Abstract
Background
Twelve populations of E. coli were serially propagated for 20,000 generations in a glucose-supplemented minimal medium in order to study the dynamics of evolution. We sought to find and characterize one of the beneficial mutations responsible for the adaptation and other phenotypic changes, including increased cell size, in one of these populations.
Results
We used transposon-tagging followed by P1-transduction into the ancestor, screening for increased cell size and fitness, co-transduction analysis, and DNA sequencing. We identified a 1-bp insertion in the BoxG1 region located upstream of glmUS, an operon involved in cell-wall biosynthesis. When transduced into the ancestor, this mutation increased competitive fitness by about 5%. This mutation spread through its population of origin between 500 and 1500 generations. Mutations in this region were not found in the other 11 evolving populations, even after 20,000 generations.
Conclusion
The 1-bp insertion in the BoxG1 region near glmUS was demonstrably beneficial in the environment in which it arose. The absence of similar mutations in the other evolved populations suggests that they substituted other mutations that rendered this particular mutation unimportant. These results show the unpredictability of adaptive evolution, whereas parallel substitutions at other loci in these same populations reveal the predictability.
Background
Evolutionary change involves a fundamental tension between chance and necessity [1]. On the one hand, mutations that produce heritable variation occur at random, although rates may be modulated by genetic and environmental factors [2]. On the other hand, natural selection tends systematically to increase the frequency of those mutants with phenotypic properties that are useful in a particular environment. How these forces play out in time has been the subject of important speculation [3,4] but is difficult to address empirically.
One approach that can rigorously address the interplay between chance and necessity is experimental evolution, especially using microorganisms. In particular, one can examine the evolution of populations founded from the same ancestral genotype and propagated under identical environmental conditions. Unlike experiments with plants and animals, an evolving population can be founded from a single haploid individual, such that there is no shared genetic variation (identical by descent) that would tend to exaggerate parallel changes. Several recent studies with viruses and bacteria have demonstrated striking examples of parallel genetic changes, affecting the same loci and sometimes even the same base-pair, that evidently confer fitness benefits [5-15]. Some of these studies have also found mutations that are present in only one of the replicate populations, but without construction and analysis of isogenic clones it is unclear whether these unique mutations are beneficial or, alternatively, are neutral or even deleterious mutations that hitchhiked with beneficial mutations.
In the longest running evolution experiment with microorganisms, 12 populations of Escherichia coli B have been serially propagated in a glucose-supplemented minimal medium for tens of thousands of generations [16-18]. During this time, the average competitive fitness increased by ~75% relative to the ancestor [18], the average cell volume substantially increased [17-21], and many other phenotypic changes occurred [11,12,18,22,23]. A number of mutations have been found in these populations by RFLP screening using IS elements as probes [24,25] as well as by random, targeted, and whole-genome sequencing [11,12,21-23,26-28]. Some of these mutations have been demonstrated to be beneficial in the experimental environment by constructing and competing clones that differ only by a single mutation [reviewed in 28]. In most cases, those genes bearing mutations that were demonstrated to be beneficial also harboured mutations in multiple replicate populations. These parallel mutations affected the same genes, but they were not usually identical at the sequence level.
The primary goals of the present paper are to identify a beneficial mutation based on its advantageous effect; and then to determine whether the same or similar mutations were substituted in the replicate evolved populations. Our strategy was as follows. Using an evolved clone, we made a pool of clones each carrying a randomly inserted marker. We then used this entire pool as donors, and transduced the marker and linked genes into the ancestor. Next, the resulting transductants were tested for increased cell size and competitive fitness. A clone with both properties served as a donor for a second round of transduction, in order to estimate the physical distance between its marker and the beneficial mutation of interest. We then sequenced the relevant regions to find any mutations. We found two mutations, one of which was previously unknown. We also sequenced the same region in clones from samples that were stored at various times to characterize the dynamics of this mutation in the focal population. Finally, we sequenced this region in clones from the replicate populations to determine whether the same or similar mutations had been substituted in them.
Results
Finding a transduced clone that carries a beneficial mutation
REL4548 is a clone that was isolated after evolving for 10,000 generations in a constant environment. From REL4548, we produced a mix of 1296 insertion mutants, each carrying a Tn10 mini-transposon that confers resistance to tetracycline. A P1 lysate was then made from that mixture of insertion mutants, and the lysate was used to transduce the ancestral strain, REL606 (see Methods for details). The total pool of transductants was propagated for 7 days (~47 generations) in the same environment in which the long-term evolution experiment was conducted. This procedure should increase the frequency of beneficial transductants that were initially present in the pool, but the period is too short to allow significant de novo evolution. In the long-term experiment itself, the first increases in fitness were discerned around generation 200, and their magnitudes were consistent with mutations having about a 10% fitness advantage [16,17]. The 7-day enrichment of a mutation that confers a 10% advantage should increase its relative frequency by about 25-fold, and smaller effect mutations would be enriched to a lesser extent. Therefore, this approach is more likely to find beneficial mutations with large effects than those with small effects. To identify beneficial transductants we first screened for larger cell size, which has been shown to be strongly correlated with higher fitness in that environment [17-19]. Measurements of cell size are easier than measurements of relative fitness, which means that more transductants can be screened for the former than for the latter. Those transductants with increased average cell size were then subsequently tested for improved fitness.
The average cell volume of the ancestral clone at stationary phase was measured as between 0.37 and 0.39 fl. Among the enriched transductant pool, there was a second mode in the cell-volume distribution centred around 0.43 fl. We initially chose ten clones that had average cell volumes of ≥0.44 fl, and we measured the competitive fitness of each relative to the ancestor. Of these, three clones were then chosen for further study because each one exhibited a statistically significant increase in fitness. Southern hybridization analyses further showed that these three candidates had the same transposon insertion and hence were progeny of the same transduced clone. One of them, designated REL10247, was chosen for further characterization.
Co-transduction analysis of the transposon and the beneficial mutation
Our next challenge was to locate the beneficial mutation linked to the mini-transposon insertion. P1-transduction can move as much as ~2% of the bacterial chromosome; thus, the beneficial mutation could lie as far away as 100,000 bp from the inserted transposon. To get some idea of the distance between the transposon-encoded Tetr marker and the beneficial mutation, we examined the co-transduction of the resistance marker and increased cell size, using REL10247 as the donor and the ancestor as the recipient.
The average cell volumes were measured for over 300 Tetr transductants, and 91.5% produced large cell sizes consistent with co-transduction. Based on the relationship between co-transduction frequency and physical distance [29], using 9.66 × 104 bp as the length of the transducing DNA [29] and 4.6 × 106 bp for the genome size [30], this frequency predicts a distance of about 2800 bp between the beneficial mutation and the inserted transposon.
Identification and fitness effect of the beneficial mutation
The second piece of information necessary to locate the beneficial mutation was to map the inserted mini-transposon. Genomic DNA from the REL10247 clone was digested with AvaI and shotgun-cloned into pUC19. A Tetr colony was chosen, the plasmid isolated, and the inserted fragment was sequenced with standard M13/pUC forward and reverse sequencing primers. The sequence corresponded to the 3' end of the glmUS operon. Another primer was then used to sequence the junction between the mini-transposon and the E. coli genome. These data showed that the transposon had inserted just beyond the glmUS operon, 20 bp downstream from the final amino acid of the GlmS protein.
