromosomes did not intermingle during the first mitosis or in two-cell embryos; a subset of condensed chromosomes (probably maternally derived) were stained by Fab313 during the second mitosis. Importantly, after transfer to the oviducts of pseudopregnant mothers, imaged embryos developed to birth (Fig. 2, C and D) like controls (Yamagata et al., 2009). Again, injection and imaging did not affect growth and development.
Figure 2.
Histone H3S10 phosphorylation in mouse preimplantation embryos. (A) Time-lapse images of an embryo injected with 100 µg/ml Fab311-488 and 5 µg/ml H2B-mRFP mRNA (maximum projection of 51 z stacks). Arrows indicate the concentration of Fab311 in the centromere-rich regions. See Video 4. (B) Maternal chromosomes are selectively detected by Fab313. Fab313-488 (red) and H2B-mRFP (cyan) images in first and second mitosis are shown. The two-cell image is contrast enhanced. See Video 5. (C) Pups developed from Fab313-488–injected embryos with their recipient mother. (D) Development of embryos injected with fluorescent Fab. Embryos generated by in vitro fertilization were injected with various concentrations (conc.) of Fab together with H2B-mRFP mRNA, imaged up to the four to eight-cell stage, and transferred to the oviduct of day 0.5 pseudopregnant mothers. The rates of normal development were similar to controls (Yamagata et al., 2009). aImaging minus (−) means the embryos were injected with Fab but not imaged. Bars, 10 µm.
Differential H3S10 phosphorylation dynamics in cancer and normal cells
To study the dynamics of H3S10ph in both cancer and normal cells, we used the fluorescent Fab technique with Fab311, which recognizes H3S10ph next to K9me0–2. In fixed cancer cell samples, missegregated chromosomes or chromosome bridges are frequently found during anaphase (Thompson and Compton, 2008), which is consistent with a higher chromosome instability, a hallmark of cancer. Such missegregated or bridged chromosomes were left near the midzone and preferentially stained with Fab311 (Fig. 3 A). This can be explained by a gradient of kinase activity of aurora B, which phosphorylates H3S10, being highest at the midzone during anaphase (Fig. 3 A; Fuller et al., 2008). Indeed, time-lapse imaging using Fab311-488 allowed us to detect chromosome missegregation more sensitively compared with a general chromatin marker, histone H2B–GFP (Fig. 3 B). Some chromosomes that segregated aberrantly generated micronuclei after cell division (Fig. 3 B and Video 6), whereas others were eventually integrated into the nucleus. As reported previously (Thompson and Compton, 2008), missegregation rates were higher in aneuploid cells (HeLa, Panc1, MDA-MB-231, and U2OS) compared with nontransformed cells (human baby skin fibroblasts, telomerase-immortalized retina pigment epithelial cells [hTERT-RPE1], and human umbilical vein endothelial cells; Fig. 3 C, left).
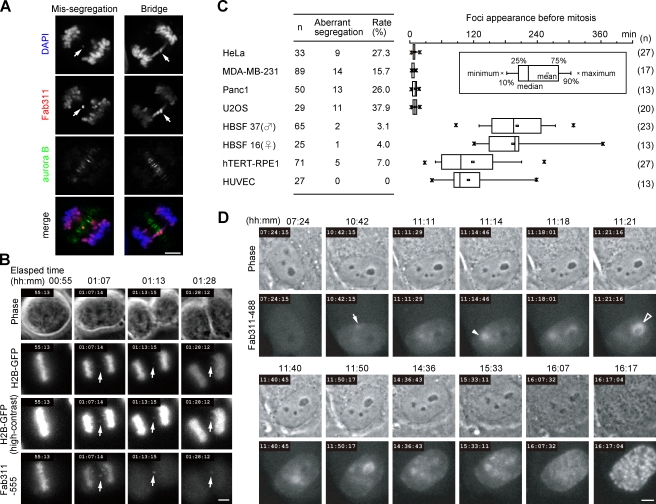
Figure 3.
Aberrant chromosome segregation and interphase phosphorylation monitored using fluorescent Fab. (A) Anaphase HeLa cells showing aberrant chromosome segregation. Fixed cells were stained with Fab311-488 (red), anti–aurora B (green), and DAPI (blue). Arrows indicate a missegregated chromosome (left) and a chromosome bridge (right). (B) Missegregated chromosomes are readily detectable in living cells using Fab311-555. HeLa cells expressing histone H2B–GFP were loaded with Fab311-555, and images were taken every 3 min. A missegregated chromosome (arrows) is easily visible by Fab311, whereas contrast enhancement is required for detection by H2B-GFP. (C) Frequency of chromosome missegregation and period of Fab311-488 foci appearance before mitosis measured using time-lapse imaging. (left) The chromosome missegregation rate. (right) The period of Fab311-488 foci appearance before mitosis. (D) Time-lapse images of a fibroblast loaded with Fab311-488. H3S10ph focus (arrow) appears >5 h before mitosis, and others appear (arrowheads) later. See Video 7. Bars, 5 µm.
