The ORP lipid-binding domain can contact two membranes simultaneously to facilitate sterol extraction or delivery at one membrane in response to the lipid composition of the other.
Abstract
Sterols are transferred between cellular membranes by vesicular and poorly understood nonvesicular pathways. Oxysterol-binding protein–related proteins (ORPs) have been implicated in sterol sensing and nonvesicular transport. In this study, we show that yeast ORPs use a novel mechanism that allows regulated sterol transfer between closely apposed membranes, such as organelle contact sites. We find that the core lipid-binding domain found in all ORPs can simultaneously bind two membranes. Using Osh4p/Kes1p as a representative ORP, we show that ORPs have at least two membrane-binding surfaces; one near the mouth of the sterol-binding pocket and a distal site that can bind a second membrane. The distal site is required for the protein to function in cells and, remarkably, regulates the rate at which Osh4p extracts and delivers sterols in a phosphoinositide-dependent manner. Together, these findings suggest a new model of how ORPs could sense and regulate the lipid composition of adjacent membranes.
Introduction
Sterols are essential membrane components that are critical for several cellular processes including membrane trafficking and signal transduction (Maxfield and Tabas, 2005; Ikonen, 2006). In mammalian cells, both the synthesis and uptake of cholesterol are regulated by sterol regulatory element-binding protein transcription factors (Espenshade and Hughes, 2007). The concentration of sterols in cellular membranes is also tightly controlled; for example, the ER has ∼5 mol% cholesterol (Lange and Steck, 1997; Radhakrishnan et al., 2008), whereas in the plasma membrane (PM), it is ∼30 mol% (van Meer et al., 2008). How this distribution is maintained is not well understood.
Sterols are moved between cellular compartments by both vesicular and less-well understood nonvesicular pathways, most of which probably use lipid transfer proteins (LTPs). These proteins reversibly bind specific lipids in a hydrophobic pocket with a 1:1 stoichiometry, a property that allows them to transfer the bound lipid between membranes. In addition to a core lipid-binding domain, many LTPs have multiple targeting motifs specific for at least two different organelles (Olkkonen, 2004). It has also been proposed that some LTPs operate at zones of tight apposition of organelle membranes, which are often called membrane contact sites (MCSs; Holthuis and Levine, 2005; Levine and Loewen, 2006). These can be directly observed in ultrastructural studies that show that the ER in particular makes contact with a wide variety of organelles (Ladinsky et al., 1999; Perktold et al., 2007). The proposal that LTPs operate at MCSs is attractive because it would explain how LTPs could efficiently move lipids between a specific pair of organelles rather than diffusing over larger distances through the cytoplasm. At an MCS, the targeting domains of LTPs may allow them to simultaneously associate with both organelles or rapidly shuttle between them (Hanada et al., 2007). However, it has been difficult to directly demonstrate how or even if LTPs function at MCSs.
The oxysterol-binding protein (OSBP)–related proteins (ORPs) comprise a large family of LTPs conserved from yeast to humans that has been implicated in vesicular trafficking, intracellular signaling, and nonvesicular sterol transport (Fairn and McMaster, 2008; Yan and Olkkonen, 2008). All ORPs contain an OSBP-related domain (ORD) that binds sterols and possibly other lipids. The structure of the ORD from the yeast ORP Osh4p (also known as Kes1p) has been solved and revealed to contain a hydrophobic binding pocket that can accommodate a single sterol and is covered by a flexible “lid” (Im et al., 2005). Some ORDs have also been shown to bind phosphoinositide phosphates (PIPs), probably at a site on the surface of the domain that is distinct from the sterol-binding pocket (Li et al., 2002; Wang et al., 2005a). Most ORPs also contain, in addition to an ORD, several N-terminal targeting domains. These usually include a pleckstrin homology (PH) domain, which binds PIPs, and an FFAT (two phenylalanines in an acid tract) motif, which binds the ER proteins called vesicle-associated membrane protein–associated proteins (Fig. 1; Fairn and McMaster, 2008; Yan and Olkkonen, 2008). Because different PIP species are enriched in various organelles but largely absent from the ER (Vicinanza et al., 2008), the PH and FFAT motifs found in most ORPs could allow them to function at multiple locations in the cell, including MCSs. Indeed, some of the ORPs in yeast have been proposed to localize to MCSs between the ER and vacuole or between the ER and PM (Levine and Munro, 2001; Loewen et al., 2003).
Figure 1.
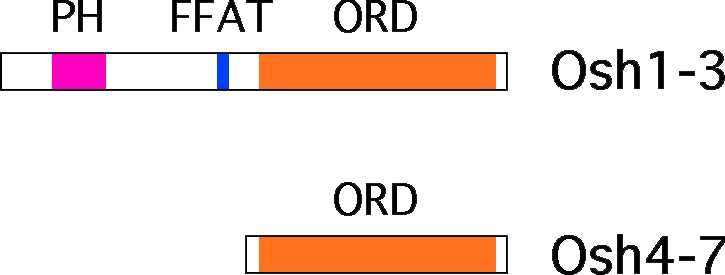
Domain structure of the seven Osh proteins (yeast ORPs).
We have previously proposed that the ORPs of the yeast Saccharomyces cerevisiae transfer sterols between intracellular membranes. The seven yeast ORPs, termed Osh proteins (Fig. 1), must have a single, shared essential function because yeast require any one of the seven for viability (Beh et al., 2001). We showed that the nonvesicular movement of exogenous sterols from the PM to the ER slows dramatically in cells lacking Osh proteins (Raychaudhuri et al., 2006). ER to PM transfer of newly synthesized sterol also slows significantly in these cells (Sullivan et al., 2006). In addition, we demonstrated that one of the Osh proteins, Osh4p, extracts and transfers sterols between liposomes in vitro. The ability to transfer sterols between membranes is not limited to yeast ORPs because it has recently been shown that two mammalian ORPs transfer sterols in vitro and perhaps in cells (Ngo and Ridgway, 2009).
Although several ORPs may transfer sterols or other lipids in cells, the functions of many ORPs remain unknown. Some ORPs may act as lipid sensors, regulating other proteins in response to binding lipids such as sterols and PIPs. For example, mammalian OSBP has been suggested to regulate the function of two other LTPs (CERT and Nir2) at the Golgi complex, perhaps at ER–Golgi contact sites (Perry and Ridgway, 2006; Peretti et al., 2008). OSBP has also been shown to function as a cholesterol-regulated scaffolding protein that modulates the activity of a phosphatase complex (Wang et al., 2005c). The yeast ORP Osh4p has been implicated in vesicular trafficking from the TGN (Fang et al., 1996; Beh and Rine, 2004; Proszynski et al., 2005), where it has been suggested to regulate a critical pool of phosphoinositol (PI)-4-phosphate (PI(4)P) needed for vesicular transport from Golgi membranes (Li et al., 2002; Fairn et al., 2007; Schaaf et al., 2008).
In this study, we demonstrate that the core lipid-binding domain present in all ORPs is able to contact two membranes simultaneously and, remarkably, that the specific lipid species associated with one membrane-binding surface can dramatically alter the probability of sterol extraction or delivery at the other binding surface. In addition, we show that four of the seven ORPs in yeast are enriched on regions of the ER that are closely associated with the PM, probably at MCSs. We conclude by presenting a model of how ORPs could transfer sterols and transmit signals at MCSs.
Results
ORDs bind two membranes simultaneously
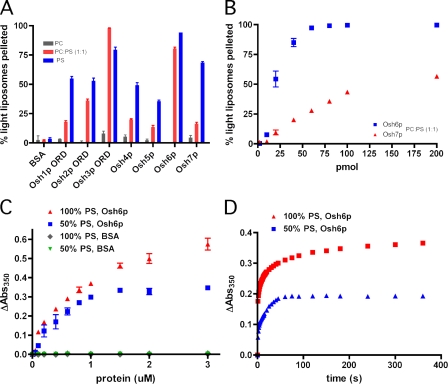
The nonvesicular transfer of sterols between the ER and PM slows dramatically in cells lacking the Osh proteins (Raychaudhuri et al., 2006; Sullivan et al., 2006). To better understand how Osh proteins interact with membranes, we purified the ORDs of all seven Osh proteins and used a pull-down assay to determine their ability to bind to and sediment with liposomes. We used liposomes containing various amounts of acidic phospholipids because many Osh proteins are known to have affinity for membranes containing these lipids (Li et al., 2002; Fairn and McMaster, 2005; Hynynen et al., 2005; Wang et al., 2005a), which is a finding we confirmed (Fig. S1 A). Surprisingly, we also found that the ORDs caused some liposomes to aggregate, particularly those containing high levels of acidic phospholipids. For this assay, we used two populations of liposomes: dense, sucrose-loaded liposomes that pellet at 16,000 g and light liposomes that contain an isoosmotic saline buffer and do not pellet at 16,000 g. We found that the ORDs of the Osh proteins caused the light liposomes to aggregate and pellet with dense liposomes (Fig. 2 A) and that the aggregation is dependent on protein concentration (Fig. 2 B). Aggregation was most efficient when the liposomes contained significant amounts of the acidic phospholipids phosphatidylserine (PS) or phosphatidic acid (Fig. S1 B). No aggregation was seen in control reactions with BSA, indicating that aggregation by Osh proteins was not the result of nonspecific interactions of proteins with liposomes.
Figure 2.
ORDs can bind two membranes simultaneously. (A) Dense, sucrose-loaded liposomes with the indicated lipid composition were incubated with light liposomes of identical lipid composition plus a trace amount of [3H]triolein (0.5 mM each) with and without purified ORD or BSA (0.4 µM) for 30 min at 30°C. Radioactivity in the supernatant after centrifugation was measured by scintillation counting. (B) Liposome pull-down assay performed as in A but with varying concentrations of Osh6p or Osh7p. (C) Turbidity assay (absorbance at 350 nm after 10 min) using 90 µM lipid and varying concentrations of Osh6p or BSA. (D) Time course of change in turbidity using 900 µM lipid and 0.4 µM Osh6p. Error bars indicate mean ± SEM (n = 3).
We confirmed these findings in three ways. First, using turbidity as an indication of liposome association, we showed that Osh6p causes liposome aggregation in a concentration-dependent fashion (Fig. 2 C). Using this assay, we found that the reaction required <5 min to reach maximal turbidity with a half-time in the range of 1–5 s (Fig. 2 D). In a second approach, we confirmed that Osh proteins cause membrane association using cryo-EM. In the absence of protein, 1:1 PS/phosphatidylcholine (PC) liposomes were almost never seen to be associated with one another; however, Osh4p and Osh6p caused dramatic association and aggregation of liposomes (Fig. S2). Finally, we ruled out the possibility that Osh proteins cause liposome fusion rather than association. Liposomes that had been aggregated by Osh proteins were treated with trypsin to digest the Osh proteins, which fully reversed tethering (Fig. S3 A), and we could not detect any liposome fusion with fluorescence-based lipid- and content-mixing assays (Fig. S3, B and C). Collectively, these findings indicate that all of the Osh ORDs can cause liposomes to associate and thus suggest that ORDs can bind two membranes simultaneously.
