Abstract
The mouse γ-aminobutyric acid (GABA) transporter mGAT1 was expressed in neuroblastoma 2a cells. 19 mGAT1 designs incorporating fluorescent proteins were functionally characterized by [3H]GABA uptake in assays that responded to several experimental variables, including the mutations and pharmacological manipulation of the cytoskeleton. Oligomerization and subsequent trafficking of mGAT1 were studied in several subcellular regions of live cells using localized fluorescence, acceptor photobleach Förster resonance energy transfer (FRET), and pixel-by-pixel analysis of normalized FRET (NFRET) images. Nine constructs were functionally indistinguishable from wild-type mGAT1 and provided information about normal mGAT1 assembly and trafficking. The remainder had compromised [3H]GABA uptake due to observable oligomerization and/or trafficking deficits; the data help to determine regions of mGAT1 sequence involved in these processes. Acceptor photobleach FRET detected mGAT1 oligomerization, but richer information was obtained from analyzing the distribution of all-pixel NFRET amplitudes. We also analyzed such distributions restricted to cellular subregions. Distributions were fit to either two or three Gaussian components. Two of the components, present for all mGAT1 constructs that oligomerized, may represent dimers and high-order oligomers (probably tetramers), respectively. Only wild-type functioning constructs displayed three components; the additional component apparently had the highest mean NFRET amplitude. Near the cell periphery, wild-type functioning constructs displayed the highest NFRET. In this subregion, the highest NFRET component represented ∼30% of all pixels, similar to the percentage of mGAT1 from the acutely recycling pool resident in the plasma membrane in the basal state. Blocking the mGAT1 C terminus postsynaptic density 95/discs large/zona occludens 1 (PDZ)-interacting domain abolished the highest amplitude component from the NFRET distributions. Disrupting the actin cytoskeleton in cells expressing wild-type functioning transporters moved the highest amplitude component from the cell periphery to perinuclear regions. Thus, pixel-by-pixel NFRET analysis resolved three distinct forms of GAT1: dimers, high-order oligomers, and transporters associated via PDZ-mediated interactions with the actin cytoskeleton and/or with the exocyst.
INTRODUCTION
The γ-aminobutyric acid (GABA) transporter, GAT1, is widely expressed in the mammalian brain. It was the first identified gene in the solute carrier 6 (SLC6) gene family (Guastella et al., 1990), which also includes transporters for dopamine, serotonin, norepinephrine, and glycine. This family is characterized by an absolute requirement for Na+ cotransport, which provides part of the driving force to accumulate neurotransmitter against an otherwise unfavorable concentration gradient. Transporters in this family also exhibit a variable and incompletely understood necessity for Cl− cotransport in the transport process (Lester et al., 1996; Rudnick, 2002; Kanner, 2006; Reith et al., 2006).
In the healthy brain, density and surface/cytoplasm distribution of GAT1 transporters are tightly regulated. Previously, we determined that presynaptic boutons and axons have GAT1 molecules at a surface density of 800–1,300 µm−2, with 61–63% of these molecules expressed on the plasma membrane (Chiu et al., 2002). This pool of transporters cycles to and from the plasma membrane, a process that plays a critical role in shaping neurotransmission and is both constitutively and functionally regulated (Whitworth and Quick, 2001; Zahniser and Doolen, 2001; Deken et al., 2003; Loder and Melikian, 2003; McHugh et al., 2004; Holton et al., 2005; Wang and Quick, 2005; Quick, 2006; Boudanova et al., 2008).
Although SLC6 transporters do function to some extent when expressed as monomers (Scholze et al., 2002; Soragna et al., 2005), constitutive transporter oligomerization is a major factor in the release of newly synthesized transporter from the ER (Scholze et al., 2002; Sorkina et al., 2003; Torres et al., 2003; Bartholomäus et al., 2008). The crystal structure of the Aquifex aeolicus leucine transporter LeuTAa (a bacterial orthologue of the mammalian SLC6 neurotransmitter transporters) reveals a dimer, with transmembrane domains (TMs) 9 and 12 probably providing the dimer interface (Yamashita et al., 2005). There may also be higher-order oligomers (Kilic and Rudnick, 2000; Hastrup et al., 2003; Just et al., 2004; Bartholomäus et al., 2008).
Sequence elements in the cytoplasmic N and C termini exert additional control over trafficking. They can serve as substrates for cytosolic regulators such as kinases or as motifs for interactions with other proteins, including postsynaptic density 95/discs large/zona occludens 1 (PDZ) domain–containing and soluble N-ethylmaleimide–sensitive fusion receptor proteins, which determine the insertion and withdrawal of transporter complexes from the cell surface (Loder and Melikian, 2003; Torres et al., 2003; Farhan et al., 2004; McHugh et al., 2004; Miranda et al., 2004; Holton et al., 2005; Quick, 2006; Boudanova et al., 2008).
We wish to understand quantitative aspects of GAT1 in synaptic transmission. To facilitate this goal, one must study transporters that are expressed in appropriate cells at normal levels and at normal locations. Most previous studies involve expression in heterologous expression systems in which the transporter is greatly overexpressed and/or has the potential to associate with incorrect partner proteins. Here, we describe the construction of 19 new fluorescent mouse GAT1 (mGAT1) transporters tagged with either cyan or yellow fluorescent proteins (CFP and YFP, respectively; XFP collectively). The majority of the designs are fusions of the XFP onto the mGAT1 C terminus with additional modifications to preserve wild-type trafficking and assembly. We report functional characterization of these constructs with a quantitative [3H]GABA uptake assay in which neither protein expression levels nor substrate transport has reached maximum when the transporter is expressed in mouse neuroblastoma 2a (N2a) cells (a neuron-like cell line); we present this expression system as an optimal one for the characterization of transiently expressed neuronal transporters. The assay revealed that several of these fluorescent mGAT1 transporters were functionally identical to wild-type mGAT1.
We applied Förster resonance energy transfer (FRET) techniques to study the assembly of the fluorescent mGAT1s. We especially evaluated, by pixel-by-pixel analysis of sensitized emission FRET, the local oligomerization of each fluorescent transporter construct in various subcellular compartments of a living cell. We conclude that pixel-by-pixel analysis of FRET has the ability to distinguish between subsurface and membrane-inserted transporter complexes, as also determined by functional [3H]GABA uptake assays. Moreover, we find that the ratio of the FRET between peripheral and perinuclear subcellular compartments correlates with each construct's function. Furthermore, the all-pixel FRET amplitude distributions have up to three components with low, intermediate, and high mean amplitudes; these correspond to mGAT1 dimers, higher-order oligomers, and mGAT1 PDZ-interaction domain–mediated events in specific subcellular compartments, respectively.
MATERIALS AND METHODS
Materials
pEYFP-C1 or pECFP-C1 vectors were purchased from Takara Bio Inc. PfuTurbo Cx Hotstart polymerase and the QuikChange II XL site-directed mutagenesis kit were purchased from Agilent Technologies. The mouse N2a (CCL-131) and the human embryonic kidney T/17 cell line (HEK 293T; CRL-11268) were obtained from American Type Culture Collection. The pcDNA3.1(+) expression vector, Dulbecco's modified Eagle's medium (DMEM) with 4 mM l-glutamine, OptiMEM 1, FBS, and Lipofectamine and PLUS reagents were purchased from Invitrogen. Penicillin/streptomycin 100× and Na pyruvate 100× solutions were purchased from Mediatech, Inc. 35-mm culture dishes with 14-mm glass bottoms of #0 thickness were procured from MatTek. Other tissue culture plasticware was purchased from Greiner Bio-One. The bicinchoninic acid protein assay, EZ-Link Sulfo-NHS-Biotin, and immobilized monomeric avidin beads were obtained from Thermo Fisher Scientific. 4-amino-n-[2,3-3H]butyric acid ([3H]GABA; 87 Ci/mmol) and ECL Plus Western blotting detection reagents were obtained from GE Healthcare. Anti-GAT1 antibody AB1570W and goat anti–rabbit peroxidase–conjugated secondary antibody were obtained from Millipore. Anti–green fluorescent protein (GFP) rabbit antiserum 132002 was obtained from Synaptic Systems GmbH. Protease inhibitor cocktail tablets were obtained from Roche. Scinti-Safe 30% liquid scintillation cocktail was purchased from Thermo Fisher Scientific. The plasmids for α4YFP and/or β2CFP nicotinic acetylcholine receptors (nAChRs) can be obtained as Addgene plasmids 15245 and 15106 (http://www.addgene.org) and were described previously, together with the cDNAs for the nonfluorescent subunits from which they were constructed (Nashmi et al., 2003; Khakh et al., 2005). The ECFP-GalT plasmid was obtained from Addgene (plasmid 11937) (Cole et al., 1996). Polyethyleneimine (PEI) 50% solution in H2O, GABA, latrunculin B, and all other reagents were purchased from Sigma-Aldrich.
Cell culture and transfections
N2a cells were cultured as described previously (Son et al., 2009). Cells were plated onto 12-well plates, 60-mm dishes, or 35-mm culture dishes with 14-mm glass bottoms that we precoated with 0.05% PEI (pH 8.4 in borate buffer; 100 mM Na2B4O7, and 100 mM H3BO3). HEK 293T cells were cultured at 37°C in 95% air, 5% CO2 in culture media composed of 88% DMEM, 10% FBS, 100 IU/ml penicillin, 100 µg/ml streptomycin, and 1 mM Na pyruvate. Transfections were performed using a modification of the manufacturer's Lipofectamine and PLUS reagent protocol to achieve expression levels of transporter that exceed neither the cells' synthetic machinery nor trafficking mechanisms (supplemental Materials and methods and Table S1). Latrunculin B incubations were performed as described previously (Imoukhuede et al., 2009).
A linear [3H]GABA uptake assay in a neuronal cell line
To evaluate the function of fluorescently modified mGAT1 constructs relative to wild type, we sought to establish an uptake assay in an appropriate expression system that faithfully reports any functional changes caused by the introduction of the fluorophore. In gene transfer experiments, quantitative analysis of protein function can be vitiated by several mechanisms that cause a “ceiling,” “maximum,” “limit,” “plateau,” “leveling off,” or “saturation” of measured uptake. The nouns in quotes have recently connoted various phenomena, but we use the verb “to exceed” for each specific effect that we sought to avoid.
(A) The quantity of template cDNA transfected into the cell may exceed the capacity of the cell's protein synthetic machinery.
(B) The quantity of overexpressed protein may exceed the capability of the cell's posttranslational processing, sorting, or trafficking mechanisms.
(C) The substrate concentration may lie outside a linear region of the dose–response relation. For transporters, Michaelis-Menten analyses describe a maximum transport velocity (Vmax) of the neurotransmitter transporter at substrate concentrations that substantially exceed the Km.
(D) Net substrate flux may exceed the capacity of the cell's efflux pumps, organelle pumps, or simple diffusional removal, leading to partial or complete equilibration.
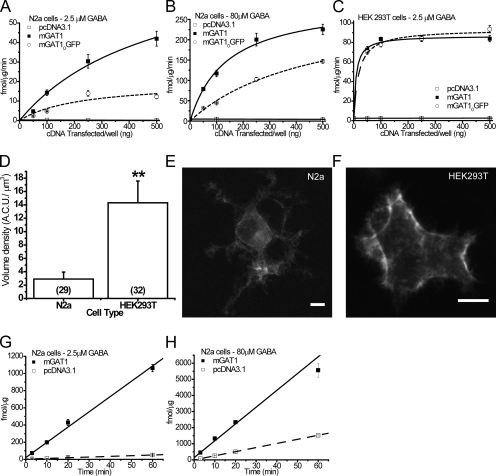
We compared the function of the wild-type mGAT1 with our original fluorescent construct, mGAT10GFP (previously called mGAT1-GFP) (Chiu et al., 2002), when expressed in the mouse N2a or human HEK 293T cell lines. Previous data show that mGAT10GFP has a major trafficking deficit in vivo (Chiu et al., 2002) because this construct design disrupts an endogenous PDZ-interacting epitope (-AYI-CO2−) at the mGAT1 C terminus of mGAT1 (Imoukhuede et al., 2009). The N2a cell line is both neuronal and murine in origin and may possess much of the necessary cellular machinery to process and traffic mGAT1 correctly; HEK 293T cells are a model human line frequently used to assess the function of plasma membrane proteins. 0, 50, 100, 250, or 500 ng of wild-type mGAT1 plasmid or an equimolar amount of mGAT10GFP or pcDNA3.1(+) plasmid was transfected in 12-well plates. 20-min [3H]GABA uptake experiments were performed in both low (2.5 µM) and high (80 µM) extracellular GABA concentrations ([GABA]O). Details of the [3H]GABA uptake assay are in the supplemental material.
[3H]GABA uptake in N2a cells transfected with wild-type mGAT1 remained in the linear range, with respect to cDNA transfected, at cDNA levels ≤100 ng cDNA/well in both 2.5 and 80 µM [GABA]O (Fig. 1, A and B). The measured [3H]GABA uptake approached a limit for transfections ≥250 ng cDNA/well. Importantly, compared with the wild-type mGAT1 control, the [3H]GABA uptake by N2a cells transfected with the mGAT10GFP construct was reduced (by 28–65%) for all cDNA transfection amounts in both 2.5 and 80 µM [GABA]O, reproducing the functional deficit reported for synaptosome preparations from mGAT10GFP knock-in mice (Chiu et al., 2002). On the other hand, HEK 293T cells did not replicate this known functional deficit when expressing the mGAT10GFP construct; [3H]GABA uptake reached a limit in 2.5 µM [GABA]O in cells transfected with as little as 100 ng cDNA/well of wild-type mGAT1 plasmid (V100 ng = 83.5 ± 1.9 fmol/µg/min; calculated maximum = 86.9 ± 2.4 fmol/µg/min; Fig. 1 C). Furthermore, when the variable parameter was the mass of cDNA transfected in HEK 293T cells, the maximum calculated mGAT10GFP [3H]GABA flux (93.5 ± 17.9 fmol/µg/min) was not significantly different from wild-type mGAT1 (Fig. 1 C).