In cancer cells, Fab311 became concentrated in foci just before mitosis (within ∼20 min; Fig. 3 C, right). In contrast, in normal cells, Fab311 foci lasted for many hours (Fig. 3 C, right), and the distribution of Fab311 presented very different patterns. In fibroblasts, Fab311 was first concentrated in foci near the nuclear periphery (Fig. 3 D, 10:42, arrow; and Video 7) but soon redistributed to new foci near nucleoli (11:11). One focus near a small nucleolus grew (Fig. 3 D, 11:14; arrowhead) and then disappeared (11:21), whereas another (around a large nucleolus; 11:21, open arrowhead) appeared and disappeared repeatedly (11:40 and 11:50) before Fab311 distribution became more stable (14:36 and 15:33) and chromosomes began to condense before mitosis (16:17). Such Fab311 redistributions from one focus to another were observed in most fibroblast nuclei imaged (e.g., Fig. 4 B). These results suggest that H3S10 phosphorylation is spatially and temporally regulated by kinase and phosphatase activities during interphase.
H3S10 phosphorylation and aurora B in living cells
As aurora B is likely to mediate H3S10 phosphorylation before mitosis (Crosio et al., 2002; Monier et al., 2007), we next visualized the dynamics of H3S10ph together with aurora B (Fig. 4). We used Alexa Fluor 488–labeled aurora B–specific Fab (FabAuB-488) to detect endogenous protein, whose expression level depends on cell type. In HeLa cells (Fig. 4 A and Video 8), FabAuB concentrated in several foci, often around nucleoli, for several hours during interphase (e.g., 00:54 and 03:51, arrows), and then Fab311 became concentrated around a few FabAuB foci ∼10–20 min before chromosome condensation (03:51 and 06:01, open arrowheads). FabAuB then localized to centromeres (Fig. 4 A, 04:01 and 04:11 [arrows]; and 04:41 [arrowhead]) on condensed chromosomes (04:01 and 04:11, asterisks) during prophase to metaphase and remained at the midzone (4:41, arrow) and midbody (06:01, arrow and arrowhead) after the chromosomes segregated, as reported previously by postfixation immunolabeling and GFP tagging (Terada et al., 1998; Murata-Hori et al., 2002). Thus, loaded FabAuB-488 appears to target aurora B in living cells without affecting its function or cell cycle progression. In contrast to such different timings of FabAuB and Fab311 foci appearance in HeLa, in fibroblasts, they were both concentrated, often close together, in foci for several hours before mitosis (typically shown in Fig. 4 B, arrowheads; and Video 9). The intensities of FabAuB and Fab311 foci fluctuated, particularly in early phase, as seen in Fig. 3 D. Whereas aurora B signals appeared more stable than H3S10ph in foci, the subtle decreases of aurora B were observed concomitantly with the highest levels of Fab311 accumulations (Fig. 4 B, green and magenta arrowheads).
To validate the timing of aurora B accumulation and H3S10 phosphorylation seen during interphase in living cells, we used postfixation immunostaining combined with BrdU labeling. Cells were treated for different periods with BrdU to label cells in S phase or those that passed through S phase. With 10 min of BrdU pulse labeling, ∼50% of aurora B–positive cells exhibited BrdU signals with patterns typically seen in late S phase (Nakayasu and Berezney, 1989), both in HeLa and fibroblasts (Fig. 4 C), which is consistent with previous studies (Crosio et al., 2002; Monier et al., 2007). As seen in living cells, H3S10ph signals were often associated with aurora B in fibroblasts (Fig. 4 C, left, arrowheads) but not in HeLa (arrows). Among H3S10ph-positive HeLa nuclei, BrdU incorporation was first detected within 2 h of treatment, and the number of BrdU-positive nuclei increased to >80% within 4 h (Fig. 4 C, right). A similar BrdU incorporation profile was seen in cells in M phase. In contrast, ∼50% of H3S10ph-positive fibroblast nuclei exhibited BrdU signals after only a 10-min pulse, as did aurora B–positive ones (Fig. 4 C, right). These results are consistent with the live cell data, indicating different H3S10 phosphorylation foci formations in HeLa and fibroblasts.
Effects of kinase and phosphatase inhibitors
We further analyzed H3S10 phosphorylation kinetics by using chemical inhibitors to disturb the balance between kinase and phosphatase activities. When fibroblasts showing FabAuB and Fab311 foci were treated with an aurora B–selective inhibitor, ZM447439 (ZM; Girdler et al., 2006) at 0.5 µM, Fab311 became diffuse within 10 min, whereas FabAuB remained concentrated in foci (Fig. 4 D); aurora B localization does not depend on its kinase activity (Girdler et al., 2006). This result indicates that H3S10 dephosphorylation was induced by inhibition of the aurora B kinase. As the counteracting phosphatase could be PP1 (protein phosphatase 1; Murnion et al., 2001; Adhvaryu and Selker, 2008; Emanuele et al., 2008), fibroblasts were also treated with a PP1-selective inhibitor, TMC (Mitsuhashi et al., 2001) at 2 ng/ml (3.3 nM). During incubation with TMC (Fig. 4 E), Fab311 became more concentrated in foci, which persisted for >20 min after the addition of ZM, which is consistent with the view that H3S10 is dephosphorylated by PP1.