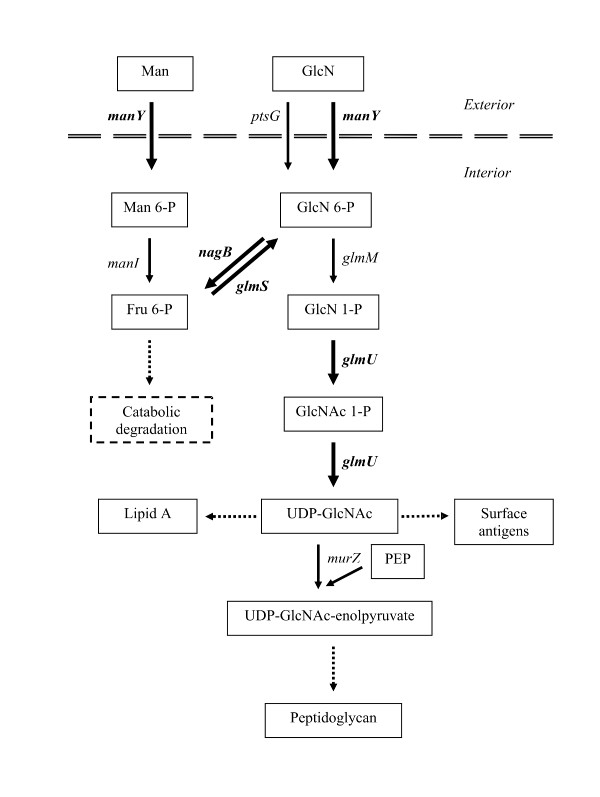
Four sets of partially overlapping primer pairs were designed to amplify ~3000 bp of DNA each. Using these primers, DNA was sequenced from both the ancestor and the 10,000-generation clone from which REL10247 was derived, and new primers were synthesized using the 3' end of the sequence to walk along each fragment. Using this approach, we sequenced ~12,000 bp total, about half on each side of the transposon insertion. Only one difference between the ancestor and evolved clone was found in this entire region, a 1-bp adenine insertion in a homopolymeric tract of seven existing adenines. This tract is in a NagC protein-binding site, called BoxG1, near the glmUS P1 promoter [31] (Figure 1). The glmUS operon encodes two proteins: GlmS converts fructose-6-phosphate (Fru6P) to glucosamine-6-phosphate (GlcN6P); GlmM (not part of the operon) converts GlcN6P to glucosamine-1-phosphate (GlcN1P); and GlmU, a bi-functional enzyme, converts GlcN1P via two reactions to uracil-diphosphate-N-acetyl-glucosamine (UDP-GlcNAc), a precursor in the syntheses of both peptidoglycan and lipopolysaccharides [32,33] (Figure 2). The 1-bp insertion mutation is 3443 bp from the inserted mini-transposon, which agrees well with the 2.8 Kb predicted by the co-transduction analysis. We refer henceforth to this mutation as BoxG18A in recognition of the insertion of an eighth adenine in the tract.
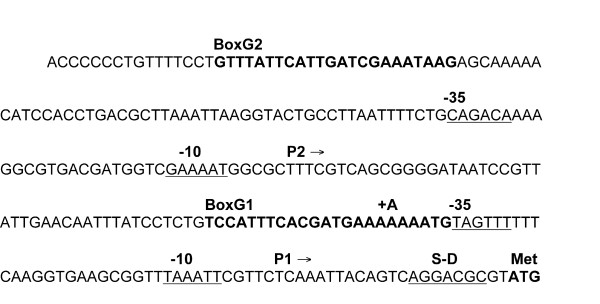
Figure 1.
Region upstream of glmUS, including site of beneficial mutation. NagC binds to two 23-bp BoxG elements shown in bold. The beneficial adenine insertion was in a tract of seven existing adenines in BoxG1; it is shown above the sequence as +A. The Shine-Dalgarno sequence (S-D) and the -10 and -35 regions of two promoters, P1 and P2, are underlined. Figure modified from Plumbridge [31].
Figure 2.
Biochemical pathways involving GlmS and GlmU. Biochemical pathways involving GlmS and GlmU, along with certain transport and biosynthetic reactions. Not all participating molecules are shown for every reaction. Genes encoding relevant enzymes are indicated next to the reactions. Genes printed in bold, and next to reactions shown as heavier arrows, are regulated by NagC. The dashed horizontal line represents the cell envelope. Dashed arrows indicate multiple steps. This figure is a composite of information from several sources [31-33,41,59].
To demonstrate conclusively that the BoxG18A mutation in glmUS was responsible for the fitness increase of the REL10247 strain, we moved that mutation into the ancestor using the gene gorging method [34] to make strain TC640. This strain had a fitness of 1.05 ± 0.01 (95% confidence interval) relative to the ancestor. This advantage was somewhat less than measured for strain REL10247 itself, suggesting that REL10247 contains one or more additional beneficial mutations. By screening REL10247 for known beneficial mutations in the vicinity of glmUS, we found that it also has a deletion of the majority of the rbs operon, which was previously shown to confer a fitness benefit of ~1.5% [11].
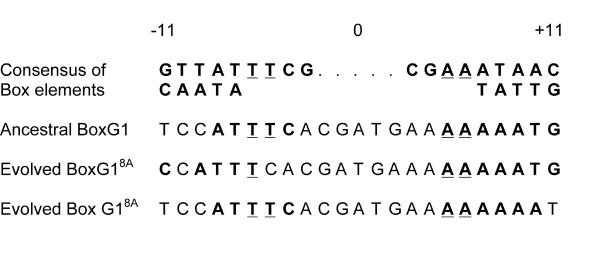
The beneficial BoxG18A mutation presumably alters binding of the transcription factor NagC to the BoxG1 region. The NagC protein acts as a transcriptional repressor for the nagE-BACD and manXYZ operons, but as a transcriptional activator for the glmUS genes [31,35,36]. NagC binds to elements associated with two promoters of the glmUS operon: BoxG1 located next to the proximal promoter P1, and BoxG2 located upstream of the distal promoter P2 [31]. P1 activity is stimulated by growth on glucose, whereas P2 activity is fairly constant across a range of conditions. NagC binding is required for activation of the P1 promoter [31]. NagC binds to a 23-bp consensus sequence [31,35,37-39] (Figure 3). Mutagenesis studies have shown that the most important bases for NagC binding to Box elements are the G or C bases at the -11 and +11 locations, and two conserved T bases at positions -5 and -6 [40]. Positions +4 thru +10 are AT rich, and the adenine insertion occurs in this region. If this insertion is viewed as shifting the BoxG1 sequence to the left, then the T at position -5 is changed to a C (Figure 3). Alternatively, if the extra adenine is viewed as shifting the sequence to the right, then the near-consensus G at position +11 is replaced by a T. In either case, it seems likely that the insertion reduces binding of the NagC activator to BoxG1, thereby reducing glmUS expression from the P1 promoter during growth on glucose.
Figure 3.
Effect of BoxG18A mutation on NagC binding sequence. The consensus sequence, including alternative bases at some sites, for a 23-bp NagC-binding Box element is shown at the top [31]. The four underlined bases are the only ones that are invariant across all such elements [31], and mutagenesis has shown that these four are also the most important for binding NagC [38]. The next row shows the ancestral BoxG1 sequence. The bottom two rows show the effect of inserting an extra adenine, as in the evolved BoxG18A allele, when viewed as shifting the sequence either to the left or to the right. The former eliminates the invariant T at position -5. The latter removes agreement with the consensus at position +11.