Figure 1.
Establishing a linearly responding [3H]GABA uptake assay in an appropriate cell type. 50, 100, 250, or 500 ng of wild-type mGAT1 plasmid or an equimolar amount of mGAT10GFP or pcDNA3.1(+) plasmid was transfected/well. 20-min uptake assays were performed on transfected N2a cells in 2.5 µM (A) or 80 µM (B) [GABA]O. (C) Identical transfections were performed in HEK 293T cells, and 20-min uptake assays were performed in 2.5 µM of extracellular [GABA]O. Each point represents the mean of six transfections ± SEM. (D) Comparison of the fluorescence volume density of mGAT10GFP expressed in N2a and HEK 293T cells. **, significant difference as determined by Student's t test; P ≤ 0.01. Summed Z-stack images of representative N2a (E) and HEK 293T cells (F) expressing mGAT10GFP. Bar, 10 µm. Time course experiments were performed in 2.5 µM (G) or 80 µM (H) [GABA]O on N2a cells transfected with 100 ng/well of wild-type mGAT1 plasmid or an equimolar amount of the pcDNA3.1(+) plasmid. Each data point represents the mean of six transfections ± SEM.
Why does the HEK 293T cell assay fail to replicate the functional deficit linked to the PDZ interaction? We suspected that this failure occurs because the HEK 293T cell assay exceeds a limit associated with one of points A–D above. We tested the relative expression of total mGAT1 protein per cell in HEK 293T cells compared with N2a cells by measuring the absolute fluorescence intensity of mGAT10GFP when expressed in each cell type. 250 ng mGAT10GFP cDNA was transfected per 35-mm dish of 3.5 × 105 cells (equivalent per cell to 100 ng cDNA per 12-well plate of 1.4 × 105 cells; Table S1). Images of complete cells (0.5-µm step Z-stack) were acquired 48 h after transfection. Integrated fluorescence measurements determined that the volume density of mGAT10GFP expressed in HEK 293T cells was 4.9-fold greater than when expressed in N2a cells (Fig. 1, D–F). We concluded (a) that N2a cells are the more suitable cell type for our experiments and (b) that one should transfect 100 ng/well of wild-type mGAT1 plasmid or its equimolar amount per unit surface in other well sizes. In addition, under these transfection conditions, [3H]GABA uptake remained linear, with respect to the assay duration, for ≤60 min in both 2.5 or 80 µM [GABA]O: linear regression analysis of the 3-, 10-, 20-, and 60-min data points revealed r2 = 0.997 and 0.989, respectively (Fig. 1, G and H).
In summary, we believe that the assay conditions faithfully reported changes caused by the mutations that we studied. We believe these conditions also rendered the optical measurements sensitive to such mutations.
Design and cloning of fluorescent mGAT1 constructs
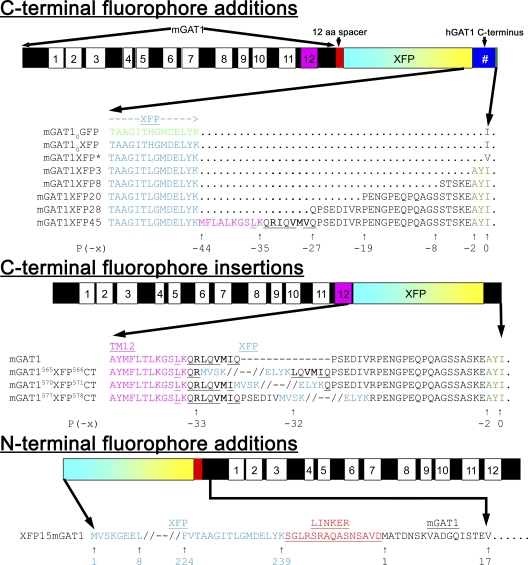
We generated 19 new fluorescent mGAT1 constructs to study trafficking and oligomerization of the GAT1 transporter. The goal was to generate fluorescent mGAT1 proteins that (a) assembled and trafficked as wild type and (b) gave robust FRET efficiencies when the transporter oligomerized. Wild-type mGAT1 and mGAT10GFP constructs have been described previously (Chiu et al., 2002). The new fluorescent mGAT1 constructs described in this study were designed to rectify trafficking deficits of mGAT10GFP; in addition, we exchanged the GFP fluorophore for ECFP or EYFP containing the “monomeric” A206K mutation to avoid distortions caused by dimerization motifs within the ECFP and EYFP proteins (Zacharias et al., 2002) and to facilitate the study of protein–protein interactions by FRET.
To generate the fluorescent mutants mGAT10XFP and mGAT1XFP* through mGAT1XFP45, the wild-type mGAT1 open reading frame (ORF) was subcloned without its original stop codon into the HindIII and EcoRI sites of the pcDNA3.1(+) expression vector multiple cloning site (MCS). XFP ORFs were then subcloned downstream from and in frame with the mGAT1 ORF at the NotI and XbaI sites of the pcDNA3.1(+) MCS. This resulted in a 12–amino acid spacer between the end of the mGAT1 sequence and the beginning of the fluorophore. We modified a method for the integration of PCR fragments without the use of restriction enzymes (Geiser et al., 2001) to add the final 3, 8, 20, 28, or 45 codons of the human GAT1 (hGAT1) ORF. These were amplified from a source plasmid using the proof-reading PfuTurbo Cx Hotstart polymerase with 5′ and 3′ extensions corresponding to the 20–22-nt regions that flanked the intended site of insertion, such that the PCR product integrated in-frame immediately after the fluorophore sequence when used as the primers in a subsequent QuikChange II XL mutagenesis PCR reaction. For mGAT1XFP*, we simply added a GTC codon for Val after the fluorophore ORF.
Fig. 2 displays the protein sequences of the modified regions of mGAT1 for each fluorescent construct. mGAT10CFP and mGAT10YFP repeated the fusion design of mGAT10GFP but with the fluorophore exchanged as annotated. The three C-terminal residues of the mGAT0XFP fusions are -YKI-CO2−, which comprises a broadly defined consensus PDZ class II–interacting motif (X-φ-X-φ, where φ designates a hydrophobic residue and X any residue) (Sheng and Sala, 2001; Hung and Sheng, 2002). We searched the Ensembl databases using Biomart (http://www.ebi.ac.uk/biomart) (Spudich et al., 2007) and applied the GO:0005886 “plasma membrane” cellular component filter. The search identified no known membrane proteins possessing the -YKI-CO2− C-terminal sequence. In the mGAT1XFP* constructs, we defined the terminal residue P(0) more narrowly, changing the terminal isoleucine residue present in mGAT10XFP to a valine in mGAT1XFP* (Fig. 2). The resulting C-terminal sequence, -YKV-CO2−, reconstituted a functional PDZ class II–interacting motif present in Ephrin B receptors, a class that relies on interactions with the PDZ domain–containing proteins for clustering (Torres et al., 1998; Brückner et al., 1999; Lin et al., 1999; Madsen et al., 2005). Other constructs in the C-terminal XFP fusion series, mGAT1XFP3, mGAT1XFP8, mGAT1XFP20, mGAT1XFP28, and mGAT1XFP45, had the most C-terminal 3, 8, 20, 28, or 45 residues of the hGAT1 appended after the mGAT1XFP fusion (Fig. 2). The differences in nucleotide sequence between the hGAT1 and mGAT1 C termini were a useful source of positive identification when we analyzed the clones during construction.
Figure 2.
Schematics of fluorescent mGAT1 construct designs and primary sequence alignments of the regions that are fused with XFP. In each schematic, the mGAT1 peptide is illustrated as a black bar, and TMs are marked as superimposed numbered boxes. TM12 is highlighted in magenta. Linkers deriving from cloning vector are shown in red, and the XFP moiety is shown as a cyan and yellow box. Additional hGAT1 sequence is shown as a dark blue box. #, a variable number of residues depending on the construct design. All schematics and their internal features are scaled according to sequence length. The three classes of florescent mGAT1 fusions, C-terminal additions, C-terminal fusions, and N-terminal additions are displayed with their names to the left. For the fusions of XFP to the mGAT1 C terminus, only the last 14 residues of the fluorophore and any appended hGAT1 or other sequence are displayed. For the mGAT1 C-terminal XFP insertions, the regions immediately adjacent to the insertion site and the first four and last four residues of the XFP are displayed. Displayed GAT1 C-terminal sequences are numbered with the terminal residue being P(0); upstream sequence positions are negative. Other important highlighted regions include: fluorophore sequence (GFP in green; XFP in light blue), the terminal hydrophobic residues of mGAT10GFP and mGAT1XFP* (purple), the endogenous GAT1 class II PDZ–interacting motif -AYI (dark green), mouse or hGAT1 TM12 sequence (magenta), nonclassical endocytic motifs homologous to FREKLAYAIT in mDAT (conserved residues underlined) (Holton et al., 2005; Boudanova et al., 2008), RL or RI ER export motif (bold) (Farhan et al., 2007), and VMI or VMV (bold) ER–Golgi intermediate compartment export motif (Farhan et al., 2008). XFP sequence position in XFP15mGAT1 constructs is displayed below the sequence in blue, and mGAT1 numbering is in black. Linker sequence between XFP and mGAT1 is shown in red.
PCR integration was applied to amplify and insert EYFP or ECFP directly between residues R565 and L566, I570 and Q571, or V577 and R578 of mGAT1 to generate the mGAT15xxXFP5xxCT constructs (Fig. 2). The site of XFP insertion in GAT1 is highlighted in the nomenclatures for these constructs by superscript residue numbers flanking the fluorophore, and the “CT” denotes that the insertion occurs within the C terminus.
The EYFP or ECFP ORFs with 15 additional codons from the downstream MCS of pEYFP-C1 or pECFP-C1 vectors were amplified and fused via PCR integration directly onto the 5′ end of the mGAT1 ORF in pcDNA3.1(+) to generate the XFP15mGAT1 N-terminally tagged constructs.
Surface biotinylation
Surface biotinylation experiments were performed as described previously (Hu and Quick, 2008). The details are in the supplemental material.
Confocal Imaging
Transfections for confocal imaging were performed as described for uptake assays but scaled up appropriately according to Table S1 for transfections in 35-mm culture dishes. Live cells grown on 14-mm glass-bottom culture dishes (MatTek) precoated with 0.05% PEI were washed once with room temperature Krebs-Ringer-HEPES buffer and observed in the same solution (Ramamoorthy et al., 1998). Live cells were imaged at room temperature on an Eclipse C1si laser-scanning confocal microscope equipped with a 63×, 1.4 numerical aperture VC Plan Apo oil objective and a 32-anode photomultiplier tube (Nikon). Where required, images were linearly unmixed using the EZ-C1 software (Nikon) for the emission spectra of the fluorophores of interest using reference spectra individually compiled for each fluorophore expressed in the same cell type and imaged under identical experimental conditions. Quantification of images was performed using ImageJ software version 1.41 (Abramoff et al., 2004). Colocalization calculations were performed using the Colocalization Finder plug-in for ImageJ.
FRET
FRET efficiency E is defined by the Förster equation (Lakowicz, 2006a). Two classes of techniques provide approximate E values based on microscopic images (Wallrabe and Periasamy, 2005; Jares-Erijman and Jovin, 2006). First, in a specimen expressing both donor- and acceptor-tagged molecules, the existence of FRET causes a decrease in the donor intensity, proportional to the number of donor-tagged molecules that interact with acceptor-tagged molecules. Thus, “acceptor photobleach” FRET directly measures E by quantifying the increase in the donor intensity after photobleach of the acceptor (although artifacts including acceptor photoconversion and donor photobleach can distort this measurement) (Rizzo et al., 2006). Second, the acceptor displays sensitized emission during excitation of the donor. Measurements of such “sensitized FRET” or “sensitized emission FRET” preserve the fluorophores in the sample and require only a single spectrally resolved image rather than a time series, but the acquired FRET index (an indirect measure of E) (Elangovan et al., 2003) responds nonlinearly to variations in the extent of interaction between fluorophore-tagged molecules (Gordon et al., 1998). This paper distinguishes between acceptor photobleach FRET and sensitized FRET measurements, as appropriate.
Acceptor photobleach FRET
Live N2a cells were visualized at room temperature in Krebs-Ringer-HEPES buffer 48 h after transfection using either an inverted LSM 510 Meta laser-scanning confocal microscope equipped with an Achroplan IR 63×, 1.4 numerical aperture oil-immersion objective and the meta array of photomultiplier tubes (Carl Zeiss, Inc.) or the Eclipse C1si LSCM. Photobleaching was performed as described previously (Son et al., 2009). FRET efficiency (E) was calculated:
| (1) |
IDA represents the normalized fluorescence intensity of CFP (100%) in the presence of nonbleached acceptor. ID represents the normalized fluorescence intensity of CFP after 100% photobleach of the acceptor, YFP. The ID value was extrapolated from a scatter plot of the percentage increase of CFP versus the percentage decrease of YFP for each cell (Nashmi et al., 2003).