We also treated HeLa cells with TMC to see the effect of PP1. In response to TMC, Fab311 concentrated at/around aurora B foci (Fig. 4 F), suggesting that PP1 may continuously inactivate aurora B by dephosphorylating autophosphorylation sites involved in activation (Sessa et al., 2005) and/or may rapidly dephosphorylate H3S10 to maintain low H3S10 phosphorylation levels in HeLa interphase nuclei. Fab311 became more concentrated during incubation with TMC; in contrast, aurora B became more diffuse, dissociating from foci (Fig. 4 F). This might mimic events during chromosome condensation; aurora B dissociates from chromosome arms after phosphorylating H3S10 (Fig. 4, A and B; Ruchaud et al., 2007).
Aberrant chromosome segregation caused by transient aurora B inhibition
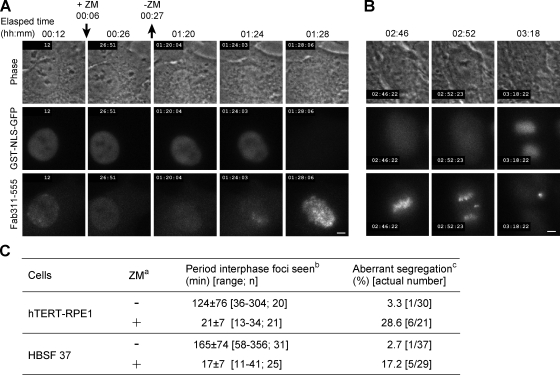
As there is a good correlation between the fidelity of chromosome segregation and H3S10 phosphorylation during interphase (Fig. 3 C), we examined whether transient inhibition with ZM during interphase affects chromosome segregation. To monitor when the nuclear membrane broke down before mitosis, GFP tagged with SV40 nuclear localization signal (NLS) and GST (GST-NLS-GFP) was loaded with Fab311-555 into hTERT-RPE1 (Fig. 5, A and B) or fibroblasts. ZM treatment led to the disappearance of Fab311 foci within several minutes (Fig. 5 A, 00:26; and Video 10). After washing away ZM, foci did not reform immediately and only appeared ∼20 min before membrane breakdown (Fig. 5, A [01:24] and C; and Video 10), as in cancer cells. Remarkably, transient ZM treatment during interphase increased chromosome missegregation (Fig. 5, B and C). All cells that showed missegregation had already exhibited Fab311 foci when treated with ZM, and we obtained no evidence that transient exposure to ZM affects cells without obvious foci.
Figure 5.
Effects of ZM treatment during the interphase. Cells were loaded with GST-NLS-GFP and Fab311-555 and treated with ZM for ∼20 min. (A) Response of Fab311 to ZM addition and removal in hTERT-RPE1. See Video 10. (B) An hTERT-RPE1 cell that exhibited chromosome missegregation after transient exposure to ZM. (C) Effects of transient exposure to ZM on the formation of Fab311 foci and chromosome segregation. aLoaded cells were incubated in the absence (−) or presence (+) of ZM. bThe period when Fab311 foci appeared before the nuclear membrane broke down (judged by the release of GST-NLS-GFP from the nucleus into the cytoplasm). Mean ± SD, range, and number of cells analyzed (n) are indicated. cThe percentage of cells showing aberrant chromosome segregation. Bars, 5 µm.
Phospho–dephospho cycle of H3S10 and chromatin reorganization
In this study, we developed a system to monitor histone H3S10 phosphorylation in growing human cells and mouse embryos. Using Fab311, we showed that the degree and timing of phosphorylation in normal and cancer cells differ profoundly, whereas aurora B accumulates within S phase in both cell types. Our data indicate that during interphase, global and local levels of H3S10ph depend on the balance between aurora B kinase and phosphatases, including PP1. Phosphatase activity appears relatively high in cancer cells, as H3S10ph levels are drastically increased by the inhibition of PP1 in HeLa. In fibroblasts, kinase and phosphatase activities appear more balanced, resulting in the dynamic redistribution of H3S10ph. The aurora B complex may initially be recruited to heterochromatin through an interaction with HP1 (heterochromatin protein 1; Ainsztein et al., 1998), which preferentially binds to methylated H3K9 (Kouzarides, 2007). The earliest H3S10 phosphorylation in heterochromatin may be analyzed using Fab313. Once H3S10 is phosphorylated, HP1 no longer binds to H3 (Fischle et al., 2005; Hirota et al., 2005), perhaps resulting in aurora B dissociation and H3S10 dephosphorylation by phosphatases. HP1 and aurora B may bind again to dephosphorylated H3S10, and repeated cycles of phosphorylation and dephosphorylation could continue as long as the balance of kinase and phosphatase activities is maintained during interphase, before aurora B becomes fully active or phosphatase activity becomes substantially reduced at the onset of mitosis.