The ability to bind and transfer sterols is likely another shared property of most ORPs; many mammalian ORPs bind cholesterol or oxysterols (Suchanek et al., 2007) and some transfer sterols in vitro (Ngo and Ridgway, 2009). We found that all of the yeast ORPs transfer cholesterol in vitro, although Osh6p and Osh7p transported very poorly with the liposomes we used in this assay (Fig. S4). It may be that they transfer sterols more efficiently in other conditions, primarily transfer other lipids, or that their main function is not lipid transfer.
Osh proteins have more than one membrane-binding surface
The ability of ORDs to bind two membranes simultaneously suggests that either they have more than one membrane-binding surface or that they bind membranes as oligomers. We used Osh4p as a model ORD because the structure of this ORP is known (Im et al., 2005). Using analytical ultracentrifugation, we confirmed that Osh4p is a monodisperse monomer in solution (Fig. S5). To determine whether Osh4p oligomerizes when binding membranes, we treated it with a variety of bifunctional cross-linkers in the presence of liposomes. Because we were unable to detect multimers after cross-linking (unpublished data), it seems likely that Osh4p interacts with membranes as a monomer and does not oligomerize on membranes. We were also unable to detect multimers of other yeast ORDs after cross-linking (unpublished data), suggesting that they all interact with membranes as monomers. Therefore, we investigated whether Osh4p has more than one membrane-binding surface, which would allow it to interact with two membranes simultaneously.
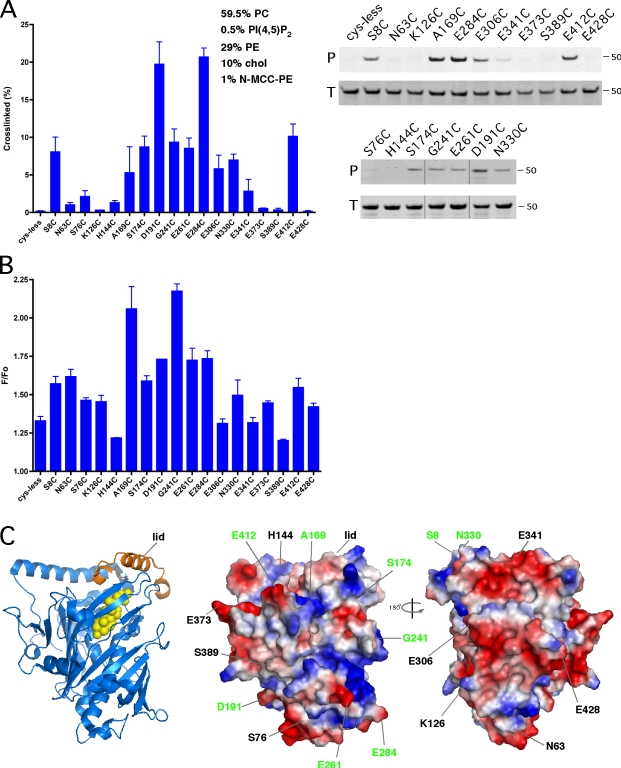
We generated a cysteine-less version of Osh4p and introduced further mutations wherein single residues were replaced with cysteines. Sites for cysteine replacement were chosen with the sole requirement that the site be accessible to the exterior surface of the protein. The cysteine-less Osh4p and the single-cysteine mutants retained the capacity to extract and transport sterol between membranes, indicating that the mutations did not substantially alter the protein structure (unpublished data). The single-cysteine mutants were mixed with liposomes containing a phospholipid with a maleimide headgroup, N-MCC-PE (1,2-di-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine-N-[4-(p-maleimidomethyl)cyclohexane-carboxamide), which can react with free sulfhydryls. Cross-linking of protein to the liposome requires the cysteine to come within ∼10 Å of the membrane surface. The cysteine replacements that resulted in the greatest degree of cross-linking were largely in two regions of the protein: near the mouth of the sterol-binding pocket (S8C, A169C, S174C, N330C, and E412C) and residues that are distal to this site on the other side of the protein (Fig. 3 A; D191C, E261C, and E284C). The wide distribution of residues that come in close contact with the membrane surface is consistent with the ability of Osh4p to interact with two membranes simultaneously.
Figure 3.
Osh4p has more than one membrane-binding surface. (A) The indicated mutations were introduced into Osh4p lacking endogenous cysteines (cys-less). The proteins were incubated with liposomes with the indicated lipid composition (N-MCC-PE contains a sulfhydryl-reactive headgroup) for 2 h at RT. The amount of protein pelleting with the membranes after washing (P) and the total amount of input protein (T) were determined by SDS-PAGE. The percentage of protein in P is shown (n = 3). Black lines indicate that intervening lanes have been spliced out. (B) Single-cysteine mutants were allowed to react with a sulfhydryl-reactive NBD derivative. The ratio of the fluorescence of the proteins before (Fo) and after (F) the addition of liposomes (99:1 PS/PI(4,5)P2) was measured (n = 6–8). (C, left) The structure of Osh4p (blue) bound to cholesterol (yellow) is shown. The flexible lid domain is shown in orange. (middle) A surface rendering of Osh4p, indicating the positively (blue) and negatively (red) charged surface. (right) The opposite side of the protein is shown. The positions of the residues changed to cysteine are indicated, and those that may interact with liposomes are shown in green. Error bars indicate mean ± SEM.
It should be noted that a few residues not in either of these two regions also cross-linked to membranes (E306C, E341C, and G241C). Interestingly, G241 is in a loop that has previously been suggested to be important for PIP binding by Osh4p (Li et al., 2002). This loop may form a third membrane-binding surface on the Osh4p.
To confirm the results of our cross-linking experiments, we used a second approach to identify membrane-interacting surfaces on Osh4p. The same single-cysteine mutants were modified with an NBD derivative that reacts with thiols and incubated with liposomes containing a physiological mix of phospholipids. Because NBD fluorescence intensity increases in a hydrophobic environment, we used this as an indicator of residues that come into close contact with the membrane. We expressed the results as the ratio of the fluorescence of the NBD-modified Osh4p after liposome addition (F) to the fluorescence intensity before liposome addition (F0). When we used liposomes with a lipid composition similar to that used for Fig. 3 A, only very low F/F0 ratios were obtained (not depicted), probably because Osh4p only transiently interacts with liposomes with physiological levels of acidic phospholipids. Substantially higher F/F0 ratios were obtained when liposomes with high amounts of acidic phospholipids were used (Fig. 3 B). Several of the same residues that had the highest F/F0 ratios also cross-linked most efficiently in Fig. 3 A, suggesting that these residues come close to the bilayer when Osh4p binds membranes.
There were also notable differences in the results between the two approaches. For example, the residues that showed the highest degree of cross-linking (D191 and E284) did not show the strongest response using the NBD approach. These differences may be caused by the different lipid compositions of the liposomes used for the two techniques. In addition, the nature of the two techniques can affect the results; cross-linking requires only that the residues come close to the surface of the membrane, whereas an increase in NBD fluorescence occurs when the fluorophore enters the hydrophobic region of the bilayer. The cross-linking efficiency is also affected by small differences in the reactivity of the thiols groups in the cysteines, which does not influence the NBD results.
Collectively, these findings indicate that Osh4p has more than one membrane-interacting surface, a property that may allow it to bind two membranes at the same time. Fig. 3 C highlights the locations of Osh4p residues, which both techniques suggest contact the bilayer. Because the ORDs of all the yeast Osh proteins can bind two membranes simultaneously, it seems likely that all ORPs have multiple membrane-binding surfaces.
The distal membrane–binding surface of Osh4p is required for PIP-regulated sterol transfer in vivo
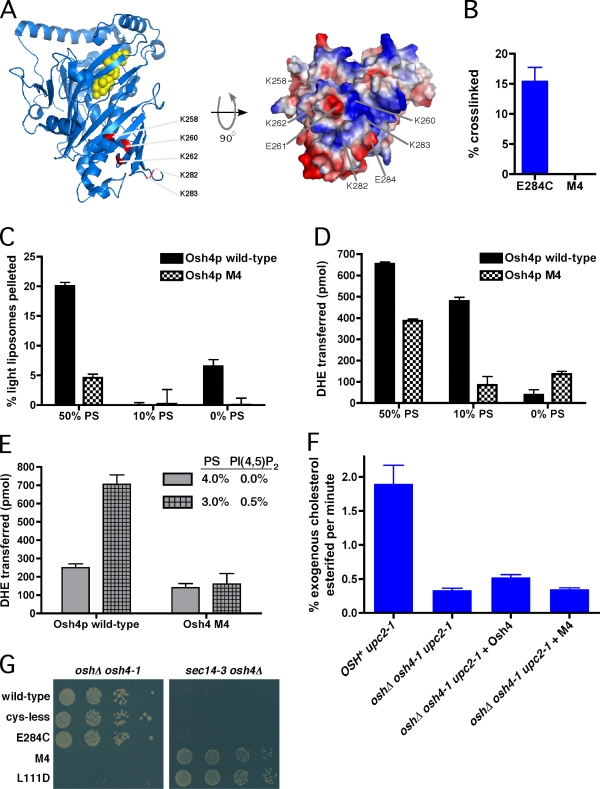
Our findings suggest that Osh4p has a membrane-binding surface near the entrance of the cholesterol-binding pocket and a second, distal membrane–binding surface. We introduced several mutations into Osh4p to ablate the distal membrane–binding surface of the protein. The results in Fig. 3 suggest that the loops containing E261 and E284 make close contact with membranes. Both loops contain several lysines that may be important for interaction with negatively charged phospholipid (Fig. 4 A; at positions 258, 260, 262, 282, and 283). We made a mutant derivative of the cysteine-less Osh4 protein called M4 in which all of these lysines were changed to glutamates. In addition, it also contains E261C and E284C. These changes did not significantly change the secondary structure of the protein, which was found to be identical with that of wild-type protein, as measured by circular dichroism (unpublished data). Thus, the mutations in M4 probably do not affect the membrane-binding surface near the mouth of the sterol-binding pocket.
Figure 4.
