mTor, acting mainly via mTORC1, controls dystrophin transcription in a raptor- and rictor-independent mechanism.
Abstract
Mammalian target of rapamycin (mTOR) is a key regulator of cell growth that associates with raptor and rictor to form the mTOR complex 1 (mTORC1) and mTORC2, respectively. Raptor is required for oxidative muscle integrity, whereas rictor is dispensable. In this study, we show that muscle-specific inactivation of mTOR leads to severe myopathy, resulting in premature death. mTOR-deficient muscles display metabolic changes similar to those observed in muscles lacking raptor, including impaired oxidative metabolism, altered mitochondrial regulation, and glycogen accumulation associated with protein kinase B/Akt hyperactivation. In addition, mTOR-deficient muscles exhibit increased basal glucose uptake, whereas whole body glucose homeostasis is essentially maintained. Importantly, loss of mTOR exacerbates the myopathic features in both slow oxidative and fast glycolytic muscles. Moreover, mTOR but not raptor and rictor deficiency leads to reduced muscle dystrophin content. We provide evidence that mTOR controls dystrophin transcription in a cell-autonomous, rapamycin-resistant, and kinase-independent manner. Collectively, our results demonstrate that mTOR acts mainly via mTORC1, whereas regulation of dystrophin is raptor and rictor independent.
Introduction
Muscle structure, mass, and composition are critical for motility, whole body metabolism, and viability. Skeletal muscle is composed of heterogeneous myofibers with distinct metabolic properties, rates of contraction, and susceptibility to fatigue (Bassel-Duby and Olson, 2006; Schiaffino et al., 2007) but exhibits remarkable metabolic and morphological adaptive capabilities in response to several physiological (e.g., exercise) and pathological (systemic diseases, myopathy, and aging) conditions. These adaptive processes include hypertrophy, atrophy, regeneration, fiber type conversion, or mitochondrial biogenesis.
As a conserved Ser/Thr kinase, the mammalian target of rapamycin (mTOR) is a central regulator of cell growth by integrating signals from nutrients, growth factors, energy status, and environmental stress. mTOR resides in two structurally and functionally distinct signaling complexes: mTOR complex 1 (mTORC1) and mTORC2. The raptor-containing complex mTORC1 regulates a vast range of cellular activities, including transcription, translation, ribosome biogenesis, and autophagy (Wullschleger et al., 2006). A key cellular substrate for mTORC1 is the ribosomal S6 kinase (S6K1), which is phosphorylated at its hydrophobic motif residue, Thr389. The rictor-containing complex mTORC2 is proposed to regulate actin organization. Moreover, mTORC2 is one of the kinases able to phosphorylate PKB/Akt on Ser473 (Jacinto et al., 2004; Sarbassov et al., 2004; Bentzinger et al., 2008).
Under acute treatment, rapamycin is thought to selectively inhibit mTORC1. Conversely, mTORC2 is considered rapamycin insensitive, although prolonged treatment disrupts mTORC2 assembly in certain cell lines (Sarbassov et al., 2006). In addition, the recent development of the ATP-competitive inhibitor Torin1, which suppresses both mTORC1 and mTORC2, allowed the identification of rapamycin-resistant mTORC1-dependent functions required for cap-dependent translation and suppression of autophagy (Thoreen et al., 2009).
Rapamycin-sensitive mTORC1 functions have been shown to be required for muscle growth (Bodine et al., 2001; Rommel et al., 2001; Ohanna et al., 2005) as well as for the maintenance of mitochondrial oxidative function by directly regulating mitochondrial gene expression through the control of a YY1 (Yin Yang 1)–PGC1α transcriptional complex (Cunningham et al., 2007). Consistent with this, muscle-specific inactivation of the mTORC1 component raptor in raptor muscle knockout (RAmKO) mice leads to muscle atrophy, impaired oxidative capacity, and increased glycogen stores, resulting in dystrophic features that were most prominent in oxidative muscles (Bentzinger et al., 2008). In contrast, the muscle-specific loss of the mTORC2 component rictor in rictor muscle knockout (RImKO) mice has minimal impact on muscle physiology (Bentzinger et al., 2008; Kumar et al., 2008), and raptor/rictor muscle double knockout (DmKO) show similar pathological changes as RAmKO mice. These studies suggested that mTOR functions in adult skeletal muscle require only mTORC1 (Bentzinger et al., 2008). However, there is evidence that some functions of mTOR, such as the activation of terminal oligopyrimidine mRNA translation, could be independent of mTORC1 and mTORC2 (Patursky-Polischuk et al., 2009).
The aim of this study was to determine the physiological functions of mTOR in differentiated mouse skeletal muscle. Because conventional mTOR knockout mice die during early embryogenesis (Gangloff et al., 2004; Murakami et al., 2004), we generated muscle-specific mTOR knockout (mTOR−) mice. In this study, we show that these animals develop a severe myopathy, displaying features of muscular dystrophy (MD) and metabolic myopathy, leading to premature death between 22 and 38 wk of age. Although the metabolic alterations are similar between mTOR− and RAmKO/DmKO mice, mTOR deficiency more severely affects muscle contractile properties. In addition, mTOR− muscles, in contrast to RAmKO and RImKO muscles, display reduced content of components of the dystrophin–glycoprotein complex (DGC). Finally, we demonstrate that mTOR controls dystrophin transcription in a cell-autonomous, rapamycin-resistant, and kinase-independent manner.
Results
Muscle-specific inactivation of mTOR leads to premature death
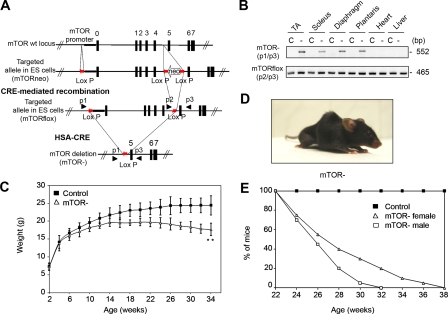
To investigate the role of mTOR in skeletal muscle development and function, we developed a conditional mTOR gene inactivation strategy based on the Cre-loxP system (Fig. 1 A). mTORflox/flox mice were crossed with human skeletal muscle α-actin (HSA)–Cre mice in which Cre-mediated recombination occurs in postmitotic myofibers but not in satellite cells (Nicole et al., 2003). At birth, Cre-positive mTORflox/flox (mTOR−) pups were viable, born at expected Mendelian ratios, and indistinguishable from their control littermates. PCR analysis confirmed the efficient deletion of the mTOR allele specifically in skeletal muscle (Fig. 1 B). At ∼4 wk of age, the growth rate of mutant animals started to decrease (Fig. 1 C). By the age of 13 wk, mutant mice started to develop spinal deformity, which increased with age in the form of kyphosis, a sign of muscle weakness, accompanied by breathing difficulties and abnormal posture of the hindlimbs (Fig. 1 D). mTOR− mice eventually died between 22 and 38 wk of age (Fig. 1 E).
Figure 1.
Muscle-specific inactivation of mTOR. (A) mTORflox and mTOR− mice were generated as described in Materials and methods. (B) PCR analysis of Cre-mediated recombination of the mTORflox allele (p2/p3) showing that the deleted mTOR allele (p1/p3) was exclusively detected in skeletal muscles of mTOR− mice. C, control; −, mTOR−. (C) Growth curves of mTOR− and control female mice (n = 15 mice). (D) Morphology of 22-wk-old female mTOR− mice. (E) Survival curve of mTOR− and control mice (n = 20). **, P < 0.01. Data indicate mean ± SD.
mTOR− mice develop a progressive MD
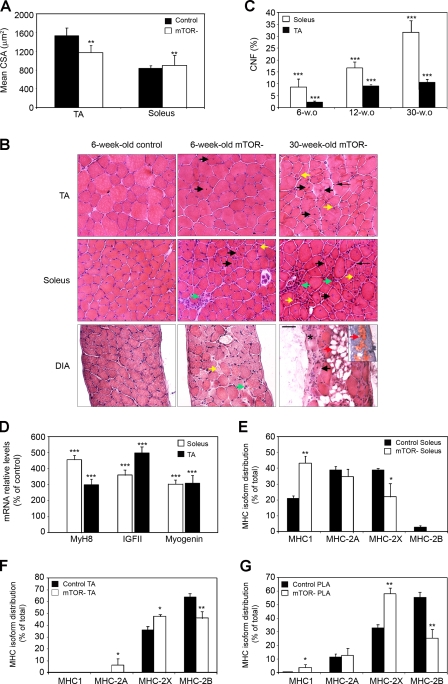
We next characterized muscles from mTOR− mice by morphometric and histological examinations. By the age of 6 wk, the body weight of mTOR− mice was reduced by ∼10% compared with controls. This reduction could be attributed to a strong decrease in the mass of fast-twitch glycolytic muscles such as tibialis anterior (TA), gastrocnemius (GC), and plantaris (PLA; Table I). In contrast, the mass of the slow-twitch oxidative soleus muscle was not significantly affected by mTOR deficiency and was even increased after normalization to body weight. Of note, the weight of other organs remained unaffected in mTOR− mice. At this age, fiber number was not significantly different between mTOR− and control muscles (unpublished data), excluding hypoplasia as the cause of reduced mass in fast-twitch muscles. However, the reduction in mTOR-TA muscle mass was associated with a 24% decrease of the mean cross-sectional area (CSA), whereas mTOR− soleus muscle displayed a 6% increase of the mean CSA (Fig. 2 A). In addition, mTOR− muscles showed characteristic dystrophic features that were exacerbated in oxidative muscles (Fig. 2 B). These included degeneration with phagocytosis and mononuclear cell infiltration (Fig. 2 B, green arrows), variation in fiber size with small atrophic fibers (Fig. 2 B, yellow arrows), interfiber connective tissue (Fig. 2 B, thin arrows), and a marked number of regenerated muscle fibers with centrally located nuclei (Fig. 2 B, black arrows) that increased with age (Fig. 2 C). Ongoing muscle regeneration was further confirmed at the molecular level by the activation of the expression of perinatal muscle myosin heavy chain (MHC) MyH8, IGF-II, and myogenin (Fig. 2 D). Dystrophic hallmarks were prominent in the diaphragm, which additionally showed fibrosis (Fig. 2 B, asterisk) and fatty infiltration (Fig. 2 B, red arrows), suggesting that respiratory failure might be the cause of premature death.
