Poc1 shores up basal bodies to support cilia formation in Tetrahymena thermophila, zebrafish, and humans; Poc1 depletion causes phenotypes commonly seen in ciliopathies.
Abstract
Centrioles are the foundation for centrosome and cilia formation. The biogenesis of centrioles is initiated by an assembly mechanism that first synthesizes the ninefold symmetrical cartwheel and subsequently leads to a stable cylindrical microtubule scaffold that is capable of withstanding microtubule-based forces generated by centrosomes and cilia. We report that the conserved WD40 repeat domain–containing cartwheel protein Poc1 is required for the structural maintenance of centrioles in Tetrahymena thermophila. Furthermore, human Poc1B is required for primary ciliogenesis, and in zebrafish, DrPoc1B knockdown causes ciliary defects and morphological phenotypes consistent with human ciliopathies. T. thermophila Poc1 exhibits a protein incorporation profile commonly associated with structural centriole components in which the majority of Poc1 is stably incorporated during new centriole assembly. A second dynamic population assembles throughout the cell cycle. Our experiments identify novel roles for Poc1 in centriole stability and ciliogenesis.
Introduction
Centrioles and basal bodies exhibit a unique, ninefold rotational symmetry of microtubules that are responsible for organizing centrosomes and cilia, respectively. The essential nature of centrioles is in their function as basal bodies for cilia formation, reflecting the predicted evolutionary origin of these complex organelles (Azimzadeh and Bornens, 2004; Basto et al., 2006; Marshall and Nonaka, 2006). Cilia defects contribute to human ciliopathies that cause a series of pathologies ranging from cystic kidneys to mental retardation. The complexity of ciliopathies reflects the vast number of cellular events that involve centriole function and are in the array of signaling pathways that act through cilia (Badano et al., 2006; Eggenschwiler and Anderson, 2007; Berbari et al., 2009; Gerdes et al., 2009). Central to these events is the proper assembly and maintenance of centrioles.
Centrioles are structural platforms that withstand the forces generated by centrosomes and ciliary beating. Centrioles are comprised of α/β-tubulin subunits that are stably incorporated so that the structure is conserved during new centriole assembly (Kochanski and Borisy, 1990; Pearson et al., 2009). Posttranslational modifications of the centriolar microtubules facilitate this stability (Piperno and Fuller, 1985; Gundersen and Bulinski, 1986; Eddé et al., 1990; Bobinnec et al., 1998b). Antibodies that recognize tubulin glutamylation disrupt centrioles and centrosomes in a manner that is exacerbated when centrosomes experience greater pushing and pulling forces. Thus, these organelles are resilient to such forces (Bobinnec et al., 1998a,b; Abal et al., 2005). Disruption of tubulin glutamylation causes defects in ciliary beating, suggesting that the stable basal bodies are necessary for normal ciliary motility (Million et al., 1999; Wloga et al., 2008). Tubulin family proteins γ- and ϵ-tubulin are necessary for centriole assembly and structural maintenance (Dupuis-Williams et al., 2002; Dutcher et al., 2002; Shang et al., 2002), whereas δ-tubulin is required for the triplet microtubule structure (Dutcher and Trabuco, 1998; Garreau de Loubresse et al., 2001). In addition, microtubule-associated components, such as CAP350, contribute to the stability of centrioles (Le Clech, 2008). Similarly, basal body fragmentation has been found in Drosophila melanogaster mutants in pericentrin-like protein (Martinez-Campos et al., 2004). The detailed mechanism for the assembly and stability that enable centrioles to withstand forces from spindle dynamics, fluid flow, and ciliary beating remain to be discovered.
A conserved series of structural centriole assembly stages have been defined predominantly by EM (for reviews see Dutcher, 2007; Pearson and Winey, 2009). Assembly begins with the generation of the ninefold symmetric cartwheel followed by microtubule triplet formation. This leads to a final cylindrical structure composed of nine triplet microtubules capped at both proximal and distal ends. The proximal end, containing the cartwheel, is critical for the assembly and maintenance of these organelles. Chlamydomonas reinhardtii Bld10 is important for the assembly of cartwheels and ultimately centrioles (Matsuura et al., 2004; Hiraki et al., 2007). In addition, Sas6 localizes to the cartwheel hub or central tubule and is essential for centriole assembly and ninefold symmetry (Dammermann et al., 2004; Leidel et al., 2005; Pelletier et al., 2006; Kilburn et al., 2007; Nakazawa et al., 2007; Rodrigues-Martins et al., 2007; Strnad et al., 2007; Vladar and Stearns, 2007; Culver et al., 2009).
Although stable molecular constituents such as tubulin contribute to centriole structure, additional molecular components transiently associate with centrioles (Kochanski and Borisy, 1990; Kirkham et al., 2003; Leidel and Gönczy, 2003, 2005; Dammermann et al., 2008; Pearson et al., 2009). In addition, some basal body components (TtCen1 and TtSas6a) exhibit both dynamic exchange through the cell cycle and stable incorporation during new basal body assembly (Pearson et al., 2009). The protein populations exhibiting these different mechanisms of association localize to specific domains within the basal body (Pearson et al., 2009). The functional significance of such incorporation is not well characterized; however, they appear to be critical for Sas4's role in proper microtubule assembly in C. elegans (Dammermann et al., 2008). Thus, centrioles are comprised of both stable and dynamic components, yet how these different populations of proteins and the centriole domains to which they localize contribute to a functionally stable organelle is not yet understood.
To determine how cartwheel proteins function in basal body assembly, maintenance, and function, we performed a proteomics screen to identify novel basal body proteins, including cartwheel proteins (Kilburn et al., 2007). One such protein, TtPoc1, is a conserved centriole component that is required for centriole assembly (Andersen et al., 2003; Avidor-Reiss et al., 2004; Li et al., 2004; Keller et al., 2005, 2009; Broadhead et al., 2006; Kilburn et al., 2007; Hames et al., 2008; Woodland and Fry, 2008). Poc1 binds to microtubules in vitro (Hames et al., 2008); however, the role for Poc1 in basal body assembly and ciliogenesis has not been studied.
Our work demonstrates that Poc1 is a stable basal body constituent that localizes to the basal body cartwheel, the site of new basal body assembly, and the microtubule cylinder walls. Poc1 is required for the stability of basal bodies, ciliary-based motility, and proper cilia formation. Poc1 provides a molecular link between the assembly and stability of centrioles for ciliary-based motility in T. thermophila and cilia formation and function in zebrafish and humans. Defects caused by Poc1 depletion generate maladies commonly associated with ciliopathies.
Results
Poc1 is required for ciliary motility
To determine the function of the basal body component, Poc1, a Poc1 deletion strain was generated in Tetrahymena thermophila (poc1Δ). poc1Δ cells incubated at 30°C grew slightly slower than wild-type controls (cell-doubling time: poc1Δ, 3.4 ± 0.3 h; wild type, 2.9 ± 0.2 h; Fig. S1 A). The growth defect in poc1Δ cells was exacerbated when cells were grown at an elevated temperature of 38°C (cell-doubling time: poc1Δ, 3.9 ± 0.5 h; wild type, 2.7 ± 0.2 h; Fig. S1 A). At this elevated temperature, poc1Δ cell populations doubled approximately twice before failing to divide any further. To determine the fate of the poc1Δ cells at 38°C, cell viability was measured (Fig. S1 B). Cell death in poc1Δ cells increased over time, reaching 91% lethality after 5 d at the restrictive temperature. poc1Δ cells maintained at 38°C for 7 d did not recover when shifted back to 30°C. The loss of POC1 causes a temperature-sensitive lethal phenotype.
Basal bodies serve as a platform for ciliary-based motility. To determine whether Poc1 is important for cilia function, the swimming rate of cells without POC1 was quantified. Cell motility in poc1Δ cells was significantly slower (poc1Δ, 281 ± 77 µm/s; wild type, 445 ± 100 µm/s; Fig. S1 C), and the decreased motility rate was exacerbated at 38°C (poc1Δ, 53 ± 51 µm/s; wild type, 481 ± 95 µm/s; Fig. S1 C). These data demonstrate that Poc1 is required for normal cilia-based motility.
Poc1 WD40 repeat domain is required for basal body frequency and organization
Basal bodies were visualized in poc1Δ cells to determine how Poc1 contributes to ciliary motility. T. thermophila basal bodies are organized into ciliary rows, and the frequency of basal bodies (number of basal bodies/unit length) is relatively uniform when normalized for cell cycle position. At 30°C, basal body organization in poc1Δ cells appeared relatively normal (Fig. 1 A). In contrast, a massive disruption of basal body organization, frequency, and loss of the oral apparatus (the basal body–rich feeding structure) was observed in poc1Δ cells that were maintained at 38°C for 24 h (Fig. 1 A). A range of defects were observed in these cells, including aberrant cell morphology with cellular extensions, binucleated cells, and cell death. The frequency of these phenotypes in poc1Δ cells at 38°C was quantified over a 5-d time course (Fig. S1 D). The reduction in basal body frequency, disruption of basal body organization, and loss of oral apparatuses were observed soon after temperature shift, suggesting that these defects are the direct result of Poc1 loss at 38°C and likely contribute to the defective cell motility. Phagocytosis is commonly lost upon oral apparatus disruption and can lead to cell inviability (Basmussen and Orias, 1975; Brown et al., 1999). Thus, the lethality may be a result of loss of the oral apparatus and phagocytosis.
Figure 1.
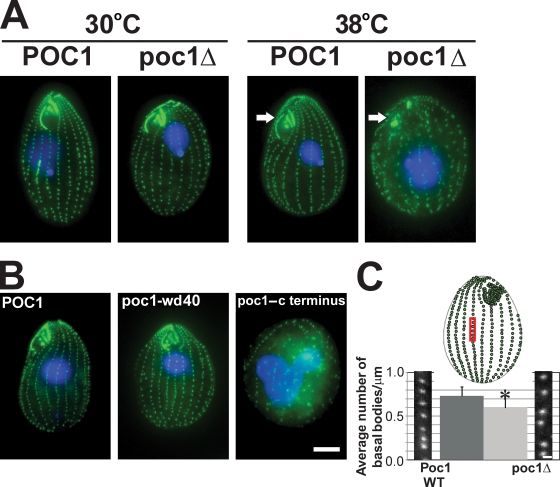
Basal body frequency and organization defects in poc1Δ. (A) Wild-type (POC1) and poc1 knockout (poc1Δ) T. thermophila cells were stained for basal bodies (green) and DNA (blue) after growing cells at either 30 or 38°C for 24 h. poc1Δ cells at 38°C exhibit a decreased frequency and disorganization of basal bodies and disruption of the oral apparatus (arrows). (B) poc1Δ cells with GFP-POC1 (aa 1–634), GFP-poc1-wd40 (aa 1–342), or GFP-poc1–c terminus (aa 343–634) at 38°C for 48 h. GFP-POC1 and the GFP-poc1-wd40 truncation rescues the lethality, oral apparatus defects, and basal body organization and frequency defects. (C) The frequency of basal bodies in poc1Δ cells is decreased in cells arrested by media starvation at 30°C for 12 h. The number of basal bodies in the medial 10 µm of cortical rows was measured to determine the mean number of basal bodies per micrometer. The red box is a representative region within the medial region of the T. thermophila cell selected for analysis. WT, wild type. *, P < 0.01; n > 150. Error bars indicate mean ± SD. Bars: (A and B) 10 µm; (C) 1 µm.