The distal membrane–binding surface of Osh4p is required for function. (A, left) Lysine residues changed to glutamate in the Osh4p mutant M4 are indicated in red, and the bound cholesterol are shown in yellow. (right) A surface rendering of Osh4p indicating the positively (blue) and negatively (red) charged surface. (B) Cross-linking experiments as in Fig. 3 A were performed with Osh4p mutant E284C or M4 proteins. The percentage of protein pelleting with the liposomes is shown. (C) Pull-down assay as described in Fig. 2 A using wild-type Osh4p or the M4 mutant and liposomes containing the indicated amount of PS. (D) DHE transport assays using 20 pmol wild-type Osh4p and liposomes containing the indicated amount of PS. The amount of DHE transferred in 1 h (30°C) was calculated by subtracting the amount of transfer in control reactions lacking protein. (E) DHE transport assays performed as in D but with the indicated liposomes. (F) [14C]cholesterol was added to cultures of strains with the indicated genotypes that contained plasmids that encoded no protein, wild-type Osh4p, or the M4 mutant. Samples were taken every 10 min for 40 min, and the rate at which the total amount of [14C]cholesterol was converted to cholesteryl ester per minute was calculated. (G) Plasmids encoding wild-type, cys-less, cys-less E284C, M4, or L111D Osh4p proteins were introduced into either oshΔ osh4-1 (left) or sec14-3 osh4Δ cells (right). Serial dilutions of the strains were incubated for 4 d at 37°C. Error bars indicate mean ± SEM (n = 3).
The mutations in M4 ablated the distal membrane–binding surface of Osh4p. Despite containing the E261C and E284C mutations, M4 did not become cross-linked to liposomes with the maleimide headgroup containing lipid N-MCC-phosphoethanolamine (PE), whereas a protein with only E284C was efficiently cross-linked (Fig. 4 B). The ability of M4 to aggregate liposomes was also significantly reduced compared with wild-type Osh4p, although it did retain the ability to aggregate liposomes with high amounts (50%) of the charged lipid PS (Fig. 4 C). Thus, the distal membrane–binding surface of M4 has a dramatically reduced affinity for membranes.
We next determined whether this reduced affinity affected the ability of M4 to transfer sterols between membranes. Liposomes with various amounts of PS were used in transport assays because we had shown that Osh4p aggregates liposomes containing high levels of PS (Fig. 2 A). We found that the ability of wild-type Osh4p to transfer sterol correlated with the amount of PS in the liposomes: it transferred most efficiently between liposomes containing 50% PS and almost not at all between liposomes lacking PS (Fig. 4 D). In comparison, M4 transferred sterols less efficiently than wild-type protein between membranes containing 10 or 50% PS but more efficiently between liposomes lacking PS, suggesting that the distal binding surface may affect sterol transfer. To more directly test this possibility, we asked whether the distal binding surface is required for Osh4p sterol transfer to be stimulated by PI-4,5-bisphosphate (PI(4,5)P2; Raychaudhuri et al., 2006). Using liposomes with relatively low PS content, we found that wild-type Osh4p was stimulated more than twofold by low amounts of PI(4,5)P2, whereas M4 was not (Fig. 4 E). These results indicate that the distal binding surface of wild-type Osh4p is responsible for regulating the efficiency of sterol transfer in response to PIP binding. They also suggest that the mutations in M4 do not indirectly affect the membrane affinity of the membrane-binding surface near the mouth of the sterol-binding pocket.
To determine whether this binding surface is also required for the function of the protein in vivo, we tested whether M4 could complement two yeast strains. Yeast requires any one of the seven Osh proteins for viability (Beh et al., 2001). The strain oshΔ osh4-1 lacks all seven OSH genes and contains a plasmid with the temperature-sensitive osh4-1 allele. M4 did not allow this strain to grow at the nonpermissive temperature, whereas wild-type Osh4p and cysteine-less Osh4p did (Fig. 4 G), indicating that M4 is not functional in vivo. As a control, we also tested L111D, a mutant that binds sterol poorly, which we previously showed does not complement this strain (Im et al., 2005). M4 was also not able to complement sec14-3 osh4Δ cells. Sec14p is an essential PI-PC LTP that is required for proper Golgi function (Mousley et al., 2007). However, cells lacking Osh4p do not require Sec14p for viability. Although the mechanism of this so-called “Sec14 bypass” phenotype is not known, it provides a way to determine whether Osh4p mutants are functional in vivo. Expression of functional Osh4p in sec14-3 osh4Δ cells renders them unable to grow at the nonpermissive temperature. In contrast, M4 and L111D did not prevent growth of this strain, indicating that they are not functional (Fig. 4 G). We ruled out the possibility that M4 fails to complement these strains because it was degraded; GFP-tagged M4 is expressed at the same level as functional Osh4p mutants (unpublished data). Therefore, M4 is not functional in vivo.
Because M4 is not functional in cells, we wondered if it has a reduced ability to mediate sterol transfer in vivo. We previously demonstrated that the rate of PM to ER sterol transfer slows significantly in oshΔ osh4-1 cells but is not affected in mutants with conditional defects in vesicular transport (Li and Prinz, 2004; Raychaudhuri et al., 2006). To measure PM to ER sterol transfer, we exploited the ER localization of the proteins that esterify free sterols (Zweytick et al., 2000). The esterification of exogenous radiolabeled cholesterol indicates that it has been transferred from the PM to the ER. To facilitate uptake of exogenous cholesterol, we used strains with an allele of a transcription factor (upc2-1) that permits cholesterol uptake under aerobic growth conditions (Crowley et al., 1998). Using this assay, we found that expression of wild-type Osh4p in oshΔ osh4-1 cells modestly restores the rate of PM to ER sterol transfer at the nonpermissive temperature, although it does not achieve the rate seen in cells expressing all seven Osh proteins (Fig. 4 F; Raychaudhuri et al., 2006). In contrast, the M4 mutant was not able to restore PM to ER cholesterol transport at all. It should be noted that the strains take up sterols with slightly different efficiencies, and to facilitate comparisons, we have expressed these results as the percentage of total cholesterol taken up that is esterified per minute. We have previously demonstrated that the rate of esterification is relatively constant over a wide range of sterol uptake rates (Li and Prinz, 2004). Collectively, these findings indicated that the M4 mutant is not functional in cells and fails to facilitate PM to ER sterol transfer in vivo. Thus, the distal membrane–binding surface of Osh4p is required to efficiently facilitate sterol transfer in cells.
PIPs in one membrane may modulate sterol extraction and delivery by Osh4p to a second membrane
Previously, we demonstrated that sterol transfer by Osh4p in vitro is enhanced by small amounts (0.5 mol%) of PI(4,5)P2 in either the donor or the acceptor liposomes (Raychaudhuri et al., 2006). We speculated that PI(4,5)P2 enhanced the dwell time of Osh4p on the liposome surface, increasing the probability that the protein would extract or deliver sterol to membranes containing PI(4,5)P2. However, our finding that the distal membrane–binding surface ablated in the M4 mutant is required for PI(4,5)P2 stimulation of sterol transfer (Fig. 4 E) led us to consider that PI(4,5)2 might stimulate sterol transfer by Osh4p by a second mechanism: Osh4p might be able to extract or deliver sterols from one bilayer while simultaneously interacting with PI(4,5)P2 in a second membrane.
To test this, we determined whether PIPs in one set of liposomes could rapidly affect the rate at which Osh4p extracts and delivers sterols to a second membrane. We mixed liposomes containing PC and [14C]cholesterol (99:1) with an equimolar amount of a second set of liposomes consisting either of 100% PC (control), 99.5:0.5 PC/PI(4,5)P2, or 99:1 PC/PI(4)P. Osh4p was added, and the sterol occupancy of Osh4p was determined over time. Remarkably, the PI(4,5)P2-containing liposomes dramatically increased the equilibration rate by approximately one order of magnitude from 130 to 18 s (Fig. 5 A). It also slightly increased the maximal binding (Bmax) from 0.13 to 0.17 pmol cholesterol/pmol Osh4p. Liposomes containing PI(4)P had the opposite effect. They did not affect the half-time of occupancy but reduced the Bmax to 0.08 pmol cholesterol/pmol Osh4p (Fig. 5 A). Because Osh4p does not measurably sediment with liposomes containing 1.0% PI(4)P (unpublished data), we were able to rule this out as an explanation for this decrease. Similar effects were seen when Osh4p was incubated with the [14C]cholesterol-containing liposomes for 15 min before addition of the PIP-containing liposomes; within 1 min of adding PIP-containing liposomes, the amount of cholesterol bound by Osh4p was altered (Fig. 5 B). Interestingly, the amount of cholesterol bound was progressively reduced with an increasing concentration of PI(4)P-containing auxiliary liposomes, but increasing the relative amount of PI(4,5)P2-containing vesicles failed to increase the maximal extraction any further (Fig. 5 C). In addition, the effects of PIP binding on the sterol occupancy of Osh4p must take place in the context of a membrane because PIP headgroups did not affect sterol binding or transport by Osh4p (unpublished data).
Figure 5.
Cholesterol extraction by Osh4p is regulated by PIPs in a second membrane. (A) 200 pmol Osh4p was incubated at 30°C with 500 µM 99:1 PC/[14C]cholesterol liposomes together with 400 µM auxiliary liposomes containing 100% PC (red squares), 99.5:0.5 PC/PI(4,5)P2 (blue triangles), or 99:1 PS/PI(4)P (gray triangles). All liposomes were sucrose filled. At the indicated times, the samples were placed on ice, and the liposomes were pelleted. The amount of [14C]cholesterol in the supernatant was determined by scintillation counting. (B) 2 µM Osh4p was incubated at 30°C with 500 µM 99:1 PC/[14C]cholesterol liposomes. After 15 min (arrow), auxiliary liposomes were added, and the amount of [14C]cholesterol extracted was determined as in A (n = 3). (C) Performed as in B except that auxiliary liposomes were added to a final concentration of 400 µM (1×), 800 µM (2×), or 1.6 mM (4×). The total incubation was 30 min at 30°C (n = 4). Error bars indicate mean ± SEM.
The effects of PIP-containing liposomes on the sterol occupancy of Osh4p in this study are consistent with the model that PIPs in one liposome can modulate the rate at which Osh4p extracts and delivers sterols to a second membrane; PIPs in one liposome may be able to interact with Osh4p while it is extracting or delivering sterols to a second liposome. These findings do not exclude the possibility that the PIP-containing liposomes alter the sterol occupancy of Osh4p by other mechanisms, including sterol exchange with the PIP-containing liposomes or binding of Osh4p to these liposomes. For example, sterol transfer to the PIP-containing liposomes could explain our findings if liposomes with PI(4,5)P2 were poor acceptors for sterols delivered by Osh4p, whereas PI(4)P-containing liposomes were good acceptors. However, this is not the case; PI(4,5)P2 increases the rate of sterol transfer when it is in acceptor liposomes, whereas PI(4)P had no effect (Raychaudhuri et al., 2006).