To evaluate how this mutation affected glmUS expression, we examined data from a previous experiment [12] that measured, with four-fold replication, genomic expression profiles for the ancestor and for evolved clones from generations 2000, 10,000 and 20,000 in the same population that we used to isolate the BoxG18A mutation. As shown below, the BoxG18A mutation arose prior to generation 2000. The expression profiles were obtained for cells that were growing exponentially in the same glucose medium and other conditions of the long-term experiment. In all 24 comparisons between ancestral and evolved clones (4 replicates × 3 generations × 2 genes), the standardized expression (adjusted for total mRNA) was lower in the evolved clone, a pattern that is extremely unlikely by chance alone. The magnitude of the reduction was ~9% for both glmU and glmS, and at all three time points. These data therefore support the hypothesis that the BoxG18A mutation caused a subtle, but significant, reduction in the expression of glmUS during growth on glucose. Such a physiological change is also consistent with the reduced demand for peptidoglycan and lipopolysaccharide, relative to other cell constituents, in larger cells based on the geometry of surface area to cell volume.
Fitness effects of the BoxG18A mutation in different media
The BoxG18A mutation is clearly beneficial in glucose, where the mutation evidently reduces binding of NagC to the BoxG1 site and thereby reduces expression of the glmUS operon. To examine further this allele's physiological effects, we performed competitions in the same minimal medium but replacing glucose with other substrates chosen for their relationships to glmUS and NagC.
First, the BoxG18A mutation has no significant effect during competition for UDP-GlcNAc, which is the end product of GlmS and GlmU activities (Figure 2). The fitness of TC640, which contains only the BoxG18A mutation on the ancestral background, relative to the ancestor in medium containing UDP-GlcNAc is 0.99 ± 0.03 (95% confidence interval). The absence of any effect is not surprising because UDP-GlcNAc is biochemically downstream of the glmUS operon.
Second, the BoxG18A allele is significantly advantageous in GlcN6P, which is a product of GlmS and is converted to GlcN1P before being a substrate for GlmU (Figure 2). The fitness of TC640 relative to the ancestor in medium containing GlcN6P is 1.15 ± 0.09 (95% confidence interval). At first glance, this advantage might suggest that expression of glmUS is increased in this case. Alternatively, GlcN6P can be converted by the nagB-encoded deaminase to F6P [41], which can be catabolised through the glycolytic pathway. GlmS acts in the opposing direction to covert Fru6P to GlcN6P for the production of cell-wall constituents. Hence, a reduction in glmUS expression caused by the BoxG18A allele may provide an advantage in GlcN6P by better balancing metabolic flux between catabolic and anabolic processes. We note, however, that this hypothesis is further complicated by the observation that GlcN6P interacts directly with NagC to reduce its affinity for at least some binding sites [42]. Third, and consistent with this second hypothesis for the advantage of BoxG18A in GlcN6P, the BoxG18A allele confers a significant benefit in competition for fructose, which is also catabolized to Fru6P. The fitness of TC640 relative to the ancestor in medium containing fructose as the sole carbon source is 1.06 ± 0.03 (95% confidence interval).
Fourth, the BoxG18A mutation confers a small but significant disadvantage during competition for glucosamine (GlcN). The fitness of TC640 relative to the ancestor in medium with GlcN as the sole source of carbon is 0.95 ± 0.02 (95% confidence interval). GlcN enters the cell via the PTS, which converts it to GlcN-6-phosphate [41]. By contrast, GlcN6P enters the cell via a permease that functions for many phosphorylated sugars [41]. The finding that the BoxG18A allele is detrimental in GlcN while beneficial in GlcN6P suggests some effect on transport, presumably mediated by NagC and its interaction with the BoxG1 site. GlcN enters the cell via two different PTS enzymes: IIGlc encoded by ptsG, and IIMan encoded by manY (ptsM) [41]. The latter route is the main one [36]. NagC is a repressor of the manXYZ operon as well as of the nagE-BACD operon [36]. Thus, NagC represses its own synthesis, although it also has constitutive promoters that stabilize its expression [36]. Given these interactions, it is unclear exactly how the feedback would impose a disadvantage to the BoxG18A allele in medium containing GlcN, but clearly the potential exists for complex regulatory effects. One possibility is that the reduced binding of NagC to the mutant BoxG1 site leaves more NagC to bind to the manXYZ operator, thereby slightly repressing it and impeding GlcN transport and catabolism. The growth rate of E. coli in GlcN has been shown to be limited by transport, and so any decrease in manY expression might reduce fitness [43]. Fifth, and consistent with this last hypothesis, the BoxG18A allele is slightly disadvantageous in competition for mannose, which is also taken up through the IIMan PTS system. The fitness of TC640 relative to the ancestor in mannose medium is 0.97 ± 0.02 (95% confidence interval).
We must emphasize that the possible explanations suggested above for the fitness effects of the BoxG18A allele in various media are hypotheses, not conclusions. We would welcome further biochemical and molecular studies of this mutation, and others like it, that might confirm or refute these hypotheses.
Substitution dynamics of the beneficial mutation
To better understand the evolutionary dynamics of the BoxG18A mutation, we sought to determine when it appeared in the evolving population and the time course of its substitution. The clone from which REL10247 was derived was isolated at generation 10,000. Preliminary sequencing of the BoxG1 region from samples taken at earlier times indicated that the mutation was spreading between 500 and 1500 generations. We therefore chose many clones at random from the population samples that had been stored at generations 500, 1000, and 1500 generations, and we sequenced the BoxG1 region in each. None of 51 clones from generation 500 had the BoxG18A mutation, whereas 14/31 (45%) clones carried it at generation 1000, and 29/30 (97%) clones had this mutation at generation 1500.
In an evolving asexual population, the actual rate of spread of a beneficial mutation is often slower than expected from its selective advantage. This discrepancy arises from a phenomenon called clonal interference, whereby a clone with a particular beneficial mutation must compete with clones bearing other beneficial mutations [27,44-46]. The decelerating effect of this interference on the mutation that ultimately prevails can be quite pronounced, because those other clones that reach high frequency tend to be strong competitors. To examine the possible effects of clonal interference on the substitution dynamics of the BoxG18A mutation, we performed pairwise competitions between ten clones that were isolated at generation 1000 from this population, five carrying the beneficial BoxG18A allele and five with the ancestral BoxG17A allele. From each clone, we then isolated a spontaneous Ara+ mutant; such mutations have been shown to be effectively neutral in the glucose-limited environment used in the long-term evolution experiment, but they allowed us to distinguish the competitors by plating on TA indicator plates. Each BoxG18A clone was competed with each BoxG17A clone, with 2-fold replication corresponding to the reciprocal Ara marker states, for a total of 50 competitions. The grand mean fitness of BoxG18A clones relative to BoxG17A clones was only 1.0075, which indicates an advantage of less than 1%. Although the BoxG18A clones were slightly more fit, this advantage is far smaller than the ~5% advantage measured relative to the ancestor. These data imply strong interference from competing clones that carry other beneficial mutations with the spread of the BoxG18A mutation.
Sequencing glmUS promoter region in other evolved lines
Having discovered the BoxG18A mutation in our focal population and shown that it was beneficial, this element and the entire glmUS promoter region became a logical candidate region for sequencing in the other 11 populations founded with the same ancestral strain and propagated in the same environment. We sequenced the glmUS promoter region in clones sampled at 10,000 and 20,000 generations from all of the populations. The focal population retained the BoxG18A allele at generation 20,000. However, all 11 other populations still had the ancestral sequence at both time points. Evidently, only one population substituted this particular mutation, although all 12 populations achieved substantial fitness gains during the experiment [17,18].