Quantification of pixel-by-pixel FRET from sensitized acceptor emission
Live cells were imaged using the Eclipse C1si LSCM. Full emission spectra images were acquired in 5-nm bins between 452 and 612 nm, linearly unmixed, and complied into donor spectral bleed-through stacks, acceptor spectral bleed-through stacks, and sample image stacks as described previously (Son et al., 2009). The Pix-FRET ImageJ plug-in was used to determine the CFP and YFP bleed-through values and to calculate the net FRET (nF) and normalized FRET (NFRET) in each pixel (Feige et al., 2005). With the background and bleed-through corrections set, the nF for each pixel as described by Eq. 2 was calculated and the data output as 32-bit images.
| (2) |
nF was normalized for donor and acceptor expression levels according to Eq. 3 to calculate NFRET and to generate a 32-bit NFRET image (Xia and Liu, 2001).
| (3) |
Normalizing nF to the square root of the product of the donor and acceptor fluorescence intensities controlled for large variations in the expression levels of each fluorophore between different cells and provided a measure of FRET that is readily comparable between different samples (Xia and Liu, 2001). Gordon et al. (1998) explained that NFRET is a relative measure of the dissociation constant (Kd) for the binding interaction between proteins that increases monotonically (but not linearly) at smaller values of Kd. Box plots were constructed showing the mean NFRET, the median NFRET, and the spread of the data for each region of interest (ROI). Extreme outliers (pixels with NFRET amplitudes greater than three times the interquartile range [IQR]) and pixels within an ROI with an NFRET amplitude of 0 were eliminated from the calculations. All-pixel NFRET amplitude distributions for a given collection of cells, or of subregions, were summarized as histograms with a bin size of 0.02 NFRET units. Distributions were best fit with two or three Gaussians. Fitting concluded when R2 ≥ 0.99.
To calculate FRET efficiency from sensitized emission experiments, one needs to modify Eq. 1 because it is not possible to directly measure ID from such samples. Because the sensitized emission of the acceptor is due to the quenching of the donor in the presence of the acceptor, ID can be substituted in Eq. 1 by adding the nF signal amplitude to the amplitude of the donor fluorescence in the presence of acceptor IDA (Elangovan et al., 2003). Thus, we calculated FRET efficiency E as
| (4) |
Radial analysis of NFRET amplitude distributions
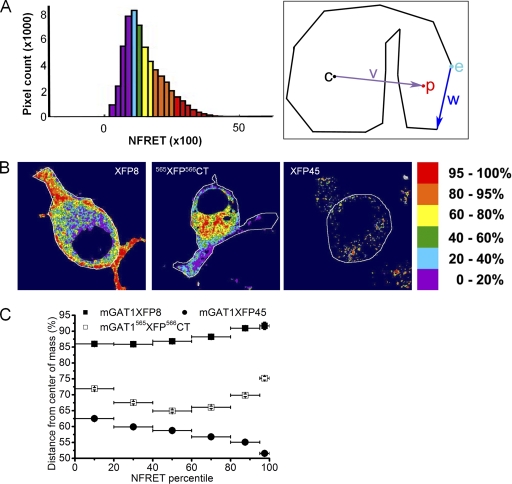
The radial NFRET amplitude distributions describe the fractional distance of pixel p to the cell edge w from the cell center c (see Fig. 13 A). Full details of the method to calculate the mean fractional displacement from c for the thousands of pixels in each NFRET amplitude percentile group are included in the supplemental material.
Figure 13.
Radial analysis of NFRET amplitude distributions. (A) Exemplar NFRET distribution with bins colored by percentile NFRET amplitude. The colors of the pixels in each bin were applied to the NFRET images as displayed in B. The right-hand panel displays how the radius of each pixel from the cell's center of mass was determined (see Materials and methods). The indentation shows that the analysis provides reliable data despite dark areas such as the nucleus or, in some cases, irregular-shaped cells. (B) NFRET images colored by NFRET amplitude percentile for exemplar cells expressing the wild-type–like functioning mGAT1XFP8, efficiently oligomerized but intracellularly retained mGAT1565XFP566CT, and the poorly oligomerized and predominantly intracellular mGAT1XFP45. The color code for the NFRET amplitude by percentile is at the right. (C) Plot of the mean distance from the center of mass of each percentile group for each construct. Each construct displayed a unique profile. Y error bars, SEM; x bars, size of each percentile group.
Data analyses
Data analyses were performed using Origin 7.0 (OriginLab) and MATLAB R2008a (The MathWorks) software.
Online supplemental material
The supplemental material includes descriptions of the methods for transfection, uptake, surface biotinylation, and radial analysis of NFRET amplitude distributions, a table of the cell densities and transfection parameters required to achieve nonsaturated transient mGAT1 expression in N2a cells (Table S1), and images of the subcellular localization of ER and trans-Golgi in N2a cells (Fig. S1). The online supplemental material is available at http://www.jgp.org/cgi/content/full/jgp.200910314/DC1.
RESULTS
Several mGAT1 C-terminal fluorophore fusions display wild-type function and surface localization
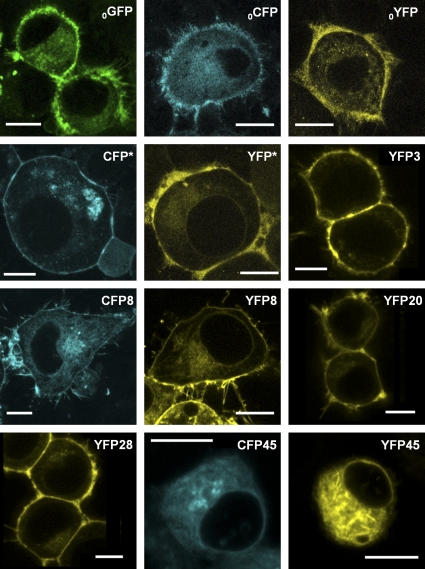
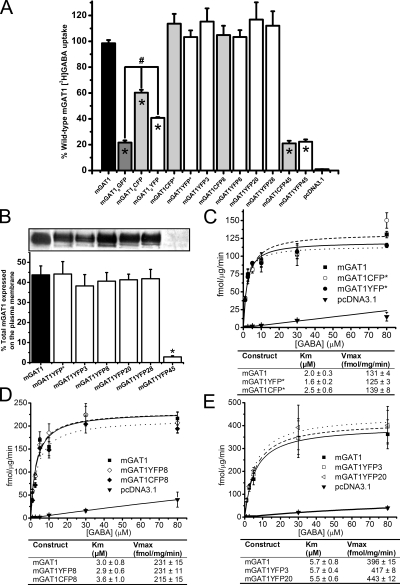
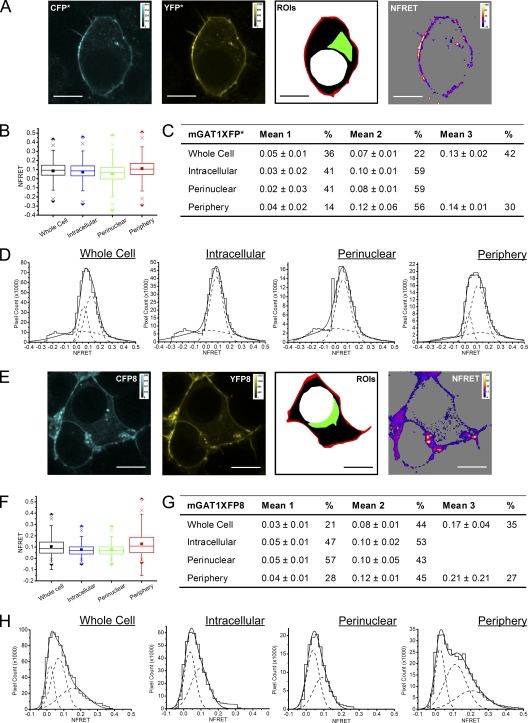
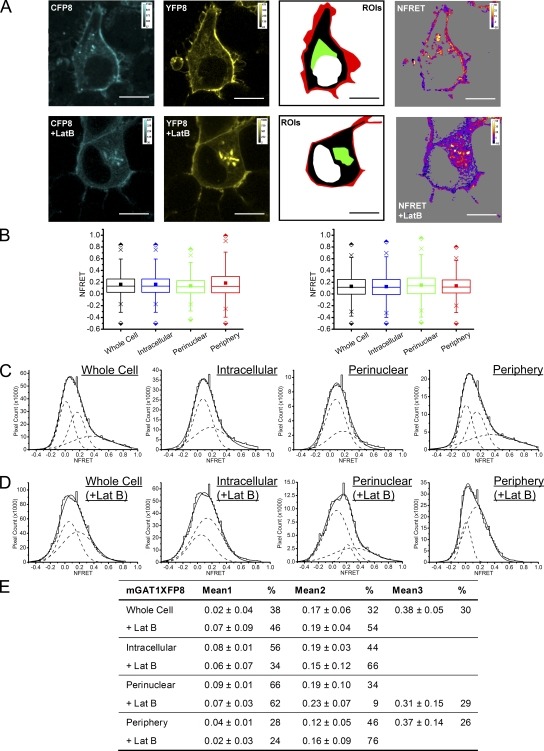
We studied the subcellular localization of the mGAT1 C-terminal fluorophore addition constructs by laser-scanning confocal fluorescence microscopy (Fig. 3) and compared their function to nonfluorescent wild-type mGAT1 in [3H]GABA uptake assays (Fig. 4 A). With the optimized [3H]GABA uptake assays described in Materials and methods, the major source of variability was apparently unavoidable variations in cell growth. Therefore, where appropriate, we used side-by-side assays (as in Figs. 4 A and 5 B); in such assays, we believe that differences in substrate transport velocity of ≥25% reflect real differences in the membrane levels of functional transporters. The mGAT10XFP constructs display fluorescence throughout the cytoplasm; at the periphery of the cells (that is, in the region near large amounts of plasma membrane), the fluorescence is brighter (Fig. 3). Fig. 4 A shows that this peripherally enhanced fluorescence does not represent complete plasma membrane insertion of mGAT10XFP constructs: [3H]GABA uptake of both mGAT10YFP and mGAT10CFP functioned less well (41 ± 1 and 60 ± 8%, respectively) compared with wild-type mGAT1. This poor function was not revealed by our previous expression system in HEK 293T cells (Chiu et al., 2002), but a functional deficiency was observed when neurons of mGAT10GFP knock-in mice exhibited reduced [3H]GABA uptake compared with wild-type mice. We therefore believe that the present assay system has good relevance to several processes that control functional levels of mGAT1 expression in neurons.
Figure 3.
Expression and subcellular localization of C-terminal XFP addition mGAT1 constructs in N2a cells. Each panel displays representative fluorescence microscopy images of live N2a cells expressing each C-terminal addition construct 48 h after transfection using the linear range parameters described in Materials and methods. The nomenclature for each construct has been abbreviated to remove the “mGAT1” because this element is common to all the constructs displayed (i.e., mGAT10GFP = 0GFP and mGAT1YFP8 = YFP8). Bars, 10 µm.
Figure 4.
Functional characterization of C-terminal fusion mGAT1XFP constructs. (A) 20-min [3H]GABA uptake from N2a cells transfected with 100 ng/well of mGAT1 wild-type plasmid, an equimolar amount of the fluorescently tagged mGAT1 plasmids, or blank pcDNA3.1(+) vector. 100% wild-type [3H]GABA uptake is 21.4 ± 1.8 fmol/µg/min. Results represent the mean ± SEM of 6–18 transfections for each construct. *, significant difference compared with wild-type; P ≤ 0.05 (ANOVA with Tukey's post-hoc test). (B) Surface biotinylation experiments comparing the plasma membrane partitioning of mGAT1XFP constructs that had displayed wild-type–like [3H]GABA uptake and one that presents an uptake deficit. Representative bands of the biotinylated mGAT1 surface fractions are displayed above the graph. Bands are arranged in the image to correspond to the appropriate column in the graph. Results represent the mean ± SEM of five to eight transfections for each construct. *, significant difference compared with wild-type; P ≤ 0.05 (one-way ANOVA with Tukey's post-hoc test). (C–E) GABA concentration dependence of six selected constructs whose uptake was closest to wild type in A. Each panel presents three groups of experiments in which the constructs were tested in pairs versus wild-type mGAT1 (100 ng cDNA transfected per 12-well plate well). The nonspecific uptake was determined from wells transfected with empty pcDNA3.1(+) vector and subtracted from the test samples, but is displayed for reference in each plot. No significant difference was found between any of these fluorescent mGAT1 constructs and wild-type mGAT1 for all concentrations, as determined by one-way ANOVA with Tukey's post hoc test (P ≤ 0.05). Data represent the mean uptake for each concentration ± SEM for six to nine transfections.
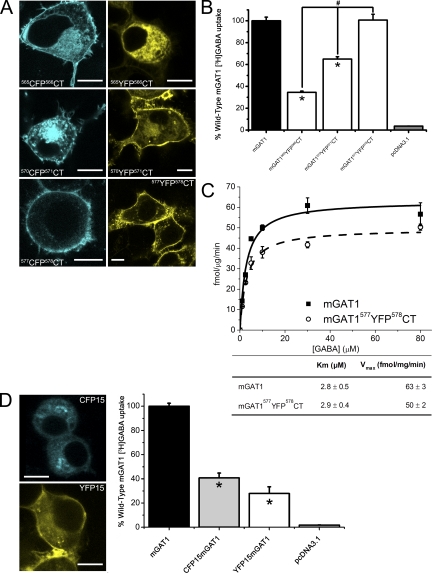
Figure 5.