Sterol transfer by Osh4p between closely apposed liposomes
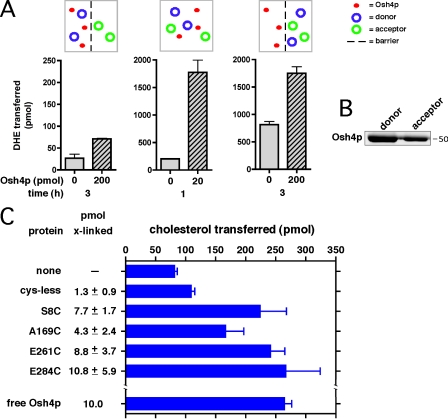
Our findings suggest that sterol extraction and delivery by Osh4p, and probably other Osh proteins, can be regulated by its interactions with a nearby membrane. One place where membranes are closely apposed is MCSs, and it is has been proposed that ORPs (and other LTPs) might work more efficiently at these sites because they have to diffuse only a short distance between membranes (Holthuis and Levine, 2005; Levine and Loewen, 2006; Hanada et al., 2007). This led us to investigate whether the sterol transfer rate by Osh4p is affected by the rate at which it diffuses between membranes. To reduce the rate of Osh4p diffusion between membranes, we set up a sterol transfer reaction in which the donor and acceptor liposomes were separated by a barrier (a Nuclepore polycarbonate membrane with 1.0-µm pores) that Osh4p can cross but the liposomes cannot. The donor liposomes contained the fluorescent sterol dehydroergosterol (DHE), and transfer to acceptor liposomes was measured fluorometrically. At the start of the assay, a 50-µl suspension containing donor liposomes and 200 pmol Osh4p were added to the side of the barrier (the input chamber); a Nuclepore membrane was laid over the mixture, and a 50-µl suspension of acceptor liposomes was added to the output chamber. Osh4p transferred sterol slowly in these conditions, moving only 0.1 pmol DHE/pmol protein/h (Fig. 6 A, left). We confirmed that the barrier did not prevent Osh4p diffusion, as substantial amounts of the protein were on both sides of the barrier at the end of the transfer assay, although it had not yet completely equilibrated across the barrier (Fig. 6 B). In contrast, sterol transfer was significantly faster in two conditions in which the donor and acceptor liposomes were not separated by a barrier. In the first, when no barrier was present, the transfer rate was 75 pmol DHE/pmol protein/h (Fig. 6 A, middle). In the second, at the start of the reaction, donor and acceptor were on the same side of the barrier, and Osh4p was on the other side. In these conditions, the transfer rate was 1.5 pmol DHE/pmol protein/h (Fig. 6 A, right). Collectively, these findings suggest that sterol transfer by Osh4p is at least partially diffusion limited, and thus, Osh4p can transfer sterols more rapidly between closely apposed membranes.
Figure 6.
Sterol transfer by Osh4p requires close contact of membranes. (A) DHE transfer from donor to acceptor liposomes was determined fluorimetrically as described in Materials and methods. Reaction components were separated by a barrier (1-µm–pore size Nuclepore membrane) where indicated. The distribution of components across the barrier at the start of the reaction is indicated. The amount of transfer with and without the indicated amount of Osh4p is shown (n = 2). (B) For the assays in A, the amount of Osh4p on the side of the barrier with the donor and acceptor liposomes at the end of the transfer assay was determined by SDS-PAGE. (C) Osh4p lacking endogenous cysteines and with the indicated mutations was covalently attached to sucrose-filled liposomes (containing 1% [14C]cholesterol) as described in Materials and methods. After a washing to remove unbound proteins, the amount of protein attached to the liposomes (picomole × linked) was determined (n = 3). These liposomes were incubated with acceptor liposomes for 1 h at 30°C, and the amount of [14C]cholesterol transferred to the acceptors was determined. (bottom) In a control reaction, the amount of transfer by free (not covalently attached) Osh4p and the same liposomes was determined. Error bars indicate mean ± SEM.
To better understand how Osh4p might transfer sterols between closely apposed membranes, we covalently attached the protein to donor liposomes and determined whether it could still transfer sterols to unattached acceptors. Some of the single-cysteine Osh4p mutants were treated with a bifunctional cross-linker that reacts with sulfhydryls and carbohydrates, thereby linking the cysteine to liposomes containing 1 mol% of the ganglioside GM1. Liposomes were washed to remove nonattached Osh4p. Remarkably, we found that the covalently attached Osh4p retained the ability to transfer cholesterol between liposomes (Fig. 6 C). The cross-linked proteins transferred cholesterol about as well as freely diffusing (non–cross-linked) wild-type protein. Interestingly, all of the cysteine mutants we tested transferred cholesterol with about the same efficiency. This is probably because the proteins were attached to the membranes with an ∼30-Å-long linker, allowing the protein some freedom of movement on the membrane. In contrast, when the proteins were attached to liposomes containing a maleimide headgroup (N-MCC-PE), which has a 10-Å linker, the proteins were not able to transfer cholesterol (unpublished data). Collectively, the findings indicate that Osh4p can transfer sterols while remaining bound to liposomes and that it can pivot between the membranes to transfer sterols between closely apposed membranes.
Four Osh proteins are enriched on ER close to the PM
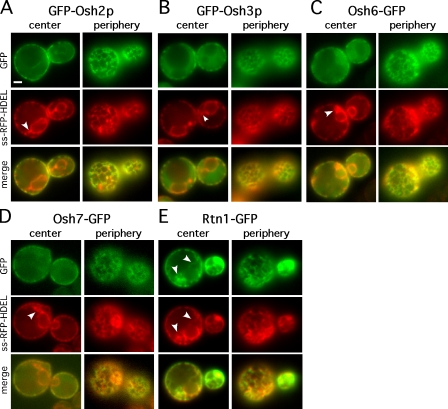
Our findings suggest that Osh proteins could transfer lipids most efficiently between closely apposed membranes, e.g., at MCSs. It has previously been shown that Osh1p is enriched at the MCS between the nucleus and the vacuole, whereas Osh2p, Osh3p, and Osh6p are enriched in patches at the cell cortex that were suggested to be ER–PM contact sites (Levine and Munro, 2001; Loewen et al., 2003; Wang et al., 2005a,b). It should be noted that these studies showed that Osh2p and Osh3p only have a mostly cortical localization in cells overexpressing Scs2p, an ER-resident FFAT-binding protein. We wanted to better understand the localization of Osh proteins in the cell cortex in these conditions. Using an RFP-labeled ER marker (ss-RFP-HDEL) along with GFP-tagged Osh proteins, we found that Osh2p, Osh3p, Osh6p, and Osh7p are enriched on regions of the ER in close proximity with the PM (Fig. 7, A–D). GFP-Osh2 and -Osh3 were visualized in cells overexpressing Scs2p. When we focused on the center of the cells, we found that the GFP fusions were largely absent from the perinuclear ER and ER tubules that are not near the PM (Fig. 7, arrowheads). Focusing on the periphery of the cells confirmed that the Osh-GFP fusions near the PM were enriched on the ER. Importantly, this localization differs from that of the reticulon Rtn1p, which is largely absent from the perinuclear ER, but unlike the Osh proteins, is found in ER tubules that are not near the PM (Fig. 7 E, arrowheads). As an additional control, we demonstrated that a typical PM protein, Hxt3p, does not colocalize with ER tubules near the PM (unpublished data).
Figure 7.
Four Osh proteins are enriched on ER near the PM. Cells expressing the indicated GFP fusions and the ER marker ss-RFP-HDEL were visualized live, focusing on either the center plane or the periphery of the cells. Images taken in the GFP and RFP channels were merged. (A and B) GFP-Osh2p (A) and GFP-Osh3p (B) were expressed in a strain that overexpressed Scs2p. (C–E) Osh6-GFP (C), Osh7-GFP (D), and Rtn1-GFP (E) were expressed in wild-type cells. Arrowheads indicate ER tubules that are not closely apposed to the PM. Bar,1 µm.
These findings indicate that Osh2p, Osh3p, Osh6p, and Osh7p are enriched only on those regions of the ER that are closely apposed to the PM. Therefore, they may be enriched at PM–ER contact sites. To obtain additional evidence that Osh proteins are enriched at ER–PM MCSs, we attempted to isolate PM-associated membranes, a subfraction of the ER that associates with the PM during PM purification (Pichler et al., 2001). However, we were unable to reproducibly isolate PM-associated membrane and could not confirm that it was enriched in Osh proteins. Collectively, these findings suggest that some Osh proteins are enriched at PM–ER contact sites.
Osh proteins can bind the PM and ER simultaneously in vitro
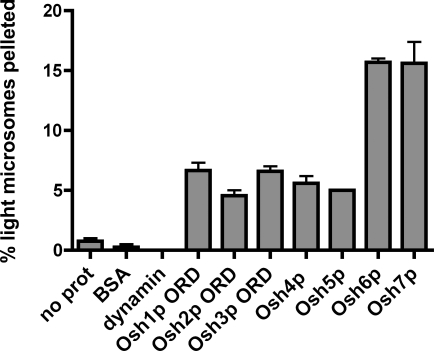
If Osh proteins localize to PM–ER contact sites, we wondered if they had affinity for both membranes and could cause them to aggregate. We found that the ORDs of all of the Osh proteins were able to cause some aggregation between inside-out PM (ISO-PM) vesicles and light microsomes, although Osh6p and Osh7p were the most efficient (Fig. 8). In control reactions, BSA and the peripheral membrane protein dynamin did not aggregate these membranes. Thus, Osh proteins cannot only bind two liposomes simultaneously, but they can also bind two cellular membranes at the same time. This could contribute to the enrichment of some Osh proteins at MCSs. However, it seems unlikely that Osh proteins are needed to create or maintain close contacts between the ER and PM. We found that most of the peripheral ER remains at the cell cortex and that the structure of the ER was not significantly altered in cells lacking Osh proteins (unpublished data).
Figure 8.
ORDs can simultaneously bind ISO-PM and ER membranes. Sucrose-loaded ISO-PM vesicles were incubated with light microsomes (that do not sediment at 16,000 g) and 0.4 µM of the indicated proteins for 30 min at 30°C. Mixtures were centrifuged for 10 min at 16,000 g, and the percentage of light microsomes pelleting was determined by quantitative immunoblotting. Error bars indicate mean ± SEM (n = 3).