Discussion
Chance and necessity
We began this paper by noting the tension between the roles of chance and necessity in evolution. We sought to address this issue by finding a beneficial mutation in one long-term experimental population of E. coli, without relying on any candidate loci, and then sequencing the region in other replicate populations to determine whether they had substituted the same or similar mutations. We found a beneficial mutation in the BoxG1 protein-binding site located near the P1 promoter of the glmUS operon. The mutation was a 1-bp insertion of an adenine in a tract of seven existing adenines. The mutation spread in its source population between generation 500, when it was rare or absent, and generation 1500, when it was present in almost every cell. The mutation persisted through 20,000 generations. It conferred a substantial fitness advantage, on the order of 5%, when transduced into the ancestor.
We also sequenced the region upstream of glmUS in 11 replicate populations. These other populations were founded by the same ancestor, and were propagated for 20,000 generations in the same environment, as the population that had evolved the BoxG18A beneficial mutation. Moreover, these replicate populations all had substantial fitness gains of similar magnitude to the gain measured in the population that substituted the BoxG18A allele [17,18]. However, none of 11 other populations evolved a change in BoxG1 nor anywhere else in the region upstream of the glmUS operon. Several other adaptive changes have occurred in only one of the replicate populations [47,48]. These findings illustrate the unpredictability of evolution, not merely at the level of random drift of neutral mutations, but instead with respect to adaptation. In striking contrast to these unique events, however, many changes in the evolving populations involve repeated beneficial mutations in the same genes [11-13,21-23,28]. Taken together, these cases nicely demonstrate that adaptation by natural selection can be both predictable and unpredictable in the same experiment.
It is interesting to consider two alternative hypotheses for why a BoxG1 mutation was substituted in only one of 12 replicate populations, despite its substantial advantage in the ancestral genetic background. According to one hypothesis, there have not been enough cell generations in each population for the same mutation to have occurred in the other populations. According to the other hypothesis, different mutations were substituted in the replicate populations that rendered this particular mutation no longer advantageous. The former hypothesis captures the idea of chance in its simplest sense of individual stochastic events. The latter explanation adds historical contingency, such that the likelihood of a particular outcome is conditional on whether some other event has already occurred [47].
The following quantitative considerations argue against the adequacy of the first and simpler hypothesis. The size of each population fluctuated between about 5 × 106 and 5 × 108 cells, owing to the daily dilution into fresh medium and re-growth. The effective population size with respect to new beneficial mutations escaping extinction by random drift is approximately equal to the minimum size multiplied by the number of generations between minimum and maximum sizes [16]: 5 × 106 × log2 100 ≈ 3 × 107 cells. The mutation rate in E. coli has been estimated to be 5 × 10-10 per bp by Drake [49], although the substitution rate of synonymous mutations in the long-term experiment yields a somewhat lower estimate of 1.5 × 10-10 [26,28]. Even using this lower rate estimate and the effective population size, and with three alternative base-pairings at each site, one expects that during 20,000 generations the average mutation has occurred (3 × 107) × (1.5 × 10-10/3) × (2 × 104) = 30 times. The 1-bp insertion of an adenine into an existing tract of seven adenines in the BoxG1 element is likely to have occurred more often, because homopolymeric tracts are very prone to insertions and deletions resulting from strand slippage during replication [50,51]. Moreover, several of the long-term populations evolved defects in their DNA repair, causing much higher mutation rates [18,26,52]. Of course, many beneficial mutations are lost by random drift while they are rare. The probability of a beneficial mutation being lost by drift is approximately 1 - 2s, where s is its selective advantage [44,53]. Thus, a mutation that confers a 5% advantage, such as the one in BoxG1, is expected to be lost by drift ~90% of the time. Even so, one would expect such a mutation to have appeared and survived drift multiple times in each population. Yet, the BoxG18A mutation was substituted in only 1 of the 12 evolving populations.
How can we explain its absence from the other populations? The best explanation, in our view, relies on the effects of clonal interference and epistasis, which together give rise to the historically contingent form of chance. In an asexual population, such as in the long-term experiment, two clones that acquire different beneficial mutations will compete with one another, and only one can ultimately prevail. Owing to this clonal interference, many beneficial mutations that escape drift nonetheless will be lost in competition with superior mutations [44,45]. We showed that clonal interference impeded the rate of spread of the BoxG18A mutation even in the population in which it was substituted. Although clonal interference might explain the failure to observe this mutation in the other populations, we doubt that it alone suffices. The likelihood that a given beneficial mutation will be eliminated by clonal interference depends on its advantage relative to other beneficial mutations still being generated in the evolving population [44]. Early in the long-term evolution experiment, a number of mutations with benefits on the order of 10% spread through all of the populations, and these could have interfered with the survival of the BoxG18A mutation [16,17]. However, the rate of fitness improvement declined sharply after the first 2000 generations [17,18,54]. Therefore, later appearances of the BoxG18A mutation would not have encountered many superior beneficial mutations, assuming that the BoxG18A mutation itself would still have conferred a ~5% advantage in these evolved genetic backgrounds. Yet, these BoxG18A mutations were not seen, even much later, in the other 11 populations. This absence suggests, therefore, that mutations in other genes were substituted that rendered the BoxG18A mutation less advantageous, neutral, or even disadvantageous. That is, some other mutations may interact epistatically with the BoxG18A mutation, such that their combined benefits are less than expected from their individual effects. After one of these other mutations had been substituted in a population, the likelihood of a BoxG18A mutation being substituted would therefore be reduced or eliminated.
Mode of action of the BoxG1 beneficial mutation
Our experiments demonstrate that the BoxG18A allele is beneficial in the ancestral background in the glucose-limited medium. The mutation is located in the upstream regulatory region of the glmUS operon, which encodes two proteins involved in the synthesis of peptidoglycan and other cell-wall components. The BoxG1 motif has been shown previously to be a NagC protein-binding site that affects the transcription of nearby genes [31].
Individual E. coli cells become larger as they grow faster [55-57], and the glucose concentration used in the long-term experiment permitted growth at a rate that was extravagant compared with the natural environments in which this species evolved for millions of years prior to our evolution experiment [58]. Given the geometric fact that the ratio of surface area to volume is reduced in larger cells, it may have been beneficial for cells to produce reduced amounts of cell-wall constituents in this experiment. And given the role of GlmS and GlmU in producing these components, one might expect the BoxG18A allele to be associated with reduced expression of this operon. Indeed, a comparison of the BoxG18A sequence with the consensus Box sequence suggests reduced binding by the NagC activator of glmUS expression. Moreover, whole-genome expression profiles showed that evolved clones from this population, when growing exponentially in the same conditions of the long-term experiment, had consistently reduced mRNA levels for both glmU and glmS relative to the ancestor. Thus, the BoxG18A allele in the long-term evolution experiment evidently leads to reduced expression of the glmUS operon, which might provide a better balance between the synthesis of cell-wall and other cellular components.