Table I.
mTOR− mice exhibit reduced body weight associated with a strong reduction in the mass of fast-twitch glycolytic muscles
| Variable | Control | mTOR− | % Control |
| Body weight (g) | 17.41 ± 1.05 | 15.85 ± 1.37a | 91.0 |
| Soleus weight (mg) | 4.93 ± 0.64 | 5.00 ± 0.61 | 101.4 |
| Soleus/body weight (mg/g) | 0.28 ± 0.03 | 0.32 ± 0.03b | 114.3 |
| TA weight (mg) | 30.93 ± 3.56 | 24.54 ± 2.29a | 79.3 |
| TA/body weight (mg/g) | 1.78 ± 0.22 | 1.55 ± 0.04a | 87.1 |
| GC weight (mg) | 71.13 ± 5.23 | 56.27 ± 4.79a | 79.1 |
| GC/body weight (mg/g) | 4.09 ± 0.26 | 3.55 ± 0.07a | 86.8 |
| PLA weight (mg) | 10.81 ± 0.53 | 8.37 ± 0.85a | 77.4 |
| PLA/body weight (mg/g) | 0.62 ± 0.03 | 0.53 ± 0.06a | 85.5 |
| Liver (mg) | 868.26 ± 53.77 | 851.84 ± 102.75 | 98.1 |
| Liver/body weight (mg/g) | 50.05 ± 4.50 | 53.69 ± 3.40 | 107.3 |
| Fat pad weight (mg) | 295.78 ± 40.15 | 286.73 ± 30.12 | 96.9 |
| Fat pad/body weight (mg/g) | 16.98 ± 1.90 | 18.08 ± 1.15 | 106.9 |
| Heart weight (mg) | 120.62 ± 9.49 | 114.17 ± 9.07 | 94.7 |
| Heart/body weight (mg/g) | 6.95 ± 0.68 | 7.22 ± 0.50 | 103.9 |
Values are for 6-wk-old control and mTOR− female mice. Fat data includes ovarian, uterine, and retroperitoneal fat pads. Data indicate mean ± SD (n > 5 mice/group).
P < 0.01.
P < 0.05.
Figure 2.
mTOR− mice develop a progressive MD. (A) Analysis of the mean fiber CSA in TA and soleus muscles from 6-wk-old mice (n = 4). (B) H&E-stained transverse sections of TA, soleus, and diaphragm (DIA) muscles from control and mTOR− mice. Degeneration with phagocytosis and mononuclear cell infiltration (green arrows), variation in fiber size with small atrophic fibers (yellow arrows), interfiber connective tissue (thin arrows), regenerated muscle fibers with centrally located nuclei (black arrows), fibrosis (asterisk), and fatty infiltration (red arrows) are shown. Adipogenic differentiation was confirmed by oil red O staining as shown in the inset. Bar, 50 µm. (C) Percentage of centrally nucleated fibers (CNF) increases with age in mTOR− muscles. (D) Relative mRNA levels of MyH8, IGF-II, and myogenin in mTOR− muscles from 6-wk-old mice. (E–G) Graphs show the percentage distribution of MHC isoforms I, 2B, 2X, and 2A in soleus (E), TA (F), and PLA (G) muscles from 6-wk-old control and mTOR− mice. (C–G) n = 5 sample sets. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data indicate mean ± SEM.
To examine a potential relationship between the prominent dystrophic features in oxidative muscles and fiber types, we analyzed the composition of MHC isoforms in whole muscles from 6-wk-old mice. In soleus the shift was from MHC-2X toward MHC-1 (Fig. 2 E and Fig. S1), in TA from MHC-2B toward MHC-2X and -2A (Fig. 2 F), and in PLA from MHC-2B toward MHC-2X and MHC-1 (Fig. 2 G). Therefore, loss of mTOR induces a significant shift toward the expression of slower MHC isoforms, indicating contractile dysfunctions in all types of muscles.
mTOR deficiency more severely affects muscle functional properties than loss of raptor
To assess the functional consequences of mTOR inactivation in skeletal muscle, the muscle contractile performance was analyzed in response to nerve and/or muscle stimulation in 9-wk-old mice (Table II). Both absolute maximum tetanic (Po) and twitch (Pt) forces were significantly decreased in mTOR− soleus and TA muscles compared with control muscles. Stimulation of mTOR− TA directly or via its nerve gave rise to a similar reduction in absolute Po, indicating that neurotransmission was not altered in mTOR− mice. The reduction in force production was more pronounced in mTOR− TA because of the large decrease in muscle mass observed in fast-twitch muscles. Indeed, when Po was normalized for TA muscle mass or for soleus muscle CSA, the specific maximum tetanic force (sPo) was reduced to the same extent (by ∼30%) in both mTOR− TA and soleus muscles, further indicating that muscle weakness of mTOR− mice is not only caused by a loss of contractile muscle mass. In contrast, loss of raptor in muscles from age-matched RAmKO mice did not significantly alter sPo.
Table II.
Comparison of muscle contractile properties between mTOR− and RAmKO mice
| Mice | Control | mTOR−/RAmKO | % Control |
| Soleus muscle | |||
| mTOR | |||
| Mass (mg) | 7.6 ± 0.4 | 7.5 ± 0.3 | 98.7 |
| Po (mN)a | 141.6 ± 33.2 | 104.3 ± 24.0c | 73.6 |
| sPo (mN/mm2)a | 187.9 ± 42.0 | 134.5 ± 37.1c | 71.6 |
| Pt (mN)a | 26.4 ± 3.7 | 16.2 ± 4.1d | 61.4 |
| TTP (ms)a | 82.7 ± 11.1 | 171.5 ± 73.8d | 207.4 |
| RT50% (ms)a | 131.7 ± 45.3 | 375.4 ± 228.2c | 285.0 |
| F20% (ms)a | 85.1 ± 14.5 | 41.1 ± 18.9d | 48.3 |
| Raptor | |||
| Mass (mg) | 7.1 ± 0.9 | 6.5 ± 0.9 | 90.9 |
| Po (mN)a | 155.6 ± 24.6 | 141.7 ± 11.0 | 91.0 |
| sPo (mN/mm2)a | 211.5 ± 38.7 | 212.4 ± 19.3 | 100.4 |
| TA muscle | |||
| mTOR | |||
| Mass (mg) | 37.5 ± 1.2 | 32.7 ± 2.2d | 87.2 |
| Po (mN)a | 536.3 ± 100.7 | 332.0 ± 55.4d | 61.9 |
| Po (mN)b | 567.1 ± 79.1 | 359.7 ± 34.8d | 63.4 |
| sPo (mN/mg)b | 15.1 ± 2.2 | 11.0 ± 1.4c | 73.0 |
| Pt (mN)b | 106.4 ± 25.8 | 60.5 ± 9.5d | 56.7 |
| TTP (ms)b | 31.5 ± 2.4 | 42.4 ± 9.3 | 134.6 |
| RT50% (ms)b | 33.2 ± 3.8 | 58.0 ± 10.9d | 174.7 |
| Deficit (%)b | −12.7 ± 5.0 | −56.0 ± 12.9d | −340.9 |
| Raptor | |||
| Mass (mg) | 41.0 ± 2.4 | 36.2 ± 1.7d | 88.5 |
| Po (mN)b | 607.1 ± 35.7 | 497.0 ± 55.2d | 81.9 |
| sPo (mN/mg)b | 14.9 ± 1.4 | 13.8 ± 1.8 | 92.5 |
| Deficit (%)b | −12.0 ± 3.5 | −12.2 ± 7.3 | 1.5 |
Muscle contractile properties of 9-wk-old mTOR and raptor mice were assessed by recording isometric forces in response to electrical stimulation. The following data were measured: muscle mass, Po, sPo, Pt, TTP, RT50%, time taken for maximal twitch tension to fall by 20% (F20%), and the percentage of force deficit following repetitive eccentric contractions (deficit). The data indicate mean ± SD (n > 4 mice/group).
Muscle electrical stimulation.
Nerve electrical stimulation.
P < 0.05.
P < 0.01.
Moreover, mTOR− muscles contracted and relaxed slowly as shown by the increased time to peak twitch tension (TTP) and one-half relaxation time (RT50%), which is consistent with the MHC distribution toward slower isoforms. Furthermore, mTOR− soleus muscle was twofold less resistant to fatigue (F20%) than control muscle, whereas RAmKO muscles were more resistant to fatigue (Bentzinger et al., 2008).
We next tested whether mTOR or raptor deficiency impacts resistance to contraction-induced injury in situ by subjecting TA muscles to a series of high stress muscle-eccentric contractions. Importantly, loss of mTOR but not of raptor resulted in increased force deficit, indicating a higher susceptibility to injury of mTOR− muscle. Collectively, these data show that mTOR deficiency more severely affects muscle contractile properties than loss of raptor.
mTOR− muscles display reduced levels of DGC components and induction of utrophin
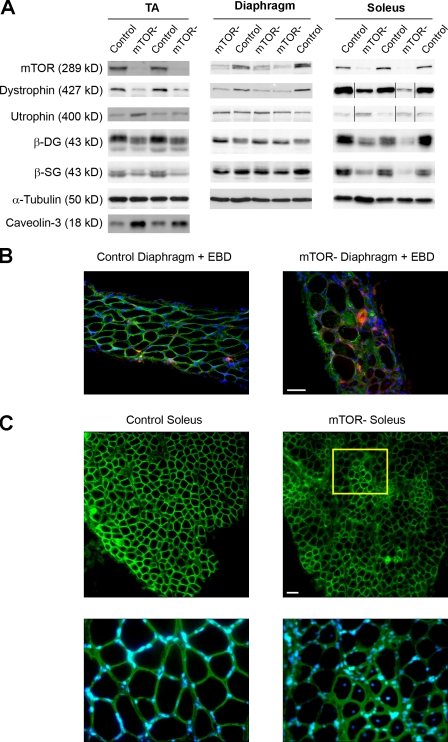
Most forms of MD arise from mutations in the DGC, which provides a physical link between the extracellular matrix and the intracellular cytoskeleton (Durbeej and Campbell, 2002). Force deficits after eccentric contractions are a functional hallmark of dystrophin-deficient muscle (Watchko et al., 2002). This observation prompted us to examine dystrophin levels in mTOR− muscles. Western blot analysis revealed a strong reduction in muscle dystrophin content in 6-wk-old mTOR− mice (Fig. 3 A and Table S1). In Duchenne MD (DMD) and the mdx mouse model for DMD, loss of dystrophin is associated with the concurrent down-regulation of other DGC members (Ohlendieck and Campbell, 1991) and with a compensatory up-regulation of its autosomal homologue utrophin (Tinsley et al., 1998). mTOR− muscles also showed decreased expression of several DGC components, including β-sarcoglycan and β-dystroglycan, and induction of utrophin (Fig. 3 A and Table S1). Dystrophin content was further reduced in mutant soleus and TA muscles from 11-wk-old mice, reaching 13 and 17% of the levels in control muscles (Fig. S2 and Table S2). In contrast, expression of caveolin-3 and calpain 3, whose deficiency leads to additional forms of MD (Watchko et al., 2002), was significantly up-regulated.