To verify that the defects are the result of Poc1 loss, GFP-POC1 was introduced into poc1Δ cells, and the cells were shifted to 38°C. These cells were viable and exhibited normal basal body organization and frequency and normal oral apparatuses (Fig. 1 B and Fig. S3). Expression of GFP-POC1 up to 7 d after poc1Δ cells without GFP-POC1 were shifted to 38°C also rescued the temperature-sensitive lethality, suggesting that basal bodies could be repaired by the reintroduction of Poc1. Truncated GFP fusions were introduced into poc1Δ cells at 38°C to determine whether specific domains of the Poc1 protein were responsible for its localization and function in normal basal body number and organization. Surprisingly, cells exhibited a wild-type phenotype when the N-terminal fragment containing only the WD40 repeat (aa 1–342) was expressed (Fig. 1 B). Rescue was not observed with the C terminus of the protein (aa 343–634). Consistent with these results, the full-length POC1 (aa 1–634) and the WD40 repeat truncation localized to basal bodies, whereas the C-terminal domain did not (Fig. S2 A). The fluorescence intensity of the GFP-poc1-wd40 truncation was reduced compared with full-length GFP-Poc1 at 30°C (Fig. S2 A). These results indicate that the WD40 repeat domain is critical for Poc1's function.
Proper basal body organization and polarity is required for efficient ciliary beating from ciliates to humans (Marshall and Kintner, 2008). In particular, the formation and organization of accessory structures determine the proper orientation of basal bodies. One prominent structure that contributes to this polarity in T. thermophila is the kinetodesmal fiber at the proximal end of basal bodies that becomes disrupted in specific positioning and orientation mutants (Jerka-Dziadosz et al., 1995; Beisson and Jerka-Dziadosz, 1999). To determine whether such organizational defects occur in poc1Δ cells, kinetodesmal fibers were immunolocalized. poc1Δ cells, at both 30 and 38°C, displayed intact kinetodesmal fibers for most of the basal bodies in the ciliary rows (Fig. S2 B), suggesting that the loss in basal body organization in poc1Δ cells at 38°C did not result from the loss of kinetodesmal fibers.
The density of basal bodies per ciliary row in T. thermophila is maintained by the assembly of new basal bodies before cell division (Nanney, 1975; Kaczanowski, 1978). To determine whether the frequency of basal bodies is altered in the absence of Poc1, poc1Δ cells were arrested in G1 by media starvation for 12 h, and the frequency of basal bodies was quantified within the medial region of the cell (number of basal bodies/µm [bb/µm]). The frequency of basal bodies was significantly reduced in poc1Δ cells at 30°C (poc1Δ, 0.60 ± 0.14 bb/µm; POC1, 0.73 ± 0.11 bb/µm; Fig. 1 C). The decreased basal body frequency was not a result of altered cell length (unpublished data). The decreased basal body frequency in the absence of Poc1 reveals a phenotype at 30°C that may be caused by either a cessation or decreased efficiency in new basal body assembly or an assembly defect causing decreased stability of basal bodies.
Poc1 loss causes basal body structural defects
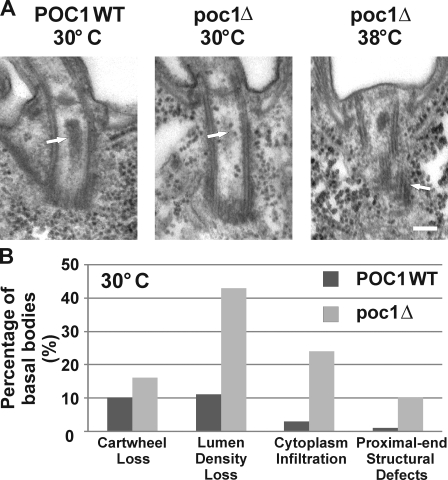
The decreased frequency of centrin-stained basal bodies in poc1Δ cells suggests that structural defects may be associated with the loss of Poc1. The ultrastructure of basal bodies in poc1Δ cells grown at 30 and 38°C was visualized using transmission EM (TEM). We found that ∼60% of basal bodies observed in poc1Δ cells at 30°C exhibited structural defects in contrast to only ∼20% of wild-type basal bodies. At 30°C, basal bodies in poc1Δ cells lacked the electron-dense material normally present in the central lumen, and cytoplasm infiltrated into this luminal region (Fig. 2). Although the cartwheel structure remained intact, a subtle increase in structural defects within the proximal end of the basal body was observed at 30°C (Fig. 2). These defects are generally characterized by anomalies in microtubule continuity or breaks in the walls of the cylinder structure. Such basal body proximal-end structural defects were exacerbated at an increased temperature (38°C; Fig. 2 A). Fewer basal bodies were found in poc1Δ samples at 38°C, suggesting that poc1Δ basal bodies at 38°C are less stable under the TEM fixation conditions. Furthermore, the cylinder walls were significantly disrupted in the observed basal bodies (Fig. 2 A).
Figure 2.
Poc1 loss causes basal body structural defects. (A) Basal body structural defects in poc1Δ cells at either 30 or 38°C for 24 h. Basal bodies in poc1Δ cells at 30°C exhibited a loss in the lumen electron density (arrows) and an infiltration of cytoplasm into the lumen (indicated by the ribosomal-like structures). (right) Defects in the triplet microtubules at the proximal end of basal bodies were observed at 30°C and were more frequently observed in the limited basal bodies at 38°C. Bar, 100 nm. (B) The relative frequency of the defects observed at 30°C. WT, wild type. n > 100 basal bodies.
Poc1 is required for basal body stability and maintenance
The basal body structural defects in poc1Δ cells suggest that Poc1 is required for the proper assembly of a mature basal body structure. To assess basal body assembly, a strategy to visualize newly assembled (immature) basal bodies was used (Shang et al., 2005; Pearson et al., 2009). Old basal bodies were labeled (K antigen; Williams et al., 1990) relative to all basal bodies (anticentrin; Stemm-Wolf et al., 2005). New basal bodies with centrin staining and no K antigen signal were found for both wild-type and poc1Δ cells at both 30 and 38°C (Fig. 3 A), suggesting that Poc1 is not required for new basal body assembly. However, some of the new basal bodies assembled in poc1Δ cells at 38°C contained variable levels of centrin fluorescence. Thus, even though new basal bodies form in the absence of Poc1, the new basal bodies are not completely mature or contain defective structures (Fig. 3 A), which is consistent with the structural defects shown in Fig. 2.
Figure 3.
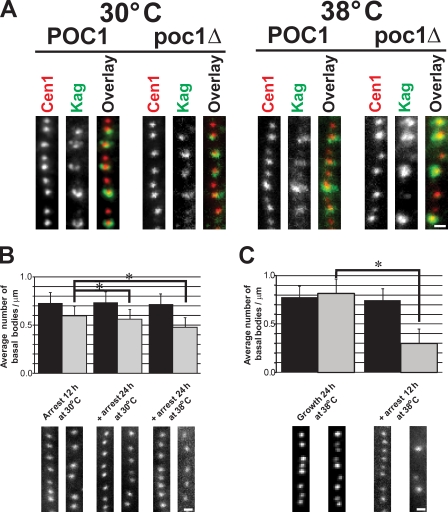
Poc1 is required for basal body stability. (A) New basal bodies assemble in the absence of Poc1. All basal bodies (centrin, red) were observed relative to mature basal bodies (K antigen, green). Newly formed basal bodies (no K antigen) were found at all conditions. (B) Basal bodies are less stable in poc1Δ cells. Cells were arrested at 30°C for 12 h (left) to create a uniform cell cycle population, and the arrest was maintained either at 30 (middle) or 38°C (right) for an additional 24 h. Basal body frequency was quantified for each condition by counting the number of centrin-stained basal bodies per micrometer. (C) Basal bodies assembled in poc1Δ cells at 38°C are less stable than at 30°C. Cells were grown at 38°C for 24 h before arrest for 12 h at 38°C. Black bars, wild-type POC1; gray bars, poc1Δ. *, P < 0.01; n > 150. Error bars indicate mean ± SD. Bars, 1 µm.
Because poc1Δ cells assemble basal bodies even at 38°C, we asked whether Poc1 was required for their stability/maintenance. poc1Δ cells were arrested by nutrient starvation so that new basal body assembly does not occur. The arrest was maintained for an additional 24 h to test whether the frequency of basal bodies decreased (indicative of a loss in stability). poc1Δ cells exhibited significantly reduced numbers of basal bodies per micrometer when arrested for an additional 24 h (Fig. 3 B). Wild-type cells did not demonstrate an appreciable difference in basal body frequency. Finally, to determine whether the basal bodies assembled without Poc1 at increased temperatures (38°C) were less stable than those assembled at 30°C, T. thermophila cells were grown at 38°C and arrested at 38°C overnight. Unlike wild-type cells, the poc1Δ cells exhibited severely decreased numbers of basal bodies per micrometer (poc1Δ, 0.30 ± 0.14 bb/µm; POC1, 0.75 ± 0.11 bb/µm). Before cell cycle arrest, the mean frequency of basal bodies was slightly higher in poc1Δ compared with wild type at 38°C, suggesting that the reduced number of basal bodies found in poc1Δ knockout cells is a result of the arrest and decreased basal body stability (poc1Δ, 0.82 ± 0.16 bb/µm; POC1, 0.78 ± 0.12 bb/µm; Fig. S3 C). In the absence of Poc1, new basal bodies are assembled, but they are less stable, especially at elevated temperatures.
Consistent with a role in basal body stability, basal bodies were more sensitive to the microtubule-depolymerizing drug nocodazole. Basal bodies were less organized in poc1Δ cells treated with 10 µg/ml nocodazole at 30°C for 7 h, and the frequency of basal bodies was decreased (Fig. S2 D).
Poc1 exhibits both stable and dynamic protein incorporation at basal bodies
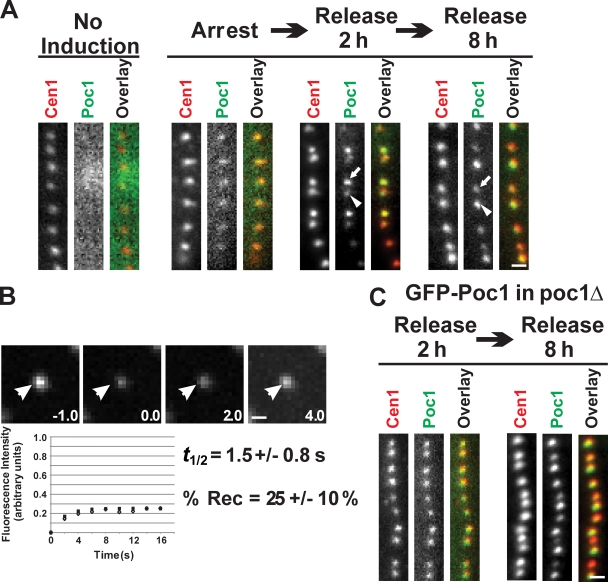
The role of Poc1 in the assembly of a stable basal body suggests that Poc1 is incorporated early during assembly. Poc1 protein deposition was monitored during new basal body assembly to understand how and when Poc1 becomes incorporated. Cells containing a transcriptional regulated version of GFP-Poc1 in the presence of endogenous, untagged Poc1 were arrested by media starvation to inhibit new basal body assembly. GFP-Poc1 expression was induced for 2 h, and a low level of GFP-Poc1 fluorescence was observed in all basal bodies (Fig. 4 A, arrest). These results suggest that Poc1 exhibits a low level of dynamic protein incorporation at basal bodies that is independent of new basal body assembly (Pearson et al., 2009). Upon release from the cell cycle arrest, GFP-Poc1 incorporated at newly assembled basal bodies. In contrast to centrin antibody staining that uniformly labeled all basal bodies, newly formed basal bodies (those assembled in the anterior position relative to its closely positioned mother basal body) were brightly labeled with GFP-Poc1 relative to their dimly labeled parent (Fig. 4 A, release 2 h). Similar results were also found when comparing the old oral apparatus and the newly forming oral apparatus. These results suggest that the majority of Poc1 is stably incorporated during new basal body assembly, whereas a smaller proportion of the Poc1 protein is dynamic and incorporated independent of new basal body assembly.