As an added benefit, the end product of GlmS and GlmU catalysis is UDP-GlcNAc, a precursor to peptidoglycan (as well as other cell-wall components). The next step in peptidoglycan production is the transfer of enolpyruvate from phosphoenolpyruvate (PEP) to UDP-GlcNAc [32,33]. PEP is important not only as a precursor metabolite but also as the source of the high-energy phosphate that drives the phosphotransferase system (PTS), by which glucose is actively transported into the cell [59]. All else equal, reductions in the expression of glmUS and synthesis of UDP-GlcNAc should leave more PEP available for driving the PTS and thereby acquiring glucose, the sole available source of carbon and energy in the long-term experiment. Consistent with the importance of PEP in this experiment, the population that evolved the BoxG18A allele later also substituted an IS150 insertion into pykF, which encodes an enzyme catalyzing the conversion of PEP to pyruvate [25]. This insertion presumably disrupts PykF function, causing the accumulation of PEP that could be used to drive the PTS-mediated acquisition of glucose. In addition to its advantage in medium containing glucose, the BoxG18A mutation has significant effects on competitiveness in media containing fructose, GlcN6P, GlcN, and mannose, possibly through NagC-mediated effects on sugar transport and catabolism genes.
All 12 populations in the long-term experiment evolved larger cell size, but only the Ara-1 population substituted a mutation anywhere in the upstream regulatory region of the glmUS operon. However, several populations, including Ara-1, substituted mutations either in the pbpA gene or near the promoter of pbpA-rodA, another operon involved in peptidoglycan production and cell-wall elongation [13,19,21]. Moreover, mutations in two additional genes involved in cell-wall synthesis, mrdB and mreB, rose to intermediate frequency in the Ara-1 population in the first 2000 generations, before they were eliminated, presumably by clonal interference [27]. Thus, although the substitution in glmUS is unique to the Ara-1 population, beneficial mutations in other genes encoding related functions arose in many of the replicate populations.
In contrast to our explanation, Graña and Acerenza [20] proposed a model in which the parallel increases in cell size and growth rate of these long-term populations were both consequences of diverting intracellular resources from unnecessary physiological functions to those that enhanced performance in the glucose-only regime. According to the model, beneficial mutations need not have a direct role in cell-wall synthesis in order to impact cell size. However, their model and our explanation are not mutually exclusive. In fact, the evolution of faster growing and larger cells caused by loss or repression of unneeded functions should intensify the selection to reduce the relative production of cell-wall components, owing to the lower surface-to-volume ratio.
Conclusions
In this study, we sought to discover and characterize one of the beneficial mutations responsible for the evolutionary adaptation in one of 12 E. coli populations grown for 20,000 generations in glucose-supplemented minimal medium. We then wanted to see whether the same or a similar mutation had occurred in the other populations, in order to examine the roles of chance and necessity in adaptive evolution. Our strategy employed transposon-tagging an evolved clone's genome followed by transduction into the ancestor, screening for increased cell size and fitness, co-transduction analysis, and sequencing the relevant region in the focal and other populations. Our main conclusions are as follows:
1) We found a 1-bp insertion in the BoxG1 region located in one of two operators of glmUS, an operon involved in synthesizing peptidoglycan and other components of the cell wall. The BoxG1 element has been shown previously to bind NagC, which activates this promoter during growth on glucose.
2) This mutation spread through the focal population between generations 500 and 1500. When transduced into the ancestral background, the BoxG18A allele increased competitive fitness by about 5%.
3) Expression profiles of ancestral and evolved clones from the focal population show a reduction of almost 10% in glmUS expression associated with the substitution of the BoxG18A allele. The insertion of the extra base-pair into the BoxG1 site apparently reduces NagC binding and thereby the activity of the P1 promoter during growth on glucose.
4) Faster growing cells are larger and, consequently, have reduced surface-to-volume ratios; therefore, they have proportionately less need for cell-wall components. The BoxG1 mutation and reduced expression of glmUS may thus confer an advantage by reducing excess synthesis of these components. Also, the end-product of GlmS and GlmU catalysis is UDP-GlcNAc, a precursor to peptidoglycan. The next step in the production of peptidoglycan consumes PEP, which would otherwise be available to drive PTS-mediated glucose transport.
5) Despite the large fitness gain conferred by the BoxG18A allele in the environment in which it arose, no similar mutations were found upstream of the glmUS operon in any of the other 11 replicate populations, even after 20,000 generations. A possible explanation is that those populations substituted mutations at other loci that interact epistatically with the glmUS operon, so that mutations in the BoxG1 region would no longer confer a substantial benefit. In fact, mutations in another operon involved in cell-wall synthesis arose in a number of replicate populations, including the focal population with the glmUS mutation. In any case, the unique substitution at the BoxG1 site illustrates the unpredictability of adaptive evolution, whereas parallel substitutions at other loci in these same populations illustrate its predictability.
Methods
Bacterial strains and culture conditions
The bacteria used in this study are part of a long-term evolution experiment that has been described in detail elsewhere [16-18]. Briefly, 12 populations of Escherichia coli B were started, six from each of two genetically marked variants of the ancestor. The populations were propagated by 1:100 daily serial transfer in Davis minimal medium supplemented with glucose at 25 μg per ml (DM25) at 37°C for 20,000 generations (3000 days). The two ancestral variants, REL606 and REL607, are phenotypically Ara- and Ara+, respectively. These variants are selectively neutral in DM25 medium, but Ara- and Ara+ genotypes can be distinguished by their red and white colonies, respectively, on tetrazolium-arabinose (TA) indicator agar. At 500-generation intervals, samples from each population were stored at -80°C.
The source of the beneficial BoxG18A mutation that is the focus of this paper is a clone, REL4548, that was isolated from population Ara-1 at generation 10,000. This population retained functional DNA repair throughout the first 20,000 generations of the evolution experiment, whereas several other populations became hypermutable as a consequence of mutations that disrupted repair functions [18,52]. The Ara-1 population also subsequently evolved a mutator phenotype, but did so only after the 20,000 generations examined in this study [27,28]. REL10247 is the designation of a P1-transductant clone that was derived using REL4548 as the donor and REL606 as the recipient. The REL10247 genome has a mini-transposon bearing a tetracycline resistance (Tetr) marker inserted 20 bp downstream from the last codon in the glmUS operon; and a 1-bp adenine insertion in a tract of seven pre-existing adenines in the BoxG1 site of the glmUS promoter region. It has no other mutational differences from the ancestor within ~6,000 bp on either side of the mini-transposon. However, it has a deletion of the rbs operon [11] that begins ~18,000 bp from the BoxG1 site (in the direction opposite from the mini-transposon), which was co-transduced from REL4548. We describe later the construction of another strain, TC640, that carries the evolved BoxG18A allele but is otherwise isogenic to the ancestor.
When sequencing BoxG1 and the rest of the region upstream of the glmUS operon, we chose clones at random from the frozen samples of the Ara-1 population taken at various time points, as well as from samples of the other 11 populations at generations 10,000 and 20,000.
Strain JM109 and plasmid pUC19 were obtained from New England Biolabs and used in certain genetic manipulations described below.
Mini-transposon tagging
The protocol that we used to produce a library of clones carrying mini-transposons and associated markers follows the one developed by Kleckner et al. [60]. Phage λNK1323 was used as the delivery vector for inserting mini-Tn10 transposons that confer tetracycline resistance into REL4548. We made 1296 independent insertion mutants. These mutants were used to inoculate individual 2-ml cultures of LBC supplemented with 15 μg/ml Tet, which were grown overnight at 37°C. This number of insertion mutants is such that, in a genome of ~4.6 × 106 bp [30], a marker should occur, on average, every ~3600 bp. The 1296 cultures were then mixed in equal volume to provide a proportional representation of each marked clone. Glycerol was added to 16%, and this mixture, designated MIX-1, was frozen at -80°C for later use in producing the P1-transducing lysate.