Discussion
In this study, we demonstrate that yeast ORPs can interact with two membranes simultaneously and facilitate regulated sterol transfer by a novel mechanism. Using Osh4p as a representative Osh protein, we identified a membrane-binding site on Osh4p that is distal to the membrane-binding surface near the mouth of the sterol-binding pocket. Our findings suggest that lipid content of the liposome interacting with the distal membrane–binding surface affects the rate of sterol extraction from and delivery to the second liposome. We confirmed the functional importance of the distal membrane–binding surface of Osh4p by altering it and showing that the mutant protein cannot replace the wild-type protein or facilitate sterol transfer in cells and that it is insensitive to the stimulatory effect of PI(4,5)P2 on sterol transfer in vitro. Because all yeast ORDs can bind two membranes simultaneously, they likely all have similar regulatory distal membrane–binding surfaces, although they almost certainly differ in terms of the preference for PIPs and other lipids.
The ability of the ORD's distal membrane–binding surface to regulate sterol extraction and delivery to a second membrane suggests a new mechanism for sterol transfer by ORPs. We have previously proposed that ORPs transport sterols by extracting a sterol from one membrane, diffusing through the aqueous phase, and delivering the bound sterol to a second membrane (Im et al., 2005; Raychaudhuri et al., 2006). Although our findings do not rule out this model, we now propose that ORPs extract or deliver sterols to a membrane most efficiently when they simultaneously interact with a second membrane via the distal membrane–binding surface of the ORD, which could occur at an MCS or any place where two membranes are closely apposed. Consistent with this model, we show that Osh4p transfers sterols most efficiently when the donor and acceptor membranes can come in close contact. In addition, we found that Osh4p transfers sterols as efficiently when it is covalently attached to a liposome as when it is free in solution. This was surprising because Osh4p probably cannot transfer a sterol between two membranes without detaching from both membranes; the movement of a sterol between a membrane and the hydrophobic binding pocket of an ORP probably only occurs when the entrance to the pocket is very close to or partially buried in the membrane. Thus, sterol transfer requires that ORPs detach from membranes. This is supported by our cross-linking data, which show that Osh4p still transfers sterols efficiently while covalently attached to one set of liposomes, but only if a cross-linker with a large arm length was used, indicating that the protein must be able to pivot between membranes to move sterols between them.
The ability of the distal membrane–binding surface of ORPs to regulate sterol extraction and deliver to a second membrane suggests how sterol transfer might be driven primarily in one direction between pairs of membranes at an MCS. The distribution of various PIP species in cellular membranes is highly regulated and membrane specific (Vicinanza et al., 2008). For example, PI(4,5)P2 is highly enriched in the PM but largely absent from the ER. Thus, an asymmetric distribution of PIPs across the two membranes of an MCS could result in net sterol transport primarily in one direction because the net result would be a differential probability of sterol extraction/delivery at each organelle. However, this is difficult to demonstrate in vitro because both the donor and acceptor liposomes in a transfer assay can associate with the distal binding surface of ORDs; thus, PIPs in either donor or acceptor liposomes could regulate transfer. Interestingly, a recent study has demonstrated PIP-induced directional transport in vitro by the mammalian ORP9L and OSBP; transport was stimulated only when PI(4)P was in the acceptor but not donor liposomes (Ngo and Ridgway, 2009). This ORP contains a PH domain that is not found in Osh4p. PI(4)P binding by the PH domain may position the ORD of these ORPs so that sterol transport is favored in one direction only.
We also found that four of the seven Osh proteins are enriched on regions of the ER that are closely apposed to the PM, which suggests that they may be enriched at PM–ER contact sites as has been previously reported (Levine and Munro, 2001; Loewen et al., 2003; Wang et al., 2005a,b). Other yeast and mammalian ORPs have also been found at MCSs, suggesting that this may be a common feature of this protein family (Levine and Munro, 2001; Rocha et al., 2009). Because the ORD of Osh4p and probably other ORPs is ∼6 nm in diameter, ORDs could only directly contact both membranes at an MCS when the bilayers are very closely apposed. This raises the question of whether the PM and ER can come close enough for ORDs to interact with both membranes simultaneously. The mean distance between the PM and ER in yeast is not well established. In Jurkat cells, the mean distance between these organelles is 17 ± 10 nm (Wu et al., 2006), which is seemingly too large to be readily bridged by an ORD. However, other findings suggest that the PM and ER are not held at a fixed distance from each other at contact sites but are dynamic and can come within at least 6 nm. Expression of putative PM–ER cross-bridging proteins, the junctophilins, in embryonic amphibian cells induced the formation of structures in which PM and ER approached within a mean distance of ∼7.6 nm (Takeshima et al., 2000). Additionally, Várnai et al. (2007) were able to cross-link proteins in the PM and ER in live mammalian cells using cross-linkers that require the membranes to be only ∼4–6 nm apart. Thus, the PM and ER can come close enough, at least transiently, for ORDs to interact with both membranes simultaneously.
What determines the localization of Osh6p and Osh7p, which contain only ORDs, is unclear. Perhaps not surprisingly, we found that the ER-localized FFAT-binding protein Scs2p is not required to enrich these proteins at PM–ER junctions (unpublished data). We also found that depleting cells of acidic phospholipids PI(4)P, PI(4,5)P2, or PS did not affect the localization of the proteins (unpublished data). The ability of ORDs to interact with two membranes simultaneously could help promote the enrichment of Osh6p and Osh7p (and perhaps other ORPs) at MCSs.
The enrichment of ORPs at MCSs may serve several functions in the cell. First, it might facilitate the bulk transfer of sterols or perhaps other lipids between organelles. Although the transfer rate of Osh proteins in vitro suggests that they may not transfer sterols rapidly enough to significantly contribute to bulk sterol transfer, our findings indicate that they probably transfer sterols much more efficiently at MCSs. Second, it is also possible that the primary function of some ORPs is not bulk lipid transfer between organelles but rather a fine tuning of the sterol concentration of organelles or subdomains of organelles such as MCSs. Thus, they might transiently alter the sterol content of one organelle in response to a signal, such as a change in PIP levels, in the second organelle. Third, ORPs could also function as lipid sensors at MCSs rather than lipid transporters. Because ORPs can interact with two membranes simultaneously, they could sense not just a single lipid but respond to differences in the lipid composition of two organelles. Such a form of coincidence detection could be useful at MCSs, which may be highly specialized structures that are defined by their lipid composition as much as their protein components. ORPs could also more directly function in a signaling pathway by transmitting a signal directly between two organelles. For example, when positioned at MCSs, they might add or remove a signaling lipid from one membrane in response to binding PIPs in a second membrane.
In summary, we demonstrate a novel mechanism of interorganellar communication and lipid exchange between closely apposed membranes. The core lipid-binding ORD domain of ORPs can sense the lipid composition of one membrane and simultaneously modify the sterol content of a second membrane. Identifying other proteins that work in concert with ORPs will lead to a better understanding of how they transfer lipids and signals between cellular compartments or subdomains of organelles.
Materials and methods
Strains and plasmids
The strains and plasmids used in this study are listed in Table I.
Table I.
Strains and plasmids used in this study
| Strain/plasmid | Genotype or description | Source |
| SEY6210 | MATa ura3-52, his3-Δ200, leu2-3, -112 trp1-Δ901, suc2-Δ9, lys2-801 | C. Beha |
| CBY926 | SEY6210 osh1Δ::kan-MX4 osh2 Δ::kan-MX4 osh3Δ::LYS2 osh4Δ::HIS3 osh5Δ::LEU2 osh6Δ::LEU2 osh7Δ::HIS3 / osh4-1, CEN-TRP1 | C. Beh |
| SEY6218 | MATa ura3-52, his3-Δ200, leu2-3, -112 trp1-Δ901, suc2-Δ9, lys2-801 | S. Emrb |
| AAY102 | SEY6218 stt4::HIS3 / pRS415 CEN-LEU2-stt4-4 | S. Emr |
| SD102 | MATa leu2 ura3 rme1 trp1 his3Δ GAL+ TOF3 mss4::HIS3MX6 / YCplac111::mss4-2ts | C. Jacksonc |
| CVY215 | Mat a leu2-3 -112 his 3-11-15 trp1-1 ura3-1 ade2-1 /ss-RFP-HDEL:TRP1 | This study |
| WPY979 | CVY215 HXT3-GFP:HIS3 | This study |
| WPY1009 | CVY215 RTN1-GFP:HIS3 | This study |
| CVY433 | CVY215 cho1::kan-MX4 | This study |
| NDY93 | MATa sec14-3 osh4::kan-MX4 ura3-1 his 3-11, -15 leu2-5, 112 | Laboratory collection |
| NDY75 | CBY926 upc2-1:URA3 | Laboratory collection |
| pJK59 | Sec63-GFP, SEC63 promoter (URA3/CEN) | Laboratory collection |
| pCV19 | Rtn1-GFP, RTN1 promoter (URA3/CEN) | Laboratory collection |
| pTL312 | PHO5 promoter, GFP- Osh2p (URA3/CEN) | T. Levined |
| pTL313 | PHO5 promoter, GFP-Osh3p (URA3/CEN) | T. Levine |
| YIplac204/TKC-DsRed-HDEL | Encodes ss-RFP-HDEL (TRP) | B. Glicke |
Simon Frasier University, Vancouver, British Columbia, Canada.
Cornell University, Ithaca, NY.
Centre National de la Recherche Scientifique, Gif-sur-Yvette, France.
Institute of Ophthalmology, University College London, London, England, UK.
University of Chicago, Chicago, IL.
Recombinant protein expression and purification
Proteins were expressed in Escherichia coli as GST fusions. The customized plasmid (pGST-1) containing GST-OSH4 was provided by Y.-J. Im (Laboratory of J. Hurley, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health). All cysteine-less and single-cysteine mutants of OSH4 were generated with a site-directed mutagenesis kit (Agilent Technologies) using that plasmid as template. ORDs of the remaining six yeast ORPs were expressed in E. coli BL21 cells as N-terminal GST fusions in the pGEX-4T-3 vector (Invitrogen). Osh4p, Osh5p, Osh6p, and Osh7p were expressed as full-length proteins. The ORDs of other Osh proteins were Osh1 (727–1,189), Osh2 (794–1,284), and Osh3 (524–997).
To induce expression of fusion protein, BL21 cells containing an expression plasmid (pGEX-4T-3 or pGST-1; see previous paragraph) were grown overnight at 37°C in 100 ml Luria broth containing 100 µg/ml ampicillin (LB-Amp). The next morning, 1 liter LB-Amp was inoculated with 100 ml preculture and allowed to grow for 1.5 h at 37°C. Protein expression was subsequently induced by adding IPTG to a final concentration of 100–200 µM and incubating the cells for ∼4 h at 30°C. Cells were harvested by centrifugation and lysed by sonication for 2.5 min on ice in a buffer containing PBS, 1 mM EDTA, 0.5% Triton X-100, and protease inhibitor cocktail (Roche). Lysate was exposed to glutathione agarose resin (Sigma-Aldrich) to isolate the fusion protein, and the resin was extensively washed with lysis buffer. To cleave the GST tag, the resin with bound GST fusion protein was exposed to either AcTEV protease in proprietary buffer (for all plasmids derived from pGST-1; Invitrogen) or thrombin (Sigma-Aldrich) in PBS at 23°C for 2 h. The eluate containing the GST-free recombinant protein was removed and concentrated followed by adjustment to a concentration of 1 mg/ml.