Figure 4.
Poc1 exhibits both dynamic and stable incorporation at basal bodies. (A) GFP-Poc1 protein incorporation during basal body assembly. Noninduced cells exhibited no fluorescence signal. GFP-Poc1 expressed for 2 h in cells arrested for 12 h to inhibit basal body assembly. Low fluorescence uniformly incorporated at all basal bodies. Cells were released from the arrest for 2 h in the presence of GFP-Poc1. Dim GFP-Poc1 signal was found at existing, mature basal bodies (arrowhead), whereas newly assembled basal bodies contained bright GFP-Poc1 (arrow). Uniform anticentrin staining was observed at all basal bodies. 8 h after release, the majority of the basal bodies were brightly labeled. In contrast to the 2-h release, at 8 h, new basal bodies in dividing cells were dim (arrow) compared with old basal bodies (arrowhead). (B) FRAP to quantify the dynamic fraction of GFP-Poc1 in poc1Δ cells. After photobleaching, fluorescence recovery was followed. (bottom) Open diamonds indicate the mean fluorescence recovery relative to the best-fit single exponential recovery (filled squares). Arrowheads indicate the photobleached basal body. (C) GFP-Poc1 decorates all basal bodies uniformly upon expression in cycling cells for 2 h in a poc1Δ strain. Bars, 1 µm.
The relative fraction of the stable and dynamic Poc1 at basal bodies was determined by measuring the ratio between the old (dim; dynamic only) and new (bright; dynamic and stable) basal bodies. The ratio of fluorescence intensities of old to new basal bodies was 0.18 ± 0.08 (n = 541), suggesting that 18% of the protein population is dynamic, whereas the remaining 82% is stably assembled only during new basal body assembly. This ratio, showing that the majority of the protein population is incorporated only during new basal body assembly, is similar to that found for other structural basal body and centriole proteins (Kochanski and Borisy, 1990; Pearson et al., 2009). As expected, cells grown for multiple cell cycles so that most basal bodies were assembled in the presence of GFP-Poc1 lead to more bright and uniform GFP-Poc1 labeling (Fig. 4 A, release 8 h). However, by this time point, new basal bodies of closely associated pairs were often dim compared with the older basal bodies (Fig. 4 A, release 8 h). We predict that the dim signal is a result of the lack of additional Poc1 incorporation during later stages of assembly and maturation. As the basal body matures early after assembly, new Poc1 continues to be incorporated at the daughter basal body until the levels equalize with the mother basal body. Cells either in interphase or arrested by starvation, where new basal body assembly is low or halted, contained uniform Poc1 fluorescence (Fig. S2 E). This indicates that Poc1 incorporation continues for a short time period after new basal body assembly.
The dynamic and stable populations of Poc1 at basal bodies were measured using FRAP. Photobleaching of brightly labeled GFP-Poc1 basal bodies (without endogenous Poc1) resulted in 25 ± 10% Poc1 recovery, indicating that ∼75% of Poc1 is stable. The dynamic population of Poc1 exhibited rapid recovery (t1/2 = 1.5 ± 0.8 s; Fig. 4 B). These results, combined with the aforementioned ratio experiments, indicate that the majority of Poc1 is stable and incorporates only during new basal body assembly.
In the presence of endogenous Poc1, induced GFP-Poc1 incorporates into the basal body structure during a specific event within the assembly process. To determine whether Poc1 incorporation at existing basal bodies increases when the Poc1-binding sites are not occupied (poc1Δ), GFP-Poc1 was expressed in poc1Δ cells for 2 h. Surprisingly, bright and uniform GFP-Poc1 signal was observed at both new and old basal bodies, indicating that unoccupied binding sites are accessible to Poc1 incorporation throughout the cell cycle (Fig. 4 C). This is in contrast to the bright and dim fluorescent protein distribution found when GFP-Poc1 was expressed in the presence of unlabeled, endogenous Poc1 (Fig. 4 A). However, once Poc1 incorporates, the protein remains stably bound to these sites (Fig. 4 C).
Stable Poc1 assembles at cartwheels and the cylinder walls
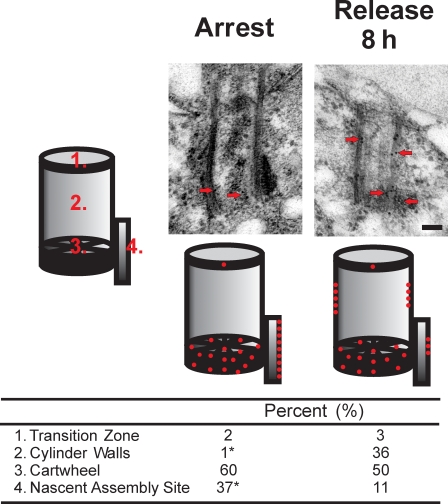
Identical basal body localization was observed by both light and immuno-EM (IEM) when all copies of the POC1 gene were replaced with either an N- or C-terminal fusion of mCherry or GFP to POC1 (Fig. S2 E; Kilburn et al., 2007; unpublished data). Endogenous Poc1 localizes to three major domains within the basal body ultrastructure: the cartwheel, the medial region cylinder walls, and the site of new basal body assembly. We studied the incorporation profile and stability of Poc1 at each of the three basal body domains. T. thermophila cells containing a regulated copy of GFP-POC1 were arrested by media starvation to inhibit new basal body assembly. GFP-Poc1 expression was induced for 2 h, and cells were fixed for IEM. Under these conditions, only dynamic Poc1 incorporation at basal bodies is observed (Fig. 4 A). This population localized to the cartwheel (60%) and the nascent site of basal body assembly (37%) (Fig. 5, arrest). These populations represent the dynamic fraction that was measured by our FRAP experiments to be only 25% of the total Poc1 population. Based on the IEM distribution, we predict that ∼15% of the total Poc1 is dynamic at the cartwheel and ∼10% is dynamic at the site of nascent basal body assembly.
Figure 5.
Dynamic and stable populations of Poc1 incorporation. IEM of GFP-Poc1 was visualized in cells arrested by starvation (arrest) and normally cycling cells (release 8 h). In arrested cells, GFP-Poc1 incorporates at only the cartwheel and the nascent site of assembly. In contrast, GFP-Poc1 assembles at basal body cylinder walls in addition to the cartwheel and nascent site when expressed in cycling cells with new basal body assembly in the presence of GFP-Poc1. The relative frequency of gold label at the nascent site of basal body assembly decreases in cycling cells compared with arrested cells. Red dots indicate the relative distribution of gold particles (n > 150). *, P < 0.01 by χ2 test. Bar, 100 nm.
Cells grown for 8 h in the presence of GFP-Poc1 have both stable and dynamic populations of Poc1 at basal bodies (Fig. 4 A). In this study, the cylinder walls were strongly decorated with Poc1 in addition to the cartwheel and nascent site of assembly (Fig. 5, release 8 h). Because 75% of the total Poc1 at basal bodies is stable and because the ratios in growing cells of antibody labeling between the cartwheel, nascent site, and microtubule walls cannot be explained only by dynamic incorporation into the cartwheel and nascent site, we calculate that ∼35% of the total stable Poc1 pool localized to the cylinder walls, whereas ∼35% assembled at the cartwheel. These results suggest that the stable Poc1 population assembles at the basal body cylinder walls and the cartwheel. A smaller dynamic fraction is also present at the cartwheel and the nascent site of assembly.
Human Poc1B is required for primary cilia formation
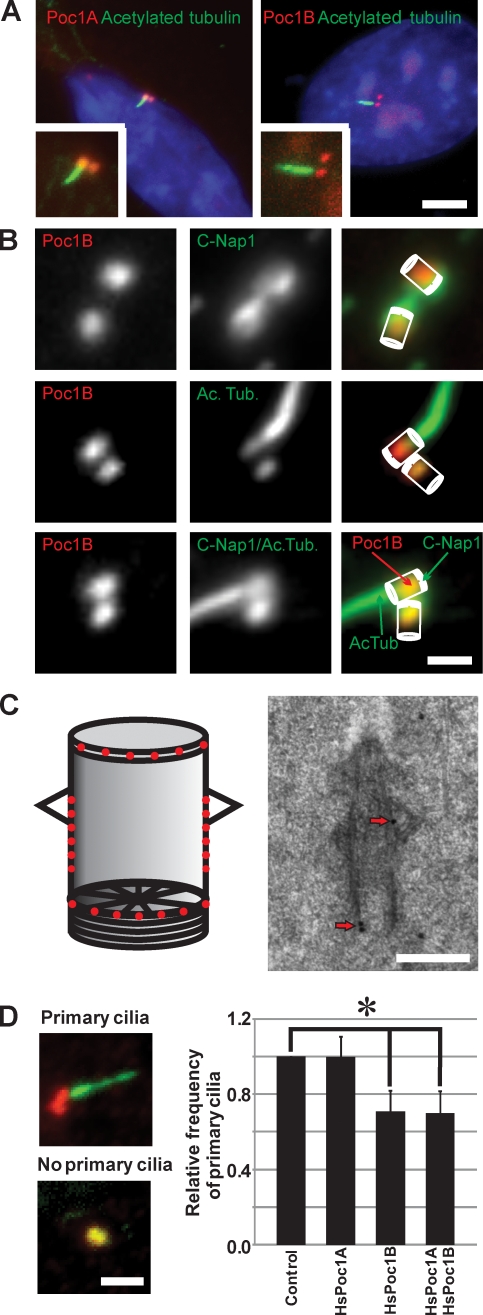
There are two Poc1-like proteins in humans; however, beyond localization of Poc1 to basal bodies (Hames et al., 2008; Blachon et al., 2009; Keller et al., 2009), the role of Poc1 in ciliogenesis has not been studied. We localized mCherry fused to the two Poc1-like proteins in ciliated human RPE-1 cells (HsPoc1A and HsPoc1B). Both Poc1-like proteins exhibited identical localization to basal bodies of primary cilia (Fig. 6 A). The same localization was observed using antibodies directed against either Poc1B (CO200; Fig. 6 B and Fig. S3) or both Poc1A and Poc1B (Hames et al., 2008). As found for TtPoc1 (Fig. S2 A), HsPoc1B and HsPoc1B truncations containing the WD40 repeat domain localized to basal bodies; however, the C-terminal domain containing the conserved Poc1 domain did not localize to centrioles or basal bodies (Keller et al., 2009; unpublished data).
Figure 6.
Human Poc1B is required for ciliogenesis. (A) Basal body localization of mCherry-Poc1A or mCherry-Poc1B (red) was visualized relative to ciliary axonemes labeled with anti-acetylated tubulin (green) in ciliated human RPE-1 cells. Insets indicate a magnified view. Bar, 5 µm. (B) HsPoc1B localized predominantly to the proximal end of basal bodies. Poc1B (red; CO200) was colocalized relative to C-Nap1 (green; basal body proximal end marker) and acetylated tubulin (ac tub; green; basal body and cilia marker). Bar, 1 µm. (C) IEM localized HsPoc1B to the transition zone (23%), cylinder walls (49%), and cartwheel (28%). (left) Schematic showing the relative distribution of gold particles. (right) A representative localization image is shown. Arrows indicate immunogold label. Bar, 200 nm. (D) Depletion of HsPoc1B inhibits primary cilia formation. Cilia axonemes were labeled with anti-acetylated tubulin (green), and basal bodies were labeled with γ-tubulin (red). The presence or absence of primary cilia was scored to determine the relative frequency of primary cilia relative to control siRNA. Bar, 2 µm. P < 0.01; n > 800 cells; n = 4. Error bars indicate mean ± SD.