P1-mediated transduction and co-transduction
Following a standard protocol [61], phage P1 (ATCC25404-B1) was grown on MIX-1 to produce a lysate stock (LYS-1) that was then used to transduce the ancestral strain REL606. An aliquot of the transduced culture was plated on LB agar containing 15 μg/ml Tet to estimate the total number of Tetr transductants. The remaining culture was added to 200 ml LB supplemented with 15 μg/ml Tet, allowed to grow overnight, then frozen at -80°C in 16% glycerol. This frozen culture, designated MIX-2, thus contained a diverse mix of genotypes. These genotypes consist of randomly-tagged DNA from the evolved clone REL4548 bearing a mini-transposon along with any physically linked DNA sequences, inserted into the ancestral REL606 background. P1 phage transfers only ~2% of the E. coli genome between donor and recipient strains. Another clone sampled from the same population and generation has a total of 28 mutations distributed throughout its sequenced genome [28], and so one would expect that most transductants in the MIX-2 culture carry no more than 1 or 2 mutations.
We also performed a second round of transduction to estimate the physical distance between the beneficial mutation and the mini-transposon marker, based on their co-transduction. A P1-transducing lysate was prepared from REL10247 using the same methods as above. This lysate, designated LYS-2, was used to transduce the ancestral strain REL606. A random sample of the Tetr transductants was screened for cell size, and some transductants were also used in fitness assays.
Enrichment procedure to find beneficial transductant
To facilitate finding a clone that carried a beneficial mutation linked to a transposon-encoded marker, we serially propagated the MIX-2 mixture of transductants in the same DM25 medium and other culture conditions as used in the long-term evolution experiment. This procedure was performed for 7 daily cycles, corresponding to ~47 cell generations. Glycerol was then added to the resulting culture, designated MIX-3, which was stored at -80°C.
Construction of isogenic mutant strain by gene gorging
To determine the phenotypic effects caused specifically by the BoxG18A mutation, we used the gene-gorging method [34] to move this mutation into the ancestral genome, producing a new strain designated TC640. Primers were designed to amplify a 1280-bp product centered on the BoxG18A mutation, one of which also contained the I-SceI homing endonuclease recognition site required for the gene gorging protocol. These primers were used to amplify a product from REL4548, the 10,000-generation clone from which REL10247 was derived. This product was cloned into pCR2.1 to make a gene-gorging mutagenesis plasmid. Subsequent manipulations followed the previous protocol [34], except LB was used instead of rich defined medium. Successful transfer of the BoxG18A mutation was confirmed by direct sequencing. To ensure that no other mutations were inadvertently introduced during construction of TC640, we then also replaced the BoxG18A mutation with the ancestral operator sequence. The fitness of this reverted strain in glucose medium was indistinguishable from the ancestor (data not shown), which strongly implies that no secondary mutations affecting fitness were present in the intermediate strain, TC640.
Screening for cell size
Previous research has shown a strong, positive correlation between cell volume and relative fitness in the evolving population studied here [17,19]. Moreover, cell size can be measured more easily and precisely than relative fitness. Therefore, we chose to focus on clones from MIX-3 that produced cells with significantly larger volumes than those produced by the ancestor REL606. Clones from MIX-3 were isolated on LB plates containing 15 μg/ml Tet, then propagated for two daily transfers in DM25 at 37°C. Average cell volumes were measured using a Coulter particle counter (model ZM and channelyzer model 256). The ancestor produces an average cell size of 0.37-0.39 fl, depending on the experiment, whereas the clones we chose for further study had average cell sizes of at least 0.44 fl. For the co-transduction analysis using REL10247 as donor and the ancestor as recipient, we scored Tetr clones with average cell volumes ≥0.44 fl as having the beneficial allele, and those with averages ≤0.39 fl as having the ancestral allele; intermediate values were ignored when estimating the co-transduction frequency.
Fitness assays
Competition experiments were performed to estimate the relative fitness of various clones. The procedures for the assays have been described in detail elsewhere [16]. In brief, two competing clones, one Ara- and the other Ara+, are separately acclimated to the conditions in which they will compete, and then mixed at a 1:1 volumetric ratio in the competition environment. Samples are plated on TA agar immediately after mixing and again at the end of the competition experiment. The competitions ran for one complete serial-transfer cycle, which encompasses the same lag, growth, and stationary phases that occur in the long-term evolution experiment. Relative fitness is defined simply as the ratio of the net growth rates realized by the two strains during their competition [16].
Unless otherwise indicated, competitions involving particular clones of interest were performed against REL607, the Ara+ ancestral strain. The competitions performed to study clonal interference used pairs of evolved clones, with spontaneous Ara+ mutants having been obtained for each clone (on minimal arabinose agar) and the competition experiments balanced with respect to the Ara marker. Also unless otherwise noted, experiments were performed in DM25 medium, in which glucose at 25 μg/ml is the only usable source of carbon. To investigate the physiological effects of the BoxG18A mutation, additional competitions of TC640 against REL607 were performed in the same minimal medium except replacing the glucose with fructose, GlcN, GlcN6P, mannose, or UDP-GlcNAc. For UDP-GlcNAc only, the amount of resource was reduced to 8.75 μg/ml to equalize the available carbon.
Southern blots
Plasmid pNKtet [60] was digested with BglI and XbaI, and the fragment containing the tetracycline-resistance gene was used as a probe. Genomic DNA of the clone designated REL10247 was isolated from an overnight LB culture using the Qiagen Genomic-tip system. A number of restriction enzymes were then used to digest this genomic DNA, including AvaI, BamHI, PvuI, and PvuII. Five μg of DNA were cut with 40 units of each restriction enzyme. The samples were run on a 0.5% agarose TBE gel, and then blotted onto a positively charged nylon membrane (Boehringer Mannheim) using a vacuum. Southern hybridization was performed using the DIG DNA Labelling and Detection Kit (Boehringer Mannheim) and the manufacturer's recommendations. The lane containing the AvaI-digested genomic DNA produced a band of ~5500 bp containing the entire functional tetracycline-resistance marker [60] as well as flanking DNA. The size of this fragment made it suitable to clone it and then sequence the genomic DNA flanking the mini-transposon.
Mapping and sequencing
To map the chromosomal location of the mini-transposon inserted in the REL10247 genome, the ~5500 bp AvaI fragment was excised, purified, cloned into pUC19, and used to transform JM109. Prior to ligation, the AvaI-digested vector was treated with alkaline phosphatase (Gibco BRL) following the manufacturer's recommendations for 5' overhangs. The ligation was performed at 16°C for 16 h after a 5' treatment at 45°C. Following transformation of JM109, a Tetr clone was isolated on an LB plate containing 15 μg/ml Tet. The DNA insert was sequenced using M13/pUC forward and reverse primers. A BLAST search was used to locate the site of the transposon insertion. The RL100 primer (5'-CGGATCCGATCATATGACAAGATGTGTA-3') was used specifically for the junction between the transposon and the E. coli genome.
Primers and PCR
We sequenced regions around the mini-transposon insertion via chromosome walking, in order to find the beneficial mutation that was co-transduced with the insertion in REL10247. To that end, genomic DNA was isolated from REL606 and REL10247. PCRs were then used to generate four partially overlapping fragments, ranging in size from ~3400 to ~4100 bp, which were purified using an Ultrafree-MC filter (Millipore) or an Elu-Quik DNA Purification Kit (Schleicher & Schuell). The PCR fragments were sequenced on each end, and new sequencing primers were designed at the 3' end of the prior sequencing run. Sequencing continued until both ends overlapped. The glmUS promoter region was sequenced on both strands using primer RL36 (positive strand: 5'-ATTTTCTGCAGACAAAAGGCGTGAC-3') and RL51 (negative strand: 5'-GTTCTCGGTGGTGCGGATAACAAT-3').