Figure 3.
Reduced DGC amount and utrophin induction in mTOR− muscles. (A) Soleus, diaphragm, and TA muscle extracts from 6-wk-old mice were immunoblotted with the indicated antibodies to examine dystrophy-related protein levels. Black lines indicate that intervening lanes have been spliced out. (B) Rare Evans blue–positive fibers can be detected in mTOR− diaphragms from 30-wk-old mice. (C) Dystrophin immunostaining (green) and Hoechst staining (blue) on soleus muscle sections from 11-wk-old control and mTOR− mice showing sarcolemmal localization of residual dystrophin. A higher magnification view (inset) shows that dystrophin expression is reduced in noncentronucleated fibers, whereas it is normal in regenerating centronucleated fibers. Bars, 50 µm.
The DGC plays a major role in regulating membrane integrity, and its loss leads to sarcolemmal fragility and permeability (Straub et al., 1997). However, the overall sarcolemmal integrity of quiescent mTOR− muscle fibers was maintained because rare muscle fibers exhibited Evans blue dye uptake (Fig. 3 B), and elevated creatine kinase serum levels were not detected in mTOR− mice (not depicted). Importantly, immunofluorescence analysis showed that residual dystrophin was still present along the sarcolemma of the noncentronucleated fibers (Fig. 3 C). Thus, it is likely that residual DGC may modulate membrane damage in mTOR− muscles.
mTOR controls dystrophin expression in a cell-autonomous, rapamycin-resistant, and kinase-independent manner
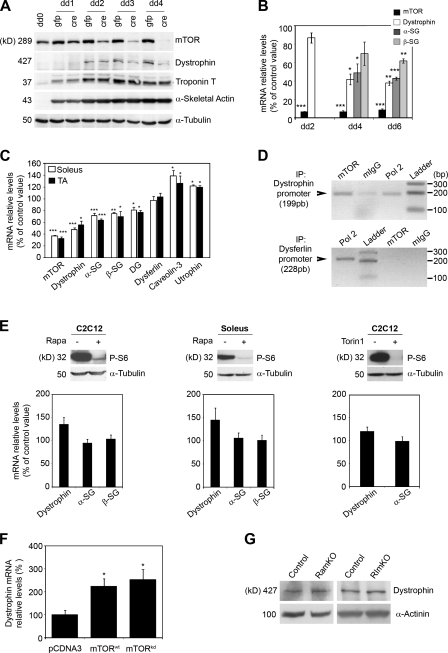
To gain better insight into the regulation of dystrophin by mTOR, we performed mTOR knockdown in primary myotubes derived from mTORflox mice infected with Cre-expressing adenovirus (Fig. 4 A). Although mTOR inactivation in cultured myotubes did not prevent terminal differentiation as shown by the induction of the late myogenic differentiation markers α-skeletal actin and troponin T, it led to a rapid and strong down-regulation of the dystrophin protein. These results demonstrate that the mTOR cell autonomously regulates dystrophin levels.
Figure 4.
mTOR controls dystrophin expression. (A) Time course for mTOR inactivation, dystrophin down-regulation, and induction of terminal differentiation markers in cultured mTORflox myotubes. The myotubes were transduced by CMV-GFP or CMV-Cre adenovirus on differentiation day 1 (dd1), harvested at a different time point after differentiation, and immunoblotted with the indicated antibodies. (B) Relative mRNA levels of mTOR and the indicated DGC components in mTORflox myotubes transduced by Ad-Cre (n = 3). (C) Relative mRNA levels of mTOR and the indicated dystrophy-related genes in muscles from 6-wk-old mice (n = 5). (D) Detection of mTOR on the dystrophin promoter by ChIP assay. Antibodies against mTOR (lane 1), normal mouse IgG (lane 2), or RNA polymerase 2 (lane 3) were used to immunoprecipitate (IP) a mouse TA chromatin extract. The precipitated DNA was analyzed by PCR using primers for the dystrophin (top) or dysferlin (bottom) promoter. (E, top) Western blot analysis showing the phosphorylation status of S6 protein in C2C12 myotubes in the absence (−) or presence (+) of 20 nM rapamycin or 250 nM Torin1 for 48 h from differentiation day 4. For soleus muscle, C57BL/6 mice were daily injected intraperitoneally with 2.5 mg/kg rapamycin or vehicle for 8 d. (bottom) mRNA levels for the indicated DGC components in C2C12 myotubes and soleus muscle in absence or presence of the indicated mTOR inhibitors. (F) Relative mRNA levels of dystrophin in the TA muscle fibers of mTOR− mice 12 d after in vivo coelectroporation of rat wild-type (wt) or kinase-dead (kd) mTOR and pRNAT-GFP vector. Graphs show means versus pcDNA3-electroporated control (n = 5 mice/group). (G) Western blot analysis showing dystrophin protein levels in soleus muscle from 14-wk-old RAmKO and RImKO mice. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data indicate mean ± SEM.
We next assessed whether reduced dystrophin content reflects changes at the mRNA levels. In cultured mTOR− myotubes and muscles, mRNA levels for several DGC components were significantly reduced as compared with control levels, indicating possible transcriptional defects (Fig. 4, B and C). In contrast, no change was detected for dysferlin, whereas caveolin-3 and utrophin were up-regulated.
Because mTOR was shown to interact and positively regulate the activity of the transcription factor YY1 in myotubes (Cunningham et al., 2007) and YY1 was shown to bind the dystrophin promoter (Galvagni et al., 1998), we hypothesized that mTOR should bind to the dystrophin promoter. To test our hypothesis, we performed chromatin immunoprecipitation (ChIP) analysis on TA muscle from control mice (Fig. 4 D). We found that mTOR is bound to the promoter of the dystrophin gene at the same extent as RNA polymerase II, whereas it was not bound to the dysferlin promoter. These results strongly suggest that mTOR directly regulates the dystrophin promoter.
We next determined whether dystrophin regulation involved rapamycin-sensitive mTOR functions and mTOR kinase activity. In cultured C2C12 myotubes and mouse soleus muscle, neither rapamycin nor Torin1 treatment reduced dystrophin mRNA levels, although both mTOR inhibitors efficiently inhibited mTORC1 signaling (Fig. 4 E). Consistently, overexpression of a rat mTOR wild type or mTOR kinase dead into the TA muscle of mTOR− mice by in vivo DNA electroporation significantly increased dystrophin mRNA levels by 2.1 and 2.5 times, respectively, as compared with control (Fig. 4 F). In agreement with these observations, dystrophin levels were not reduced in muscles lacking raptor and rictor (Fig. 4 G). Collectively, these findings demonstrate that mTOR controls dystrophin expression in a rapamycin-resistant and kinase-independent manner independently of raptor and rictor.
MD in mTOR− mice is associated with a down-regulation of mTORC1 signaling and hyperactivation of PKB/Akt
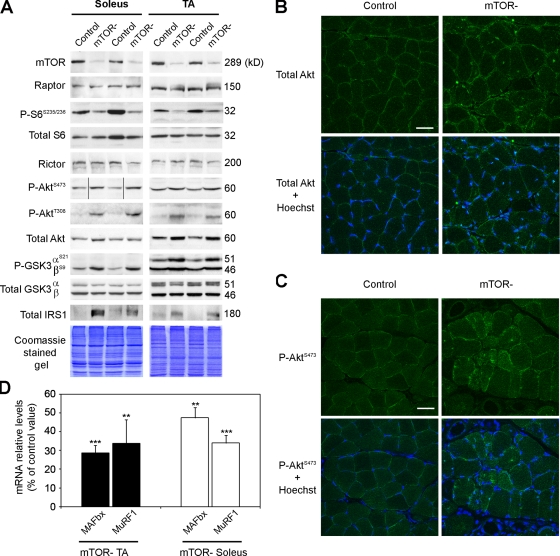
We next investigated the consequences of mTOR depletion on signal transduction in mutant muscles. Western blot analysis revealed that mTOR protein level was strongly decreased in mutant muscles (Fig. 5 A and Table S3). Residual mTOR expression likely arose from nonmuscle cells as well as from cells in which the mTOR allele was not recombined, including newly repaired muscle fibers and satellite cells. In contrast, expression of the specific mTORC1 and mTORC2 components, raptor and rictor, respectively, remained unchanged. mTORC1 and mTORC2 activity in mutant muscles was next assessed by monitoring phosphorylation of S6 and PKB/Akt, respectively. Decreased phosphorylation of S6 was consistent with down-regulation of mTORC1. Surprisingly, phosphorylation of the mTORC2 downstream target PKB/Akt was strongly increased on Ser473. This observation implies the existence of a distinct kinase from mTOR able to phosphorylate PKB/Akt at this residue as previously suggested (Bentzinger et al., 2008). In addition, PDK1-dependent phosphorylation of PKB/Akt on Thr308 was also increased. Increased PKB/Akt activation in mTOR− muscle could result from the suppression of the inhibitory feedback from S6K on insulin signaling. The negative feedback involves S6K phosphorylation of IRS-1 leading to its degradation, thereby suppressing insulin signaling (Um et al., 2006). Consistent with the loss of negative feedback, IRS-1 protein levels were strongly increased in mTOR− muscles (Fig. 5 A). Interestingly, hyperactivation of PKB/Akt was accompanied by an up-regulation of its protein level, a feature that has been previously reported in both DMD and mdx muscles, irrespective of the muscle pathogenic state (Dogra et al., 2006). Consistently, immunostaining for PKB/Akt (Fig. 5 B) and P-PKB/AktS473 (Fig. 5 C) was stronger in regenerating centronucleated and nonregenerating mTOR− muscle fibers compared with control fibers.
Figure 5.
Signal transduction in mTOR− muscles. (A) Muscle extracts from 6-wk-old mice were immunoblotted with the indicated antibodies to examine mTORC1 and mTORC2 signaling in soleus and TA muscles. Black lines indicate that intervening lanes have been spliced out. (B and C) Immunofluorescence staining for total PKB/Akt (B) and P-PKB/Akt (C; green) and Hoechst staining (blue) on control and mTOR− soleus muscle sections. Bars, 50 µm. (D) Relative mRNA levels of MuRF1 and MAFbx in mTOR− soleus and TA muscles. **, P < 0.01; ***, P < 0.001. Data indicate means versus control ± SEM (n = 5 sample sets).
PKB/Akt has been shown to phosphorylate and inactivate the forkhead box transcription factors FoxO1 and FoxO3, thereby blocking the transcriptional up-regulation of the atrophy-related genes MAFbX and MuRF1 (Stitt et al., 2004). Consistent with PKB/Akt hyperactivation, both MaFbX and MuRF1 mRNA were down-regulated in mTOR− muscles (Fig. 5 D), indicating that the atrophy observed in mutant muscle is not associated with activation of the ubiquitin proteasome pathway. In addition, PKB/Akt phosphorylates and inactivates GSK3, an inhibitory kinase of glycogen synthase (Cross et al., 1997). Accordingly, phosphorylation of the regulatory residues Ser9 in GSK3β and/or Ser21 in GSK3α was increased in mTOR− muscles (Fig. 5 A).