Expression and functional characterization of C-terminal insertion mGAT1XFPCT constructs. (A) Each panel displays exemplar fluorescence microscopy images of live N2a cells expressing each of the C-terminal insertion constructs 48 h after transfection. Note that the subcellular distributions of mGAT1565CFP566CT and mGAT1565YFP566CT differ in the displayed panels. These variations are not caused by the different fluorophores but are displayed to exemplify each of the two types of subcellular distributions observed for this construct design. Bars, 10 µm. (B) 20-min uptake of 2.5 µM GABA/25 nM [3H]GABA from N2a cells transfected with 100 ng/well of mGAT1 wild-type plasmid, an equimolar amount of the fluorescently tagged mGAT1 plasmids, or blank pcDNA3.1(+) vector. For these experiments, 100% wild-type [3H]GABA uptake is 5.3 ± 0.6 fmol/µg/min. Results represent the mean ± SEM of 6–12 transfections for each construct. *, significant difference compared with wild-type; P ≤ 0.05 (ANOVA with Tukey's post-hoc test). (C) GABA concentration dependence of mGAT1577YFP578CT compared with wild-type mGAT1. Each data point represents the mean ± SEM of six transfections of each construct at each [GABA]O. (D) Left-hand panels display representative expression pattern of XFP15mGAT1 constructs in N2a cells (bar, 10 µm) 48 h after transfection. Right-hand graph displays the results of 20-min [3H]GABA uptake from N2a cells transfected with 100 ng/well of mGAT1 wild-type plasmid, an equimolar amount of the XFP15mGAT1 plasmids, or blank pcDNA3.1(+) vector. 100% wild-type [3H]GABA uptake is 21.4 ± 1.8 fmol/µg/min. Results represent the mean ± SEM of 18 wild-type mGAT transfections and 6 transfections of CFP15mGAT1 and YFP15mGAT1. *, significant difference compared with wild-type; P ≤ 0.05 (ANOVA with Tukey's post-hoc test).
We include data for mGAT10GFP for primarily historical reasons. Its poor function in a knock-in mouse strain (Chiu et al., 2002) provided a major motivation for the present study. Surprisingly, the mGAT10GFP functioned even less well (22 ± 2% of wild type) than the corresponding YFP and CFP constructs. We have not systematically explored the source of the differences among the functional data for the three fluorophore fusions (YFP, CFP, and GFP), but as noted, the generally poor function of these mGAT10XFP constructs was expected from the functional deficits of the knock-in mouse strain. Because the YFP-CFP FRET pair forms the basis of most measurements, we have not constructed additional GFP-containing mGAT1 mutants.
The constructs mGAT1XFP*, mGAT1XFP3, mGAT1XFP8, mGAT1XFP20, and mGAT1XFP28 exhibited strong localization in the cell periphery (Fig. 3). We determined that a large part of this fluorescence distribution arose from surface membrane expression. Three complementary assay types provide evidence of this point. (1) For each of these five constructs, [3H]GABA uptake from transfected N2a cells did not differ significantly from wild-type mGAT1 (Fig. 4 A). (2) Surface biotinylation experiments determined that 38–44% of the total expressed mGAT1XFP*, mGAT1XFP3, mGAT1XFP8, mGAT1XFP20, or mGAT1XFP28 protein inserted into the plasma membrane, which again did not differ significantly from wild-type mGAT1 (43%; Fig. 4 B). (3) Concentration dependence of [3H]GABA uptake for mGAT1XFP*, mGAT1XFP8, mGAT1YFP3, and mGAT1YFP20 was, again, identical to wild-type mGAT1 (Fig. 4, C–E). Each panel of Fig. 4 (C–E) presents the concentration dependence data for two fluorescent constructs versus wild-type mGAT1 transfections performed in side-by-side assays. Within each set of experiments, the Km and Vmax for the fluorescently tagged transporters did not differ significantly from those of wild-type mGAT1.
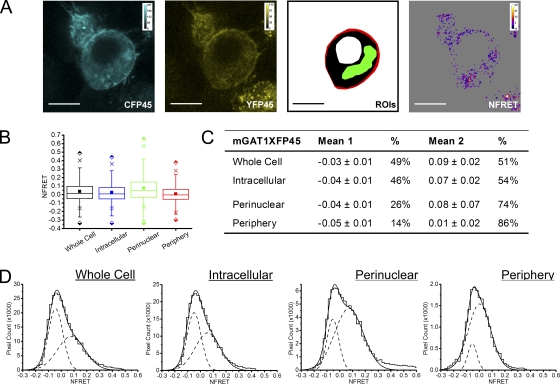
On the other hand, mGAT1XFP45 constructs were expressed primarily inside the cell (Fig. 3). This point is reported by markedly impaired [3H]GABA uptake compared with wild-type mGAT1 (mGAT1CFP45, 21 ± 2%; mGAT1YFP45, 22 ± 2%; Fig. 4 A) and by the very small fraction of the total mGAT1XFP45 protein that partitioned into the plasma membrane in biotinylation assays (2.8 ± 0.6% compared with 44 ± 5% for wild type; Fig. 4 B). It should be noted that mGAT1XFP45 was particularly challenging to process during surface biotinylation experiments because the majority of the protein sample aggregated in the well of the acrylamide gels and did not separate under electrophoresis. Apparently, the mGAT1XFP45 constructs suffer from one or more major deficits.
Diminished mGAT1 function results if XFP is incorporated close to TM12
In the mGAT1565XFP566CT and mGAT1570XFP571CT constructs, the XFP insertion sites interrupted motifs that are important in the regulation of mGAT1 trafficking (Holton et al., 2005; Farhan et al., 2007, 2008; Boudanova et al., 2008). However, the C terminus region interrupted by our mGAT1577XFP578CT construct has little influence on mGAT1 oligomerization and trafficking, according to a previous report (Farhan et al., 2004). Fig. 5 A displays the subcellular localization of the mGAT1565XFP566CT, mGAT1570XFP571CT, and mGAT1577XFP578CT constructs. These three construct designs expressed varying degrees of concentrated fluorescence at the periphery of the cells close to the plasma membrane as well as in intracellular compartments. The intracellular localization was greatest in the mGAT1565XFP566CT constructs, and in many instances, expression was confined entirely to intracellular compartments. This observation is exemplified by the mGAT1565YFP566CT image presented in Fig. 5 A (21 of out of 79 cells imaged). Notably, all cells expressing mGAT1570XFP571CT or mGAT1577XFP578CT constructs exhibited some apparent plasma membrane localization, with mGAT1577XFP578CT showing strong fluorescence concentrated in the cell periphery (Fig. 5 A), similar to that observed for mGAT1XFP3-mGAT1XFP28. We studied the [3H]GABA uptake properties of the C-terminal YFP insertions (Fig. 5 B). mGAT1565YFP566CT exhibited a marked deficit in uptake compared with wild-type mGAT1. Uptake from N2a cells expressing mGAT1570YFP571CT was intermediate between mGAT1565YFP566CT and wild-type mGAT1. Uptake from mGAT1577YFP578CT-expressing cells did not differ significantly from wild-type mGAT1. In concentration–response experiments, the Km for mGAT1577YFP578CT also did not differ significantly from wild-type mGAT1, and its Vmax was 78% that of wild type (Fig. 5 C). Thus, the presence of the XFP fluorophores between the C-terminal R565 and Q571 residues inhibited interactions required for normal trafficking of mGAT1 to the plasma membrane. Locating the XFP six residues more distally in the C terminus avoided such inhibitions and maintained mGAT1577XFP578CT transporters at the plasma membrane in numbers almost equivalent to the wild-type nonfluorescent mGAT1.
N-terminal fluorophore additions impair mGAT1 function and trafficking
The N-terminally labeled XFP15mGAT1 fusions were primarily retained within intracellular regions (Fig. 5 D) and displayed significantly reduced [3H]GABA uptake compared with wild-type mGAT1 (CFP15mGAT1 35 ± 2% and YFP15mGAT1 28 ± 6%; Fig. 5 E). This extended our previous experience with an N-terminally labeled GFPmGAT1 fusion that had only eight amino acids in its linker between the fluorophore and the mGAT1; the previous construct also failed to localize appropriately to the plasma membrane (Chiu et al., 2002).
Acceptor photobleach FRET with fluorescent mGAT1 constructs shows that oligomerization is required for wild-type function
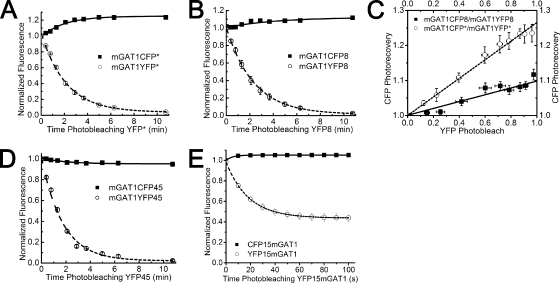
Dimerization/oligomerization of SLC6 transporter protomers is required for export of functional transporter from the ER to the plasma membrane (Farhan et al., 2006; Bartholomäus et al., 2008). Therefore, fluorescent mGAT1 constructs that behaved functionally like wild-type mGAT1 were expected to form oligomers of mGAT1 protomers. We studied the assembly of our fluorescent GAT1 constructs using the acceptor photobleach FRET technique (Nashmi et al., 2003). We first investigated acceptor photobleach FRET between mGAT1XFP* pairs and between mGAT1XFP8 pairs, which are C-terminal GAT1–XFP fusion constructs that functioned like wild-type mGAT1 in both uptake and surface biotinylation experiments (Fig. 6). To quantify acceptor photobleach FRET between the mGAT1 oligomers, we constructed a scatter plot of the photobleach-induced changes in CFP and YFP fluorescence of each cell during the entire bleaching time, fitted a regression line to the data, and extrapolated to complete YFP photobleach (Fig. 6 C). The extrapolated values of CFP photorecovery for the mGAT1CFP*-mGAT1YFP* and the mGAT1CFP8-mGAT1YFP8 transfection pairs were 1.10 ± 0.01 and 1.26 ± 0.01, respectively (Fig. 6 C). Eq. 1 yields calculated FRET efficiencies of 9% for mGAT1CFP8/mGAT1YFP8 and 21% for mGAT1CFP*/mGAT1YFP*. For the C-terminal mGAT1XFP45 fusions, which have poor function, we detected no measurable FRET between mGAT1CFP45-mGAT1YFP45 coexpressed in N2a cells (Fig. 6 D). Likewise, no detectable acceptor photobleach FRET signal was recorded from cells coexpressing the CFP15mGAT1-YFP15mGAT1 constructs (Fig. 6 E).
Figure 6.
FRET assessed by acceptor photobleach. (A) Plots of acceptor photobleach and donor photorecovery for mGAT1CFP*/mGAT1YFP* (n = 10) or (B) mGAT1CFP8/mGAT1YFP8 (n = 7) coexpressed in N2a cells. Data for each fluorophore were normalized to absolute fluorescence at time zero and fitted to a single exponential plus finite baseline: . (C) A scatter plot of CFP photorecovery versus YFP photobleach for the FRET pairs in A and B. FRET efficiency for each transfection was calculated by extrapolating to the linear regression fits to YFP photobleach = 1.0. The Y-intersect (ID) was then entered into Eq. 1. (D) Plot of acceptor photobleach and donor photorecovery for mGAT1CFP45/mGAT1YFP45 (n = 12). (E) Mean acceptor photobleach and donor photorecovery for the CFP15mGAT1/YFP15mGAT1 cotransfection in N2a cells (n = 5).
Pixel-based FRET reveals several components to the total FRET signal amplitude distribution
The acceptor photobleach FRET data gave a reasonable report of the average FRET signal from confocal images of whole N2a cells expressing fluorescent GAT1 protomers. However, acceptor photobleaching is not optimal for measurement of FRET at subcellular resolution in live cells. Attempting acceptor photobleach FRET in subcellularly differentiated ROIs would introduce error in the temporal resolution because imaging would be far slower than the dynamics of transporter trafficking. To determine FRET in specific ROIs, we used a spectrally resolved, sensitized emission FRET approach that determines FRET in a sample on a pixel-by-pixel basis and during the few microseconds required to measure each pixel (pixels are 69-nm squares). The spectrally unmixed images provide data similar to the three-filter method reported by Xia and Liu (2001), but using an optically more efficient, simultaneous detection of photons. Furthermore, we included the NFRET amplitude of each pixel as a datum in the analysis rather than averaging the signal amplitudes of all the pixels in an ROI. As shown below, this technique revealed that (a) FRET varies among subcellular compartments as defined by each ROI, and (b) within every ROI, the total NFRET distribution consists of two or three subcomponents, each with a distinct mean NFRET amplitude. For each fluorescent mGAT1 construct design studied, we analyzed the number of NFRET components, their amplitudes, and the proportion of the total NFRET distribution represented by each component. Comparing these data with the measured functional properties of each construct suggested that the subcomponents of NFRET distributions may represent mGAT1 dimers, higher-order mGAT1 oligomers, and mGAT1 oligomers interacting with PDZ domain–containing complexes, as explained in the Discussion and Appendix.