Some of our experiments involved sequencing the glmUS promoter region from many clones. These PCRs used primers RL32 (5'-GTTCTGGCCGACACCGCAAT-3') and RL35 (5'-GCAGTTTTTCAGCCTGTTCGGACT-3').
Sequence data were managed using the DNASTAR software (Madison, Wisconsin).
Abbreviations
CI: confidence interval; Fru: fructose; GlcN: glucosamine; IS: insertion sequence; LPS: lipopolysaccharide; PEP: phosphoenolpyruvate; PTS: phosphotransferase system; TetR: tetracycline resistant; UDP-GlcNAc: uracil-diphosphate-N-acetyl-glucosamine.
Authors' contributions
MTS and TC performed the genetic and competition experiments reported here. REL was responsible for the overall direction of these experiments as well as the long-term evolution experiment. All authors contributed substantially to writing this paper, and approved the final manuscript.
Contributor Information
Mark T Stanek, Email: mstanek@ltu.edu.
Tim F Cooper, Email: tfcooper@uh.edu.
Richard E Lenski, Email: lenski@msu.edu.
Acknowledgements
We thank Neerja Hajela for technical assistance, Mike Thomashow for strategic advice, and Dom Schneider and Bob Woods for comments and discussion. This work was supported by grants from the US National Science Foundation to REL and from the DARPA FunBio Program to TFC and REL.
References
- Monod J. Chance and Necessity. New York, Knopf; 1971. [Google Scholar]
- Sniegowski PD, Lenski RE. Mutation and adaptation: the directed mutation controversy in evolutionary perspective. Ann Rev Ecol Syst. 1995;26:553–578. doi: 10.1146/annurev.es.26.110195.003005. [DOI] [Google Scholar]
- Wright S. Evolution in Mendelian populations. Genetics. 1931;16:97–159. doi: 10.1093/genetics/16.2.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. Wonderful Life: The Burgess Shale and the Nature of History. New York, Norton; 1989. [Google Scholar]
- Bull JJ, Badgett MR, Wichman HA, Huelsenbeck JP, Hillis DM, Gulati A, Ho C, Molineux IJ. Exceptional convergent evolution in a virus. Genetics. 1997;147:1497–1507. doi: 10.1093/genetics/147.4.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham CW, Jeng K, Husti J, Badgett M, Molineux IJ, Hillis DM, Bull JJ. Parallel molecular evolution of deletions and nonsense mutations in bacteriophage T7. Mol Biol Evol. 1997;14:113–116. doi: 10.1093/oxfordjournals.molbev.a025697. [DOI] [PubMed] [Google Scholar]
- Treves DS, Manning S, Adams J. Repeated evolution of an acetate cross-feeding polymorphism in long-term populations of Escherichia coli. Mol Biol Evol. 1998;15:789–797. doi: 10.1093/oxfordjournals.molbev.a025984. [DOI] [PubMed] [Google Scholar]
- Wichman HA, Badgett MR, Scott LA, Boulianne CM, Bull JJ. Different trajectories of parallel evolution during viral adaptation. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, Ferenci T. Adaptive mgl regulatory mutations and genetic diversity evolving in glucose-limited Escherichia coli populations. Environ Microbiol. 1999;1:33–43. doi: 10.1046/j.1462-2920.1999.00002.x. [DOI] [PubMed] [Google Scholar]
- Notley-McRobb L, Ferenci T. Experimental analysis of molecular events during mutational periodic selections in bacterial evolution. Genetics. 2000;156:1493–1501. doi: 10.1093/genetics/156.4.1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Schneider D, Blot M, Lenski RE. Mechanisms causing rapid and parallel losses of ribose catabolism in evolving populations of E. coli B. J Bacteriol. 2001;183:2834–2841. doi: 10.1128/JB.183.9.2834-2841.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF, Rozen DE, Lenski RE. Parallel changes in gene expression after 20,000 generations of evolution in E. coli. Proc Natl Acad Sci USA. 2003;100:1072–1077. doi: 10.1073/pnas.0334340100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods R, Schneider D, Winkworth CL, Riley MA, Lenski RE. Tests of parallel molecular evolution in a long-term experiment with Escherichia coli. Proc Natl Acad Sci USA. 2006;103:9107–9112. doi: 10.1073/pnas.0602917103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiers AJ, Kahn SG, Bohannon J, Travisano M, Rainey PB. Adaptive divergence in experimental populations of Pseudomonas fluorescens. I. Genetic and phenotypic bases of wrinkly spreader fitness. Genetics. 2002;161:33–46. doi: 10.1093/genetics/161.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski EA, Woods R, Lenski RE. The genetic basis of parallel and divergent phenotypic responses in evolving populations of Escherichia coli. Proc R Soc Lond B. 2008;275:277–284. doi: 10.1098/rspb.2007.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Rose MR, Simpson SC, Tadler SC. Long-term experimental evolution in Escherichia coli. I. Adaptation and divergence during 2,000 generations. Am Nat. 1991;138:1315–1341. doi: 10.1086/285289. [DOI] [Google Scholar]
- Lenski RE, Travisano M. Dynamics of adaptation and diversification: a 10,000-generation experiment with bacterial populations. Proc Natl Acad Sci USA. 1994;91:6808–6814. doi: 10.1073/pnas.91.15.6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper VS, Lenski RE. The population genetics of ecological specialization in evolving E. coli populations. Nature. 2000;407:736–739. doi: 10.1038/35037572. [DOI] [PubMed] [Google Scholar]
- Lenski RE, Mongold JA. In: Scaling in Biology. Brown JH, West GB, editor. Oxford, Oxford University Press; 2000. Cell size, shape, and fitness in evolving populations of bacteria; pp. 221–235. [Google Scholar]
- Graña M, Acerenza L. A model combining cell physiology and population genetics to explain Escherichia coli experimental evolution. BMC Evol Biol. 2001;1:12. doi: 10.1186/1471-2148-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe N, Pelosi N, Lenski RE, Schneider D. Evolution of penicillin-binding protein 2 concentration and cell shape during a long-term experiment with Escherichia coli. J Bacteriol. 2009;191:909–921. doi: 10.1128/JB.01419-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat E, Philippe N, Lenski RE, Geiselmann J, Schneider D. Long-term experimental evolution in Escherichia coli. XII. DNA topology as a key target of selection. Genetics. 2005;169:523–532. doi: 10.1534/genetics.104.035717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelosi L, Kühn L, Guetta D, Garin J, Geiselmann J, Lenski RE, Schneider D. Parallel changes in global protein profiles during long-term experimental evolution in Escherichia coli. Genetics. 2006;173:1851–1869. doi: 10.1534/genetics.105.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadopoulos D, Schneider D, Meier-Eiss J, Arber W, Lenski RE, Blot M. Genomic evolution during a 10,000-generation experiment with bacteria. Proc Natl Acad Sci USA. 1999;96:3807–3812. doi: 10.1073/pnas.96.7.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider D, Duperchy E, Coursange E, Lenski RE, Blot M. Long-term experimental evolution in Escherichia coli. IX. Characterization of insertion sequence-mediated mutations and rearrangements. Genetics. 2000;156:477–488. doi: 10.1093/genetics/156.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenski RE, Winkworth CL, Riley MA. Rates of DNA sequence evolution in experimental populations of Escherichia coli during 20,000 generations. J Mol Evol. 2003;56:498–508. doi: 10.1007/s00239-002-2423-0. [DOI] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harbor Symposia on Quantitative Biology. 2009. in press . [DOI] [PMC free article] [PubMed]