T. thermophila Poc1 localizes to multiple basal body domains. Consistent with the T. thermophila cartwheel localization, we found that HsPoc1B localizes to the proximal end of both basal bodies of primary cilia and centrioles (Figs. 6 B). HsPoc1B was found to reside between C-Nap1, which localizes to the base and fibers linking basal bodies (Fry et al., 1998), and Cetn2, which predominantly localizes to the distal end of centrioles (Baron et al., 1992; Paoletti et al., 1996). IEM localization of HsPoc1B to centrioles and basal bodies exhibited localization to the cylinder walls, cartwheel, and the transition zone. Consistent with the fluorescence localization, most HsPoc1B was found at the proximal end of the basal body. In addition, a significant level of gold label (23%) was found at the transition zone.
To determine whether HsPoc1 contributes to primary ciliogenesis, HsPoc1 was depleted using siRNA in cells induced to undergo primary cilia formation. We found that siRNA of Poc1A did not have an effect on cilia formation or length, whereas there was a significant decrease in ciliogenesis in Poc1B siRNA-treated cells (∼30% decreased primary ciliogenesis; Fig. 6 C). In addition, the HsPoc1B knockdown cells that were cilia positive commonly exhibited shorter (HsPoc1B siRNA, 3.3 ± 1.3 µm; control, 4.6 ± 1.2 µm; P < 0.001) and bent cilia compared with wild-type and HsPoc1A siRNA. The depletion of HsPoc1A was not as efficient as that found for HsPoc1B (Fig. S4).
Zebrafish Poc1B morphants exhibit defects commonly found in ciliopathies
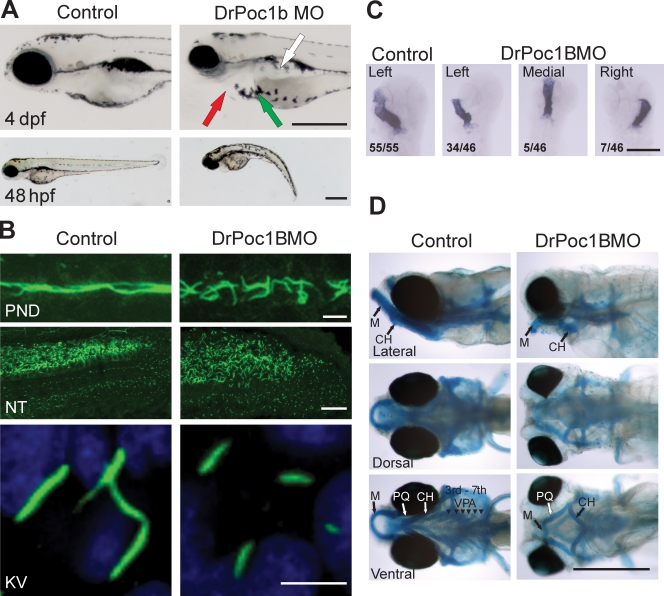
The morphological anomalies associated with human ciliopathies can be reproduced in zebrafish with ciliary defects. To establish whether Poc1B-depleted zebrafish exhibit ciliopathy-associated defects, Dr.poc1b morpholinos (MO) were used.
Dr.poc1b morphants exhibited smaller eyes, pericardial edemas, shorter and curved bodies, and mislocalized melanocytes (Fig. 7 A and Fig. S5). Furthermore, at 120 h post fertilization (hpf), ∼20% of the MO-injected fish displayed kidney cysts (Fig. S5 D). The zebrafish pronephros is lined with motile cilia that permit the movement of filtrate through the pronephric ducts for excretion from the body via the cloaca. To determine whether the kidney cysts were associated with impairment of renal filtration, a rhodamine–dextran clearance assay was performed (Hentschel et al., 2005; Tobin and Beales, 2008). The fraction of rhodamine–dextran cleared from the pericardium 24 h post injection in the Dr.poc1b morphants was significantly reduced (Dr.poc1b MO, 48%; control, 66%; Fig. S5 E). The decreased clearance suggests that the efficiency of renal clearance is reduced in the absence of Poc1B.
Figure 7.
Zebrafish Dr.poc1B morphants exhibit ciliopathy defects. (A) Dr.poc1B morphants (DrPoc1B MO) display smaller eyes, heart edemas (red arrow), mislocalized melanocytes (green arrow), truncated bodies, and kidney cysts (white arrow) at 4 d post fertilization (dpf). Bars, 0.5 mm. (B) Cilia (green) of Dr.poc1B morphant embryos at 48 hpf are disorganized and appear shorter within the pronephretic duct (PND) and are expanded within the posterior neural tube (NT). Short cilia were visible in the KV at 12 hpf (Dr.poc1BATG MO, 3.6 ± 0.9 µm; Dr.poc1BSPL MO, 3.6 ± 0.9 µm; control, 6.2 ± 0.8 µm; n > 100; P < 0.001). Bars: (top) 10 µm; (middle) 25 µm; (bottom) 5 µm. (C) Heart laterality defects were observed in Dr.poc1B morphants at 48 hpf. Using in situ hybridization to cmlc2, heart loops were visualized in flat-mounted sections. The ratios indicate the relative frequency of heart positioning. Bar, 0.5 mm. (D) Fish were stained with Alcian blue to visualize cartilage and bone. Morphants displayed craniofacial abnormalities consistent with the ciliopathy phenotype. Dr.poc1B morphants exhibit pharyngeal arch and jaw defects at 6 d post fertilization. PQ, palatoquadrate; M, Meckel's cartilage; CH, ceratohyal; VPA, visceral pharyngeal arches. Bar, 0.5 mm.
Consistent with inefficient ciliary beating and/or flow in the pronephric ducts, cilia stained for acetylated tubulin were less organized and shorter compared with the noninjected control embryos (Fig. 7 B). Cilia in the posterior neural tube were expanded and disorganized (Fig. 7 B). The short cilia were more apparent in the Kupffer's vesicle (KV), in which the ciliary length in Dr.poc1b MO–injected zebrafish was approximately half that of the controls (Fig. 7 B).
Situs inversus is a common developmental defect observed in ciliopathies. The deregulation of morphogen flow within the ciliated node or the zebrafish KV is considered to be a major cause of laterality disorders. The short KV cilia in Dr.poc1b morphants suggests that laterality defects may result from DrPoc1B loss. We quantified the positioning of heart looping by in situ hybridization using a heart marker, cmlc2 (cardiac myosin light chain 2; Peterkin et al., 2007). 26% of Dr.poc1B MO–injected embryos exhibited laterality defects with 15% of embryos displaying right-sided heart looping and 11% with medially located hearts (Fig. 7 C).
Ciliopathy patients commonly display facial gestalt dysmorphology as a result of aberrant neural crest cell (NCC) migration, which is necessary to form the craniofacial skeleton. Zebrafish ciliopathy gene morphants exhibit analogous craniofacial defects (Tobin et al., 2008). Consistent with DrPoc1B having a role in proper NCC migration, Dr.poc1B morphants exhibit jaw and pharyngeal arch defects (Fig. 7 D). Both the dorsal (palatoquadrate) and ventral (Meckel's cartilage) components of the mandibular arch were short and malformed in the morphants, whereas the pharyngeal arches were absent. Morphants also displayed abnormal polarity of the ceratohyal cartilage, which formed in the opposite direction to control embryos. Furthermore, neural crest–derived melanocytes displayed mislocalization, which is congruent with disruption of NCC migration.
Discussion
Poc1 is a conserved WD40 repeat–containing protein that incorporates within multiple domains of the basal body to function in centriole stability and cilia biogenesis. Poc1's role in cilia function is conserved from T. thermophila to vertebrates. Poc1B loss in zebrafish causes maladies found in ciliopathies. Our work suggests an important role for Poc1 function in both motile and immotile cilia. We propose that Poc1's role in maintaining the structural integrity of basal bodies is critical for proper cilia formation.
Basal body structural integrity
Poc1 protein localizes to three major domains within the basal body: the cartwheel, the cylinder walls, and the site of nascent basal body assembly. The cylinder wall localization of Poc1 is consistent with that found in C. reinhardtii and humans (Fig. 7; Hames et al., 2008; Keller et al., 2009). However, these studies did not identify the cartwheel localization. In T. thermophila, the total population of Poc1 at basal bodies is represented by a dynamic fraction that makes up the Poc1 at the site of new assembly and a small fraction at the cartwheel population. The stable and most abundant population of basal body–localized Poc1 is at the cartwheel and the microtubule cylinder walls. Our analyses of Poc1 assembly dynamics defines how and when Poc1 protein localizes to discrete structural elements within the basal body. During new basal body assembly, the cartwheel and cylinder walls acquire the stable population of Poc1, whereas a smaller fraction of Poc1 is dynamically exchanging at basal bodies throughout the cell cycle.
The complex localization of Poc1 to basal body domains for proper assembly and stability, such as the cartwheel and the cylinder walls, suggested that Poc1 is critical for these events. New basal bodies assemble in poc1Δ cells; however, the basal bodies contain morphological defects. The central lumen density was disrupted, as were the microtubules of the proximal end. Cytoplasm also infiltrated the luminal space. Consistent with the stable incorporation of Poc1 assembly at the cylinder walls, we predict that this population of Poc1 is required for stabilizing the microtubule cylinder walls to maintain the central lumen integrity. The loss of lumen electron density was also observed in T. thermophila Sas6a knockouts (Culver et al., 2009), and analogous to Sas6a loss, the loss of the stable Poc1 population at the cartwheel may also perturb the cartwheel structure. However, we did not observe an appreciable difference in cartwheel structure in poc1Δ cells. Structural defects in the basal body cartwheel and/or the cylinder walls likely result in the decreased basal body stability.
Basal bodies disassemble in poc1Δ cells that are maintained under conditions that either block new assembly or destabilize microtubules, nocodazole. The nocodazole sensitivity suggests that Poc1's basal body stability role is at the cylinder walls. Furthermore, the structural and stability defects are exacerbated by increased temperature. At high temperatures, in the absence of Poc1, centrin staining of new basal bodies is decreased, and the ultrastructural defects are increased. Consistent with the structural defects, the stability of basal bodies is reduced. Finally, basal bodies assembled in poc1Δ cells are unstable and do not withstand a cell cycle arrest, suggesting that cell cycle–dependent maturation events must occur before basal bodies assemble into a stable structure. Such stability is facilitated at least in part by Poc1.
We predict that a source of basal body instability is the forces that are exerted on basal bodies or centrioles by attached microtubule arrays. The temperature-sensitive lethality of poc1Δ cells may be the result of the loss of organization and stability of basal bodies in T. thermophila. Elevated temperatures increase cell swimming rates, causing greater forces on basal bodies (Fig. S1 C). This increased force may contribute to the instability of basal bodies, ultimately leading to diminished cell swimming rates in the absence of Poc1. In the case of centrioles, both HsPoc1A and HsPoc1B are required for centriole over duplication during a prolonged S phase arrest (Keller et al., 2009; unpublished data). Drosophila Poc1, along with Sas6 and Plk4, is required for the early formation of a proximal centriole-like structure at the single spermatid centriole (Blachon et al., 2009). We predict that Poc1-dependent stabilization is required to maintain newly assembled centrioles in the S phase arrest over-duplication assay in human cells and at the Drosophila proximal centriole-like structure. Consistent with a model for centriole stabilization, Poc1 overexpression leads to elongated centriole-like structures, whereas Poc1 depletion causes shorter centrioles, suggesting that microtubule stability is facilitated by Poc1 family members (Blachon et al., 2009; Keller et al., 2009).