We also showed that transient inhibition of aurora B during interphase affects chromosome segregation. This suggests that H3S10 phosphorylation during interphase or moderate aurora B activity is critically required to maintain the fidelity of chromosome segregation. Although phosphorylation of other proteins may also be required, genetic knockouts point to the importance of H3S10 phosphorylation by aurora B in Tetrahymena thermophila (Wei et al., 1999) and fission yeast (Mellone et al., 2003). Recent studies (Giménez-Abián et al., 2004; Dai et al., 2006; Yamagishi et al., 2008) also show that timely aurora B activity is required to displace HP1 and cohesins that tie sister chromatids together, along with its regulator shugoshin (Sgo1), from chromosome arms to the centromere. Indeed, we found little colocalization of H3S10ph with the cohesin subunit Rad21/Scc1 or Sgo1 in G2 cells (Fig. S2). In normal cells, the phospho–dephospho cycle of H3S10 during interphase may stimulate chromatin reorganization and ensure complete removal of Sgo1 and sister chromatid cohesion from chromosome arms. However, in transformed cells associated with chromosome instability, aurora B activity and/or H3S10 phosphorylation appears to be actively suppressed by PP1 during interphase; the rapid phosphorylation during prophase to prometaphase may not be sufficient for full chromatin reorganization. The significance and exact mechanism of the interphase phospho–dephospho cycle on chromosome instability should be investigated in future studies. As H3S10 phosphorylation is also involved in gene activation and epigenetic regulation (Johansen and Johansen, 2006; Sabbattini et al., 2007; Adhvaryu and Selker, 2008), we speculate that this phosphorylation may generally be associated with chromatin reorganization through displacing HP1 and cohesin (Peters et al., 2008).
Conclusions
The method developed in this study is a powerful and straightforward technique for visualizing the endogenous proteins and their modifications in various cell types and is applicable to any modifications when specific antibodies are available. We anticipate that our approach will prove useful in understanding the important role that other histone modifications play in regulating the life of the cell.
Materials and methods
Antibodies and Fab preparation
Mice were immunized with synthetic peptides, ARTKQTARK(phospho-S)TGGKAPRKQC and ARTKQTAR(trimethyl-K) (phospho-S)TGGKAPRKQC for CMA311 and CMA313, respectively, and hybridomas were screened by ELISA using the peptides listed in Kimura et al. (2008). CMA311 and CMA313 were isotyped as IgG1-κ using a kit (AbD Serotec). After purifying IgG through a protein G column (GE Healthcare), Fab fragments were prepared using a kit (Thermo Fisher Scientific) and then conjugated with an Alexa Fluor dye (Invitrogen), yielding the labeling ratio of dye for Fab of ∼3 (mol/mol), according to the manufacturers’ instructions. The sample was concentrated, and the buffer was replaced with PBS using an Ultrafree 0.5 filter (10 k-cut off; Millipore). Fab was also prepared from an AIM/aurora B–specific mAb (BD) and labeled with Alexa Fluor 488 (Invitrogen).
Imaging of culture cells
Human umbilical vein endothelial cells were purchased from Takara Bio Inc. and cultured according to the manufacturer’s instructions. Human baby skin fibroblasts (obtained from M. Naito and S. Suzuki, Kyoto University, Kyoto, Japan), hTERT-RPE1, HeLa, MDA-MB-231 (obtained from N. Matsuura, Osaka University, Suita, Osaka, Japan), Panc1 (obtained from N. Matsuura), and U2OS (obtained from M. Tsuchiya, Osaka University) were grown in DME and high glucose (Sigma-Aldrich) supplemented with 10 U/ml penicillin, 50 µg/ml streptomycin, and 10% fetal calf serum. Cells plated on a glass-bottom dish (Mat-Tek) were loaded with 4 µl of 0.5 mg/ml Fab311-488 using glass beads (Manders et al., 1999) and regrown for >4 h in phenol red–free DME (Nacalai Tesque, Inc.) supplemented with 10 U/ml penicillin, 50 µg/ml streptomycin, and 10% fetal calf serum. The dish was placed on an inverted microscope (Ti-E; Nikon) with a Plan-Apochromat VC 60× NA 1.2 water immersion objective lens with Immersol W (Carl Zeiss, Inc.), featuring a culture system (Tokai Hit) at 37°C under 5% CO2. Fluorescence images were captured under the operation of NIS Elements version 3.0 (Nikon) using an EM charge-coupled device (iXon+; Andor; 100 ms; gain 300; typically 3-min intervals) with a GFP-3035B filter set (Semrock) and a 75-W Xenon lump attenuated through two neutral-density (4× and 16×) and 440-nm long-pass filters. Phase-contrast images were also collected (50 ms) using an external phase ring. In most cases, three different focal planes were imaged with 2-µm intervals. For Figs. 1 D and 4, a Plan-Apochromat VC 100× NA 1.4 oil immersion lens was used. For double labeling, cells were loaded with 2 µl of 0.5 mg/ml Fab311-555 in combination with 2 µl of 0.5 mg/ml Fab313-488, 1 µl of 1.3 mg/ml FabAuB-488, or 2 µl of 0.2 mg/ml GST-NLS-GFP (gift from M. Hieda, Osaka University; Yokoya et al., 1999), and images were captured sequentially using GFP-3035B (100 ms) or LF488-A (200–400 ms; Semrock) and TRITC-A (300–400 ms; Semrock) filters. When necessary, 0.5 µM of chemical inhibitor ZM (Tocris Bioscience) and 3.3 nM TMC (Sigma-Aldrich) were added during the imaging.