Fluorescence microscopy
Yeast strains were grown in synthetic complete medium (0.67% yeast nitrogen base and 2% glucose) and imaged live in medium at room temperature using a microscope (BX61; Olympus) with a UPlan Apo 100×/1.35 NA lens, a camera (Retiga EX; QImaging), and IVision software (version 4.0.5). The brightness and contrast of the images were adjusted with Canvas software (version 10.4.9; ACD Systems). The GFP fusions to the Osh proteins were expressed on CEN plasmids under the PHO5 promoter (GFP-Osh2 and -Osh3) or under the endogenous promoter (Osh6- and Osh7-GFP).
Preparation of liposomes
Most phospholipids were obtained from Avanti Polar Lipids, Inc. and, unless otherwise noted, are all dioleoyl (di-18:1). Lissamine rhodamine (Rho) DHPE (Rho B 1,2-dihexadecanoyl-sn-glycero-3-PE) was obtained from Invitrogen. DHE and cholesterol were obtained from Sigma-Aldrich. [14C]cholesterol and [3H]triolein were obtained from American Radiolabeled Chemicals. Liposomes were prepared essentially as described previously (Raychaudhuri et al., 2006). Most liposomes were hydrated in standard vesicle buffer (20 mM Hepes, pH 7.3, 100 mM NaCl, and 1 mM EDTA) followed by at least five freeze-thaw cycles (1 min in liquid nitrogen followed by 3 min in warm water) and subsequent extrusion through a 0.4-µm–pore size track-etched Nuclepore membrane (Whatman) using a mini extruder (Avanti Polar Lipids, Inc.). Sucrose-loaded liposomes were created by resuspending the dried lipid in sucrose vesicle buffer (20 mM Hepes, pH 7.3, 180 mM sucrose, and 1 mM EDTA). After extrusion, the sucrose-containing liposomes were diluted 1:5 in standard vesicle buffer and centrifuged at 16,000 g for 10 min. The lipid pellet was gently resuspended in standard (sucrose free) vesicle buffer.
Liposome association and tethering assays
Sucrose-loaded liposomes were prepared as described in the previous paragraph. 950 µM liposomes of varying composition were incubated with 6 µg (∼120 pmol) of protein for 30 min at 30°C. Liposomes were pelleted at 16,000 g for 10 min, and the top 90 µl of supernatant was separated. Proteins were purified from the pellet and supernatant fractions by cold acetone precipitation, dissolved in SDS sample buffer, and separated by SDS-PAGE on a 12-well NuPage 4–12% Bis-Tris polyacrylamide gel (Invitrogen). Gels were stained with Coomassie blue. For sedimentation-based tethering assays, 450 µM sucrose-loaded liposomes were mixed with 450-µM tracer liposomes (made in standard vesicle buffer) containing trace amounts of [3H]triolein. After incubation with protein, sucrose-loaded liposomes were pelleted by centrifugation, and the radioactivity remaining in 50 µl supernatant was measured by scintillation counting in Bio-Safe II fluid (Research Products International) in a counter (LS 6500; Beckman Coulter). Turbidity assays were performed in a spectrophotometer (UV160U; Shimadzu) set to read absorbance values at 350 nm. Liposomes (50:50 PC/PS or 100% PS) were prepared in standard vesicle buffer as described in the previous paragraph but extruded through a 0.1-µm–pore size membrane (Whatman). The time-based assay used 0.4 µM protein and 900 µM lipid, whereas the protein concentration curves used 90 µM lipid. Total volume in all cases was 200 µl, and readings were measured in a quartz cuvette.
Organelle tethering assays
Microsomes and ISO-PM membranes were purified exactly as previously described (Zinser and Daum, 1995; Fritz et al., 1999). For the ISO-PM tethering experiments, 400 µg ISO-PM membranes (purified as sucrose-loaded vesicles) was mixed with 100 µg microsomal membranes and 80 pmol of the indicated proteins in a total volume of 200 µl 20 mM Hepes, pH 7.3, 180 mM NaCl, and 1 mM EDTA. After 30 min at 30°C, the dense ISO-PM were pelleted at 16,000 g. Proteins from the pellet and supernatant were separated by SDS-PAGE, and the amount of the ER protein Dpm1p was determined by quantitative immunoblotting using anti-Dpm1p (Invitrogen).
Radiolabeled sterol extraction and transfer assays
Extraction and transfer assays were performed as previously described (Raychaudhuri et al., 2006) with the following modifications. For the extraction assay, a 50-µl suspension of prewarmed (30°C) 99:1 PC/[14C]cholesterol liposomes was mixed with 40 µl prewarmed auxiliary liposomes containing 100% PC, 99.5:0.5 PC/PI(4,5)P2, or 99:1 PC/PI(4)P. All liposomes were sucrose filled. Prewarmed Osh4p (200 pmol in 10 µl) was immediately added to the donor/auxiliary liposomes mixture. Reactions were performed at 30°C and terminated on wet ice. Mixtures were centrifuged for 10 min at 16,000 g to clear the sucrose-loaded liposomes, and 50 µl supernatant was removed for scintillation counting. The amount of radioactivity in control reactions without protein was subtracted to calculate the amount of [14C]cholesterol extracted.
For transfer assays, donor liposomes were sucrose loaded and contained 60:20:10:9:1 PC/PE/PS/cholesterol/[14C]cholesterol. Acceptor liposomes were created in standard vesicle buffer and were composed of 70:20:10 PC/PE/PS. After incubation of liposomes (450 µM donors and 450 µM acceptors) with protein (0.4 µM) at 30°C in a total reaction volume of 100 µl, donor liposomes were cleared from supernatant by centrifugation (16,000 g for 10 min). Radioactivity in 50 µl supernatant was measured by scintillation counting.
DHE transfer assay
All fluorometric readings were taken using a photomultiplier (detector voltage set to 750 W; model 814; Photon Technology International [PTI]) with a short arc lamp (75W Xenon; Ushio) as a light source. Data were collected and analyzed using the FeliX software (version 32; PTI). All readings were taken as 100 µl samples in a 3-mm quartz cell (Starna Cells). All liposomes were prepared in standard vesicle buffer. For the sterol transfer assay, donor liposomes incorporated 9 mol% DHE, and acceptor liposomes include 2.5 mol% dansyl-PE. Liposomes contained the stated amount of PS, 20% PE, and the remainder PC. Sterol transport assays were performed at the same protein and lipid concentrations as described in the previous paragraph, terminated by placing on ice, and diluted 1:1 immediately before fluorescence measurement. Energy transfer between DHE and dansyl-PE in the acceptor liposomes was expressed as the ratio of the excitation peaks at 330 and 344 nm (constant 498-nm emission wavelength and 2-mm slit sizes), a slight modification of a previously described method (John et al., 2002). A standard curve was generated using liposomes containing 2.5% dansyl-PE and varying concentrations of DHE.
DHE transfer with a barrier
DHE donor liposomes (10% PS and 0.5% PI(4,5)P2) and dansyl-PE acceptor liposomes (10% PS and 1% PI(4)P) were made as described in Preparation of liposomes and extruded through 1.0-µm Nuclepore track-etched membranes. Liposomes were loaded into the input chamber of a reusable Teflon standard dialyzer (Harvard Apparatus) in which the barrier membrane was a 1.0-µm–pore size Nuclepore track-etched membrane. Liposomes were dialyzed against 1 liter of vesicle buffer for 3 h at room temperature to remove liposomes that could pass freely through the membranes. The final liposome concentration was estimated by fluorimetrically assaying for DHE or dansyl content as appropriate. For the transfer assay, the input chamber of the standard dialyzer (50-µl capacity) was loaded with 10 µl of vesicle buffer or 200 pmol of Osh4p (10 µl of 1 mg/ml stock) and the predialyzed DHE donor liposomes (∼45 nanomoles of total lipid) and vesicle buffer (if necessary) to a total volume of 50 µl. A 1.0-µm Nuclepore track-etched membrane was laid over the input mixture, and the membrane was secured by the open-top screw lid. A 50-µl suspension of predialyzed dansyl-PE acceptor liposomes was gently laid over the membrane. The dialyzers were capped with parafilm and incubated for 3 h at 30°C. After the incubation period, the acceptor liposome solution was gently collected from the top of the membrane, and the membrane was briefly washed with 50 µl of vesicle buffer. The acceptor and wash fractions were pooled and subjected to a DHE-dansyl FRET assay as described in the previous paragraph. All samples that included Osh4p were saved and subjected to cold acetone precipitation for SDS-PAGE. The contents of the input chamber (protein + DHE donors) were collected for SDS-PAGE by puncturing the membrane with a 200-µl pipette tip and removing the input mixture. The input chamber was briefly rinsed with 50 µl of vesicle buffer, and the input and wash fractions were combined and subjected to cold acetone precipitation.
NBD labeling
Osh4p (10 µg) was labeled with 0.5 mM IANBD amide (Invitrogen) in a final reaction volume of 100 µl. After 2 h at room temperature, the mixture was applied to a CentriSep column to separate the labeled protein from free NBD. The protein was mixed with liposomes for fluorimetric measurements. NBD emission intensity was measured from 40 µl NBD-Osh4p mixed with membranes (40 µl of 1 mM stock) in a final volume of 200 µl. NBD was excited at 478 nm, and emission was monitored at 541 nm.
Reaction of proteins with NEM-containing liposomes
The NEM-containing phospholipid N-MCC-PE was obtained from Avanti Polar Lipids, Inc. 10 µg Osh4p was incubated with sucrose-loaded liposomes containing PC/PE/MPB-PE/cholesterol/PI(4,5)P2 (59.5:29:1:10:0.5) for 2 h at room temperature. The membranes were pelleted at 16,000 g for 10 min and washed three times. Protein amounts in the pellet were determined by Coomassie staining and subsequent quantification with an infrared imaging system (Odyssey; LI-COR Biosciences).