Loss of mTOR leads to increased muscle glucose uptake and glycogen accumulation
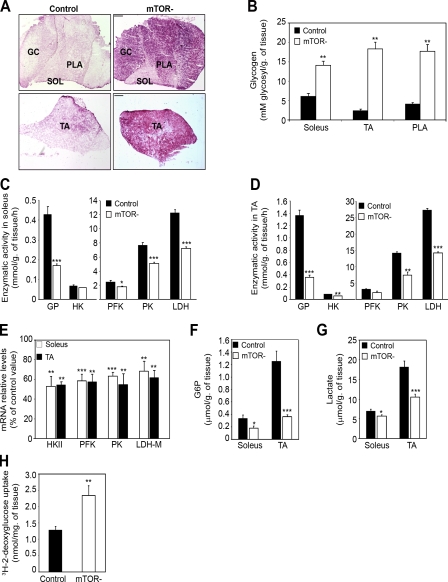
Inactivation of GSK3 by PKB/Akt was shown to promote glycogen synthesis (Cross et al., 1997), suggesting that glycogen synthesis might be increased in mTOR− muscles. Consistent with this, periodic acid Schiff staining demonstrated that glycogen content was strongly increased in mTOR− muscles (Fig. 6 A). Quantification of glycogen levels showed a 2.3-, 7.5-, and 4.3-fold increase in mTOR− soleus, TA, and PLA muscles, respectively, as compared with control muscles (Fig. 6 B). We next assessed whether increased glycogen content in mTOR− muscles could also result from diminished glycogenolysis and glycolytic capacity. Indeed, the activity of glycogen phosphorylase (GP), hexokinase, phosphofructokinase (PFK), pyruvate kinase (PK), and lactate dehydrogenase (LDH) was consistently reduced in mTOR− soleus (Fig. 6 C) and TA muscles (Fig. 6 D). In addition, reduced activity of glycolytic enzymes in mTOR− muscles was associated with a significant reduction in their expression (Fig. 6 E). Consistent with diminished glycolytic flow, reduced levels of glucose-6-phosphate (Fig. 6 F) and lactate (Fig. 6 G) were measured in mTOR− muscles. Importantly, this decrease was not caused by defects in glucose uptake because basal 2-deoxyglucose uptake was even increased 1.8-fold into isolated mTOR− soleus muscle compared with controls (Fig. 6 H).
Figure 6.
Altered glucose usage and glycogen accumulation in mTOR− muscle. (A) Representative periodic acid Schiff staining of soleus (SOL), PLA, GC, and TA muscle sections from 6-wk-old mice showing glycogen accumulation in mTOR− muscles. Bar, 400 µm. (B) Quantification of muscle glycogen content (n = 5 sample sets). (C and D) GP and glycolytic enzyme activity in soleus (C) and TA (D) muscles (control, n = 5; mTOR−, n = 4). (E) Relative mRNA levels of the indicated glycolytic enzymes in mTOR− muscles (n = 5 sample sets). (F) Quantification of intramuscular glucose-6-phosphate (G6P) in control and mTOR− mice. (G) Quantification of intramuscular lactate in control and mTOR− mice. (H) Enhanced basal glucose uptake in isolated soleus muscle from 5-wk-old control and mTOR− mice (n = 5–6 mice). (F and G) n = 8 sample sets. HK, hexokinase. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Data indicate mean ± SEM.
mTOR deficiency in muscle alters mitochondrial regulation and oxidative capacity
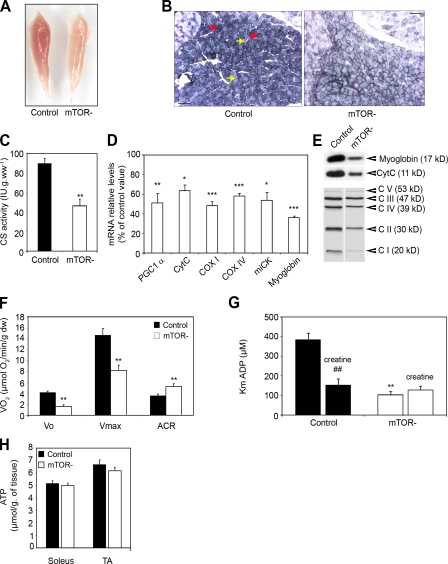
At the macroscopic level, mTOR− muscles appeared paler than control muscles. This was particularly striking in mTOR− soleus muscles, which lacked the red color characteristic of oxidative muscles (Fig. 7 A). Moreover, staining for succinate dehydrogenase, a marker of mitochondrial complex II, was weak in mTOR− soleus fibers to the extent that the difference in oxidative activity classically observed between fiber types was barely detectable (Fig. 7 B). These observations suggested changes to have taken place in the metabolic properties of mTOR− soleus muscle. Recent evidence indicates that mTORC1 controls mitochondrial biogenesis and respiration, notably via YY1 and PGC1α (Cunningham et al., 2007; Bentzinger et al., 2008). In agreement, measurement of citrate synthase activity indicated a twofold reduction in mitochondrial content (Fig. 7 C), and the expression of several genes involved in oxidative metabolism was markedly reduced in mTOR− soleus muscle (Fig. 7, D and E).
Figure 7.
Impaired oxidative metabolism in mTOR− soleus muscle. (A) mTOR− soleus muscle is paler than control muscle. (B) Succinate dehydrogenase histochemical staining demonstrating alterations of oxidative metabolism in mTOR− soleus muscle from 6-wk-old mice. Oxidative and glycolytic fibers are indicated by red and yellow arrows, respectively. Bar, 100 µm. (C) Decreased citrate synthase (CS) activity in mTOR− soleus muscle (n = 5 sample sets). (D) Relative mRNA levels of genes involved in oxidative energy production in mTOR− soleus muscle (n = 5 sample sets). (E) Western blot analysis showing reduced protein levels for myoglobin, cytochrome c (cytc), and respiratory chain components (CI–CV) in mTOR− soleus muscle. Equal protein loading was controlled by measuring total protein content and Coomassie blue staining. (F) Effect of mTOR depletion on mitochondrial respiration in saponin-skinned fibers from soleus muscles from 6-wk-old mice with glutamate and malate as substrates. The following data were measured: basal rate of mitochondrial oxygen consumption in the absence of ADP (Vo), maximal rate of oxygen consumption in the presence of 2 mM ADP (Vmax), and acceptor control ratio (ACR; control, n = 15; mTOR−, n = 8). (G) Effect of mTOR depletion on mitochondria sensitivity for ADP in saponin-skinned fibers from soleus muscles. The apparent Km for ADP was measured in the absence and presence of 20 mM creatine (control, n = 8; mTOR−, n = 5). (H) Intramuscular ATP levels in muscles from control and mTOR− mice (n = 8 sample sets). ##, P < 0.01 (for control with creatine versus control without creatine); **, P < 0.01 (for mTOR− without creatine versus control without creatine). Data indicate mean ± SEM.
To investigate the consequences of mTOR inactivation on intrinsic oxidative capacities in soleus muscles, we analyzed mitochondrial respiration and function in saponin-skinned fibers with nonlimiting amounts of substrate and oxygen. Consistent with decreased mitochondrial content in mTOR− soleus, the rate of oxygen consumption in muscle was diminished both in the absence (basal) or presence (maximal) of ADP, whereas the ratio of maximal to basal respiration (acceptor control ratio) was increased (Fig. 7 F). These results indicate that the lack of mTOR in soleus muscles reduced the maximal capacity of oxidative phosphorylation, whereas the coupling efficiency between oxidation and phosphorylation was improved. Mitochondrial function depends on its sensitivity to ADP and creatine, which differs in oxidative and glycolytic muscle fibers (Kuznetsov et al., 1996). In oxidative fibers, the apparent Km (Michaelis–Menten constant) for ADP is high and is decreased by creatine as a consequence of the functional coupling of mitochondrial creatine kinase to ATP production and energy transfer (Fig. 7 G). Surprisingly, in mTOR− soleus muscle fibers, the apparent Km for ADP was unusually low and remained unaffected by creatine. Altogether, our findings demonstrate that loss of mTOR not only affects mitochondrial biogenesis and oxygen consumption, but additionally leads to profound changes into the mitochondria metabolic properties of oxidative fibers, which share characteristics with those of glycolytic fibers. However, intramuscular ATP concentrations were not significantly reduced in mTOR− muscles (Fig. 7 H).
mTOR deficiency in muscle does not impair whole body glucose homeostasis
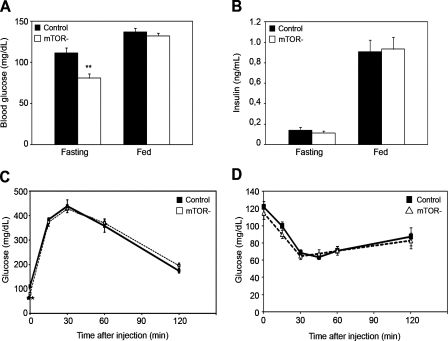
We next investigated the effect of mTOR deficiency in skeletal muscle on whole body glucose homeostasis. In the fasting state, mTOR− mice had significantly lower glucose levels (Fig. 8 A), which is possibly the result of increased basal glucose uptake in skeletal muscle (Fig. 6 H). However, the glucose and insulin concentrations in the fed state and the insulin level in fasting mice were not statistically different between mTOR− and control mice (Fig. 8, A and B), suggesting that glucose tolerance and insulin sensitivity were not affected. Consistent with this, mTOR− and control mice showed similar responses during the glucose tolerance test (Fig. 8 C) and the insulin tolerance test (Fig. 8 D). Collectively, these results indicate that mTOR functions in skeletal muscle are not crucial in controlling whole body glucose homeostasis.
Figure 8.
Glucose homeostasis and insulin sensitivity in mTOR− mice. (A) Fasting and fed blood glucose concentrations in mTOR− and control male mice. (B) Fasting and fed serum insulin concentrations in mTOR− and control male mice (n = 7 mice/group). (C) Glucose tolerance test on male mice (n = 15 mice/group). (D) Insulin tolerance test on mTOR− and control mice (n = 8 mice/group). **, P < 0.01. Data indicate mean ± SEM.