Images were deconvolved (four iterations; Fig. 4 B), the contrast was stretched linearly using the same values throughout a time sequence, and the cropped area and planes were converted to avi using NIS Elements or MetaMorph version 7 (MDS Analytical Technologies). Avi videos were converted to QuickTime files with high quality Sorenson or H.264 compression using ImageJ version 1.41 (National Institutes of Health). Graphs in Figs. 1 and 3 were drawn using Origin version 7 (OriginLab).
Measurements of chromosome missegregation rate and H3S10 foci formation period
Chromosome missegregation was scored using the criteria that a few chromosomes were highlighted with Fab311-488 in at least two frames in time-lapse images, whereas others showed weak staining after anaphase. The time when interphase foci appeared was judged by obvious concentrations of fluorescence over background, and timing of chromosome condensation was defined by nucleolar deformation and clear changes in chromosome optical density seen in phase-contrast images.
FRAP
The mobility of Fab311-488 was analyzed using a confocal microscope (FV-1000; Olympus; operated by the built-in software version 1.7) with a PlanS-Apochromat 60× NA 1.35 oil immersion lens (Kimura et al., 2006). To analyze rapid diffusion, images were collected using 488-nm argon-ion laser excitation (0.4% transmission; 132 ms/frame; 2 µs/pixel; 256 × 64 pixels; pinhole 800 µm; 10×), and a 2-µm diameter spot was bleached using the second scanner (75% 405-nm laser transmission; 10 ms). To analyze binding kinetics, 10 images were collected (0.4% 488-nm laser transmission; 428 ms/frame; 2 µs/pixel; 256 × 256 pixels; pinhole 800 µm; 6×), a 2-µm diameter spot was bleached (100% 488-nm laser transmission; four iterations), and a further 90 images were collected using the original settings.
Imaging of mouse preimplantation embryos
Synthesis and purification of mRNA for injection have been described previously (Yamagata et al., 2009). Gametes were collected from BDF1 mice (7–12 wk old) and fertilized in vitro. Anaphase II/telophase II stage one-cell embryos were injected with a mixture of 5 µg/ml H2B-mRFP mRNA (cDNA encoding H2B and mRFP was provided by T. Kanda [Aichi Cancer Center, Chikusa-ku, Nagoya, Japan] and R. Tsien [University of California, San Diego, La Jolla, CA]; Kanda et al., 1998; Campbell et al., 2002) and 25–100 µg/ml Fab311-488 or Fab313-488 using a piezo manipulator (Prime Tech Ltd.). The embryos were transferred to drops of Chatot-Ziomek-Bavister medium on a glass-bottom dish and placed in a chamber stage (Tokai Hit) at 37°C under 5% CO2 on an inverted microscope (IX-71; Olympus) equipped with a spinning disk confocal (CSU10; Yokogawa), EM charge-coupled device (iXon BV-887; Andor Technology), and a filter wheel and z motor (Mac5000; Ludl). Fluorescence images in 51 different focal planes with 2-µm intervals were captured every 10–15 min using a UPlan-Apochromat 20× NA 0.8 or a UPlan-Apochromat 40× NA 1.0 (for Fig. 2 B) oil immersion lens with 488- and 568-nm laser lines (argon-krypton; CVI Melles Griot) under the operation of MetaMorph version 7. After imaging for 44 h at 15-min intervals using a UPlan-Apochromat 20× NA 0.8 oil immersion lens, four- to eight-cell stage embryos were transferred to the oviduct of day 0.5 pseudopregnant mothers. 176 time points, 51 in z axis, two colors, and a total of 17,952 fluorescent images were acquired. All animals were maintained in accordance with the Animal Experiment Handbook at the Kobe Center for Developmental Biology (RIKEN).