This function is consistent with the stable Poc1 population localized to the cylinder walls (Fig. 5). A similar nocodazole sensitivity of centrioles was observed in human RPE-1 cells depleted of CAP350 (Le Clech, 2008) and with over-duplicated centrioles formed during an S phase arrest or overexpression of Plk4 (Balczon et al., 1999; Le Clech, 2008). Poc1 may function in a class of protein components along with CAP350 (Le Clech, 2008) to form stable centrioles.
The role of Poc1 in cilia formation and function
The formation of both motile and immotile cilia can occur by several distinct mechanisms, but they all require the presence of a functional basal body. Thus, we would expect that Poc1 functions at both motile and immotile cilia. However, it has been postulated, based on evolutionary studies, that Poc1's function is specific to undulating cilia (Woodland and Fry, 2008). To determine if this is the case, we tested whether human Poc1A and Poc1B are required for normal ciliogenesis of immotile primary cilia. Poc1B had a distinct function in primary cilia formation in human RPE-1 cells (Fig. 6), suggesting that Poc1 family members have roles in both types of cilia. This is consistent with localization showing that Poc1 resides at the basal bodies of both motile and immotile cilia (Avidor-Reiss et al., 2004; Keller et al., 2005, 2009; Kilburn et al., 2007; Hames et al., 2008; this study).
We found that only HsPoc1B is required for primary cilia formation and ciliary length. This suggests that the two HsPoc1 proteins have divergent functions in which both proteins have a role in centriole duplication (Keller et al., 2009; unpublished data), whereas HsPoc1B is specifically required for cilia formation. Alternatively, although the siRNA treatment conditions with HsPoc1A were sufficient to inhibit centrosome reduplication (unpublished data), it is possible that a further depletion of HsPoc1A is required to manifest a primary ciliogenesis defect.
The role of HsPoc1B in ciliogenesis remains unclear. We predict that Poc1-stabilized basal bodies may be required for normal ciliogenesis. Consistent with a stability function, the cartwheel protein Bld10 in Drosophila exhibits similar roles in ciliogenesis (Blachon et al., 2009; Mottier-Pavie and Megraw, 2009). However, it is worth noting that Bld10 in Drosophila localizes to domains other than the cartwheel. Similarly, HsPoc1B also localizes near the transition zone that links the basal body to the cilia (Fig. 6 C). This fraction of HsPoc1B protein in human cells may be required for proper cilia assembly and function.
The inhibition of primary ciliogenesis and shorter cilia in human cells suggests that the Poc1 family is required for proper cilia formation and function. Cilia participate in an extensive number of processes required for normal embryonic development, and cilia defects cause a wide range of morphological anomalies (Gerdes et al., 2009). To determine whether Poc1 family members are required for such critical ciliary-based roles, we examined the phenotypes of DrPoc1B morphants. Zebrafish DrPoc1B morphants exhibit ciliopathy-like defects with truncated bodies, curved tails, small eyes, heart edemas, short KV cilia and laterality defects, neural tube cilia defects, and renal cilia defects with associated kidney cysts. These defects are not observed with all mutant centrosomal proteins, indicating that Poc1B function may be specific to basal body assembly and function in ciliogenesis (Graser et al., 2007; Wilkinson et al., 2009).
Laterality defects or a disruption of the left–right asymmetry of heart placement was discovered in Dr.poc1b morphants. This is commonly associated with improper assembly or motility of the nodal cilia or the zebrafish equivalent, KV, during embryogenesis. Motility is required for leftward flow of determinants during early development, leading to proper organ placement (Essner et al., 2005). KV cilia in Dr.poc1b morphants are significantly shorter, and these defects are likely the cause of the asymmetric heart-positioning anomalies.
In the absence of DrPoc1B, the motile cilia of the zebrafish pronephric duct appear shorter and, as found for T. thermophila, appear disorganized and decreased in frequency. Such defects were also found for the centrosomal proteins Cep70 and Cep131 (Wilkinson et al., 2009). As expected for defects in renal cilia formation and organization, the frequency of kidney cysts is increased in Dr.poc1b morphants. Cystic kidney diseases are among the most common genetic diseases resulting in human death (for review see Hildebrandt and Otto, 2005). Kidney cysts may develop as a result of improper polarity and mitotic spindle positioning in the lumen of the pronephric ducts. Such polarity cues are facilitated by the noncanonical Wnt–planar cell polarity signaling pathway either through proper ciliogenesis or maintenance of cilia.
Consistent with a role in polarity and spatial cues to direct cell migration, Dr.poc1b MO–injected embryos display facial defects analogous to those found with Bardet–Biedl syndrome MO–injected fish. Such anomalies are likely the result of improper NCC migration. For proper migration, NCCs are regulated by and receive cues from both the noncanonical Wnt and Sonic hedgehog signaling pathways, both of which rely on cilia for their proper function. As found for Bardet–Biedl syndrome genes (Tobin et al., 2008), Poc1B may play a role in these critical signaling pathways for NCC migration and patterning by enabling the proper formation and stabilization of basal bodies and functional cilia.
Finally, we identify the importance of basal body components in basal body stability enabling their function as a structural platform for both motile and immotile cilia. We propose that defects in basal body stability can lead to partially functional basal bodies and cilia and ultimately a more subtle defect than the complete loss of the structure. These subtle defects may be a key principle and novel mechanism for the onset of human ciliopathies.
Materials and methods
T. thermophila cell culture
T. thermophila cells were grown at 30°C in 2% SPP (2% proteose peptone, 0.2% glucose, 0.1% yeast extract, and 0.003% Fe-EDTA) unless otherwise indicated. Strains CU427, CU428, B2086, and B*VII were used in this study (T. thermophila Stock Center, Cornell University). For G1 arrest to inhibit new basal body assembly, cells were starved by washing into starvation media (SM; 10 mM Tris-HCl, pH 7.4) for 12 h. Cell densities were quantified using a Counter Z1 (Beckman Coulter).
To measure cell viability, cells were collected at 24-h intervals, washed with NaPO4 buffer, and resuspended into phosphate buffer with 25 µg/ml propidium iodide for 30 min at RT. The cell lethality (n > 5,000 cells) was quantified using a flow cytometer (FACSCan; BD). For nocodazole treatments, T. thermophila cells were grown at log phase in the presence of either 10 µg/ml nocodazole or DMSO for 7 h at 30°C before fixation and immunofluorescence. The basal body frequency of cells with early oral primordium (stages 1 and 2) was quantified for the nocodazole sensitivity experiments. This is distinct from the starvation experiments in which cells were arrested in G1.
Plasmids
The T. thermophila POC1 gene (NCBI Protein Database accession no. XP_001029778.1) knockout cassette that replaced the entire POC1 ORF with the NEO2 paromomycin resistance gene was constructed by PCR amplification and insertion of a 1.3-kb sequence upstream of POC1 flanked by SacII and BamHI sites and a 1.3-kb sequence downstream of POC1 flanked by XhoI and KpnI into p4T2-1 (Gaertig et al., 1994). pBSmttGFPPOC1 was constructed by cloning the entire POC1 cDNA into the Gateway pENTR-D (Invitrogen). This was subcloned into pBSmttGFPgtw using Gateway LR Clonase (Invitrogen). Truncated forms of POC1 were similarly constructed by cloning the coding sequence for aa 1–342 to create pBSmttGFPpoc1-wd40 and aa 343–end (634) to generate pBSmttGFPpoc1–c terminus. A C-terminal mCherry fusion (p4T21-POC1-mCherryLAP) with POC1 was constructed by amplifying and inserting flanking sequences immediately upstream (flanked by Kpn1 and EcoR1) and downstream (flanked by BamH1 and Sac1) of the POC1 termination codon and into p4T21-MTTUTR-CherryLAP.
mCherry-tagged fusion cassettes were created for both HsPoc1A (NCBI Protein database accession no. AAI19694.1) and HsPoc1B (NCBI Protein Database accession no. NP_758440.1; pIC194-HsPoc1A and pIC194-HsPoc1B). The cDNA sequence for HsPOC1A was amplified from HeLa total RNA using an RT-PCR kit (Superscript One-step; Invitrogen) with flanking XhoI and BamHI restriction sites for cloning into the pIC194 LAP vector (Cheeseman and Desai, 2005). HsPOC1B cDNA was PCR amplified from American Type Culture Collection clone MGC-26753 with flanking XhoI and BamHI restriction sites to clone into the pIC194. The cassettes were confirmed by DNA sequencing. Truncations of HsPoc1A were generated by subcloning the coding sequence for aa 1–299 and 300–552 flanked by XhoI and BamHI restriction sites to generate pIC194-Hspoc1a-wd40 and pIC194-Hspoc1a–c terminus fragments, respectively. Truncations of HsPoc1B were generated by cloning the coding sequence for aa 1–298 and 299–478 flanked by XhoI and BamHI restriction sites to generate pIC194-Hspoc1b-wd40 and pIC194-Hspoc1b–c terminus, respectively.
Generation of T. thermophila poc1Δ
A plasmid containing poc1::NEO2 was digested with SacI and KpnI to release the knockout cassette and was introduced into the micronucleus of conjugating cells (B2086 and CU428) by biolistic bombardment (Bruns and Cassidy-Hanley, 2000). Proper integration and replacement of NEO2 into the POC1 locus was confirmed by PCR. Crosses to confirm that the transformation occurred in the micronucleus were performed, and the knockout heterokaryon was constructed as described previously (Hai et al., 2000). Knockout strains were generated by mating star and nonstar strains containing the homozygous poc1::NEO2 in the micronucleus (poc1Δ). The NEO2 insert was eliminated during mating of the micronuclear knockout heterokaryons so that the knockout could not be detected by paromomycin resistance. This elimination of exogenous sequences into the micronucleus has been previously described (Liu et al., 2005). Complete elimination of POC1 was confirmed using PCR and RT-PCR and by wild-type POC1 rescue of the knockout phenotypes by reintroduction at the RPL29 locus (pBSmttGFPPOC1).
Macronuclear transformation
The GFP fusions were transformed into the T. thermophila macronucleus by biolistic bombardment (Bruns and Cassidy-Hanley, 2000). Paromomycin- or cycloheximide-resistant cells (depending on the experiment) were selected and confirmed by basal body fluorescence.
Light microscopy
Fluorescence microscopy, FRAP, and the quantification of basal body fluorescence intensities were performed as previously described (Pearson et al., 2009). Imaging was performed using an upright microscope (DMRXA; Leica) with a 63× Plan Apo NA 1.32 objective and a charge-coupled device camera (CoolSNAP HQ2; Photometrics). MetaMorph imaging software (MDS Analytical Technologies) was used to collect and analyze the images. All images were acquired at RT. Acquisition exposure times between 100 and 500 ms were used. Basal body fluorescence intensities were determined by subtracting the background fluorescence (Fbackground). 5 × 5 (inner fluorescence intensity [Fi])–and 9 × 9 (outer fluorescence intensity [Fo])–pixel regions were symmetrically placed around each basal body, and the total integrated fluorescence intensity was determined for each region. Fbackground was calculated based on the surrounding fluorescence that is not contained in the inner fluorescence intensity. The background fluorescence intensity was corrected for the total region size of Fi (Fbackground = [Fo − Fi] × 25/56). The basal body fluorescence intensity was determined by subtracting Fbackground from Fi.