Discussion
To determine the physiological role of mTOR in postnatal skeletal muscle, we generated muscle-specific mTOR knockout mice (mTOR−) using HSA-Cre mice expressing Cre in postmitotic myofibers but not in satellite cells (Nicole et al., 2003). Our study reveals that mTOR deficiency in skeletal muscle leads to metabolic changes resulting in glycogen accumulation. Specifically, these changes include an increase in glucose uptake and glycogen synthesis associated with reduced glycogen breakdown through glycogenolysis and glycolytic and oxidative pathways. Our in vivo data support and extend findings of previous in vitro studies (Hardwick et al., 1999; Peng et al., 2002; Edinger et al., 2003; Schieke et al., 2006; Cunningham et al., 2007). Indeed, mammalian cells transfected with mTOR or raptor short hairpin RNAs or treated with rapamycin demonstrate altered glycolysis and oxidative metabolism associated with a parallel change in gene expression. In addition, mTOR inhibition by rapamycin abrogated the PKB/Akt-mediated induction of glycolytic enzymes in a constitutive PKB/Akt murine model of prostate intraepithelial neoplasia (Majumder et al., 2004). Finally, mTOR and raptor have been shown to directly regulate mitochondrial biogenesis and oxidative functions through the control of a YY1–PGC1α transcriptional complex (Cunningham et al., 2007). Our study further reveals that deficiency in intracellular energy transfer could be an additional mechanistic basis for the mitochondrial alterations in mTOR− muscle.
However, intramuscular ATP levels were preserved in mTOR− mice. Similarly, ATP concentration was not changed in cells treated with rapamycin, despite lower rates of oxygen consumption and glycolysis (Cunningham et al., 2007). Although glucose metabolism was altered, basal glucose uptake and glycogen synthesis were increased in mTOR− muscle. These effects are probably mediated by the hyperactivation of PKB/Akt in mutant muscles. Increased basal glucose uptake in muscle could possibly contribute to the lower basal glucose levels in mTOR− mice. However, glucose tolerance and insulin sensitivity were not affected in mTOR− mice, indicating that muscle mTOR does not play a crucial role in controlling whole body glucose homeostasis.
Decreased oxidative capacity, altered mitochondrial regulation, glycogen accumulation, and hyperactivation of PKB/Akt were also reported in RAmKO but not in RImKO muscles (Bentzinger et al., 2008), indicating that mTORC1 disruption likely accounts for these changes in mTOR− muscles. Although glycolytic pathways were not investigated in RAmKO mice, our observations strongly suggest that glycolytic capacity is also reduced in this model. Hyperactivation of PKB/Akt in mTOR− muscle is associated with reduced mTORC1 signaling and increased IRS-1 levels, indicating that this feature results from the loss of the well-documented S6K-mediated inhibitory feedback on IRS-1 (Um et al., 2006) as observed in RAmKO muscle (Bentzinger et al., 2008). In addition, the persistence of PKB/Akt phosphorylation on Ser473 in mTOR− muscle is reminiscent of muscle lacking both raptor and rictor but still containing mTOR. Thus, our observations confirm and extend the findings by Bentzinger et al. (2008) that skeletal muscles express a kinase distinct from mTOR that is able to phosphorylate PKB/Akt on this site. In line with this, additional kinases, designated as PDK2 kinases, have been proposed to phosphorylate Akt on Ser473 (Bayascas and Alessi, 2005). Despite the hyperactivation of PKB/Akt in raptor-deficient muscles, RAmKO mice exhibited altered glucose tolerance (Bentzinger et al., 2008). It is not clear at present how raptor and rictor (Kumar et al., 2008) but not mTOR inactivation in muscle generates glucose intolerance.
In addition, we show that loss of mTOR in muscle results in a progressive muscle myopathy similar to that observed in RAmKO mice (Bentzinger et al., 2008). However, muscle pathology is exacerbated and extended to all types of muscles in mTOR− mice. For example, mTOR− muscles contain up to 35% of regenerating fibers, whereas the proportion of regenerating fibers remained very low in RAmKO mice. Moreover, mTOR deficiency severely affects the functional properties of slow- and fast-twitch skeletal muscles, whereas the contractile properties of the fast glycolytic extensor digitorum longus muscle were mainly spared in RAmKO mice. Finally, the sPo remained unaltered in muscles from RAmKO mice, whereas it is reduced by almost 30% in mTOR− muscles, thus demonstrating that muscle weakness of mTOR− mice is not only caused by a loss of muscle mass.
A possible explanation for the more severe myopathy in mTOR− muscle is our finding that these muscles additionally exhibit reduced levels of both dystrophin and the entire DGC, a feature that is observed neither in RAmKO nor RImKO muscles. Consistently with this finding, mTOR− muscles display several features reminiscent of mdx and DMD muscles. At the molecular level, these include the up-regulation of utrophin (Tinsley et al., 1998), caveolin-3 (Repetto et al., 1999), and PKB/Akt protein levels (Acharyya et al., 2005; Dogra et al., 2006; Peter and Crosbie, 2006). Finally, the functional characteristics of mTOR− muscles are similar to mdx muscle, as they also exhibit a contraction-induced force deficit (Watchko et al., 2002). In addition, skeletal muscle from mdx mice generally shows increased mass attributed to the ongoing cycle of degeneration and regeneration, producing larger but weaker muscle fibers (Watchko et al., 2002).
However, mTOR− muscles do not display increased sarcolemmal permeability, one of the most characteristic and early features of muscles lacking dystrophin (Straub et al., 1997), although sarcolemmal fragility of mTOR− muscle is demonstrated by the great force deficits associated with eccentric contractions. Importantly, dystrophin levels in mTOR− muscles reach ∼10–20% of those in control muscles, and levels of DGC components are only mildly decreased, whereas the entire protein complex is lost in DMD and mdx muscle (Ohlendieck and Campbell, 1991). Therefore, it is likely that residual sarcolemmal DGC or utrophin–glycoprotein complex modulate membrane damage of mTOR− muscles (Peter et al., 2009). In line with this, studies on mdx mice treated with gentamicin have demonstrated that 10–20% of dystrophin levels are sufficient to restore the assembly of DGC and can afford significant but not complete protection against injury (Barton-Davis et al., 1999).
We also provide evidence that the reduction of dystrophin is based on a reduction of transcripts. Moreover, we show that mTOR binds the dystrophin promoter, strongly suggesting that this reduction is controlled at the transcriptional level. mTOR controls the transcription of many genes (Peng et al., 2002). In particular, mTOR interacts with and positively regulates YY1 transcriptional activity to activate mitochondrial gene expression in myotubes (Cunningham et al., 2007). Interestingly, YY1 has also been shown to bind and regulate the dystrophin promoter (Galvagni et al., 1998). Therefore, it is tempting to speculate that mTOR may positively modulate YY1 function in myotubes to drive dystrophin expression. In addition, our findings that mTOR regulatory function on dystrophin transcription is insensitive to rapamycin are consistent with the observation that mTOR interacts with YY1 independently of rapamycin and that dystrophin is not down-regulated in RAmKO muscles. Interestingly, using rapamycin, Akt activation was shown to prevent the force deficit induced in mdx muscles by eccentric contractions independently of mTOR (Blaauw et al., 2008). Our finding that mTOR regulates dystrophin expression in a rapamycin-resistant manner may provide an explanation for this discrepancy.
We also provided evidence that mTOR kinase activity is not required to regulate dystrophin, thus further extending the list of catalytic independent mTOR functions. Indeed, mTOR was shown to regulate IGF-II expression and myogenesis via a kinase-independent mechanism (Erbay and Chen, 2001; Erbay et al., 2003).
Collectively, our data point toward a new and specific role of mTOR in the regulation of dystrophin expression in a way that is not strictly dependent on mTORC1 and mTORC2. Of note, down-regulation of dystrophin in mTOR− muscle cannot be attributed to the loss of both mTORC1 and mTORC2 functions because DmKO mice neither display a more severe phenotype than RAmKO nor the characteristic Akt protein up-regulation observed in dystrophin-deficient muscles (Dogra et al., 2006). Evidence for mTOR functions that do not involve the contribution of these mTORCs is supported by a recent study showing that mTOR regulates terminal oligopyrimidine mRNA translation in a raptor- and rictor-independent manner (Patursky-Polischuk et al., 2009).
In conclusion, we demonstrate that the loss of mTOR in skeletal muscle leads to a severe myopathy, thereby further extending the list of disorders related to mTOR dysfunction. Our findings underscore the critical role of mTOR in regulating the genetic repertoire required for the appropriate maintenance of skeletal muscle integrity. About 40% of all myopathies have not yet been genetically characterized, whereas mTOR− muscles recapitulate many of the features of a broad range of these disorders, suggesting that alterations of mTOR-mediated processes could contribute to these disorders. Therefore, the importance of the mTOR pathway in maintaining muscle integrity provides new avenues of investigation in the diagnosis and therapy of skeletal muscle pathologies.
Materials and methods
Animals
The generation of the mTORneo embryonic stem (ES) clones used to functionally inactivate the mTOR gene was described previously (Gangloff et al., 2004). In brief, a targeting vector was produced by introducing a loxP site upstream of the mTOR promoter region and a neo cassette flanked by two loxP sites in the intron preceding exon 6. To generate mTORneo ES cells, the construct was electroporated into 129SVJae ES cells, and G418-resistant clones were screened for homologous recombination by Southern blotting. For excision of the floxed PGKneo cassette, the mTORneo ES cells were transfected with 0.8 µg pEGFP-N1 (Takara Bio Inc.) and 0.2 µg pMC-Cre using transfection reagent (Effectene; QIAGEN). ES cells expressing EGFP were cultured in the presence and absence of G418. G418-sensitive colonies were analyzed by PCR. ES cells showing the selective deletion of the neomycin resistance cassette and containing the mTOR floxed allele were injected into C57BL/6J blastocysts to generate chimeric mice. Chimeric mice were mated with C57BL/6J mice to generate the heterozygous floxed mutant (mTOR+/flox) through germline transmission. Positive F1 offspring were identified by PCR and further backcrossed with C57BL/6J mice during five generations (F5 offspring). mTOR+/flox mice were interbred to obtain a homozygous mutant (mTORflox/flox). mTORflox/flox (control) mice were bred with HSA-Cre mice to generate Cre-positive mTORflox/flox (mTOR−) mice. ES cells and mice were genotyped by PCR.
The primers used to identify the mTOR flox allele were p2 (5′-GCTCTTGAGGCAAATGCCACTATCACC-3′) and p3 (5′-TCATTACCTTCTCATCAGCCAGCAGTT-3′), and the primers to identify the recombined mTOR− allele were p1 (5′-TTCATTCCCTTGAAAGCCAGTCTCACC-3′) and p3. The animals were provided with mouse chow and water ad libitum in a restricted-access, specific pathogen–free animal care facility at the Ecole Normale Superieure of Lyon (Plateau de Biologie Experimentale de la Souris). All procedures were performed in accordance with national and European legislation on animal experimentation.