Immunofluorescence
Secondary antibodies (Jackson ImmunoResearch Laboratories, Inc.), anti-AIM/aurora B (BD), and CMA311 were conjugated with Alexa Fluor dyes (Invitrogen) according to the manufacturer’s instructions. For Fig. 1 B and Fig. S1, cells grown on 24-well glass-bottom plates (Iwaki) were fixed with 1% formaldehyde (Electron Microscopy Sciences) in 250 mM Hepes-NaOH, pH 7.4, containing 0.1% Triton X-100 for 10 min, permeabilized with 1% Triton X-100 in PBS for 20 min, and washed three times with PBS (Kimura et al., 2006). After blocking in Blocking-One P (Nacalai Tesque, Inc.) for 30 min, cells were incubated with 1:10,000 guinea pig anti–human CENP-C (gift from K. Yoda, Nagoya University, Chikusa-ku, Nagoya, Japan; Ando et al., 2002) for 2 h and then with 1 µg/ml Alexa Fluor 647–conjugated anti–guinea pig Ig, 1 µg/ml Fab311-488, and 1 µg/ml Fab313-555 overnight. For Fig. 1 F and Fig. S2, cells were fixed with 4% formaldehyde in 250 mM Hepes-NaOH, pH 7.4, for 10 min before permeabilization, blocking, and immunolabeling. Cells were stained for 2 h with 1 µg/ml Fab311-555 (Fig. 1 F) or, in Fig. S2, with 1:250 mouse anti-Rad21 (Millipore) or 1:3,000 anti-SgoL1 (Abnova) and then with 1 µg/ml Fab311-488 and 1:500 Cy3-conjugated goat anti–mouse IgG, Fc-γ fragment specific (Jackson ImmunoResearch Laboratories, Inc.). For Fig. 4 C, cells were incubated in 50 µM BrdU for 10 min to 5 h before fixation (4% formaldehyde; 10 min) and staining for 2 h with 3.5 µg/ml Alexa Fluor 488–conjugated anti–aurora B and 3.5 µg/ml Alexa Fluor 555–conjugated CMA311. After refixation, DNA was denatured in 2 N HCl for 20 min, and BrdU was detected with 1:150 rat anti-BrdU (2 h; BU1/75[ICR1]; Funakoshi) and 1.5 µg/ml Alexa Fluor 647–conjugated anti–rat Ig (2 h). DNA was counterstained with DAPI. Fluorescence images of single optical sections were sequentially collected using a confocal microscope (FV-1000; Olympus) with a PlanS-Apochromat 60× NA 1.35 oil immersion lens (512 × 512 pixels; 10 µs/pixel; 4 line Kalman; pinhole 120 µm) as described previously (Kimura et al., 2008).
Online supplemental material
Fig. S1 shows the staining patterns of Fab311, Fab313, and CENP-C on chromosomes in fixed cells at different phases of the cell cycle. Fig. S2 shows the localization of Rad21 and Sgo1 in G2 cells showing foci of H3S10 phosphorylation. Videos 1 and 3 show HeLa cells loaded with Fab311-488. Video 2 shows a HeLa cell loaded with Fab313-488 and Fab311-555. Videos 4 and 5 show mouse preimplantation embryos injected with Fab311-488 and Fab313-488, respectively, in combination with H2B-mRFP mRNA. Video 6 shows a HeLa cell that exhibited chromosome missegregation. Video 7 shows a normal fibroblast loaded with Fab311-488. Videos 8 and 9 show HeLa cells and fibroblasts, respectively, loaded with FabAuB-488 and Fab311-555. Video 10 shows an hTERT-RPE1 cell loaded with Fab311-488 and treated with ZM. Table S1 estimates how many Fab311-488 molecules are loaded into a HeLa cell. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200904137/DC1.
Acknowledgments
We thank M. Hieda, T. Kanda, N. Matsuura, M. Naito, S. Suzuki, R. Tsien, M. Tsuchiya, and K. Yoda for materials and P.R. Cook, T. Fukagawa, H. Funabiki, K. Morris, and C. Obuse for comments on the manuscript. We are also grateful to Y. Hiraoka and T. Wakayama for their support.
This work was supported by grants in aid and the Genome Network Project from the Ministry of Education, Culture, Sports, Science and Technology of Japan. Y. Hayashi-Takanaka was supported by a Japan Society for the Promotion of Science Restart Postdoctoral fellowship. N. Nozaki is a founder of MAB Institute Inc.
Footnotes
Abbreviations used in this paper:
- H3S10ph
- phosphorylated H3S10
- mRFP
- monomeric RFP
- NLS
- nuclear localization signal
- TMC
- tautomycetin
References
- Adhvaryu K.K., Selker E.U. 2008. Protein phosphatase PP1 is required for normal DNA methylation in Neurospora. Genes Dev. 22:3391–3396 10.1101/gad.1738008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ainsztein A.M., Kandels-Lewis S.E., Mackay A.M., Earnshaw W.C. 1998. INCENP centromere and spindle targeting: identification of essential conserved motifs and involvement of heterochromatin protein HP1. J. Cell Biol. 143:1763–1774 10.1083/jcb.143.7.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando S., Yang H., Nozaki N., Okazaki T., Yoda K. 2002. CENP-A, -B, and -C chromatin complex that contains the I-type α-satellite array constitutes the prekinetochore in HeLa cells. Mol. Cell. Biol. 22:2229–2241 10.1128/MCB.22.7.2229-2241.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell R.E., Tour O., Palmer A.E., Steinbach P.A., Baird G.S., Zacharias D.A., Tsien R.Y. 2002. A monomeric red fluorescent protein. Proc. Natl. Acad. Sci. USA. 99:7877–7882 10.1073/pnas.082243699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowell I.G., Aucott R., Mahadevaiah S.K., Burgoyne P.S., Huskisson N., Bongiorni S., Prantera G., Fanti L., Pimpinelli S., Wu R., et al. 2002. Heterochromatin, HP1 and methylation at lysine 9 of histone H3 in animals. Chromosoma. 111:22–36 10.1007/s00412-002-0182-8 [DOI] [PubMed] [Google Scholar]