FRAP of GFP-Poc1 was performed as previously described (Pearson et al., 2009). Basal bodies were photobleached using a targeted 13 × 13–pixel region with a 500-ms laser exposure from a digital diaphragm system (Mosaic; Photonics Systems). Images were collected using an inverted microscope (TE2000U; Nikon) fitted with a confocal system (Solamere), confocal head (CSU-10; Yokogawa), Plan Apo 60× NA 1.4 violet-corrected objective (Nikon), and a charge-coupled device camera (Cascade II-512B; Photometrics). 350-ms exposures were acquired, the time intervals ranged from 1 s to 1 min, and a maximum experimental length of 5 min was performed. Fluorescence intensities for each time point were determined using the aforementioned background subtraction, and the fluorescence recovery was analyzed using Excel software (Microsoft). A single exponential recovery curve was fit to each recovery profile of the short time interval experiments and averaged for 16 experiments. The percentage of recovery, rate constant (k), and recovery t1/2 were calculated. To verify that the recovery for each protein exhibited single exponential recovery kinetics, the natural logarithm of the fluorescence recovery was plotted relative to time to determine whether a single, straight line (predicting a single rate constant) was observed. An r2 value was calculated (0.98) for a regression line through the dataset. A theoretical recovery curve based on the k value and final percentage of recovery was displayed relative to the mean recovery plots for comparison.
Cell motility was measured by washing log phase growing cells into SM for 30 min. Samples were diluted to 1.0 × 104 cells/ml in SM and transferred into an imaging chamber (CoverWell perfusion chamber; Invitrogen). Cells were imaged for 0.45 s at 0.045-s intervals. The cell motility rates were measured using the Track Points function in MetaMorph. A minimum of 200 traces were analyzed for each condition.
To identify new basal body assembly, old and new basal bodies were followed as previously described (Shang et al., 2005; Pearson et al., 2009). Cells growing in log phase were starved for 12 h before they were resuspended in fresh SPP media. After 2 h of growth, when a fraction of the cells were dividing, cells were fixed and stained for centrin and K antigen. T. thermophila cells duplicate their basal bodies anterior to their parent basal bodies within the medial region of cells before cell division.
The frequency of T. thermophila basal bodies per unit length was quantified in either starved or log phase growing cells to ensure that a consistent cell cycle stage was analyzed in growing cells in either interphase (no oral primordium) or early cell division (oral primordium stages 1–2; Bakowska et al., 1982), depending on the experiment. Anticentrin staining was used to mark all basal bodies (Stemm-Wolf et al., 2005). The number of basal bodies in a 10-µm length of each ciliary row at the cell median was counted (Fig. 1 C, top). A minimum of 150 rows were counted for each condition. The mean cell length was measured to ensure that changes in basal body frequency were not a result of changes in cell length.
TEM
IEM in T. thermophila was performed as previously described (Kilburn et al., 2007; Pearson et al., 2009). Images were collected using an electron microscope (CM10; Philips) equipped with a digital camera (BioScan2; Gatan) and digital micrograph software (Gatan). IEM in human RPE-1 cells was performed by inducing primary cilia formation by starving cells in DME with 0.25% FBS for 48 h. Cells were trypsinized, and the PBS-washed cell pellet was resuspended in 150 mM mannitol and 5% dextran (19 kD; Sigma-Aldrich) in DME with 0.25% FBS and gently centrifuged. Aliquots of the loose pellet were loaded into the 100-µm well of a type A specimen carrier (Technotrade International) and high pressure frozen (HPM-010; BAL-TEC). Frozen samples were freeze substituted in 0.1% uranyl acetate in acetone using an EM automatic freeze substitution system (AFS; Leica). Initial freeze substitution was performed at −85°C for 3 d and gradually warmed to −50°C for infiltration and embedding in Lowicryl HM20 (Electron Microscopy Sciences). Serial 60-nm-thin sections were collected on formvar-coated nickel slot grids. Grids were exposed to a blocking solution consisting of 1.0% nonfat dry milk in PBS with 0.05% Tween 20 for 30 min and floated on droplets containing the affinity purified HsPoc1B antibody diluted to either 1:5 or 1:10 in blocking reagent for 2 h at RT. After thorough rinsing with PBST, grids were incubated on 15-nm gold goat anti–rabbit IgG secondary antibody for 1 h at RT followed by rinsing with PBST and water. After drying, grids were stained with 2% uranyl acetate in 70% methanol and lead citrate for 3 min each. 87 gold particles were localized in a total of nine centrioles and basal bodies from serial 60-nm sections. Gold particles were localized to the transition zone, cylinder wall, and cartwheel. The relative gold distribution pattern using 25 gold particles is shown in Fig. 6 C (left).
Structural analysis of basal bodies in poc1Δ was performed using chemically fixed T. thermophila cells. T. thermophila cultures were prepared for conventional TEM using a modified protocol from Orias et al. (1983). Cells were fixed at RT for 3 min in 2% glutaraldehyde, 1% sucrose, and 5 mM sodium phosphate buffer, pH 7.0. An equal volume of 2% osmium tetroxide in 20 mM phosphate buffer was added, and the cells were left in the combined fixative for 10 min. The cells were centrifuged, and the fixative mixture was replaced with fresh 2% osmium tetroxide in 20 mM phosphate buffer for 30 min. Samples were partially dehydrated in 50% ethanol then stained en bloc with 1% uranyl acetate in 70% ethanol. Dehydration was completed with ethanol followed by propylene oxide, and samples were embedded in epon/araldite epoxy resin. 70-nm-thin sections were stained with 2% uranyl acetate and lead citrate and imaged using a transmission electron microscope (CM10; Philips).
Immunofluorescence
T. thermophila were fixed and stained with rabbit anti-Cen1 (Stemm-Wolf et al., 2005), mouse anti–K antigen (10D12; Williams et al., 1990; Pearson et al., 2009), or anti–kinetodesmal fiber antibodies (striated rootlet; 5D8; Jerka-Dziadosz et al., 1995) provided by N. Williams and J. Frankel (University of Iowa, Iowa City, IA). A modified ethanol fixation procedure was used (Pearson et al., 2009). 10 ml cultured cells were pelleted and resuspended in 2.0 ml fixative (70% ethanol and 0.2% Triton X-100) on ice for 10 min. Cells were subsequently washed and stained for immunofluorescence in suspension. Samples were incubated with anti-Cen1 (1:1,000) and anti–K antigen (1:50) for 1 h at RT or overnight at 4°C before washing and incubating with secondary antibodies for 1 h at RT (Alexa Fluor 488– and Texas red–conjugated anti–rabbit or –mouse secondary antibodies; 1:1,000). Cells were washed, incubated on poly-l-lysine–coated coverslips, and mounted in Citifluor (Ted Pella, Inc.).
Human cells were stained with mouse anti–C-Nap1 (1:1,000; BD), mouse anti-acetylated tubulin (1:2,000; 6–11-B1; Sigma-Aldrich), mouse anti–γ-tubulin (1:500; GTU-88; Sigma-Aldrich), mouse anti-Cetn2 (1:2,000; 20H5; Salisbury et al., 1988), rabbit anti–γ-tubulin (1:1,000; DQ-19; Sigma-Aldrich), and rabbit anti-HsPoc1B (1:250; CO200).
RPE-1 or human osteosarcoma (U2OS) cells were fixed with 100% methanol (−20°C) for 2 min, washed once with PBS/Mg (PBS + 1 mM MgCl2), and permeabilized for 10 min with PBS + 0.5% Triton X-100. Cells were washed in PBS/Mg, blocked for 1 h in blocking buffer (1× PBS, 0.5% bovine serum albumin, 0.5% NP-40, 1 mM MgCl2, and 1 mM NaN3), and incubated for at least 2 h in primary antibody diluted in blocking buffer. Cells were washed 4× with PBS/Mg and incubated in diluted secondary antibodies before a final wash and mounting in Citifluor.
Primary cilia lengths were quantified in human RPE-1 cells. At least 110 cilia were measured for each condition (lamin A/C or HsPoc1BUS siRNA) for three total experiments. Cilia lengths were measured using MetaMorph.
Transfections
For transient expression of mCherry, mCherry-HsPoc1A, mCherry-HsPoc1B, and mCherry-HsPoc1A and -HsPoc1B truncations, either RPE-1 or U2OS cells were transfected with each expression plasmid using Effectene reagent (QIAGEN). Transfected cells were identified by fluorescence microscopy.
Human siRNA
siRNA nucleotide duplexes were synthesized by Proligo (Sigma-Aldrich). HsPoc1AMID (5′-CTGGGTCCGCTGTGCCAAG-3′), HsPoc1ADS (5′-GGGACCAGCCACCACTGTT-3′), HsPoc1BUS (5′-CTTCTCAGATTCCGTTGGA-3′), and HsPoc1BDS (5′-AAAGTTGAGACTGTAGAAA-3′) were used to knockdown the two human Poc1 proteins. Transfections were performed using Oligofectamine transfection reagent as described by Invitrogen. For controls, cells were either treated with Oligofectamine without siRNA or lamin A/C siRNA (Applied Biosystems). siRNA knockdown was assessed using RT-PCR to amplify a region of each mRNA with GAPDH (glyceraldehyde 3-phosphate dehydrogenase) as a negative control (Fig. S4 A). The knockdown of HsPoc1B was also confirmed by measuring the decrease in anti-HsPoc1B (CO200) staining at basal bodies (Fig. S4 B). Both decreased primary cilia length and frequency were found for the two HsPoc1B siRNA duplexes.
Human primary ciliogenesis
To measure primary cilia formation, RPE-1 cells were seeded in DME with 10% FBS for 24 h. Cells were transfected with siRNAs for 4 h and resuspended in DME with 0.25% FBS. The frequency of primary cilia was monitored by staining for acetylated tubulin 48 h after refeeding.
Poc1 antibodies
The HsPOC1B (NCBI Protein Database accession no. NP_758440.1) coding sequence for aa 312–406 was PCR amplified from American Type Culture Collection cDNA clone MGC-26753 and cloned into pQE10 to generate a 6× His fusion antigen for HsPoc1B. This fusion was injected into rabbits for antibody production (Covance). Antibodies (anti-HsPoc1B; CO200) were purified by binding to antigen (6× His-Hspoc1; aa 312–406) isolated on a Western blot. The bound antibody was washed and eluted using low pH glycine. This antibody was not successful for Western blot analysis (no detectable bands); however, it was productive for immunofluorescence of centrioles and basal bodies. Localization was identical to that of the mCherry-tagged versions of HsPoc1A and HsPoc1B (Fig. S3). The recognition of centrioles and basal bodies by anti-HsPoc1B could be blocked by prebinding the antibody with the antigen before incubation with cells (Fig. S3 A, bottom), indicating the specificity for HsPoc1B.
Zebrafish
Wild-type zebrafish from AB × Tup LF strain were used for DrPoc1 knockdown experiments. Staging and husbandry were performed as previously described (Westerfield, 1995). Antisense MO oligonucleotides (GeneTools, LLC) were designed against the Drpoc1 (NCBI Protein database accession no. CAM15113.1) transcript start codon (Drpoc1ATG, 5′-GATCCTCCATTACAGACGCCATGAT-3′) and the intron 1–exon 2 splice site (Drpoc1Spl, 5′-TCCTCCTGAATAAGACAGAGAACTT-3′).
The rhodamine–dextran clearance and renal filtration assay was performed as previously described (Tobin and Beales 2008) by injecting 1 nl of 10 kD rhodamine–dextran (Invitrogen) into the pericardium of 72-hpf embryos. Embryos were mounted at 3 and 24 h post injection in 3% methylcellulose, and images were acquired under identical parameters using a stereomicroscope (Lumar V12; Carl Zeiss, Inc.) equipped with a camera (AxioCam HRC; Carl Zeiss, Inc.) and AxioVision software (version 4.6; Carl Zeiss, Inc.). Relative cardiac fluorescent intensity was measured using a 100 × 100–pixel region of interest with ImageJ software (National Institutes of Health). Between time points, embryos were kept in fish water maintained at 28.5°C.