In vivo electroporation of mTOR mutants
Expression vectors were injected into TA muscles (7 µg total DNA in 30 µl 0.9% NaCl) of 6-wk-old mTOR− female mice. Injected muscles were electroporated with 1-cm2 plaque electrodes as described previously (Méjat et al., 2005). The electroporated DNA mixtures contained 5 µg of the rat mTOR expression vectors (wild type or kinase dead) and 2 µg pRNAT-GFP, allowing the visualization and microdissection of electroporated fibers for further gene expression analysis by quantitative real-time PCR. pcDNA3/mTOR eukaryotic expression plasmids encoding wild-type or kinase-dead (D2338A) rat mTOR were generated previously (Brunn et al., 1997).
ChIP assay
Mouse TA muscles were incubated for 15 min on ice in buffer RBI (100 mM KCl, 5 mM MgCl2, 5 mM EDTA, 5 mM Na pyrophosphate, pH 6.8, and protease inhibitors) then transferred to a 2% formaldehyde solution for 15 min at 37°C. Reactions were stopped by addition of glycine at 0.125 M final and incubation at 25°C for 5 min. After a PBS wash, muscles were crushed in lysis buffer A (10 mM Tris-HCl, pH 7.9, 85 mM KCl, 0.5% NP-40, and protease inhibitors). The mixture was homogenized by 20 strokes of Dounce followed by a 5-min spin at 5,000 rpm at 4°C. The pellet was resuspended in lysis buffer B (50 mM Tris-HCl, pH 8, 10 mM EDTA, 1% SDS, and protease inhibitors) incubated on ice for 10 min followed by a 5-min centrifugation at 5,000 rpm at 4°C. The supernatant was sonicated in a Bioruptor (Diagenode) for 15 min (medium; 30 s/30 s cycles). The size of chromatin fragment was checked to be in the range of 300–500-bp length on an agarose gel after de–cross-linking. ChIP was performed using the EZ Magna ChIP kit (Millipore) with 200 µg chromatin and 20 µg antibody.
The antibodies used were mTOR (Cell Signaling Technology), normal mouse IgG (12–371; Millipore), and anti-RNA Pol2 CTD4H8 (05-623B; Millipore). PCR conditions were optimized for each couple of primers. The primers used were 5′-AGTTGAACATTATTTGAACT-3′ and 5′-ACTGAGTGAGTCAACATAGT-3′ for the dystrophin promoter and 5′-ACAGTCAGGGAAGGAAGCAA-3′ and 5′-CCTTCCACCTCTGTTCAGGA-3′ for the dysferlin promoter.
Muscle histology, immunohistochemistry, morphometric measurements, and imaging
Skeletal muscles were collected, embedded in tragacanth gum, and quickly frozen in isopentane cooled in liquid nitrogen. 10-µm-thick sections were obtained from the middle portion of frozen muscles and processed for histological and immunohistochemical analysis according to standard protocols. The fiber CSA and the number of centrally nucleated fibers were determined for three consecutive H&E-stained sections from at least four animals using Metamorph software (version 6.3; MDS Analytical Technologies). For immunostaining, cross sections were fixed in acetone at −20°C permeabilized with 0.1% Triton X-100/PBS, and incubated with specific primary antibody followed by Alexa Fluor 488–labeled secondary antibodies (Invitrogen).
The following primary antibodies were used: dystrophin (Dys2; Novocastra), Akt (Cell Signaling Technology), and P-Akt (Cell Signaling Technology). For vital staining with Evans blue dye, the dye was resuspended in PBS at 1 mg/0.1 ml/10 g body wt and injected intraperitoneally. Muscles were collected after 24 h, and frozen sections were rinsed in PBS and examined by fluorescence microscopy.
Confocal microscopy was performed using a spectral confocal laser-scanning microscope (TCS SP5; Leica) on an upright microscope (DM6000 B; Leica). Confocal software (LAS AF; Leica) was used for acquisition with a Plan Apo 63× NA 1.4 oil objective (HCX; Leica). Fluorescence and light microscopy were performed using an upright microscope (Axioplan2; Carl Zeiss, Inc.) and 10× NA 0.3 (Ph1), 20× NA 0.5, or 40× NA 0.75 Plan NeoFluor objectives (Carl Zeiss, Inc.). Images were captured using a charge-coupled device monochrome camera (Coolsnap HQ; Photometrics) and MetaMorph software. For all imaging, exposure settings were identical between compared samples. All samples for microscopy were viewed at room temperature.
Quantitative real-time PCR and immunoblot analysis
Total RNA was isolated from cultured cells or muscle tissues using Trizol (Invitrogen). RNA was analyzed by quantitative real-time PCR using SYBR Green (Roche). The data were normalized to cyclophilin B, β2-microglobulin, and hypoxanthine–guanine phosphoribosyltransferase mRNA levels, which gave similar results. Western blots were performed as described previously (Ohanna et al., 2005). Primary antibodies used to study mTOR signaling were obtained from Cell Signaling Technology. Primary antibodies against dystrophy-related proteins were obtained from Novocastra, and antibodies against troponin T were obtained from Sigma-Aldrich. Antibodies against respiratory chain components, myoglobin, and cytochrome c were obtained from Mitosciences, Santa Cruz Biotechnolgoy, Inc., and BD, respectively.
Cell cultures
Primary cultures were derived from GC and TA muscles of 4-wk-old mTORflox mice as described previously (Ohanna et al., 2005). mTORflox myoblasts were differentiated in DME/Ham F12 containing 2% horse serum. The myotubes were transduced by cytomegalovirus (CMV)-GFP or CMV-Cre adenovirus (Genethon) at 100 MOI on differentiation day 1 and harvested at different time points for gene expression analysis by quantitative real-time PCR or Western blotting. C2C12 myoblasts were differentiated in DME with 2% horse serum and treated at differentiation day 4 with vehicle (DMSO), 20 nM rapamycin, or 250 nM Torin1 for 48 h and harvested for gene expression analysis by quantitative real-time PCR or Western blotting.
Enzyme analysis
Frozen muscle samples were homogenized in a stabilizing medium containing 50% glycerol, 20 mM phosphate buffer, pH 7.4, 5 mM β-mercaptoethanol, 0.5 mM EDTA, and 0.02% BSA at a dilution of 1:50 based on wet weight. All of the enzyme activities were measured at 22°C as described previously (Passonneau and Lowry, 1993). Hexokinase activity was determined in a two-step assay. The glucose-6-phosphate accumulated for 60 min in the first step and reacted with NADP and glucose-6-phosphate dehydrogenase in a second step. The NADPH generated was detected by fluorescence spectroscopy with excitation/emission wavelengths of 350/460 nm. GP, PFK, PK, and LDH activities were measured in a one-step spectrophotometer assay by directly measuring the rate of appearance or disappearance of NADPH/NADH at 340 nm for 10 min. For GP activity measurement, glucose-1-phosphate formation was coupled to phosphoglucomutase and glucose-6-phosphate dehydrogenase reactions. For PFK activity measurement, fructose-1,6-bisphosphate formation was coupled to aldolase, triosephosphate isomerase, and glycerol-3-phosphate dehydrogenase reactions. For PK activity measurement, pyruvate formation was coupled to LDH reaction. LDH activity was measured in the direction of lactate formation. Citrate synthase activity was determined by using Ellman's reagent with acetyl-CoA and oxaloacetate (Leek et al., 2001). Standards were carried out through the entire procedure.
Determination of intramuscular glucose-6-phosphate, lactate, and ATP concentrations
Muscles were quickly removed from anesthesized animals, weighted, and directly frozen in liquid nitrogen. Muscles were homogenized in 0.65 M perchloric acid (1:40 wt/vol) and incubated for 15 min at 4°C. Muscle homogenates were centrifuged at 5,000 g for 10 min at 4°C. Resulting acid supernatants were neutralized with 2 N KOH in 0.4 M TEA/KCl and used for the fluorometric determination of lactate, ATP, and glucose-6-phosphate contents by coupled enzyme assays (Passonneau and Lowry, 1993).
Glucose and insulin tolerance tests
For glucose tolerance tests, 12-wk-old male mice were fasted overnight and injected intraperitoneally with 3 mg glucose/g body wt. For the insulin tolerance test, 4-h-fasted male mice were injected intraperitoneally with insulin (0.75 mUI/g body wt; Sigma-Aldrich). Blood glucose levels were determined from tail venous blood using an automatic glucose monitor (Roche). Serum levels of insulin were determined with murine ELISA kit (Linco Research).
Glucose uptake assays
After cervical dislocation of the mice, the soleus muscle was rapidly isolated and tied to stainless steel clips by the tendons. All incubations were performed at 37°C under an atmosphere of 95% O2/5% CO2 in 1 ml Krebs-Ringer bicarbonate buffer, pH 7.3, supplemented with 1% BSA (fraction V; pH 7; Intergen) and 2 mM sodium pyruvate. Glucose transport was measured as follows: after a preincubation period of 15 min, muscles were incubated for 10 min in the same medium supplemented with 2-[3H]deoxyglucose (0.1 mM and 0.5 µCi/ml). Afterward, muscles were washed for 30 min in ice-cold saline buffer and dissolved in 1 N NaOH before scintillation counting for 3H-labeled radioactivity was performed. Sample aliquots were used for protein determination.
Functional properties of mitochondria
Respiratory parameters of the total mitochondrial population were studied in situ in fresh saponin-skinned fibers (50 µg/ml saponin for 30 min) and determined with a Clark electrode (Hansatech Instruments) in an oxygraphic cell at 22°C as described previously (Kuznetsov et al., 2008). Respiration rates were expressed as µmol O2/min/g dry weight. Respiration solution, pH 7.1, contained 10 mM EGTA-CaEGTA buffer (free Ca2+ concentration 100 nM), 1 mM free Mg2+, 20 mM taurine, 0.5 mM dithiothreitol, 20 mM imidazole, ionic strength 160 (potassium methane sulfonate), 5 mM glutamate, 2 mM malate, 3 mM phosphate, and 2 mg/ml BSA. Basal oxygen consumption without ADP (V0) was recorded in the presence or absence of 20 mM creatine, and increasing amounts of ADP were added until maximal respiration was reached. After measurements, fibers were carefully removed, dried, and weighed. The ADP-stimulated respiration (VADP) above basal oxygen consumption (V0) was plotted as a function of ADP with and without creatine. The apparent Km values for ADP in the presence or absence of creatine and VADP were calculated with a nonlinear fit of the Michaelis–Menten equation. The maximal respiration rate (Vmax) = VADP + V0. The acceptor control ratio = Vmax/V0. Three determinations were made for each muscle sample.