Cholesterol transport assays with covalently attached Osh4p
10 µg Osh4p was labeled with 0.4 mM of the cross-linker 4-(4-N-maleimidophenyl)butyric acid hydrazide hydrochloride (Thermo Fisher Scientific) in a final reaction volume of 50 ul in PBS, pH 7. After 2 h at room temperature, the mixture was applied to Centri Sep column (Applied Biosystems) to remove excess cross-linker. 1 mM sucrose-loaded liposomes containing DOPC/PE/GM1/cholesterol/PI(4,5)P2 (59.5:29:0.1:10:0.5) were oxidized with 10 mM periodate for 30 min on ice in the dark. Excess periodate was removed by centrifugation at 16,000 g for 10 min at 4°C. The oxidized liposomes were briefly washed, resuspended, and mixed with the labeled protein in a final reaction volume of 100 µl. After 2 h at room temperature, the conjugation reactions were terminated by centrifugation. After the pellet was washed three times by pelleting at 16,000 g, the conjugated proteins were analyzed by SDS-PAGE.
For the transport assay, donor liposomes were prepared as described in the previous paragraph but containing [14C]cholesterol (10%) and mixed with 1 mM acceptor liposomes containing DOPC/PE/PI(4,5)P2 (79.5:20:0.5) for 1 h at 30°C. The amount of radiolabeled sterol transferred to acceptor membranes was determined by scintillation counting.
Cholesterol uptake and esterification
Quantification of the uptake and esterification of exogenous [14C]cholesterol was performed as described previously (Li and Prinz, 2004) with few modifications. Strains were grown in synthetic complete medium without uracil to which was added 6.0 µM [14C]cholesterol in Tween 80/ethanol (1:1) so that the final Tween 80 concentration was 0.5%. Samples were removed after 5, 10, 20, and 30 min and added to an equal volume of ice-cold 20 mM NaN3. The cells were washed two times with ice-cold 10 mM NaN3 and lysed in a mini beadbeater-8 (BioSpec Products). Lipids were extracted from 1.8 ml lysate by adding 4 ml methanol, 2 ml chloroform, and 2 ml 0.9% NaCl. The chloroform phase was dried under N2 and spotted onto thin-layer chromatography plates (Silica Gel 60; EMD), and the plates were developed with hexanes/diethyl ether/acetic acid (70:30:1). The amounts of free and esterified cholesterol were quantitated with a phosphorimager (FLA-5100; Fujifilm).
Cryo-EM
PC/PS (1:1) at 1 mM was mixed with 20–40 pmol Osh protein and applied to glow-discharged 200 mesh Cu grids (R3.5/1; Quantifoil Micro Tools GmbH) and rapidly frozen in liquid ethane using the Vitrobot system (FEI). They were imaged with a field emission gun–scanning microscope (CM200; FEI) fitted with a cryoholder (626; Gatan) and operating at an accelerating voltage of 120 kV. Micrographs were recorded at 0.8–1.5 µm underfocus and were later digitized by a scanner (Leafscan 45; LaserSoft Imaging) at a step size of 12.5 µm.
Analytical ultracentrifugation
Osh4p was purified by size exclusion chromatography in PBS and diluted in PBS as required for sedimentation experiments. Sedimentation velocity experiments were conducted at 20.0°C on an analytical ultracentrifuge (ProteomeLab XL-I; Beckman Coulter). Samples of Osh4p (loading volume of 400 µl) were analyzed at loading concentrations of 4.0, 4.8, and 12.1 µM and a rotor speed of 50 krpm. 80 scans were collected at 6.2-min intervals with data acquired using both absorbance and interference detection systems. Absorbance data were collected as single measurements at 280 nm using a radial spacing of 0.003 cm. Data were analyzed in SEDFIT 11.71 (Schuck, 2003) in terms of a continuous c(s) distribution covering an s20,w range of 1.0 – 7.0 S (Svedberg units) with a resolution of 100 and a confidence level (F ratio) of 0.68. Excellent fits were obtained with root/mean/square distance values of 0.0032–0.0047 absorbance units and 0.0042–0.0053 fringes. Solution densities (ρ) and viscosities (η) were calculated based on the solvent composition using SEDNTERP 1.09. Osh4p partial specific volumes (v) were calculated based on the amino acid sequence in SEDNTERP 1.09, and sedimentation coefficients s were corrected to s20,w.
Sedimentation equilibrium experiments were conducted at 20.0°C on an analytical ultracentrifuge (Optima XL-A; Beckman Coulter). Samples (loading volume of 135 µl) were studied at loading concentrations of 3.4, 5.9, and 12.3 µM. Data were acquired at various rotor speeds ranging from 14,000 to 30,000 rpm as a mean of four absorbance measurements at a wavelength of 280 nm and a radial spacing of 0.001 cm. Equilibrium was achieved within 30 h. Data were analyzed globally using SEDPHAT 6.21 (Lebowitz et al., 2002) in terms of a single ideal solute with excellent data fits.
Lipid-mixing and fusion assays
FRET-based lipid-mixing assays were performed using the donor/acceptor pair NBD and Rho. Liposomes composed of 50:20:28:1:1 DOPS/DOPE/DOPC/NBD-PS/Rho-PE (labeled) and 50:20:30 DOPS/DOPE/DOPC (unlabeled) were prepared as described in Preparation of liposomes and extruded through 0.4-µm Nuclepore filters. Equal amounts of labeled and unlabeled liposomes were mixed at a final concentration of 900 µM with 40 pmol of Osh protein for 30 min at 30°C then diluted 1:1 into vesicle buffer. NBD fluorescence emission was measured at 530 nm with the excitation wavelength set to 467 nm (2-mm slits). For PEG fusion, 50 µl of a 900 µM mixture of labeled and unlabeled liposomes was mixed with 50 µl 50% PEG in vesicle buffer, incubated for 30 min at 30°C, and NBD emission was measured. To determine the expected fluorescence increase for full lipid mixing, 50:20:29:0.5:0.5 DOPS/DOPE/DOPC/NBD-PS/Rho-PE liposomes were prepared and measured at 450 µM concentration.
Content-mixing assays were performed as previously described (Kreye et al., 2008) using the fluorophore 8-hydroxypyrene-1,3,6-trisulfonic acid (HPTS or pyranine) and its quencher p-Xylene bis-(N-pyridinium bromide) (DPX). In brief, 1 µmol DOPS was rehydrated in 1 ml of a buffer containing 20 mM Hepes, pH 7.3, 30 mM HPTS, and 45 mM DPX, subjected to freeze-thaw cycles, and extruded through a 0.4-µm Nuclepore filter. Intact liposomes were separated from external HPTS/DPX by gel filtration using a Hepes/NaCl buffer to elute. HPTS/DPX-loaded liposomes were incubated in a 1:9 mixture with unlabeled DOPS liposomes and either buffer (control), 40 pmol Osh protein, or 5 mM Ca2+. HPTS fluorescence was monitored over time at 520 nm with an excitation wavelength of 460 nm. Reactions always took place with 50 mM DPX in the external medium to prevent any fluorescence increase by HPTS leakage.
Online supplemental material
Fig. S1 shows that yeast ORPs bind to liposomes containing acidic phospholipids. Fig. S2 shows aggregation of liposomes by Osh6p and Osh4p visualized with cryo-EM. Fig. S3 shows that Osh-mediated tethering is reversible. Fig. S4 shows cholesterol transport by yeast ORPs. Fig. S5 shows that Osh4p is a monodisperse monomer. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905007/DC1.
Acknowledgments
We thank R. Chung for technical assistance; Y.-J. Im, T. Levine, and B. Glick for strains and plasmids, P. McPhie for performing circular dichroism on recombinant proteins and mutants, and L. Chernomordik, J. Hanover, and J. Hurley for advice and critique.
This work was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases.
Footnotes
Abbreviations used in this paper:
- DHE
- dehydroergosterol
- ISO-PM
- inside-out PM
- LTP
- lipid transfer protein
- MCS
- membrane contact site
- ORD
- OSBP-related domain
- ORP
- OSBP-related protein
- OSBP
- oxysterol-binding protein
- PC
- phosphatidylcholine
- PE
- phosphoethanolamine
- PH
- pleckstrin homology
- PI
- phosphoinositol
- PI(4,5)P2
- PI-4,5-bisphosphate
- PI(4)P
- PI-4-phosphate
- PIP
- phosphoinositide phosphate
- PM
- plasma membrane
- PS
- phosphatidylserine
- Rho
- rhodamine
References
- Beh C.T., Rine J. 2004. A role for yeast oxysterol-binding protein homologs in endocytosis and in the maintenance of intracellular sterol-lipid distribution. J. Cell Sci. 117:2983–2996 10.1242/jcs.01157 [DOI] [PubMed] [Google Scholar]
- Beh C.T., Cool L., Phillips J., Rine J. 2001. Overlapping functions of the yeast oxysterol-binding protein homologues. Genetics. 157:1117–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley J.H., Leak F.W., Jr., Shianna K.V., Tove S., Parks L.W. 1998. A mutation in a purported regulatory gene affects control of sterol uptake in Saccharomyces cerevisiae. J. Bacteriol. 180:4177–4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espenshade P.J., Hughes A.L. 2007. Regulation of sterol synthesis in eukaryotes. Annu. Rev. Genet. 41:401–427 10.1146/annurev.genet.41.110306.130315 [DOI] [PubMed] [Google Scholar]
- Fairn G.D., McMaster C.R. 2005. The roles of the human lipid-binding proteins ORP9S and ORP10S in vesicular transport. Biochem. Cell Biol. 83:631–636 10.1139/o05-064 [DOI] [PubMed] [Google Scholar]
- Fairn G.D., McMaster C.R. 2008. Emerging roles of the oxysterol-binding protein family in metabolism, transport, and signaling. Cell. Mol. Life Sci. 65:228–236 10.1007/s00018-007-7325-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairn G.D., Curwin A.J., Stefan C.J., McMaster C.R. 2007. The oxysterol binding protein Kes1p regulates Golgi apparatus phosphatidylinositol-4-phosphate function. Proc. Natl. Acad. Sci. USA. 104:15352–15357 10.1073/pnas.0705571104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang M., Kearns B.G., Gedvilaite A., Kagiwada S., Kearns M., Fung M.K., Bankaitis V.A. 1996. Kes1p shares homology with human oxysterol binding protein and participates in a novel regulatory pathway for yeast Golgi-derived transport vesicle biogenesis. EMBO J. 15:6447–6459 [PMC free article] [PubMed] [Google Scholar]
- Fritz F., Howard E.M., Hoffman M.M., Roepe P.D. 1999. Evidence for altered ion transport in Saccharomyces cerevisiae overexpressing human MDR 1 protein. Biochemistry. 38:4214–4226 10.1021/bi981929n [DOI] [PubMed] [Google Scholar]