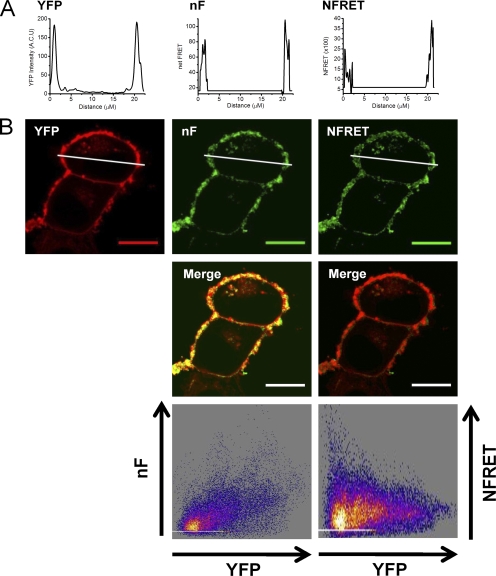
Negative controls with noninteracting membrane proteins
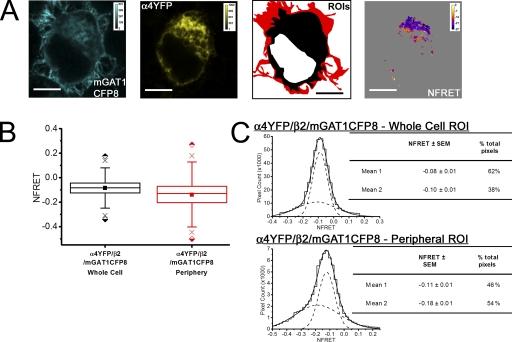
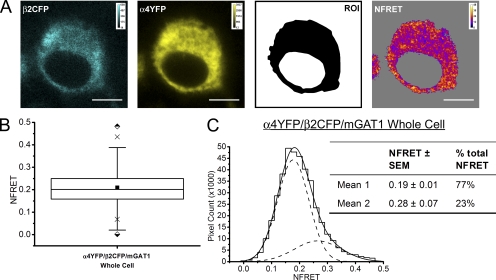
For the negative control, we expressed two fluorescently tagged plasma membrane proteins, mGAT1 and the α4β2 nAChR, which do not interact in N2a cells (Drenan et al., 2008). The mGAT1CFP8 construct was cotransfected with plasmids that assemble α4YFP/β2 nAChRs (Nashmi et al., 2003; Khakh et al., 2005; Drenan et al., 2008; Son et al., 2009). Control transfections of mGAT1CFP8/α4/β2 and mGAT1/α4YFP/β2 plasmids (250 ng of each plasmid) were also performed (a) to generate reference spectra and (b) to determine spectral bleedthrough as described previously. Both fluorescently tagged proteins maintained their normal expression pattern when coexpressed in N2a cells (Fig. 7 A): the fluorescence pattern for fluorescent α4YFPβ2 nAChRs was uniform with little enhancement at the plasma membrane (Fig. 7 A), as described previously in N2a cells (Drenan et al., 2008; Son et al., 2009), and the fluorescence pattern for mGAT1CFP8 resembled that of Fig. 3.
Figure 7.
Pixel-by-pixel quantification of sensitized emission FRET between mGAT1CFP8 and α4YFPβ2 nAChRs. A negative control experiment. (A; from left to right) mGAT1CFP8 fluorescence and α4YFP nAChR subunit fluorescence unmixed from an N2a cell coexpressing mGAT1CFP8, α4YFP nAChR subunit, and wild-type nonfluorescent β2 nAChR subunit (calibration bars in arbitrary calibration units [ACUs]). ROIs were used to determine FRET. The red-shaded area described the “peripheral” ROI, and the combined red and black areas correspond to the whole cell ROI. The fourth panel displays the NFRET image (calibration bar, NFRET × 100). Pixels with signal amplitude below threshold are shaded gray. Bars, 10 µm. (B) Box plots displaying the range of NFRET detected from these negative control data. The box highlights the IQR (Q1–Q3), the center line in the box indicates the median, and the closed square symbol represents the mean. The whiskers' ends represent the boundaries of the lower and upper inner fences (1.5 × IQR). The × marks the first and 99th percentiles. The half-shaded diamond symbols indicate the absolute maximum and minimum data point in each set. The plots from the whole cell and peripheral ROIs are colored black and red, respectively, corresponding to the ROI color codes in A. The mean and median NFRET amplitudes for all pixels in each ROI are displayed in Table I. (C) Histograms displaying the distribution of pixel NFRET amplitudes for each condition (bin width, 0.02). Distributions for each ROI were fit to two Gaussian components. The individual components are shown as dashed lines, and the sum of the fit is shown as a solid line. The tables in each panel report the means of each component and the percentage of the pixels comprising each component.
We defined a whole cell ROI that encompassed all the fluorescent pixels in the cell minus the nonfluorescent cell nucleus. The mean NFRET for all pixels within this ROI for cells coexpressing mGAT1CFP8 and α4YFP/β2 nAChR was negative, indicating that no FRET occurred and that there was some overcorrection for donor and acceptor bleedthrough when there was no FRET (Fig. 7 B and Table I).
TABLE I.
Average NFRET for all pixels measured in each ROI
| Transfection | ROI NFRET | |||||||||||||||
| Whole Cell | Intracellular | Perinuclear | Periphery | |||||||||||||
| Meana | Median | IQR | Pixels (×105) | Meana | Median | IQR | Pixels (×105) | Meana | Median | IQR | Pixels (×105) | Meana | Median | IQR | Pixels (×105) | |
| α4YFP/β2CFP/mGAT1 | 0.20 | 0.20 | 0.09 | 3.9 | ||||||||||||
| α4YFP/β2/mGAT1CFP8 | −0.08 | −0.08 | 0.31 | 4.3 | −0.14 | −0.13 | 0.12 | 0.7 | ||||||||
| mGAT1XFP* | 0.10 | 0.10 | 0.09 | 7.3 | 0.10 | 0.09 | 0.09 | 4.7 | 0. 07 | 0.07 | 0.10 | 1.8 | 0. 11 | 0.11 | 0.11 | 2.2 |
| mGAT1XFP8 | 0.10 | 0.09 | 0.10 | 7.7 | 0.09 | 0.07 | 0.07 | 4.0 | 0.08 | 0.07 | 0.06 | 1.2 | 0.13 | 0.11 | 0.14 | 3.3 |
| mGAT1XFP45 | 0.04 | 0.02 | 0.14 | 3.1 | 0.03 | 0.01 | 0.13 | 2.5 | 0.08 | 0.05 | 0.18 | 0.9 | 0.01 | −0.004 | 0.12 | 0.2 |
| mGAT1565XFP566CT | 0.18 | 0.17 | 0.11 | 4.3 | 0.17 | 0.16 | 0.11 | 3.1 | 0.17 | 0.16 | 0.11 | 1.7 | 0.18 | 0.17 | 0.11 | 0.7 |
| mGAT1570XFP571CT | 0.12 | 0.11 | 0.09 | 4.5 | 0.11 | 0.10 | 0.09 | 3.0 | 0.11 | 0.11 | 0.07 | 1.5 | 0.12 | 0.11 | 0.09 | 0.9 |
| mGAT1577XFP578CT | 0.16 | 0.14 | 0.16 | 2.6 | 0.12 | 0.10 | 0.13 | 1.3 | 0.12 | 0.10 | 0.12 | 0.7 | 0.20 | 0.18 | 0.17 | 1.2 |
| mGAT10XFP | 0.13 | 0.11 | 0.11 | 23 | 0.13 | 0.12 | 0.11 | 15 | 0.12 | 0.11 | 0.10 | 3.9 | 0.11 | 0.09 | 0.10 | 7.6 |
Standard error of the mean NFRET values range from 6 × 10−5 to 7 × 10−4.
Because the mGAT1CFP8 construct localized strongly in the cell periphery, we used this fluorescence to define a second ROI that contributed “peripheral NFRET.” The peripheral ROI encompassed both the plasma membrane and a narrow annulus (∼700 nm) of immediately adjacent cytoplasm. We use this description because constructs such as mGAT10XFP exhibit concentrated fluorescence in the cell periphery due to pooling of transporter-containing vesicles within ∼500 nm of the outer lipid bilayer of the cell, rather than because of efficient insertion into the plasma membrane (Chiu et al., 2002). The calculated peripheral NFRET for coexpressed mGAT1CFP8 and α4YFP/β2 nAChR was also negative (Fig. 7 B and Table I).
Fig. 7 C introduces frequency distributions of NFRET amplitudes from each pixel of several dozen cells. These data are binned to form all-pixel NFRET amplitude distributions: NFRET amplitude on the x axis and number of pixels on the y axis. Although the NFRET distributions for both ROIs for these negative control transfections were best fit with two Gaussians, both components had negative mean NFRET amplitudes (Fig. 7 C). These data confirm that the method detects no interaction between the α4β2 nAChR and GAT1 in intracellular regions or in the cell periphery.
Region-specific FRET quantification reveals high FRET efficiency in the periphery of cells expressing fluorescent GAT1 constructs that exhibit wild-type function
To investigate FRET between fluorescent GAT1 fusions, we defined four ROIs for each cell imaged. These were named the “whole cell ROI,” the “intracellular ROI,” the “perinuclear ROI,” and the “peripheral ROI.” The whole cell and peripheral ROIs were defined in the control experiments. Intracellular ROI is the space within the concentrated fluorescence at the cell periphery, but subtracting the dark space occupied by the cell nucleus, and is densely filled by ER (Fig. S1 A). The perinuclear ROI describes a concentrated region of fluorescence in cells expressing the fluorescent mGAT1 constructs adjacent to the cell nucleus; according to organelle markers, this ROI comprises mainly ER and Golgi (Fig. S1 B). Fluorescence images of cells coexpressing the CFP and YFP variants of fluorescent GAT1 fusions were acquired and processed as described above.
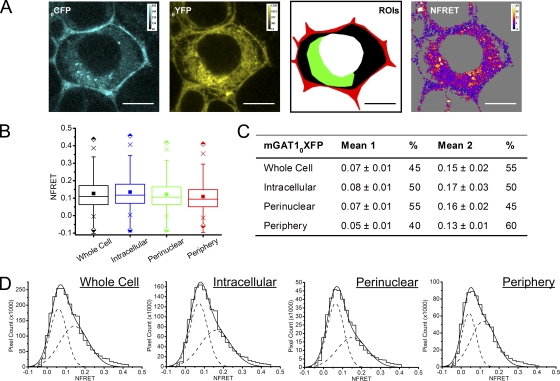
Sensitized FRET from cells expressing the wild-type–like C-terminal fusion constructs mGAT1XFP* and mGAT1XFP8 confirmed earlier acceptor photobleach FRET findings that oligomerization was required to observe wild-type–like function from our fluorescent constructs (Fig. 8). In addition, for mGAT1XFP* and mGAT1XFP8, respectively, NFRET images showed that 30.1 and 43.3% of the whole cell ROI NFRET signal came from the peripheral ROI (Fig. 8, A and E, and Table I). The mean NFRET amplitudes for all pixels in an ROI for N2a cells expressing mGAT1XFP* or mGAT1XFP8 constructs were greatest in the peripheral ROI compared with the intracellular or the perinuclear ROIs (Fig. 8, B and F, and Table I). Specifically, the ratios of NFRET in the peripheral ROI were 1.6- and 1.7-fold greater than in the perinuclear ROIs for mGAT1XFP* and mGAT1XFP8, respectively. Also, the reported NFRET from pixels in the peripheral ROI of mGATXFP8-expressing cells had a much broader IQR than those from the perinuclear ROI (Fig. 8 F and Table I). mGAT1 oligomerization was therefore detected in all ROIs, but an additional molecular event, specifically localized to the periphery of cells expressing mGAT1XFP* and mGAT1XFP8, resulted in elevated FRET in this region. Analysis of the NFRET distributions from each ROI determined that the NFRET from the intracellular and perinuclear ROIs of mGAT1XFP*- or mGAT1XFP8-expressing cells were best fit with two Gaussians, both reporting positive mean NFRET. However, the whole cell ROIs and the peripheral ROIs were best fit with three Gaussians (Fig. 8, D and H). The highest-amplitude NFRET component (1.2- to 1.8-fold greater mean NFRET than from the intermediate-amplitude component) represented 27–30% of the pixels in the peripheral ROIs (Fig. 8, C, D, G, and H). Among the three subcellular ROIs, this highest-amplitude component appeared only in the peripheral ROI and was accompanied by an ∼30% reduction in the peripheral ROI NFRET signal contributed by the lowest-amplitude component when compared with the intracellular or perinuclear ROIs (Fig. 8, C, D, G, and H). Thus, for the wild-type–like functioning mGAT1XFP* and mGAT1XFP8, we infer that two different oligomerization events were described by the low- and intermediate-amplitude components that were common to the NFRET distributions of all examined ROIs (see Discussion and Appendix). In addition, elevated mean NFRET in the peripheral ROI versus intracellular regions corresponded to the third high-amplitude component in the peripheral ROI NFRET distribution. We infer that a specific mGAT1 oligomerization or interaction event—the molecular correlate for the third high-amplitude NFRET component—is highly localized to the cell periphery.
Figure 8.
Pixel-by-pixel quantification of sensitized emission FRET between mGAT1XFP* and mGAT1XFP8. (A; left two panels) mGAT1CFP* fluorescence and mGAT1YFP* fluorescence (calibration bars, ACUs). In the third panel, ROIs were used to determine NFRET. The red-shaded area described the “peripheral” ROI, the green-shaded area corresponds to the “perinuclear” ROI, the black and green region with the red area describes the “intracellular ROI,” and the combined red, green, and black areas correspond to the whole cell ROI. The fourth panel displays the NFRET image of the same cell (color calibration bar, NFRET × 100). The color code in the NFRET image includes negative and positive signal amplitudes. Pixels with signal amplitude below threshold are shaded gray. Bars, 10 µm. (B) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT1XFP*. The box, whiskers, and other data points are represented as in Fig. 7 B. Box plots for each ROI are colored according to the code described for A (ROI), and the intracellular data are colored blue. (C) Table of results for mGAT1XFP* transfections displaying the mean of each Gaussian component of the summed fit of the distributions in D and the percentage of the pixels comprising each component. (D) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1XFP*-expressing cells. The individual components are shown as dashed lines, and the sum of the fit is shown as a solid line. (E) Representative fluorescence images of mGAT1XFP8-expressing N2a cells, the ROIs, and corresponding NFRET image. Calibration and color coding are as for A. (F) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT1XFP8. Color coding and data are represented as in B. (G) Table of results for mGAT1XFP8 transfections displaying the mean of each Gaussian component of the summed fit of the distributions in H and the percentage of the pixels comprising each component. (H) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1XFP8-expressing cells. The individual components are shown as dashed lines, and their sum is shown as a solid line.