Measurements of muscle contractile properties and lengthening contractions
Mice were anesthetised with 60 mg/kg pentobarbital. The knee and foot were fixed using clamps and pins. The isometric contractile properties of TA muscles were studied in situ as previously described (Mouisel et al., 2006). The distal tendon of the TA muscle was attached to a lever arm of a servomotor system (305B Dual-Mode Lever; Aurora Scientific). All data provided by the servomotor system were recorded and analyzed on a microcomputer using the PowerLab system (4SP; ADInstruments) and software (Chart 4; ADInstruments). The sciatic nerve (proximally crushed) was stimulated by a bipolar silver electrode using a supramaximal (10 V) square wave pulse of 0.1-ms duration. In some instances, muscles were directly stimulated (80 V). All isometric contraction measurements were made at an initial muscle length of L0 (length at which maximal tension, Pt, was obtained during the twitch). Po was measured during isometric contractions in response to electrical stimulation (frequency of 25–150 Hz; train of stimulation of 500 ms). sPo was calculated by dividing Po by muscle weight. Pt, TTP, and RT50% were also measured in response to a single stimulation. The in situ contraction–induced injury protocol for the TA muscle used in this study was similar to that described previously (Dellorusso et al., 2001). The sciatic nerve was stimulated for 300 ms (frequency of 125 Hz). An isometric contraction of the TA muscle was initiated during the first 100 ms. Then, muscle lengthening (3 mm; ∼45% fiber length [Lf]) at a velocity of 15 mm/s (∼2 Lf/s) was imposed during the last 200 ms. Two lengthening contractions of the TA muscle were performed, each separated by a 60-s rest period. The force deficit after contraction-induced damage was determined by calculating the difference between the Po measured 1 min after the lengthening contractions and the Po determined before lengthening contractions and was expressed as a percentage of Po determined before lengthening contractions.
The isometric contractile properties of soleus muscles were studied in vitro as previously described (Agbulut et al., 2009). Soleus muscles were soaked in an oxygenated Krebs solution (95% O2 and 5% CO2) containing 58.5 mM NaCl, 24 mM NaHCO3, 5.4 mM KCl, 1.2 mM KH2PO4, 1.8 mM CaCl2, 1 mM MgSO4, and 10 mM glucose, pH 7.4, and maintained at a temperature of 22°C. One of the muscle tendons was attached to a lever arm of a servomotor system (300B Dual-Mode Lever; Aurora Scientific). After equilibration (30 min), electrical stimulation was delivered through electrodes running parallel to the muscle. 1-ms pulses were generated by a high power stimulator (701B; Aurora Scientific). Po was measured during isometric contractions in response to electrical stimulation (frequency of 50–125 Hz; train of stimulation of 1,500 ms). sPo was calculated by dividing the force by the estimated CSA of the muscle. Assuming muscles have a cylindrical shape and a density of 1.06 mg mm−3, the CSA corresponds to the volume of the muscle divided by Lf. The Lf to L0 ratio of 0.70 was used to calculate Lf. Fatigue resistance was determined after a 5-min rest period. The muscles were stimulated at 50 Hz during 500 ms every second for 1 min. The time taken for initial force to fall by 20% (F20%) was calculated.
Myosin isoform identification
Muscles were subjected to the analysis of MHC isoforms as described previously (Talmadge and Roy, 1993). Myosin was extracted from small sections of muscles in 7 vol of buffer solution (0.3 M NaCl, 0.1 M NaH2PO4, 0.05 M Na2HPO4, 0.01 M Na4P2O7, 1 mM MgC12-6H2O, 10 mM EDTA, and 1.4 mM 2β-mercaptoethanol, pH 6.5). Myosin was separated in acrylamide gel solution containing 30% glycerol, 8% acrylamide-bis (50:1), 0.2 M Tris, 0.1 M glycine, and 0.4% SDS using a Mini Protean II system (Bio-Rad Laboratories). Gels were run at 72 V for 31 h and silver stained. The bands were scanned and quantified using a densitometer equipped with an integrator (GS-800; Bio-Rad Laboratories).
Statistical analysis
A two-tailed Student's t test was used for statistical analysis. All data are expressed as means ± SEM or SD, and significance was established at the P < 0.05 level.
Online supplemental material
Fig. S1 shows the distribution of the different fiber types on soleus muscle transverse sections from control and mTOR− mice. Fig. S2 shows Western blot analysis for dystrophy-related proteins in 11-wk-old mTOR− muscles. Table S1 presents the quantification of Western blot analysis for DGC components in muscles from 6-wk-old control and mTOR− mice. Table S2 presents the quantification of Western blot analysis for dystrophy-related proteins in muscles from 11-wk-old control and mTOR− mice. Table S3 presents the quantification of Western blot analysis for signal transduction in mTOR− muscles. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200903131/DC1.
Acknowledgments
We thank R.D. Roy and C. Scutt for critical reading of the manuscript and helpful discussions, J. Melki for providing the HSA-Cre line, and B. Serrurier and A. Tran-Van-Minh for technical assistance. We thank the Novartis Foundation and the animal (Le Plateau de Biologie Expérimental de la Souris) and microscopy (Plateau Technique Imagerie/Microscopie) facilities of the IFR128 Biosciences. We also thank the Whitehead Institute and Dana-Farber Cancer Institute for supplying Torin1 inhibitor and Dr. G. Chiang from the Brockhouse Institute for Materials Research for supplying the rat mTOR plasmids.
This work was supported by an EMBO fellowship (to Y.-G. Gangloff), the Association Française contre les Myopathies (to L. Mazelin, L. Schaeffer, and Y.-G. Gangloff), the Agence Nationale de la Recherche (to M. Pende and Y.-G. Gangloff), the Association pour la Recherche contre le Cancer (to Y.-G. Gangloff), and the Ministère de la Recherche et de la Technologie (ACI BDPI to L. Schaeffer).
Footnotes
Abbreviations used in this paper:
- ChIP
- chromatin immunoprecipitation
- CMV
- cytomegalovirus
- CSA
- cross-sectional area
- DGC
- dystrophin–glycoprotein complex
- DMD
- Duchenne MD
- DmKO
- raptor/rictor muscle double knockout
- ES
- embryonic stem
- GC
- gastrocnemius
- GP
- glycogen phosphorylase
- HSA
- human skeletal muscle α-actin
- LDH
- lactate dehydrogenase
- MD
- muscular dystrophy
- MHC
- myosin heavy chain
- mTOR
- mammalian target of rapamycin
- mTORC
- mTOR complex
- PFK
- phosphofructokinase
- PK
- pyruvate kinase
- PLA
- plantaris
- RAmKO
- raptor muscle knockout
- RImKO
- rictor muscle knockout
- TA
- tibialis anterior
- TTP
- time to peak twitch tension
References
- Acharyya S., Butchbach M.E., Sahenk Z., Wang H., Saji M., Carathers M., Ringel M.D., Skipworth R.J., Fearon K.C., Hollingsworth M.A., et al. 2005. Dystrophin glycoprotein complex dysfunction: a regulatory link between muscular dystrophy and cancer cachexia. Cancer Cell. 8:421–432 10.1016/j.ccr.2005.10.004 [DOI] [PubMed] [Google Scholar]
- Agbulut O., Vignaud A., Hourde C., Mouisel E., Fougerousse F., Butler-Browne G.S., Ferry A. 2009. Slow myosin heavy chain expression in the absence of muscle activity. Am. J. Physiol. Cell Physiol. 296:C205–C214 10.1152/ajpcell.00408.2008 [DOI] [PubMed] [Google Scholar]
- Barton-Davis E.R., Cordier L., Shoturma D.I., Leland S.E., Sweeney H.L. 1999. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J. Clin. Invest. 104:375–381 10.1172/JCI7866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby R., Olson E.N. 2006. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75:19–37 10.1146/annurev.biochem.75.103004.142622 [DOI] [PubMed] [Google Scholar]
- Bayascas J.R., Alessi D.R. 2005. Regulation of Akt/PKB Ser473 phosphorylation. Mol. Cell. 18:143–145 10.1016/j.molcel.2005.03.020 [DOI] [PubMed] [Google Scholar]
- Bentzinger C.F., Romanino K., Cloëtta D., Lin S., Mascarenhas J.B., Oliveri F., Xia J., Casanova E., Costa C.F., Brink M., et al. 2008. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 8:411–424 10.1016/j.cmet.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Blaauw B., Mammucari C., Toniolo L., Agatea L., Abraham R., Sandri M., Reggiani C., Schiaffino S. 2008. Akt activation prevents the force drop induced by eccentric contractions in dystrophin-deficient skeletal muscle. Hum. Mol. Genet. 17:3686–3696 10.1093/hmg/ddn264 [DOI] [PubMed] [Google Scholar]
- Bodine S.C., Stitt T.N., Gonzalez M., Kline W.O., Stover G.L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J.C., Glass D.J., Yancopoulos G.D. 2001. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3:1014–1019 10.1038/ncb1101-1014 [DOI] [PubMed] [Google Scholar]
- Brunn G.J., Hudson C.C., Sekulić A., Williams J.M., Hosoi H., Houghton P.J., Lawrence J.C., Jr., Abraham R.T. 1997. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 277:99–101 10.1126/science.277.5322.99 [DOI] [PubMed] [Google Scholar]
- Cross D.A., Watt P.W., Shaw M., van der Kaay J., Downes C.P., Holder J.C., Cohen P. 1997. Insulin activates protein kinase B, inhibits glycogen synthase kinase-3 and activates glycogen synthase by rapamycin-insensitive pathways in skeletal muscle and adipose tissue. FEBS Lett. 406:211–215 10.1016/S0014-5793(97)00240-8 [DOI] [PubMed] [Google Scholar]
- Cunningham J.T., Rodgers J.T., Arlow D.H., Vazquez F., Mootha V.K., Puigserver P. 2007. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 450:736–740 10.1038/nature06322 [DOI] [PubMed] [Google Scholar]
- Dellorusso C., Crawford R.W., Chamberlain J.S., Brooks S.V. 2001. Tibialis anterior muscles in mdx mice are highly susceptible to contraction-induced injury. J. Muscle Res. Cell Motil. 22:467–475 10.1023/A:1014587918367 [DOI] [PubMed] [Google Scholar]