- Crosio C., Fimia G.M., Loury R., Kimura M., Okano Y., Zhou H., Sen S., Allis C.D., Sassone-Corsi P. 2002. Mitotic phosphorylation of histone H3: spatio-temporal regulation by mammalian Aurora kinases. Mol. Cell. Biol. 22:874–885 10.1128/MCB.22.3.874-885.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J., Sullivan B.A., Higgins J.M. 2006. Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev. Cell. 11:741–750 10.1016/j.devcel.2006.09.018 [DOI] [PubMed] [Google Scholar]
- Emanuele M.J., Lan W., Jwa M., Miller S.A., Chan C.S., Stukenberg P.T. 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181:241–254 10.1083/jcb.200710019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W., Tseng B.S., Dormann H.L., Ueberheide B.M., Garcia B.A., Shabanowitz J., Hunt D.F., Funabiki H., Allis C.D. 2005. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 438:1116–1122 10.1038/nature04219 [DOI] [PubMed] [Google Scholar]
- Fuller B.G., Lampson M.A., Foley E.A., Rosasco-Nitcher S., Le K.V., Tobelmann P., Brautigan D.L., Stukenberg P.T., Kapoor T.M. 2008. Midzone activation of aurora B in anaphase produces an intracellular phosphorylation gradient. Nature. 453:1132–1136 10.1038/nature06923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giménez-Abián J.F., Sumara I., Hirota T., Hauf S., Gerlich D., de la Torre C., Ellenberg J., Peters J.M. 2004. Regulation of sister chromatid cohesion between chromosome arms. Curr. Biol. 14:1187–1193 10.1016/j.cub.2004.06.052 [DOI] [PubMed] [Google Scholar]
- Girdler F., Gascoigne K.E., Eyers P.A., Hartmuth S., Crafter C., Foote K.M., Keen N.J., Taylor S.S. 2006. Validating Aurora B as an anti-cancer drug target. J. Cell Sci. 119:3664–3675 10.1242/jcs.03145 [DOI] [PubMed] [Google Scholar]
- Hendzel M.J., Wei Y., Mancini M.A., Van Hooser A., Ranalli T., Brinkley B.R., Bazett-Jones D.P., Allis C.D. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 106:348–360 10.1007/s004120050256 [DOI] [PubMed] [Google Scholar]
- Hirota T., Lipp J.J., Toh B.H., Peters J.M. 2005. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 438:1176–1180 10.1038/nature04254 [DOI] [PubMed] [Google Scholar]
- Jenuwein T., Allis C.D. 2001. Translating the histone code. Science. 293:1074–1080 10.1126/science.1063127 [DOI] [PubMed] [Google Scholar]
- Johansen K.M., Johansen J. 2006. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 14:393–404 10.1007/s10577-006-1063-4 [DOI] [PubMed] [Google Scholar]
- Kanda T., Sullivan K.F., Wahl G.M. 1998. Histone-GFP fusion protein enables sensitive analysis of chromosome dynamics in living mammalian cells. Curr. Biol. 8:377–385 10.1016/S0960-9822(98)70156-3 [DOI] [PubMed] [Google Scholar]
- Kanno T., Kanno Y., Siegel R.M., Jang M.K., Lenardo M.J., Ozato K. 2004. Selective recognition of acetylated histones by bromodomain proteins visualized in living cells. Mol. Cell. 13:33–43 10.1016/S1097-2765(03)00482-9 [DOI] [PubMed] [Google Scholar]
- Kimura H., Cook P.R. 2001. Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J. Cell Biol. 153:1341–1353 10.1083/jcb.153.7.1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Takizawa N., Allemand E., Hori T., Iborra F.J., Nozaki N., Muraki M., Hagiwara M., Krainer A.R., Fukagawa T., Okawa K. 2006. A novel histone exchange factor, protein phosphatase 2Cγ, mediates the exchange and dephosphorylation of H2A–H2B. J. Cell Biol. 175:389–400 10.1083/jcb.200608001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura H., Hayashi-Takanaka Y., Goto Y., Takizawa N., Nozaki N. 2008. The organization of histone H3 modifications as revealed by a panel of specific monoclonal antibodies. Cell Struct. Funct. 33:61–73 10.1247/csf.07035 [DOI] [PubMed] [Google Scholar]
- Kouzarides T. 2007. Chromatin modifications and their function. Cell. 128:693–705 10.1016/j.cell.2007.02.005 [DOI] [PubMed] [Google Scholar]
- Lin C.W., Ting A.Y. 2004. A genetically encoded fluorescent reporter of histone phosphorylation in living cells. Angew. Chem. Int. Ed. Engl. 43:2940–2943 10.1002/anie.200353375 [DOI] [PubMed] [Google Scholar]