Sensitized NFRET from mGAT1XFP45, which reported little function and no FRET by acceptor photobleach analysis, was studied pixel by pixel (Fig. 9). This method did detect some mGAT1XFP45 oligomerization in small regions. These pixels were too few to influence the whole cell averaging algorithms that calculated FRET by acceptor photobleaching. The perinuclear ROI contained most of these small regions highlighted by the sensitized NFRET approach in mGAT1XFP45-expressing specimens. The mean NFRET amplitude for all pixels in the perinuclear ROI resembled that recorded for mGAT1XFP* or mGAT1XFP8 (Fig. 9 B and Table I). For mGAT1XFP45, the FRETing pixels in the peripheral ROI contributed only 6.4% of the whole cell ROI NFRET signal, nonetheless indicating that some oligomerized mGAT1XFP45's were exported from the perinuclear region and could eventually contribute to the small but significant [3H]GABA uptake for this construct in the functional assays. The NFRET distributions for all ROIs of mGATXFP45-expressing cells were best fit with two Gaussians, the first of which reported a negative mean NFRET (Fig. 9, C and D). The positive population made up >75% of the NFRET signal in the peripheral and perinuclear ROIs. We observed that the mean NFRET of the positive Gaussian component was 1.4-fold larger in the peripheral ROI compared with the same component of the mGAT1XFP45 perinuclear ROI NFRET. Even though very few mGAT1XFP45 oligomers inserted into the plasma membrane, it appeared that those that did insert were subject to the same molecular event that caused increased FRET in the periphery of cells expressing wild-type functioning fluorescent mGAT1 constructs.
Figure 9.
Pixel-by-pixel quantification of sensitized emission FRET between mGAT1XFP45. (A; from left to right) mGAT1CFP45 fluorescence and mGAT1YFP45 fluorescence (calibration bars, ACUs), ROIs used to determine NFRET (color coding as in Fig. 8), and the NFRET image (color calibration bar, NFRET × 100) of the same cell. Pixels with amplitude below threshold are shaded gray. Bars, 10 µm. (B) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT1XFP45. The box, whiskers, and other data points are represented as in Fig. 7 B. Box plots for each ROI are colored according to the code in A (ROI), and the intracellular data are colored blue. (C) Table of results for mGAT1XFP45 transfections displaying the mean of each Gaussian component of the summed fit of the distributions in Fig. 8 D and the percentage of the pixels that comprise each component. (D) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1XFP45-expressing cells. The individual components and their sum are shown as dashed and solid lines, respectively.
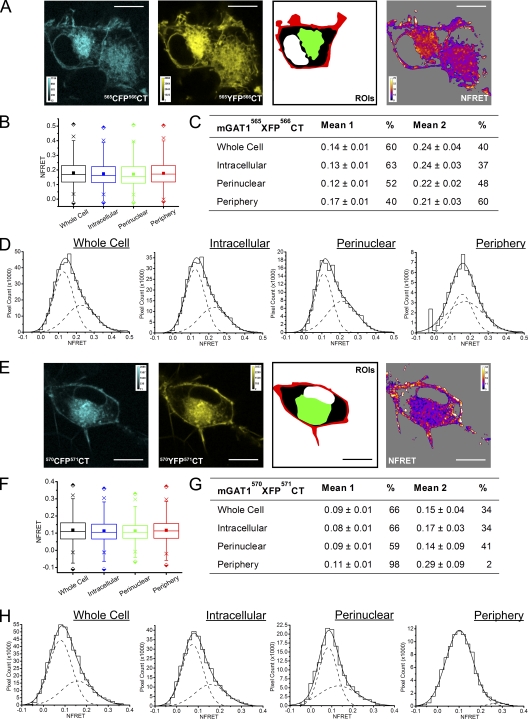
Next, we examined the FRET of the mGAT15xxXFP5xxCT constructs, which show a graded improvement in function as the XFP moiety is moved downstream within the C terminus away from TM12 (Fig. 5). mGAT1565XFP566CT reported a robust mean NFRET amplitude for all pixels (≥0.17) in all ROIs examined (Fig. 10, A and B, and Table I), indicating that significant transporter oligomerization was occurring. However, the poor function of this construct, the completely internal distribution of fluorescence in 27% of cells expressing these constructs, and the minor component (16% of the whole cell ROI NFRET) contributed from the peripheral ROI lead us to conclude that the NFRET signal was mainly due to assembled intracellular mGAT1565XFP566CT rather than plasma membrane–inserted transporters, even in the peripheral ROI. The ratio of the mean NFRET for all pixels in the peripheral ROI compared with the perinuclear ROI was 0.99. mGAT1565XFP566CT NFRET distributions were best fit with two Gaussians in all ROIs examined (Fig. 10, C and D). In the peripheral and perinuclear ROIs, the two subpopulations contributed approximately equally to the total NFRET signal. In the intracellular ROI, the lower mean NFRET signal predominated.
Figure 10.
Pixel-by-pixel quantification of sensitized emission FRET between mGAT1565XFP566CT and mGAT1570XFP571CT. (A; from left to right) mGAT1565CFP566CT fluorescence and mGAT1565YFP566CT fluorescence (calibration bars, ACUs), ROIs used to determine NFRET (color coding as in Fig. 8), and the NFRET image (color calibration bar, NFRET × 100) of the same cell. Pixels with signal amplitude below threshold are shaded gray. Bars, 10 µm. (B) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT1565XFP566CT. The box, whiskers, and other data points are represented as in Fig. 7 B. Box plots for each ROI are colored according to the code in Fig. 8 A (ROI), and the intracellular data are colored blue. (C) Table of results for mGAT1565XFP566CT transfections displaying the mean of each Gaussian component of the summed fit of the distributions in D and the percentage of the pixels comprising each component. (D) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1565XFP566CT-expressing cells. The individual components and the sum of the fit are shown as dashed and solid lines, respectively. (E) Representative fluorescence images of mGAT1570XFP571CT-expressing N2a cells, the regions of interest, and corresponding NFRET image. Calibration and color coding are as for Fig. 8 A. (F) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT1570XFP571CT. Color coding and data are represented as in B. (G) Table of results for mGAT1570XFP571CT transfections displaying the mean of each Gaussian component of the summed fit of the distributions in H and the percentage of the pixels that comprise each component. (H) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1570XFP571CT-expressing cells. The individual components and their sum are shown as dashed and solid lines, respectively.
The mean NFRET amplitude from mGAT1570XFP571CT-expressing cells (Fig. 10, E and F, and Table I) was less than that from those expressing mGAT1565XFP566CT. This probably arose in part from the increased fluorophore separation in this construct. The XFP moiety is fused five residues more distal from the end of the TM12 helix in the mGAT1570XFP571CT constructs. Exact intermolecular distances cannot be calculated from FRET efficiencies of proteins fused to GFP derivatives (Rizzo et al., 2006). However, side-by-side FRET efficiency calculations (using Eq. 4) suggested that in the perinuclear ROIs, the apparent fluorophore separation in mGAT1570XFP571CT oligomers (E = 7.9 ± 1.9%) is on average 1.25-fold greater than in mGAT1565XFP566CT oligomers (E = 25 ± 3.5%). As for mGAT1565XFP566CT, the mean NFRET amplitude from all pixels in cells expressing mGAT1570XFP571CT was similar between the peripheral and perinuclear ROIs (ratio 1.0). Analysis of the NFRET distributions for each ROI determined that all were best fit with two Gaussians (Fig. 10, G and H). However, in the periphery, the two components did not appear to describe the same populations reported by the components of the other ROIs. The mean amplitude of the major peripheral NFRET component (98% of the signal) lay approximately halfway between the amplitudes of the two NFRET components of the perinuclear, intracellular, or whole cell ROIs. We suggest that two Gaussian components merged into one with an intermediate NFRET amplitude in the periphery because the populations mix at a level too fine to resolve in a 69-nm2 pixel (see Appendix). The second component of the mGAT1570XFP571CT peripheral NFRET signal had a mean amplitude (0.29 ± 0.09) twice that observed for the second Gaussian component of the perinuclear ROI NFRET (Fig. 10, G and H). Although contributing only a small fraction (2.3%) of the total signal for the peripheral ROI, the ratio of mean NFRET amplitude of this component versus the second component of the perinuclear or intracellular ROIs (2.1 and 1.7, respectively) resembled that of the highest-amplitude component of mGAT1XFP* or mGAT1XFP8 NFRET (Fig. 8). The small but measureable high-amplitude NFRET component in the mGAT1570XFP571CT peripheral NFRET signal presumably reflects its impaired plasma membrane insertion, as determined by GABA uptake assays (Fig. 5 B), and is also consistent with the observation that mGAT1570XFP571CT functions better than mGAT1565XFP566CT (Fig. 5 B).
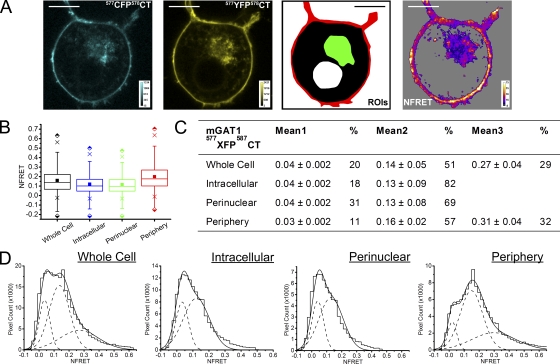
mGAT1577XFP578CT, which exhibited almost wild-type function (Fig. 5, B and C), displayed strong mean NFRET for all pixels in the peripheral ROI, 1.7-fold greater than that observed in the perinuclear ROI (Fig. 11, A and B, and Table I). This NFRET amplitude was 1.5- to 1.8-fold stronger than that for the mean NFRET amplitude from all pixels in the peripheral ROI of mGAT1XFP*- or mGAT1XFP8-expressing cells (Fig. 8). As observed for those other wild-type functioning C-terminal fusions, the NFRET amplitude in the intracellular and whole cell ROIs was significantly less. The NFRET distributions from intracellular and perinuclear ROIs of mGAT1577XFP578CT-expressing N2a cells could be fitted with two Gaussians with low and intermediate mean NFRET amplitudes, the latter component providing approximately two thirds of the NFRET signal in the perinuclear ROI (Fig. 11, C and D). Importantly, in the peripheral ROI, a third Gaussian component appeared; this component was also detectable in the whole cell ROI (Fig. 11 D). This component had a mean NFRET amplitude (0.31 ± 0.04) twice that of the second intermediate-amplitude NFRET component and comprised ∼30% of the peripheral NFRET signal. The relative distribution of transporter subpopulations for this construct, as determined by the components of NFRET in each ROI, resembled that for the wild-type functioning mGAT1XFP* and mGAT1XFP8 constructs (Fig. 8).
Figure 11.
Pixel-by-pixel quantification of sensitized emission FRET between mGAT1577XFP578CT. (A; from left to right) mGAT1577CFP578CT fluorescence and mGAT1577YFP578CT fluorescence (calibration bars, ACUs), ROIs used to determine NFRET (color coding as in Fig. 8), and the NFRET image (color calibration bar, NFRET × 100) of the same cell. Pixels with amplitude below threshold are shaded gray. Bars, 10 µm. (B) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT1577XFP578CT. The box, whiskers, and other data points are represented as in Fig. 7 B. Box plots for each ROI are colored according to the code in A (ROI), and the intracellular data are colored blue. (C) Table of results for mGAT1577XFP578CT transfections displaying the mean of each Gaussian component of the summed fit of the distributions in D and the percentage of the pixels that comprise each component. (D) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1577XFP578CT-expressing cells. The individual components and the sum of the fit are shown as dashed and solid lines, respectively.
High NFRET in the peripheral ROI is independent of variance of YFP intensity
We examined whether high NFRET amplitudes measured in this study, particularly those at the periphery of cells expressing the wild-type functioning mGAT1XFP8 constructs, were large because of high local concentrations of YFP-tagged protomers in some domains. Line profile plots through cells expressing mGAT1XFP8 emphasized that the pixels with the greatest YFP signal intensity were usually located at the periphery of the cell (Fig. 12 A), as were the pixels with the highest amplitude net FRET (nF) or NFRET. We measured the colocalization of the YFP signal with both the nF signal and the NFRET signals. As a positive control, we determined that the mean Pearson product-moment correlation (r) for the colocalization of the CFP and YFP signals coexpressing mGAT1CFP8 and mGAT1YFP8 was 0.9 ± 0.01 (n = 31 cells) (not depicted). Colocalization analysis of the YFP fluorescence and the nF signal in the same group of cells reported that r = 0.5 ± 0.05 (Fig. 12 B). The square of r indicated that 25 ± 1.5% of the variance of nF in mGAT1XFP8-expressing cells is associated with changes in YFP intensity. When the relative contributions of the CFP and YFP intensities to the FRET signal in each pixel were normalized to generate the NRET images from the same cells, r = 0.01 ± 0.05 for IYFP versus NFRET colocalization (Fig. 12 B). This indicated that the normalization step in NFRET corrects for the influence of IYFP variance. One concludes that calculated NFRET was not artificially large due to high local IYFP in some ROIs.
Figure 12.
Colocalization of IYFP with net FRET and NFRET signals. (A) Line profile analysis of YFP fluorescence. nF and NFRET measurements through the mGAT1XFP8-expressing cell in B illustrate the peripheral localization of the signals for all parameters. (B) Images report the colocalization of YFP-expressing pixels with the nF or NFRET signals. Correlation of the nF or NFRET signal with YFP intensity is plotted in the bottom panels. Note that the normalization routine allows NFRET to measure FRET independently of YFP intensity.