- Barrick JE, Yu D-S, Yoon SH, Jeong H, Oh KT, Schneider D, Lenski RE, Kim JF. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 2009;461:1243–1247. doi: 10.1038/nature08480. [DOI] [PubMed] [Google Scholar]
- Low KB. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC, ASM Press; 1996. Genetic mapping; pp. 2511–2517. [Google Scholar]
- Jeong H, Barbe V, Lee CH, Vallenet D, Yu DS, Choi S-H, Couloux A, Lee S-W, Yoon SH, Cattolico L, Hur C-G, Park H-S, Ségurens B, Kim SC, Oh TK, Lenski RE, Studier FW, Daegelen P, Kim JF. Genome sequences of Escherichia coli B strains REL606 and BL21(DE3) J Mol Biol. 2009;394:644–652. doi: 10.1016/j.jmb.2009.09.052. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. Co-ordinated regulation of amino sugar biosynthesis and degradation: the NagC repressor acts as both an activator and a repressor for the transcription of the glmUS operon and requires two separated NagC binding sites. EMBO J. 1995;14:3958–3965. doi: 10.1002/j.1460-2075.1995.tb00067.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz CRH. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC, ASM Press; 1996. Bacterial lipopolysaccharides: a remarkable family of bioactive macroamphiphiles; pp. 1035–1063. [Google Scholar]
- van Heijenoort J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC, ASM Press; 1996. Murein synthesis; pp. 1025–1034. [Google Scholar]
- Herring CD, Glasner JD, Blattner FR. Gene replacement without selection: regulated suppression of amber mutations in Escherichia coli. Gene. 2003;331:153–163. doi: 10.1016/S0378-1119(03)00585-7. [DOI] [PubMed] [Google Scholar]
- Plumbridge J, Kolb A. CAP and Nag repressor binding to the regulatory regions of the nagE-B and manX genes of Escherichia coli. J Mol Biol. 1991;217:661–679. doi: 10.1016/0022-2836(91)90524-A. [DOI] [PubMed] [Google Scholar]
- Plumbridge J. How to achieve constitutive expression of a gene within an inducible operon: the example of the nagC gene of Escherichia coli. J Bacteriol. 1996;178:2629–2636. doi: 10.1128/jb.178.9.2629-2636.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumbridge J, Kolb A. DNA loop formation between Nag repressor molecules bound to its two operator sites is necessary for repression of the nag regulon of Escherichia coli in vivo. Mol Microbiol. 1993;10:973–981. doi: 10.1111/j.1365-2958.1993.tb00969.x. [DOI] [PubMed] [Google Scholar]
- Plumbridge J, Kolb A. Nag repressor-operator interactions: protein-DNA contacts cover more than two turns of the DNA helix. J Mol Biol. 1995;249:890–902. doi: 10.1006/jmbi.1995.0346. [DOI] [PubMed] [Google Scholar]
- Plumbridge J, Kolb A. DNA bending and expression of the divergent nagE-B operons. Nucleic Acids Res. 1998;26:1254–1260. doi: 10.1093/nar/26.5.1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Quadi S, Plumbridge J. Switching control of expression of ptsG from the Mlc regulon to the NagC regulon. J Bacteriol. 2008;190:4677–4686. doi: 10.1128/JB.00315-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ECC. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC, ASM Press; 1996. Dissimilatory pathways for sugars, polyols, and carboxylates; pp. 307–342. [Google Scholar]
- Plumbridge J. Repression and induction of the nag regulon of Escherichia coli K12: the roles of nagC and nagA in maintenance of the uninduced state. Mol Microbiol. 1991;5:2053–2062. doi: 10.1111/j.1365-2958.1991.tb00828.x. [DOI] [PubMed] [Google Scholar]
- Alverez-Añorve LI, Calcagno ML, Plumbridge J. Why does Escherichia coli grow more slowly on glucosamine than N-acetylglucosamine? Effects of enzyme levels and allosteric activation of GlcN6P deaminase (NagB) on growth rates. J Bacteriol. 2005;187:2974–2982. doi: 10.1128/JB.187.9.2974-2982.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish PJ, Lenski RE. The fate of competing beneficial mutations in an asexual population. Genetica. 1998;102/103:127–144. doi: 10.1023/A:1017067816551. [DOI] [PubMed] [Google Scholar]
- Shaver AC, Dombrowski PG, Sweeney JY, Treis T, Zappala RM, Sniegowski PD. Fitness evolution and the rise of mutator alleles in experimental Escherichia coli populations. Genetics. 2002;162:557–566. doi: 10.1093/genetics/162.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper TF. Recombination speeds adaptation by reducing competition between beneficial mutations in populations of Escherichia coli. PLoS Biology. 2007;5:e225. doi: 10.1371/journal.pbio.0050225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount ZD, Borland CZ, Lenski RE. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc Natl Acad Sci USA. 2008;105:7899–7906. doi: 10.1073/pnas.0803151105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen DE, Lenski RE. Long-term experimental evolution in Escherichia coli. VIII. Dynamics of a balanced polymorphism. Am Nat. 2000;155:24–35. doi: 10.1086/303299. [DOI] [PubMed] [Google Scholar]
- Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser JN, Love JM, Moxon ER. The molecular mechanism of phase variation of H. influenzae lipopolysaccharide. Cell. 1989;59:657–665. doi: 10.1016/0092-8674(89)90011-1. [DOI] [PubMed] [Google Scholar]
- Moxon RE, Rainey PB, Nowak MA, Lenski RE. Adaptive evolution of highly mutable loci in pathogenic bacteria. Curr Biol. 1994;4:24–33. doi: 10.1016/S0960-9822(00)00005-1. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Haldane JBS. A mathematical theory of natural and artificial selection. V. Selection and mutation. Proc Camb Phil Soc. 1927;23:838–844. doi: 10.1017/S0305004100015644. [DOI] [Google Scholar]
- De Visser JAGM, Lenski RE. Long-term experimental evolution in Escherichia coli. XI. Rejection of non-transitive interactions as cause of declining rate of adaptation. BMC Evo Biol. 2002;2:19. doi: 10.1186/1471-2148-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Åkerlund T, Nordström K, Bernander R. Analysis of cell size and DNA content in exponentially growing and stationary-phase batch cultures of Escherichia coli. J Bacteriol. 1995;177:6791–6797. doi: 10.1128/jb.177.23.6791-6797.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC, ASM Press; 1996. Modulation of chemical composition and other parameters of the cell by growth rate; pp. 1553–1569. [Google Scholar]
- Mongold JA, Lenski RE. Experimental rejection of a nonadaptive explanation for increased cell size in Escherichia coli. J Bacteriol. 1996;178:5333–5334. doi: 10.1128/jb.178.17.5333-5334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savageau MA. Escherichia coli habitats, cell types, and molecular mechanisms of gene control. Am Nat. 1983;122:732–744. doi: 10.1086/284168. [DOI] [Google Scholar]
- Postma PW, Lengeler JW, Jacobson GR. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. Neidhardt FC, editor. Washington, DC, ASM Press; 1996. Phosphoenolpyruvate: carbohydrate phosphotransferase systems; pp. 1149–1174. [Google Scholar]
- Kleckner N, Bender J, Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. full_text. [DOI] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics. Cold Spring Harbor, New York, Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]