- Hanada K., Kumagai K., Tomishige N., Kawano M. 2007. CERT and intracellular trafficking of ceramide. Biochim. Biophys. Acta. 1771:644–653 [DOI] [PubMed] [Google Scholar]
- Holthuis J.C., Levine T.P. 2005. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6:209–220 10.1038/nrm1591 [DOI] [PubMed] [Google Scholar]
- Hynynen R., Laitinen S., Käkelä R., Tanhuanpää K., Lusa S., Ehnholm C., Somerharju P., Ikonen E., Olkkonen V.M. 2005. Overexpression of OSBP-related protein 2 (ORP2) induces changes in cellular cholesterol metabolism and enhances endocytosis. Biochem. J. 390:273–283 10.1042/BJ20042082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonen E. 2006. Mechanisms for cellular cholesterol transport: defects and human disease. Physiol. Rev. 86:1237–1261 10.1152/physrev.00022.2005 [DOI] [PubMed] [Google Scholar]
- Im Y.J., Raychaudhuri S., Prinz W.A., Hurley J.H. 2005. Structural mechanism for sterol sensing and transport by OSBP-related proteins. Nature. 437:154–158 10.1038/nature03923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John K., Kubelt J., Müller P., Wüstner D., Herrmann A. 2002. Rapid transbilayer movement of the fluorescent sterol dehydroergosterol in lipid membranes. Biophys. J. 83:1525–1534 10.1016/S0006-3495(02)73922-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreye S., Malsam J., Söllner T.H. 2008. In vitro assays to measure SNARE-mediated vesicle fusion. Methods Mol. Biol. 440:37–50 10.1007/978-1-59745-178-9_3 [DOI] [PubMed] [Google Scholar]
- Ladinsky M.S., Mastronarde D.N., McIntosh J.R., Howell K.E., Staehelin L.A. 1999. Golgi structure in three dimensions: functional insights from the normal rat kidney cell. J. Cell Biol. 144:1135–1149 10.1083/jcb.144.6.1135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange Y., Steck T.L. 1997. Quantitation of the pool of cholesterol associated with acyl-CoA:cholesterol acyltransferase in human fibroblasts. J. Biol. Chem. 272:13103–13108 10.1074/jbc.272.20.13103 [DOI] [PubMed] [Google Scholar]
- Lebowitz J., Lewis M.S., Schuck P. 2002. Modern analytical ultracentrifugation in protein science: a tutorial review. Protein Sci. 11:2067–2079 10.1110/ps.0207702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine T., Loewen C. 2006. Inter-organelle membrane contact sites: through a glass, darkly. Curr. Opin. Cell Biol. 18:371–378 10.1016/j.ceb.2006.06.011 [DOI] [PubMed] [Google Scholar]
- Levine T.P., Munro S. 2001. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell. 12:1633–1644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Prinz W.A. 2004. ATP-binding cassette (ABC) transporters mediate nonvesicular, raft-modulated sterol movement from the plasma membrane to the endoplasmic reticulum. J. Biol. Chem. 279:45226–45234 10.1074/jbc.M407600200 [DOI] [PubMed] [Google Scholar]
- Li X., Rivas M.P., Fang M., Marchena J., Mehrotra B., Chaudhary A., Feng L., Prestwich G.D., Bankaitis V.A. 2002. Analysis of oxysterol binding protein homologue Kes1p function in regulation of Sec14p-dependent protein transport from the yeast Golgi complex. J. Cell Biol. 157:63–77 10.1083/jcb.200201037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen C.J., Roy A., Levine T.P. 2003. A conserved ER targeting motif in three families of lipid binding proteins and in Opi1p binds VAP. EMBO J. 22:2025–2035 10.1093/emboj/cdg201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxfield F.R., Tabas I. 2005. Role of cholesterol and lipid organization in disease. Nature. 438:612–621 10.1038/nature04399 [DOI] [PubMed] [Google Scholar]
- Mousley C.J., Tyeryar K.R., Vincent-Pope P., Bankaitis V.A. 2007. The Sec14-superfamily and the regulatory interface between phospholipid metabolism and membrane trafficking. Biochim. Biophys. Acta. 1771:727–736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo M., Ridgway N.D. 2009. Oxysterol binding protein-related protein 9 (ORP9) is a cholesterol transfer protein that regulates Golgi structure and function. Mol. Biol. Cell. 20:1388–1399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olkkonen V.M. 2004. Oxysterol binding protein and its homologues: new regulatory factors involved in lipid metabolism. Curr. Opin. Lipidol. 15:321–327 10.1097/00041433-200406000-00013 [DOI] [PubMed] [Google Scholar]
- Peretti D., Dahan N., Shimoni E., Hirschberg K., Lev S. 2008. Coordinated lipid transfer between the endoplasmic reticulum and the Golgi complex requires the VAP proteins and is essential for Golgi-mediated transport. Mol. Biol. Cell. 19:3871–3884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perktold A., Zechmann B., Daum G., Zellnig G. 2007. Organelle association visualized by three-dimensional ultrastructural imaging of the yeast cell. FEMS Yeast Res. 7:629–638 10.1111/j.1567-1364.2007.00226.x [DOI] [PubMed] [Google Scholar]
- Perry R.J., Ridgway N.D. 2006. Oxysterol-binding protein and vesicle-associated membrane protein-associated protein are required for sterol-dependent activation of the ceramide transport protein. Mol. Biol. Cell. 17:2604–2616 10.1091/mbc.E06-01-0060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichler H., Gaigg B., Hrastnik C., Achleitner G., Kohlwein S.D., Zellnig G., Perktold A., Daum G. 2001. A subfraction of the yeast endoplasmic reticulum associates with the plasma membrane and has a high capacity to synthesize lipids. Eur. J. Biochem. 268:2351–2361 10.1046/j.1432-1327.2001.02116.x [DOI] [PubMed] [Google Scholar]
- Proszynski T.J., Klemm R.W., Gravert M., Hsu P.P., Gloor Y., Wagner J., Kozak K., Grabner H., Walzer K., Bagnat M., et al. 2005. A genome-wide visual screen reveals a role for sphingolipids and ergosterol in cell surface delivery in yeast. Proc. Natl. Acad. Sci. USA. 102:17981–17986 10.1073/pnas.0509107102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhakrishnan A., Goldstein J.L., McDonald J.G., Brown M.S. 2008. Switch-like control of SREBP-2 transport triggered by small changes in ER cholesterol: a delicate balance. Cell Metab. 8:512–521 10.1016/j.cmet.2008.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Im Y.J., Hurley J.H., Prinz W.A. 2006. Nonvesicular sterol movement from plasma membrane to ER requires oxysterol-binding protein–related proteins and phosphoinositides. J. Cell Biol. 173:107–119 10.1083/jcb.200510084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha N., Kuijl C., van der Kant R., Janssen L., Houben D., Janssen H., Zwart W., Neefjes J. 2009. Cholesterol sensor ORP1L contacts the ER protein VAP to control Rab7–RILP–p150Glued and late endosome positioning. J. Cell Biol. 185:1209–1225 10.1083/jcb.200811005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaf G., Ortlund E.A., Tyeryar K.R., Mousley C.J., Ile K.E., Garrett T.A., Ren J., Woolls M.J., Raetz C.R., Redinbo M.R., Bankaitis V.A. 2008. Functional anatomy of phospholipid binding and regulation of phosphoinositide homeostasis by proteins of the sec14 superfamily. Mol. Cell. 29:191–206 10.1016/j.molcel.2007.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuck P. 2003. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal. Biochem. 320:104–124 10.1016/S0003-2697(03)00289-6 [DOI] [PubMed] [Google Scholar]
- Suchanek M., Hynynen R., Wohlfahrt G., Lehto M., Johansson M., Saarinen H., Radzikowska A., Thiele C., Olkkonen V.M. 2007. The mammalian oxysterol-binding protein-related proteins (ORPs) bind 25-hydroxycholesterol in an evolutionarily conserved pocket. Biochem. J. 405:473–480 10.1042/BJ20070176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan D.P., Ohvo-Rekilä H., Baumann N.A., Beh C.T., Menon A.K. 2006. Sterol trafficking between the endoplasmic reticulum and plasma membrane in yeast. Biochem. Soc. Trans. 34:356–358 10.1042/BST0340356 [DOI] [PubMed] [Google Scholar]
- Takeshima H., Komazaki S., Nishi M., Iino M., Kangawa K. 2000. Junctophilins: a novel family of junctional membrane complex proteins. Mol. Cell. 6:11–22 10.1016/S1097-2765(00)00003-4 [DOI] [PubMed] [Google Scholar]
- van Meer G., Voelker D.R., Feigenson G.W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9:112–124 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Várnai P., Tóth B., Tóth D.J., Hunyady L., Balla T. 2007. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 complex. J. Biol. Chem. 282:29678–29690 10.1074/jbc.M704339200 [DOI] [PubMed] [Google Scholar]
- Vicinanza M., D'Angelo G., Di Campli A., De Matteis M.A. 2008. Phosphoinositides as regulators of membrane trafficking in health and disease. Cell. Mol. Life Sci. 65:2833–2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Duan W., Munn A.L., Yang H. 2005a. Molecular characterization of Osh6p, an oxysterol binding protein homolog in the yeast Saccharomyces cerevisiae. FEBS J. 272:4703–4715 10.1111/j.1742-4658.2005.04886.x [DOI] [PubMed] [Google Scholar]
- Wang P., Zhang Y., Li H., Chieu H.K., Munn A.L., Yang H. 2005b. AAA ATPases regulate membrane association of yeast oxysterol binding proteins and sterol metabolism. EMBO J. 24:2989–2999 10.1038/sj.emboj.7600764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.Y., Weng J., Anderson R.G.W. 2005c. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 307:1472–1476 10.1126/science.1107710 [DOI] [PubMed] [Google Scholar]
- Wu M.M., Buchanan J., Luik R.M., Lewis R.S. 2006. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J. Cell Biol. 174:803–813 10.1083/jcb.200604014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D., Olkkonen V.M. 2008. Characteristics of oxysterol binding proteins. Int. Rev. Cytol. 265:253–285 10.1016/S0074-7696(07)65007-4 [DOI] [PubMed] [Google Scholar]
- Zinser E., Daum G. 1995. Isolation and biochemical characterization of organelles from the yeast, Saccharomyces cerevisiae. Yeast. 11:493–536 10.1002/yea.320110602 [DOI] [PubMed] [Google Scholar]
- Zweytick D., Leitner E., Kohlwein S.D., Yu C., Rothblatt J., Daum G. 2000. Contribution of Are1p and Are2p to steryl ester synthesis in the yeast Saccharomyces cerevisiae. Eur. J. Biochem. 267:1075–1082 10.1046/j.1432-1327.2000.01103.x [DOI] [PubMed] [Google Scholar]