Radial distance analysis of NFRET signal amplitude correlates to functional phenotype
The presence of the three NFRET components from only the fluorescent mGAT1 constructs with wild-type–like function suggested that one component arose from a mechanism that operates only in the cell periphery for mGAT1 oligomers, which are appropriately inserted into the plasma membrane. Apparently, this plasma membrane–associated component is also the component with the highest NFRET amplitude. We have already qualitatively observed that the largest amplitude NFRET signal occurred in the periphery of the cell (Fig. 12 A). As described in Materials and methods, we further analyzed NFRET images by grouping pixels into NFRET amplitude percentile groups (Fig. 13 A; look-up table in far right panel of Fig. 13 B). The pixels contained within each bin or group are highlighted with the same color in the NFRET image (Fig. 13 B). We calculated each pixel's percent radial distance from the cell's center to the cell periphery. We analyzed the localization of NFRET amplitude for three representative constructs: mGAT1XFP8, which oligomerized efficiently as determined by acceptor photobleach and sensitized FRET analysis and functioned like wild-type nonlabeled mGAT1; mGAT1565XFP566CT, which showed strong FRET but compromised function; and mGAT1XFP45, which demonstrated weak function and weak FRET. The results of the analyses plotted in Fig. 13 C determined that the NFRET signals from pixels in cells expressing mGAT1XFP8 were the most peripherally located; the minimum percent distance from the center of mass for any NFRET amplitude percentile group was >85%. The pixels in the top fifth percentile of NFRET amplitude for mGAT1XFP8 were located a mean distance of 93% of the total radius from the center to mass to the edge of the cell, quantifying the statement that the highest-amplitude NFRET pixels were also the most peripherally located in the cell. Overall, the localization of NFRET signals of all amplitudes from pixels expressing mGAT1565XFP566CT trended toward the cell periphery rather than the center of mass, but significantly less so than from mGAT1XFP8. The trend followed a U-shaped pattern, with the bottom and top 20% NFRET pixel amplitudes being the most peripherally located (>70%), and the medium amplitude NFRET pixels residing in more intracellular locations. Localization of mGAT1XFP45 pixels that reported positive NFRET trended to more random distributions with increasing NFRET amplitude.
Correlation between function of fluorescent mGAT1 transporters and their NFRET properties
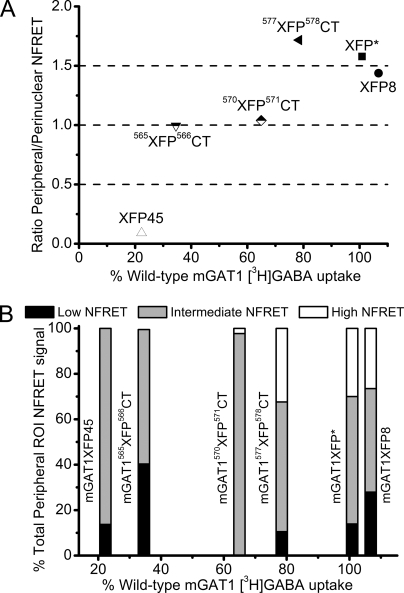
Fig. 14 provides two ways to summarize the trends in NFRET and their relationship to function ([3H]GABA uptake) of mGAT1 constructs. Both panels of Fig. 14 highlight the set of three correctly oligomerized, correctly trafficked, and correctly functioning fluorescent constructs: mGAT1XFP*, mGAT1XFP8, and mGAT577XFP578CT. In Fig. 14 A, we compare the function of each fluorescent construct (relative to nontagged wild-type mGAT1) versus the ratio of peripheral to perinuclear NFRET. The peripheral/perinuclear ratio was calculated from the mean NFRET for all pixels in each ROI. In this analysis, the fluorescent transporters could be classified into three groups:
Figure 14.
The relationship between transporter function and NFRET. (A) Scatter plot of the ratio and mean (peripheral NFRET)/(perinuclear NFRET) for all pixels in the ROIs versus the percent function of each construct as compared with nonfluorescent wild-type mGAT1 in [3H]GABA uptake assays. Data points for constructs are individually labeled and omit the mGAT1 prefix, which is common to all. Constructs with ≥75% wild-type function are represented by solid symbols. Constructs that showed ≤65% wild-type function but good whole cell NFRET are represented by half-filled symbols. mGAT1XFP45, which demonstrated poor function and poor whole cell NFRET, is represented by an open symbol. (B) The proportions of each Gaussian component of the total peripheral ROI NFRET signal for each construct are plotted in stacked columns. Each column represents a single construct design and appears on the x axis at the percentage of wild-type [3H]GABA uptake it exhibits in uptake assays.
(1) The three constructs with >75% wild-type function, as determined by [3H]GABA uptake, displayed a peripheral/perinuclear NFRET ratio ≥1.4.
(2) Constructs with compromised function (30–65% wild-type [3H]GABA uptake) demonstrated efficient oligomerization as determined by FRET quantification, but less efficient plasma membrane insertion, indicated by peripheral/perinuclear NFRET ratios close to 1.0.
(3) Represented by mGAT1XFP45, the constructs that display <30% of wild type function, limited oligomerization, and poor trafficking properties expressed a peripheral/perinuclear NFRET ratio of 0.1.
Wild-type functioning fluorescent transporters could therefore be clearly distinguished from constructs with poor oligomerization and/or trafficking: the former possess a peripheral/perinuclear NFRET ratio ≥1.4.
Fig. 14 B presents an additional hallmark of the three well-functioning constructs (as shown by [3H]GABA uptake levels). For these constructs, the peripheral ROI NFRET distributions could be fitted to three Gaussian components, and we have presented analyses that the highest-amplitude component arises from plasma membrane molecules. On the other hand, constructs with significantly reduced substrate transport rates (corresponding to reduced plasma membrane insertion) relative to wild-type GAT1 displayed peripheral ROI NFRET distributions comprising only two components (Fig. 14 B). The data suggest a threshold effect: if the peripheral NFRET distribution includes ≥∼30% of the high-amplitude NFRET component, fluorescent mGAT1 molecules exhibit wild-type–like function (Fig. 14 B).
Nicotinic receptors also display a two-component NFRET distribution
Data presented to this point indicate that mGAT1, a membrane protein, displays only a two-component NFRET distribution if its oligomers are not inserted into the plasma membrane at normal wild-type levels. We assessed whether this conclusion holds for another membrane protein whose oligomerization has been studied in our laboratory in the N2a system: the heteropentameric mouse α4β2 nAChR (Nashmi et al., 2003; Drenan et al., 2008; Son et al., 2009). Previous data indicate that α4β2 receptors are retained to a large extent in intracellular compartments (Nashmi et al., 2003; Kuryatov et al., 2005; Sallette et al., 2005; Drenan et al., 2008; Son et al., 2009).
As with previous studies of this channel expressed in N2a cells (Drenan et al., 2008; Son et al., 2009), HEK 293 cells (Nashmi et al., 2003), transfected neurons (Nashmi et al., 2003; Khakh et al., 2005), and α4YFP knock-in mice (Nashmi et al., 2007), the fluorescence of both α4YFP and β2CFP nAChR subunits appeared evenly throughout the ER of N2a cells (Fig. 13 A), with a high degree of colocalization (Pearson product-moment correlation coefficient: r = 0.9 ± 0.02; n = 11 cells) (not depicted). As described above (Fig. 7), the even fluorescence distribution throughout the cell provided only a single ROI per cell for this transfection: the ROI encompassed the entire cell without the nucleus. This was determined by the pixels exhibiting YFP fluorescence.
Robust FRET was observed throughout cells coexpressing α4YFPβ2CFP nAChR subunits; the mean NFRET for all pixels was 0.20 (Fig. 15, A and B, and Table I). The distribution of NFRET amplitudes for all analyzed pixels was best fit by two Gaussian components, with mean NFRET of 0.19 (representing 75% of all FRETing pixels) and 0.28 (Fig. 15 C).
Figure 15.
Pixel-by-pixel quantification of sensitized emission FRET within α4YFPβ2CFP nAChRs. (A; from left to right) Panels display an N2a cell coexpressing β2CFP and α4YFP nAChR subunits with nonfluorescent wild-type mGAT1 (calibration bars, ACUs). The whole cell ROI in which FRET was quantified is displayed in black in the third panel. The fourth panel displays the NFRET image (calibration bar, NFRET × 100). Pixels with signal amplitude below threshold are shaded gray. Bars, 10 µM. (B) Box plot displaying the range of NFRET detected from α4YFPβ2CFP-coexpressing N2a cells. The box, whiskers, and other data points are represented as in Fig. 7 B. The mean and median NFRET amplitudes for all pixels in each ROI are displayed in Table I. (C) Distribution of NFRET signal amplitudes per pixel (bin width, 0.02). The histogram was fit to two Gaussian distributions. The individual components are shown as dashed lines, and the sum of the fit is shown as a solid line. The inset table reports the mean NFRET amplitude of each component and the percentage of the total pixels that comprise each component.
C-terminal PDZ interactions are required to produce NFRET distributions comprising three Gaussian components
The PDZ type II–interacting domain at the distal C terminus of mGAT1 is necessary for at least two important transporter interactions in the cell periphery: with the exocyst complex (Farhan et al., 2004) and with the actin cytoskeleton (Imoukhuede et al., 2009). To determine whether an interaction of the C-terminal mGAT1 PDZ-interaction domain produced the observed third, high NFRET component in peripheral ROI NFRET distributions, we coexpressed mGAT10CFP and mGAT10YFP and examined the resulting subcellular NFRET profiles. As demonstrated earlier (Fig. 4 A), this construct design had compromised function. This occurs because the XFP moiety blocks access to the endogenous PDZ-interaction domain of mGAT1 (Imoukhuede et al., 2009). Fig. 16 A displays representative images of the donor and acceptor fluorescence in an mGAT10XFP-expressing cell together with the defined ROIs and calculated NFRET image. The ratio of mean NFRET amplitude for the peripheral versus the perinuclear regions for 63 cells expressing mGAT10XFP was 0.9 (Fig. 16 B and Table I), in contrast to 1.4 observed above for mGAT1XFP8 (Fig. 8 F and Table I). Furthermore, the NFRET distributions for all ROIs from mGAT10XFP-expressing cells were best fit by two Gaussian components. This indicated that mGAT1 transporters must possess a functional C-terminal PDZ-interacting domain to exhibit the third high-amplitude NFRET component in the cell periphery.
Figure 16.
Pixel-by-pixel quantification of sensitized emission FRET between mGAT10XFP constructs. (A; from left to right) mGAT10CFP fluorescence and mGAT10YFP fluorescence (calibration bars, ACUs), ROIs used to determine NFRET (color coding as in Fig. 8), and the NFRET image (color calibration bar, NFRET × 100) of the same cell. Pixels with amplitude below threshold are shaded gray. Bars, 10 µm. (B) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT10XFP. The box, whiskers, and other data points are represented as in Fig. 7 B. Box plots for each ROI are colored according to the code in A (ROI), and the intracellular data are colored blue. (C) Table of results of mGAT10XFP transfections displaying the mean of each Gaussian component of the summed fit of the distributions in D and the percentage of the pixels that each component represents. (D) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT10XFP-expressing cells. The individual components and the sum of the fit are shown as dashed and solid lines, respectively.
PDZ-mediated tethering to the cytoskeleton locates third high-amplitude NFRET component in the cell periphery
A PDZ domain interaction occurs in the ezrin-mediated tethering of mGAT1 oligomers to the actin cytoskeleton (Imoukhuede et al., 2009). We therefore investigated whether an intact cytoskeleton is required for the observed high NFRET component at the cell periphery.
We transfected N2a cells with the mGAT1XFP8 constructs and in half of the transfected dishes depolymerized the actin cytoskeleton with 5 µM latrunculin B (Fig. 17). As previously observed, mGAT1XFP8 showed detectable NFRET in all ROIs. The ratio of mean peripheral to perinuclear NFRET amplitude was 1.4 (Fig. 17 B, left; 67 cells, 4.7 and 1.8 × 105 pixels, respectively). 5 µM latrunculin B decreased this ratio to 0.9 (Fig. 17 B, right). The NFRET distributions for nontreated mGAT1XFP8-expressing N2a cells were fit with three Gaussian components in the whole cell and peripheral ROIs, with the highest mean amplitude NFRET component comprising ∼30% of the total NFRET signal (Fig. 17, C and E). The intracellular and perinuclear ROIs were best fit with two Gaussian components (Fig. 17 C). Treatment of mGATXFP8-expressing cells with latrunculin B produced NFRET distributions from whole cell and peripheral ROIs that were best fit with two Gaussian components, similar to data for the fluorescent mGAT1 constructs with less than wild-type function (Fig. 17 D). Latrunculin B treatment therefore demonstrated that the polymerized actin cytoskeleton was required for the PDZ-interacting domain–mediated high NFRET component in the cell periphery.
Figure 17.
Pixel-by-pixel quantification of sensitized emission FRET from mGAT1XFP8 when cells are treated with 5 µM latrunculin B. (A; top row, from left to right) mGAT1CFP8 fluorescence and mGAT1YFP8 fluorescence (calibration bars, ACUs), ROIs used to determine NFRET (color coding as in Fig. 8), and the NFRET image (color calibration bar, NFRET × 100) of the same cell. (Bottom row) mGAT1XFP8-expressing cells after 1-h treatment with 5 µM latrunculin B. Bars, 10 µm. (B) Box plots displaying NFRET for all pixels in each ROI of cells expressing mGAT1XFP8 (left) and mGAT1XFP8 treated with latrunculin B (right). The box, whiskers, and other data points are represented as in Fig. 7 B. Box plots for each ROI are colored according to the code in A (ROI), and the intracellular data are colored blue. (C) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1XFP8-expressing cells. The individual components and the sum of the fit are shown as dashed and solid lines, respectively. (D) Distributions of NFRET signal amplitude per pixel for each ROI (bin width, 0.02) from mGAT1XFP8-expressing cells plus latrunculin B. The individual components and the sum of the fit are shown as dashed and solid lines, respectively. (E) Table of results for mGAT1XFP8 transfections before and after latrunculin B treatment displaying the mean of each Gaussian component of the summed fit of the distributions in C and D, and the percentage of the pixels that comprise each component.