Zebrafish embryos were fixed in 4% PFA. Ciliary morphology was determined using a monoclonal anti-acetylated tubulin antibody (1:500; T6793; Sigma-Aldrich) with Alexa Fluor secondary reagents (1:1,000; Invitrogen). Embryos were flat mounted in Citifluor and imaged using a confocal microscope (SP2 [Leica] or LSM710 [Carl Zeiss, Inc.]).
Craniofacial defects were characterized using the cartilage stain Alcian blue as previously described (Neuhauss et al., 1996). Embryos were bleached in 30% hydrogen peroxide until sufficiently translucent. Before incubating overnight in Alcian blue solution, embryos were cleared in acidic ethanol until the desired clarity was resolved and specimens were stored and imaged in glycerol.
RNA was extracted from ∼30 48-hpf embryos per experimental group using TRIzol (Invitrogen) and isopropanol RNA extraction and precipitation. Embryos were ground in 50 µl TRIzol (50–100 mg tissue/ml) using a micropestle incubated for 5 min at RT before adding 10 µl chloroform (200 µl chloroform/ml TRIzol). The solution was vigorously mixed and incubated for 3 min before centrifuging for 15 min. Centrifugation was performed at 12,000 g in a 4°C microcentrifuge. 25 µl isopropanol (500 µl isopropanol/ml TRIzol) was added to the aqueous phase in a fresh tube, mixed, and incubated for 10 min at RT. The precipitate was centrifuged for 10 min, supernatant were removed, and RNA were washed in 75% ethanol (1 ml ethanol/ml TRIzol) by brief vortexing and further centrifugation for 5 min at 7,500 g. RNA was left to air dry for 5 min before redissolving in 10 µl of nuclease-free water. First-strand cDNA was synthesized using random nanomers (Sigma-Aldrich) and Omniscript transcriptase (QIAGEN) according to the manufacturer's instructions. Standard PCR was performed using primers against the forward strand of Dr.poc1b within the 5′ untranslated region (5′-GTGGCTCAACATGGAGGAGT-3′) and either the reverse strand of exon 3 (5′-TGGGTGTCCACAATCGAAC-3′) or exon 12 (5′-TCACAACGGGGGTCCCTCGGA-3′). GAPDH primers (forward, 5′-TTAAGGCAGAAGGCGGCAAA-3′; reverse, 5′-AAGGAGCCAGGCAGTTGGTG-3′) were used as control.
Online supplemental material
Fig. S1 shows the cell growth, viability, swimming, and morphological defects in poc1Δ cells. Fig. S2 shows that Poc1 localization is dependent on the WD40 repeat domain and that basal bodies are less stable in poc1Δ cells. Fig. S3 shows that endogenous HsPoc1B localizes to basal bodies and centrioles. Fig. S4 shows the level of HsPoc1A and HsPoc1B depletion by siRNA. Fig. S5 shows the defective renal clearance and increased kidney cysts in zebrafish Poc1B morphants. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200908019/DC1.
Supplementary Material
Acknowledgments
We thank A. Stemm-Wolf, J. Deluca, and M. Jones for critical comments and discussion on the manuscript. We thank I. Cheeseman, D. Chalker, and A. Stemm-Wolf for plasmids, M. Rout, J. Salisbury, J. Frankel, and N. Williams for antibodies, and J. Meehl and C. Clarissa for EM expertise.
This work was funded by the National Institutes of Health (grant GM074746) and the John Simon Guggenheim Memorial Foundation to M. Winey and by the European Union (EUCILIA contract number 201804) to P.L. Beales and D. Osborn. P.L. Beales is a Wellcome Trust Senior Fellow. C.G. Pearson is supported by the Damon Runyan Cancer Research Foundation (grant 1879-05).
Footnotes
Abbreviations used in this paper:
- hpf
- h post fertilization
- IEM
- immuno-EM
- KV
- Kupffer's vesicle
- MO
- morpholino
- NCC
- neural crest cell
- SM
- starvation media
- TEM
- transmission EM
References
- Abal M., Keryer G., Bornens M.. 2005. Centrioles resist forces applied on centrosomes during G2/M transition. Biol. Cell. 97:425–434. 10.1042/BC20040112 [DOI] [PubMed] [Google Scholar]
- Andersen J.S., Wilkinson C.J., Mayor T., Mortensen P., Nigg E.A., Mann M.. 2003. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 426:570–574. 10.1038/nature02166 [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T., Maer A.M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C.S.. 2004. Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell. 117:527–539. 10.1016/S0092-8674(04)00412-X [DOI] [PubMed] [Google Scholar]
- Azimzadeh J., Bornens M.. 2004. The centrosome in evolution. In Centrosomes in Development and Disease. Nigg E.A., editor. Wiley-VCH, Weinheim, Germany. 431 pp. [Google Scholar]
- Badano J.L., Mitsuma N., Beales P.L., Katsanis N.. 2006. The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7:125–148. 10.1146/annurev.genom.7.080505.115610 [DOI] [PubMed] [Google Scholar]
- Bakowska J., Frankel J., Nelsen E.M.. 1982. Regulation of the pattern of basal bodies within the oral apparatus of Tetrahymena thermophila. J. Embryol. Exp. Morphol. 69:83–105. [PubMed] [Google Scholar]
- Balczon R., Varden C.E., Schroer T.A.. 1999. Role for microtubules in centrosome doubling in Chinese hamster ovary cells. Cell Motil. Cytoskeleton. 42:60–72. [DOI] [PubMed] [Google Scholar]
- Baron A.T., Greenwood T.M., Bazinet C.W., Salisbury J.L.. 1992. Centrin is a component of the pericentriolar lattice. Biol. Cell. 76:383–388. 10.1016/0248-4900(92)90442-4 [DOI] [PubMed] [Google Scholar]
- Basmussen L., Orias E.. 1975. Tetrahymena: growth without phagocytosis. Science. 190:464–465. 10.1126/science.1166313 [DOI] [PubMed] [Google Scholar]
- Basto R., Lau J., Vinogradova T., Gardiol A., Woods C.G., Khodjakov A., Raff J.W.. 2006. Flies without centrioles. Cell. 125:1375–1386. 10.1016/j.cell.2006.05.025 [DOI] [PubMed] [Google Scholar]
- Beisson J., Jerka-Dziadosz M.. 1999. Polarities of the centriolar structure: morphogenetic consequences. Biol. Cell. 91:367–378. 10.1016/S0248-4900(99)80097-5 [DOI] [PubMed] [Google Scholar]
- Berbari N.F., O'Connor A.K., Haycraft C.J., Yoder B.K.. 2009. The primary cilium as a complex signaling center. Curr. Biol. 19:R526–R535. 10.1016/j.cub.2009.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blachon S., Cai X., Roberts K.A., Yang K., Polyanovsky A., Church A., Avidor-Reiss T.. 2009. A proximal centriole-like structure is present in Drosophila spermatids and can serve as a model to study centriole duplication. Genetics. 182:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Khodjakov A., Mir L.M., Rieder C.L., Eddé B., Bornens M.. 1998a. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J. Cell Biol. 143:1575–1589. 10.1083/jcb.143.6.1575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y., Moudjou M., Fouquet J.P., Desbruyères E., Eddé B., Bornens M.. 1998b. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil. Cytoskeleton. 39:223–232. [DOI] [PubMed] [Google Scholar]
- Broadhead R., Dawe H.R., Farr H., Griffiths S., Hart S.R., Portman N., Shaw M.K., Ginger M.L., Gaskell S.J., McKean P.G., Gull K.. 2006. Flagellar motility is required for the viability of the bloodstream trypanosome. Nature. 440:224–227. 10.1038/nature04541 [DOI] [PubMed] [Google Scholar]
- Brown J.M., Marsala C., Kosoy R., Gaertig J.. 1999. Kinesin-II is preferentially targeted to assembling cilia and is required for ciliogenesis and normal cytokinesis in Tetrahymena. Mol. Biol. Cell. 10:3081–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruns P.J., Cassidy-Hanley D.. 2000. Biolistic transformation of macro- and micronuclei. Methods Cell Biol. 62:501–512. 10.1016/S0091-679X(08)61553-8 [DOI] [PubMed] [Google Scholar]
- Cheeseman I.M., Desai A.. 2005. A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci. STKE. 2005:pl1. 10.1126/stke.2662005pl1 [DOI] [PubMed] [Google Scholar]
- Culver B.P., Meehl J.B., Giddings T.H., Jr., Winey M.. 2009. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol. Biol. Cell. 20:1865–1877. 10.1091/mbc.E08-08-0838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dammermann A., Müller-Reichert T., Pelletier L., Habermann B., Desai A., Oegema K.. 2004. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 7:815–829. 10.1016/j.devcel.2004.10.015 [DOI] [PubMed] [Google Scholar]
- Dammermann A., Maddox P.S., Desai A., Oegema K.. 2008. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin–mediated addition of centriolar microtubules. J. Cell Biol. 180:771–785. 10.1083/jcb.200709102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupuis-Williams P., Fleury-Aubusson A., de Loubresse N.G., Geoffroy H., Vayssié L., Galvani A., Espigat A., Rossier J.. 2002. Functional role of ϵ-tubulin in the assembly of the centriolar microtubule scaffold. J. Cell Biol. 158:1183–1193. 10.1083/jcb.200205028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K. 2007. Finding treasures in frozen cells: new centriole intermediates. Bioessays. 29:630–634. 10.1002/bies.20594 [DOI] [PubMed] [Google Scholar]
- Dutcher S.K., Trabuco E.C.. 1998. The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol. Biol. Cell. 9:1293–1308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutcher S.K., Morrissette N.S., Preble A.M., Rackley C., Stanga J.. 2002. Epsilon-tubulin is an essential component of the centriole. Mol. Biol. Cell. 13:3859–3869. 10.1091/mbc.E02-04-0205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddé B., Rossier J., Le Caer J.P., Desbruyères E., Gros F., Denoulet P.. 1990. Posttranslational glutamylation of alpha-tubulin. Science. 247:83–85. 10.1126/science.1967194 [DOI] [PubMed] [Google Scholar]
- Eggenschwiler J.T., Anderson K.V.. 2007. Cilia and developmental signaling. Annu. Rev. Cell Dev. Biol. 23:345–373. 10.1146/annurev.cellbio.23.090506.123249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner J.J., Amack J.D., Nyholm M.K., Harris E.B., Yost H.J.. 2005. Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 132:1247–1260. 10.1242/dev.01663 [DOI] [PubMed] [Google Scholar]
- Fry A.M., Mayor T., Meraldi P., Stierhof Y.D., Tanaka K., Nigg E.A.. 1998. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle–regulated protein kinase Nek2. J. Cell Biol. 141:1563–1574. 10.1083/jcb.141.7.1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J., Gu L., Hai B., Gorovsky M.A.. 1994. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 22:5391–5398. 10.1093/nar/22.24.5391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garreau de Loubresse N., Ruiz F., Beisson J., Klotz C.. 2001. Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol. 2:4. 10.1186/1471-2121-2-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J.M., Davis E.E., Katsanis N.. 2009. The vertebrate primary cilium in development, homeostasis, and disease. Cell. 137:32–45. 10.1016/j.cell.2009.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graser S., Stierhof Y.D., Lavoie S.B., Gassner O.S., Lamla S., Le Clech M., Nigg E.A.. 2007. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 179:321–330. 10.1083/jcb.200707181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundersen G.G., Bulinski J.C.. 1986. Distribution of tyrosinated and nontyrosinated alpha-tubulin during mitosis. J. Cell Biol. 102:1118–1126. 10.1083/jcb.102.3.1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B., Gaertig J., Gorovsky M.A.. 2000. Knockout heterokaryons enable facile mutagenic analysis of essential genes in Tetrahymena. Methods Cell Biol. 62:513–531. 10.1016/S0091-679X(08)61554-X [DOI] [PubMed] [Google Scholar]