- Dogra C., Changotra H., Wergedal J.E., Kumar A. 2006. Regulation of phosphatidylinositol 3-kinase (PI3K)/Akt and nuclear factor-kappa B signaling pathways in dystrophin-deficient skeletal muscle in response to mechanical stretch. J. Cell. Physiol. 208:575–585 10.1002/jcp.20696 [DOI] [PubMed] [Google Scholar]
- Durbeej M., Campbell K.P. 2002. Muscular dystrophies involving the dystrophin-glycoprotein complex: an overview of current mouse models. Curr. Opin. Genet. Dev. 12:349–361 10.1016/S0959-437X(02)00309-X [DOI] [PubMed] [Google Scholar]
- Edinger A.L., Linardic C.M., Chiang G.G., Thompson C.B., Abraham R.T. 2003. Differential effects of rapamycin on mammalian target of rapamycin signaling functions in mammalian cells. Cancer Res. 63:8451–8460 [PubMed] [Google Scholar]
- Erbay E., Chen J. 2001. The mammalian target of rapamycin regulates C2C12 myogenesis via a kinase-independent mechanism. J. Biol. Chem. 276:36079–36082 10.1074/jbc.C100406200 [DOI] [PubMed] [Google Scholar]
- Erbay E., Park I.H., Nuzzi P.D., Schoenherr C.J., Chen J. 2003. IGF-II transcription in skeletal myogenesis is controlled by mTOR and nutrients. J. Cell Biol. 163:931–936 10.1083/jcb.200307158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvagni F., Cartocci E., Oliviero S. 1998. The dystrophin promoter is negatively regulated by YY1 in undifferentiated muscle cells. J. Biol. Chem. 273:33708–33713 10.1074/jbc.273.50.33708 [DOI] [PubMed] [Google Scholar]
- Gangloff Y.G., Mueller M., Dann S.G., Svoboda P., Sticker M., Spetz J.F., Um S.H., Brown E.J., Cereghini S., Thomas G., Kozma S.C. 2004. Disruption of the mouse mTOR gene leads to early postimplantation lethality and prohibits embryonic stem cell development. Mol. Cell. Biol. 24:9508–9516 10.1128/MCB.24.21.9508-9516.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J.S., Kuruvilla F.G., Tong J.K., Shamji A.F., Schreiber S.L. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA. 96:14866–14870 10.1073/pnas.96.26.14866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto E., Loewith R., Schmidt A., Lin S., Rüegg M.A., Hall A., Hall M.N. 2004. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 6:1122–1128 10.1038/ncb1183 [DOI] [PubMed] [Google Scholar]
- Kumar A., Harris T.E., Keller S.R., Choi K.M., Magnuson M.A., Lawrence J.C. 2008. Muscle-specific deletion of rictor impairs insulin-stimulated glucose transport and enhances basal glycogen synthase activity. Mol. Cell. Biol. 28:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznetsov A.V., Tiivel T., Sikk P., Kaambre T., Kay L., Daneshrad Z., Rossi A., Kadaja L., Peet N., Seppet E., Saks V.A. 1996. Striking differences between the kinetics of regulation of respiration by ADP in slow-twitch and fast-twitch muscles in vivo. Eur. J. Biochem. 241:909–915 10.1111/j.1432-1033.1996.00909.x [DOI] [PubMed] [Google Scholar]
- Kuznetsov A.V., Veksler V., Gellerich F.N., Saks V., Margreiter R., Kunz W.S. 2008. Analysis of mitochondrial function in situ in permeabilized muscle fibers, tissues and cells. Nat. Protoc. 3:965–976 10.1038/nprot.2008.61 [DOI] [PubMed] [Google Scholar]
- Leek B.T., Mudaliar S.R., Henry R., Mathieu-Costello O., Richardson R.S. 2001. Effect of acute exercise on citrate synthase activity in untrained and trained human skeletal muscle. Am. J. Physiol. Regul. Integr. Comp. Physiol. 280:R441–R447 [DOI] [PubMed] [Google Scholar]
- Majumder P.K., Febbo P.G., Bikoff R., Berger R., Xue Q., McMahon L.M., Manola J., Brugarolas J., McDonnell T.J., Golub T.R., et al. 2004. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat. Med. 10:594–601 10.1038/nm1052 [DOI] [PubMed] [Google Scholar]
- Méjat A., Ramond F., Bassel-Duby R., Khochbin S., Olson E.N., Schaeffer L. 2005. Histone deacetylase 9 couples neuronal activity to muscle chromatin acetylation and gene expression. Nat. Neurosci. 8:313–321 10.1038/nn1408 [DOI] [PubMed] [Google Scholar]
- Mouisel E., Blondet B., Escourrou P., Chatonnet A., Molgó J., Ferry A. 2006. Outcome of acetylcholinesterase deficiency for neuromuscular functioning. Neurosci. Res. 55:389–396 10.1016/j.neures.2006.05.002 [DOI] [PubMed] [Google Scholar]
- Murakami M., Ichisaka T., Maeda M., Oshiro N., Hara K., Edenhofer F., Kiyama H., Yonezawa K., Yamanaka S. 2004. mTOR is essential for growth and proliferation in early mouse embryos and embryonic stem cells. Mol. Cell. Biol. 24:6710–6718 10.1128/MCB.24.15.6710-6718.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicole S., Desforges B., Millet G., Lesbordes J., Cifuentes-Diaz C., Vertes D., Cao M.L., De Backer F., Languille L., Roblot N., et al. 2003. Intact satellite cells lead to remarkable protection against Smn gene defect in differentiated skeletal muscle. J. Cell Biol. 161:571–582 10.1083/jcb.200210117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohanna M., Sobering A.K., Lapointe T., Lorenzo L., Praud C., Petroulakis E., Sonenberg N., Kelly P.A., Sotiropoulos A., Pende M. 2005. Atrophy of S6K1(-/-) skeletal muscle cells reveals distinct mTOR effectors for cell cycle and size control. Nat. Cell Biol. 7:286–294 10.1038/ncb1231 [DOI] [PubMed] [Google Scholar]
- Ohlendieck K., Campbell K.P. 1991. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J. Cell Biol. 115:1685–1694 10.1083/jcb.115.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passonneau J.V., Lowry O.H. 1993. Enzymatic Analysis: A Practical Guide. Humana Press, Inc., Totowa, NJ: 403 pp [Google Scholar]
- Patursky-Polischuk I., Stolovich-Rain M., Hausner-Hanochi M., Kasir J., Cybulski N., Avruch J., Rüegg M.A., Hall M.N., Meyuhas O. 2009. The TSC-mTOR pathway mediates translational activation of TOP mRNAs by insulin largely in a raptor- or rictor-independent manner. Mol. Cell. Biol. 29:640–649 10.1128/MCB.00980-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T., Golub T.R., Sabatini D.M. 2002. The immunosuppressant rapamycin mimics a starvation-like signal distinct from amino acid and glucose deprivation. Mol. Cell. Biol. 22:5575–5584 10.1128/MCB .22.15.5575-5584.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter A.K., Crosbie R.H. 2006. Hypertrophic response of Duchenne and limb-girdle muscular dystrophies is associated with activation of Akt pathway. Exp. Cell Res. 312:2580–2591 10.1016/j.yexcr.2006.04.024 [DOI] [PubMed] [Google Scholar]
- Peter A.K., Ko C.Y., Kim M.H., Hsu N., Ouchi N., Rhie S., Izumiya Y., Zeng L., Walsh K., Crosbie R.H. 2009. Myogenic Akt signaling upregulates the utrophin-glycoprotein complex and promotes sarcolemma stability in muscular dystrophy. Hum. Mol. Genet. 18:318–327 10.1093/hmg/ddn358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto S., Bado M., Broda P., Lucania G., Masetti E., Sotgia F., Carbone I., Pavan A., Bonilla E., Cordone G., et al. 1999. Increased number of caveolae and caveolin-3 overexpression in Duchenne muscular dystrophy. Biochem. Biophys. Res. Commun. 261:547–550 10.1006/bbrc.1999.1055 [DOI] [PubMed] [Google Scholar]
- Rommel C., Bodine S.C., Clarke B.A., Rossman R., Nunez L., Stitt T.N., Yancopoulos G.D., Glass D.J. 2001. Mediation of IGF-1-induced skeletal myotube hypertrophy by PI(3)K/Akt/mTOR and PI(3)K/Akt/GSK3 pathways. Nat. Cell Biol. 3:1009–1013 10.1038/ncb1101-1009 [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Kim D.H., Guertin D.A., Latek R.R., Erdjument-Bromage H., Tempst P., Sabatini D.M. 2004. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 14:1296–1302 10.1016/j.cub.2004.06.054 [DOI] [PubMed] [Google Scholar]
- Sarbassov D.D., Ali S.M., Sengupta S., Sheen J.H., Hsu P.P., Bagley A.F., Markhard A.L., Sabatini D.M. 2006. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol. Cell. 22:159–168 10.1016/j.molcel.2006.03.029 [DOI] [PubMed] [Google Scholar]
- Schiaffino S., Sandri M., Murgia M. 2007. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda). 22:269–278 [DOI] [PubMed] [Google Scholar]
- Schieke S.M., Phillips D., McCoy J.P., Jr., Aponte A.M., Shen R.F., Balaban R.S., Finkel T. 2006. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J. Biol. Chem. 281:27643–27652 10.1074/jbc.M603536200 [DOI] [PubMed] [Google Scholar]
- Stitt T.N., Drujan D., Clarke B.A., Panaro F., Timofeyva Y., Kline W.O., Gonzalez M., Yancopoulos G.D., Glass D.J. 2004. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell. 14:395–403 10.1016/S1097-2765(04)00211-4 [DOI] [PubMed] [Google Scholar]
- Straub V., Rafael J.A., Chamberlain J.S., Campbell K.P. 1997. Animal models for muscular dystrophy show different patterns of sarcolemmal disruption. J. Cell Biol. 139:375–385 10.1083/jcb.139.2.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge R.J., Roy R.R. 1993. Electrophoretic separation of rat skeletal muscle myosin heavy-chain isoforms. J. Appl. Physiol. 75:2337–2340 [DOI] [PubMed] [Google Scholar]
- Thoreen C.C., Kang S.A., Chang J.W., Liu Q., Zhang J., Gao Y., Reichling L.J., Sim T., Sabatini D.M., Gray N.S. 2009. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284:8023–8032 10.1074/jbc.M900301200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinsley J., Deconinck N., Fisher R., Kahn D., Phelps S., Gillis J.M., Davies K. 1998. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat. Med. 4:1441–1444 10.1038/4033 [DOI] [PubMed] [Google Scholar]
- Um S.H., D'Alessio D., Thomas G. 2006. Nutrient overload, insulin resistance, and ribosomal protein S6 kinase 1, S6K1. Cell Metab. 3:393–402 10.1016/j.cmet.2006.05.003 [DOI] [PubMed] [Google Scholar]
- Watchko J.F., O'Day T.L., Hoffman E.P. 2002. Functional characteristics of dystrophic skeletal muscle: insights from animal models. J. Appl. Physiol. 93:407–417 [DOI] [PubMed] [Google Scholar]
- Wullschleger S., Loewith R., Hall M.N. 2006. TOR signaling in growth and metabolism. Cell. 124:471–484 10.1016/j.cell.2006.01.016 [DOI] [PubMed] [Google Scholar]