- Manders E.M., Kimura H., Cook P.R. 1999. Direct imaging of DNA in living cells reveals the dynamics of chromosome formation. J. Cell Biol. 144:813–821 10.1083/jcb.144.5.813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Fung J.C., Sedat J.W. 1997. Deconstructing the nucleus: global architecture from local interactions. Curr. Opin. Genet. Dev. 7:259–263 10.1016/S0959-437X(97)80136-0 [DOI] [PubMed] [Google Scholar]
- Mayer W., Smith A., Fundele R., Haaf T. 2000. Spatial separation of parental genomes in preimplantation mouse embryos. J. Cell Biol. 148:629–634 10.1083/jcb.148.4.629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellone B.G., Ball L., Suka N., Grunstein M.R., Partridge J.F., Allshire R.C. 2003. Centromere silencing and function in fission yeast is governed by the amino terminus of histone H3. Curr. Biol. 13:1748–1757 10.1016/j.cub.2003.09.031 [DOI] [PubMed] [Google Scholar]
- Mitsuhashi S., Matsuura N., Ubukata M., Oikawa H., Shima H., Kikuchi K. 2001. Tautomycetin is a novel and specific inhibitor of serine/threonine protein phosphatase type 1, PP1. Biochem. Biophys. Res. Commun. 287:328–331 10.1006/bbrc.2001.5596 [DOI] [PubMed] [Google Scholar]
- Monier K., Mouradian S., Sullivan K.F. 2007. DNA methylation promotes Aurora-B-driven phosphorylation of histone H3 in chromosomal subdomains. J. Cell Sci. 120:101–114 10.1242/jcs.03326 [DOI] [PubMed] [Google Scholar]
- Murata-Hori M., Tatsuka M., Wang Y.L. 2002. Probing the dynamics and functions of aurora B kinase in living cells during mitosis and cytokinesis. Mol. Biol. Cell. 13:1099–1108 10.1091/mbc.01-09-0467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murnion M.E., Adams R.R., Callister D.M., Allis C.D., Earnshaw W.C., Swedlow J.R. 2001. Chromatin-associated protein phosphatase 1 regulates aurora-B and histone H3 phosphorylation. J. Biol. Chem. 276:26656–26665 10.1074/jbc.M102288200 [DOI] [PubMed] [Google Scholar]
- Nakayasu H., Berezney R. 1989. Mapping replicational sites in the eucaryotic cell nucleus. J. Cell Biol. 108:1–11 10.1083/jcb.108.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters A.H., Kubicek S., Mechtler K., O’Sullivan R.J., Derijck A.A., Perez-Burgos L., Kohlmaier A., Opravil S., Tachibana M., Shinkai Y., et al. 2003. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol. Cell. 12:1577–1589 10.1016/S1097-2765(03)00477-5 [DOI] [PubMed] [Google Scholar]
- Peters J.M., Tedeschi A., Schmitz J. 2008. The cohesin complex and its roles in chromosome biology. Genes Dev. 22:3089–3114 10.1101/gad.1724308 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Sabbattini P., Canzonetta C., Sjoberg M., Nikic S., Georgiou A., Kemball-Cook G., Auner H.W., Dillon N. 2007. A novel role for the Aurora B kinase in epigenetic marking of silent chromatin in differentiated postmitotic cells. EMBO J. 26:4657–4669 10.1038/sj.emboj.7601875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos F., Peters A.H., Otte A.P., Reik W., Dean W. 2005. Dynamic chromatin modifications characterise the first cell cycle in mouse embryos. Dev. Biol. 280:225–236 10.1016/j.ydbio.2005.01.025 [DOI] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L.B., Schneider T.R., Stukenberg P.T., Musacchio A. 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell. 18:379–391 10.1016/j.molcel.2005.03.031 [DOI] [PubMed] [Google Scholar]
- Terada Y., Tatsuka M., Suzuki F., Yasuda Y., Fujita S., Otsu M. 1998. AIM-1: a mammalian midbody-associated protein required for cytokinesis. EMBO J. 17:667–676 10.1093/emboj/17.3.667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G., Lens S.M.A. 2008. The Aurora kinase family in cell division and cancer. Biochim. Biophys. Acta. 1786:60–72 [DOI] [PubMed] [Google Scholar]
- Wei Y., Yu L., Bowen J., Gorovsky M.A., Allis C.D. 1999. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 97:99–109 10.1016/S0092-8674(00)80718-7 [DOI] [PubMed] [Google Scholar]
- Yamagata K., Suetsugu R., Wakayama T. 2009. Long-term, six-dimensional live-cell imaging for the mouse preimplantation embryo that does not affect full-term development. J. Reprod. Dev. 55:343–350 10.1262/jrd.20166 [DOI] [PubMed] [Google Scholar]
- Yamagishi Y., Sakuno T., Shimura M., Watanabe Y. 2008. Heterochromatin links to centromeric protection by recruiting shugoshin. Nature. 455:251–255 10.1038/nature07217 [DOI] [PubMed] [Google Scholar]
- Yamazaki T., Yamagata K., Baba T. 2007. Time-lapse and retrospective analysis of DNA methylation in mouse preimplantation embryos by live cell imaging. Dev. Biol. 304:409–419 10.1016/j.ydbio.2006.12.046 [DOI] [PubMed] [Google Scholar]
- Yokoya F., Imamoto N., Tachibana T., Yoneda Y. 1999. β-catenin can be transported into the nucleus in a Ran-unassisted manner. Mol. Biol. Cell. 10:1119–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]