- Hames R.S., Hames R., Prosser S.L., Euteneuer U., Lopes C.A., Moore W., Woodland H.R., Fry A.M.. 2008. Pix1 and Pix2 are novel WD40 microtubule-associated proteins that colocalize with mitochondria in Xenopus germ plasm and centrosomes in human cells. Exp. Cell Res. 314:574–589. 10.1016/j.yexcr.2007.10.019 [DOI] [PubMed] [Google Scholar]
- Hentschel D.M., Park K.M., Cilenti L., Zervos A.S., Drummond I., Bonventre J.V.. 2005. Acute renal failure in zebrafish: a novel system to study a complex disease. Am. J. Physiol. Renal Physiol. 288:F923–F929. 10.1152/ajprenal.00386.2004 [DOI] [PubMed] [Google Scholar]
- Hildebrandt F., Otto E.. 2005. Cilia and centrosomes: a unifying pathogenic concept for cystic kidney disease? Nat. Rev. Genet. 6:928–940. 10.1038/nrg1727 [DOI] [PubMed] [Google Scholar]
- Hiraki M., Nakazawa Y., Kamiya R., Hirono M.. 2007. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr. Biol. 17:1778–1783. 10.1016/j.cub.2007.09.021 [DOI] [PubMed] [Google Scholar]
- Jerka-Dziadosz M., Jenkins L.M., Nelsen E.M., Williams N.E., Jaeckel-Williams R., Frankel J.. 1995. Cellular polarity in ciliates: persistence of global polarity in a disorganized mutant of Tetrahymena thermophila that disrupts cytoskeletal organization. Dev. Biol. 169:644–661. 10.1006/dbio.1995.1176 [DOI] [PubMed] [Google Scholar]
- Kaczanowski A. 1978. Gradients of proliferation of ciliary basal bodies and the determination of the position of the oral primordium in Tetrahymena. J. Exp. Zool. 204:417–430. 10.1002/jez.1402040313 [DOI] [PubMed] [Google Scholar]
- Keller L.C., Romijn E.P., Zamora I., Yates J.R., III, Marshall W.F.. 2005. Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr. Biol. 15:1090–1098. 10.1016/j.cub.2005.05.024 [DOI] [PubMed] [Google Scholar]
- Keller L.C., Geimer S., Romijn E., Yates J., III, Zamora I., Marshall W.F.. 2009. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol. Biol. Cell. 20:1150–1166. 10.1091/mbc.E08-06-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilburn C.L., Pearson C.G., Romijn E.P., Meehl J.B., Giddings T.H., Jr., Culver B.P., Yates J.R., III, Winey M.. 2007. New Tetrahymena basal body protein components identify basal body domain structure. J. Cell Biol. 178:905–912. 10.1083/jcb.200703109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkham M., Müller-Reichert T., Oegema K., Grill S., Hyman A.A.. 2003. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 112:575–587. 10.1016/S0092-8674(03)00117-X [DOI] [PubMed] [Google Scholar]
- Kochanski R.S., Borisy G.G.. 1990. Mode of centriole duplication and distribution. J. Cell Biol. 110:1599–1605. 10.1083/jcb.110.5.1599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Clech M. 2008. Role of CAP350 in centriolar tubule stability and centriole assembly. PLoS One. 3:e3855. 10.1371/journal.pone.0003855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidel S., Gönczy P.. 2003. SAS-4 is essential for centrosome duplication in C elegans and is recruited to daughter centrioles once per cell cycle. Dev. Cell. 4:431–439. 10.1016/S1534-5807(03)00062-5 [DOI] [PubMed] [Google Scholar]
- Leidel S., Gönczy P.. 2005. Centrosome duplication and nematodes: recent insights from an old relationship. Dev. Cell. 9:317–325. 10.1016/j.devcel.2005.08.004 [DOI] [PubMed] [Google Scholar]
- Leidel S., Delattre M., Cerutti L., Baumer K., Gönczy P.. 2005. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat. Cell Biol. 7:115–125. 10.1038/ncb1220 [DOI] [PubMed] [Google Scholar]
- Li J.B., Gerdes J.M., Haycraft C.J., Fan Y., Teslovich T.M., May-Simera H., Li H., Blacque O.E., Li L., Leitch C.C., et al. 2004. Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell. 117:541–552. 10.1016/S0092-8674(04)00450-7 [DOI] [PubMed] [Google Scholar]
- Liu Y., Song X., Gorovsky M.A., Karrer K.M.. 2005. Elimination of foreign DNA during somatic differentiation in Tetrahymena thermophila shows position effect and is dosage dependent. Eukaryot. Cell. 4:421–431. 10.1128/EC.4.2.421-431.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Kintner C.. 2008. Cilia orientation and the fluid mechanics of development. Curr. Opin. Cell Biol. 20:48–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall W.F., Nonaka S.. 2006. Cilia: tuning in to the cell's antenna. Curr. Biol. 16:R604–R614. 10.1016/j.cub.2006.07.012 [DOI] [PubMed] [Google Scholar]
- Martinez-Campos M., Basto R., Baker J., Kernan M., Raff J.W.. 2004. The Drosophila pericentrin-like protein is essential for cilia/flagella function, but appears to be dispensable for mitosis. J. Cell Biol. 165:673–683. 10.1083/jcb.200402130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura K., Lefebvre P.A., Kamiya R., Hirono M.. 2004. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J. Cell Biol. 165:663–671. 10.1083/jcb.200402022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Million K., Larcher J., Laoukili J., Bourguignon D., Marano F., Tournier F.. 1999. Polyglutamylation and polyglycylation of alpha- and beta-tubulins during in vitro ciliated cell differentiation of human respiratory epithelial cells. J. Cell Sci. 112:4357–4366. [DOI] [PubMed] [Google Scholar]
- Mottier-Pavie V., Megraw T.L.. 2009. Drosophila bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol. Biol. Cell. 20:2605–2614. 10.1091/mbc.E08-11-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y., Hiraki M., Kamiya R., Hirono M.. 2007. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 17:2169–2174. 10.1016/j.cub.2007.11.046 [DOI] [PubMed] [Google Scholar]
- Nanney D.L. 1975. Patterns of basal body addition in ciliary rows in Tetrahymena. J. Cell Biol. 65:503–512. 10.1083/jcb.65.3.503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhauss S.C., Solnica-Krezel L., Schier A.F., Zwartkruis F., Stemple D.L., Malicki J., Abdelilah S., Stainier D.Y., Driever W.. 1996. Mutations affecting craniofacial development in zebrafish. Development. 123:357–367. [DOI] [PubMed] [Google Scholar]
- Orias J.D., Hamilton E.P., Orias E.. 1983. A microtubule meshwork associated with gametic pronucleus transfer across a cell-cell junction. Science. 222:181–184. 10.1126/science.6623070 [DOI] [PubMed] [Google Scholar]
- Paoletti A., Moudjou M., Paintrand M., Salisbury J.L., Bornens M.. 1996. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J. Cell Sci. 109:3089–3102. [DOI] [PubMed] [Google Scholar]
- Pearson C.G., Winey M.. 2009. Basal body assembly in ciliates: the power of numbers. Traffic. 10:461–471. 10.1111/j.1600-0854.2009.00885.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Giddings T.H., Jr., Winey M.. 2009. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol. Biol. Cell. 20:904–914. 10.1091/mbc.E08-08-0835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier L., O'Toole E., Schwager A., Hyman A.A., Müller-Reichert T.. 2006. Centriole assembly in Caenorhabditis elegans. Nature. 444:619–623. 10.1038/nature05318 [DOI] [PubMed] [Google Scholar]
- Peterkin T., Gibson A., Patient R.. 2007. Redundancy and evolution of GATA factor requirements in development of the myocardium. Dev. Biol. 311:623–635. 10.1016/j.ydbio.2007.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piperno G., Fuller M.T.. 1985. Monoclonal antibodies specific for an acetylated form of alpha-tubulin recognize the antigen in cilia and flagella from a variety of organisms. J. Cell Biol. 101:2085–2094. 10.1083/jcb.101.6.2085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues-Martins A., Bettencourt-Dias M., Riparbelli M., Ferreira C., Ferreira I., Callaini G., Glover D.M.. 2007. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 17:1465–1472. 10.1016/j.cub.2007.07.034 [DOI] [PubMed] [Google Scholar]
- Salisbury J.L., Baron A.T., Sanders M.A.. 1988. The centrin-based cytoskeleton of Chlamydomonas reinhardtii: distribution in interphase and mitotic cells. J. Cell Biol. 107:635–641. 10.1083/jcb.107.2.635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Li B., Gorovsky M.A.. 2002. Tetrahymena thermophila contains a conventional γ-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J. Cell Biol. 158:1195–1206. 10.1083/jcb.200205101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y., Tsao C.C., Gorovsky M.A.. 2005. Mutational analyses reveal a novel function of the nucleotide-binding domain of γ-tubulin in the regulation of basal body biogenesis. J. Cell Biol. 171:1035–1044. 10.1083/jcb.200508184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf A.J., Morgan G., Giddings T.H., Jr., White E.A., Marchione R., McDonald H.B., Winey M.. 2005. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol. Biol. Cell. 16:3606–3619. 10.1091/mbc.E04-10-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P., Leidel S., Vinogradova T., Euteneuer U., Khodjakov A., Gönczy P.. 2007. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 13:203–213. 10.1016/j.devcel.2007.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin J.L., Beales P.L.. 2008. Restoration of renal function in zebrafish models of ciliopathies. Pediatr. Nephrol. 23:2095–2099. 10.1007/s00467-008-0898-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobin J.L., Di Franco M., Eichers E., May-Simera H., Garcia M., Yan J., Quinlan R., Justice M.J., Hennekam R.C., Briscoe J., et al. 2008. Inhibition of neural crest migration underlies craniofacial dysmorphology and Hirschsprung's disease in Bardet-Biedl syndrome. Proc. Natl. Acad. Sci. USA. 105:6714–6719. 10.1073/pnas.0707057105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vladar E.K., Stearns T.. 2007. Molecular characterization of centriole assembly in ciliated epithelial cells. J. Cell Biol. 178:31–42. 10.1083/jcb.200703064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. 1995. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio). Vol. 385. University of Oregon Press, Eugene, OR. 350 pp. [Google Scholar]
- Wilkinson C.J., Carl M., Harris W.A.. 2009. Cep70 and Cep131 contribute to ciliogenesis in zebrafish embryos. BMC Cell Biol. 10:17. 10.1186/1471-2121-10-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams N.E., Honts J.E., Kaczanowska J.. 1990. The formation of basal body domains in the membrane skeleton of Tetrahymena. Development. 109:935–942. [DOI] [PubMed] [Google Scholar]
- Wloga D., Rogowski K., Sharma N., Van Dijk J., Janke C., Eddé B., Bré M.H., Levilliers N., Redeker V., Duan J., et al. 2008. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot. Cell. 7:1362–1372. 10.1128/EC.00084-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland H.R., Fry A.M.. 2008. Pix proteins and the evolution of centrioles. PLoS One. 3:e3778. 10.1371/journal.pone.0003778 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.