Interestingly, the perinuclear ROI of latrunculin B–treated cells was best fit by three Gaussian components (Fig. 17 D). A three-component perinuclear distribution was not found under other conditions in this study. The amplitude of the third component was equivalent to that seen in the periphery of nontreated cells and comprised a similar proportion of the total NFRET distribution in that ROI (∼30%; Fig. 17 E). This was not observed in the perinuclear ROI of mGAT10XFP-expressing cells, suggesting that the intact PDZ-interacting domain of mGAT1XFP8 plays a role.
DISCUSSION
FRET between GFP variant-tagged molecules reports separations of <100 Å. In live cells, this distance can be achieved only by directly interacting proteins (Sekar and Periasamy, 2003). FRET has previously demonstrated that GAT1 and other SLC6 neurotransmitter transporters oligomerize (Soragna et al., 2005; Farhan et al., 2006; Bartholomäus et al., 2008). The dimerization of transporter protomers is required for efficient export from the ER (Farhan et al., 2006). Acceptor photobleach FRET gave good whole cell measurements of mGAT1 assembly but did not differentiate between fluorescent constructs with wild-type–like or compromised plasma membrane insertion.
Pixel-by-pixel analysis of NFRET allowed the accurate measurement of FRET in various subcellular regions, and the individual components of NFRET distributions in each ROI correlate with functional phenotype. Analyzing each construct's number of Gaussian components in an NFRET distribution overcame complications from the fact that each construct has a unique separation and/or orientation between fluorophores. We simply assumed that the rank order of NFRET distribution components arises from mechanisms that are common across the entire set of constructs.
We observed at least two oligomerization states of the mGAT1 transporter in the basal state (in the absence of substrate). These oligomerization states probably bear no relation to the time-resolved conformational states that occur during the permeation cycle for the SLC6 neurotransmitter transporter family (Mager et al., 1994; Li et al., 2000; Loo et al., 2000; Kanner, 2003; Rudnick, 2006). Rather, they represent the basic oligomer formed by mGAT1 protomers and high-order complexes of basic mGAT1 oligomers (e.g., dimers of dimers). In intracellular organelles (e.g., ER or Golgi), these two oligomer types are represented by two Gaussian components of the NFRET distribution. A third mGAT1 oligomer exists in the periphery of cells expressing fluorescent transporters that are functionally indistinguishable from the nontagged wild-type mGAT1. This species is represented in NFRET distributions by a third Gaussian component that always possesses the highest mean amplitude of the three. If the high-amplitude component accounts for ∼30% of the total NFRET distribution, the fluorescent mGAT1 construct functions with >75% of the wild-type rate.
Oligomeric states may generate the components in the Gaussian fits to mGAT1 NFRET distributions
We consider three explanations for the measured NFRET between fluorescently tagged GAT1 protomers.
(1) The XFPs themselves might drive apparent oligomerization of the fused protein of interest. We rule out this possibility because the monomeric A206K mutation was introduced into every construct (Zacharias et al., 2002).
(2) The GAT1 protomers might have no tendency to associate directly and are distributed randomly in cellular membranes, with FRET occurring through stochastic interactions in domains where the protomers are clustered at high density. Thus, controlling for collisional FRET is critical (Blackman et al., 1998; Tadross et al., 2009). In this paper, we believe there is minimum contribution from this scenario: at the expression levels used in this study, for constructs that exhibit >25% wild-type mGAT1 function, very little of the total NFRET arises from stochastic interactions of high local concentrations of mGAT1XFP monomers. To support this statement, we invoke the examples of the mGAT1XFP45 and the XFP15mGAT1 constructs. Neither of these exhibited FRET by the acceptor photobleach method, despite extensive colocalization of CFP- and YFP-tagged mGAT1 protomers. In the pixel-by-pixel analysis of sensitized mGAT1XFP45 NFRET, some positively FRETing pixels were observed. However, a fraction of the mGAT1XFP45 FRETing pixels is also expected to represent the small proportion of mGAT1XFP45 protomers that was able to assemble as functional oligomers and traffic to the plasma membrane, as reported by the [3H]GABA uptake assays. Oligomerization is a prerequisite for ER export of the SLC6 family of transporters (Farhan et al., 2006; Bartholomäus et al., 2008); therefore, the functional mGAT1XFP45 construct in the plasma membrane must be at least a dimer. In summary, we believe that donor–acceptor excitations are rare occurrences unless the fluorophores are part of a biologically functional oligomeric complex.
(3) Therefore, in all likelihood, mGAT1 dimers and possibly higher-order oligomers (e.g., dimers of dimers or trimers of dimers) assemble. Even if mGAT1 can only maximally oligomerize as dimers, a high concentration of dimers in a particular domain can result in inter-dimer energy transfer, producing NFRET. In our linear expression system, we expect minimal contributions of inter-oligomer proximity to the measured NFRET amplitude, for the same reason that we expect little contribution to the NFRET amplitude from random proximity of donor- and acceptor-tagged mGAT1 monomers.
The basic oligomeric unit of mGAT1 is a dimer, and an atomic-scale structure is available for dimers of a homologous transporter (Yamashita et al., 2005). Features of this dimer render it unlikely that a trimer would form, so that the simplest likely high-order oligomer is a dimer of dimers. The Appendix presents assumptions and calculations analyzing the FRET properties of dimers and tetramers. The results are consistent with the idea that the first and second Gaussian components of the all-pixel mGAT1 NFRET amplitude distributions represent the dimer and a high-order oligomerization state (probably a square or rhomboid tetramer), respectively.
When NFRET distributions are fit to three Gaussian components, the highest NFRET component reports the specific locations of mGAT1 C-terminal PDZ interactions
The peripheral ROI includes both the plasma membrane and the immediately adjacent intracellular region. Several of the constructs display high fluorescence in the cell periphery but have major functional deficits, presumably owing to poor plasma membrane insertion. FRET determined that these constructs with trafficking deficits efficiently oligomerized, but the ratio (peripheral NFRET/perinuclear NFRET) was ∼1.0 (in contrast to the value of ∼1.4 for the best-inserted constructs) (Fig. 14 A). Furthermore, both the peripheral and perinuclear NFRET distributions were fit by two Gaussian components, again in contrast to the three components for the best-inserted constructs (Figs. 9 D, 10, D and H, and 16 D). The data are consistent with the idea that well-inserted plasma membrane transporters display an additional component with higher mean NFRET amplitude comprising ∼30% of the NFRET distribution.
In the basal state, one third of the acutely recycling pool of GAT1 transporters resides in the plasma membrane (Wang and Quick, 2005). This fraction resembles the proportion of the total peripheral NFRET represented by the highest-amplitude component in the cell periphery. The radial analysis of NFRET amplitudes (Fig. 13) suggests that the highest-amplitude component of peripheral NFRET represents most of the fluorescent GAT1 complexes inserted into the plasma membrane. Furthermore, the presence of this component and the proportion of the total NFRET that it represents serve as an indicator of transporter function; the assay could help in the future design of other fluorescent transporter constructs. Judging the generality of the pixel-by-pixel method will require examination with additional classes of transmembrane proteins. We demonstrate here that α4β2 nAChRs predominantly reside in intracellular compartments in the basal state and have NFRET distributions comprising two Gaussian components (Fig. 12). These two components may represent the two possible functional stoichiometries of α4 and β2 nAChR subunits, (α4)3(β2)2 and (α4)2(β2)3 (Son et al., 2009). Moreover, independent investigations suggested the geometry of G protein–coupled receptor oligomers by fitting FRET distributions with Gaussian components (Raicu et al., 2009).
We determined that the physical origin of the third high NFRET component amplitude for mGAT1 oligomers closely associated with the surface membrane requires (a) that the fluorescent mGAT1 construct possess an intact PDZ type II–interaction domain at the distal C terminus, and (b) that the mGAT1 C terminus be tethered to the actin cytoskeleton via this domain (Fig. 16). mGAT10XFP does not interact with PDZ proteins (Imoukhuede et al., 2009), limiting its presentation at the plasma membrane (Chiu et al., 2002), probably because of compromised interaction with the exocyst (Farhan et al., 2004). mGAT10XFP NFRET distributions are always fit to only two Gaussian components in any ROI (Fig. 16). Furthermore, the NFRET distributions from cells expressing a different class of plasma membrane oligomers, the α4β2 nAChR, which does not make direct PDZ-mediated interactions in the cell periphery, can be fit to only two components (Fig. 15). In N2a cells expressing mGAT1XFP8, which has a functional PDZ-interaction domain, actin depolymerization via latrunculin B eliminated the third high-amplitude NFRET component present in the peripheral ROI (Fig. 17). Collectively, these results support the view that, if the C terminus of mGAT1 is tethered to an intact cytoskeleton via a functional PDZ-interaction domain, one observes three NFRET components in the periphery. We suggest that this interaction produces the highest mean amplitude NFRET component of the three, and thus this component can be used as a marker of this intracellular event. Presumably, the fluorophores located at the very distal C termini of mGAT1XFP8 oligomers are constrained in a way that increases FRET. If the tethering restricts the motion of the C terminus without changing the average fluorophore separation, this might actually decrease FRET (Lakowicz, 2006b). On the other hand, if the tethering draws the C termini closer together, or restricts the rotation of the fluorescent moieties to angles that have favorable dipole–dipole coupling, this would markedly increase FRET efficiency.
In this investigation, latrunculin B treatment of mGAT1XFP8-expressing cells was the only manipulation causing three NFRET components in an intracellular ROI. The amplitude and proportion of the total NFRET for the perinuclear ROI of the third component resembled that observed in the periphery of untreated cells. We suspect that the perinuclear high-FRET component for the latrunculin treatment arose from a pathological concentration of mGAT1 oligomers in perinuclear organelles. In addition to decoupling membrane-resident mGAT1 from the actin cytoskeleton, actin depolymerization disrupts normal endocytosis and endosomal sorting and recycling (Roux et al., 2006; Gauthier et al., 2007; Doherty and McMahon, 2009; Morel et al., 2009). Significant slowing of actin-dependent endocytic traffic relative to overall mGAT1 insertion rates during latrunculin B incubation would explain our previously observed increases in [3H]GABA uptake for both mGAT1YFP8 or mGAT10GFP (Imoukhuede et al., 2009). Additionally, PDZ-associated mGAT1XFP8 oligomers internalized during the early minutes of the latrunculin B incubation could possibly get trapped inside the cell (Taunton et al., 2000; Gauthier et al., 2007; Morel et al., 2009). Nonetheless, exocyst-driven plasma membrane GAT1 insertion requires the transporter's PDZ-interacting domain (Farhan et al., 2004). Most proteins that form the exocyst complex are loaded onto vesicles in the trans-Golgi region (Munro, 2004), and their trafficking to the site of exocytosis requires an intact actin cytoskeleton (Boyd et al., 2004). In latrunculin B–treated N2a cells, mGAT1XFP8 oligomers presumably associate normally via their PDZ-interaction domains, with the vesicle-associated exocyst complex in the vicinity of the trans-Golgi. However, cytoskeletal disruption inhibits transport of the vesicles and their cargo away from this location, resulting in a third high-amplitude NFRET component being recorded in the perinuclear ROI. In contrast, despite possessing a wild-type PDZ-interaction domain, the efficiently oligomerized but intracellularly retained mGAT1565XFP566CT constructs present only two perinuclear NFRET components because the location of the fluorophore prevents the normal interaction with the COPII coat complex (Farhan et al., 2007). This mutation dominates over the PDZ interactions: transporter complexes cannot exit from the ER (Farhan et al., 2004) to the trans-Golgi, where the first exocyst/PDZ-interacting domain interactions occur.
Pixel-by-pixel resolution in NFRET
Ultimately, one would like to measure the assembly of membrane proteins at single-molecule resolution in each cellular compartment. At present, this is possible only at the plasma membrane using total internal reflection fluorescence in FRET mode (Riven et al., 2006; Bal et al., 2008a,b), although superresolution microscopy may enable further progress (Hess et al., 2007; Manley et al., 2008; Tokunaga et al., 2008). We have therefore resorted to measurements on single pixels, 69-nm square. We estimate that each pixel represents 10–150 mGAT1 molecules, based on comparisons with known samples that we have imaged at single-molecule resolution under comparable conditions (Chiu et al., 2001). A pixel's NFRET amplitude is determined by the concentration of each FRET-yielding oligomer type in that pixel (Corry et al., 2005b), not by the concentration of fluorophores. Methods to measure FRET in biological samples are continually evolving. Recently, investigators have minimized spurious FRET by judiciously selecting the fluorophores by using appropriate instrumentation, and by controlling optical variations during both an individual experiment and the entire investigation (Raicu et al., 2009; Tadross et al., 2009). Combining these enhancements with the method presented in this paper will further increase the resolution and reliability of confocal FRET data. We hope eventually to determine the supramolecular geometry of the high-order mGAT1 oligomers represented by the intermediate-amplitude component of the NFRET distributions (Raicu et al., 2009).
Trafficking and oligomerization motifs in mGAT1 and similar transporters
Appropriate mGAT1 plasma membrane expression requires a class II PDZ–interacting motif at the terminal residues of mGAT1 (Chiu et al., 2002; Farhan et al., 2004). In mGAT1, this motif mediates interactions with the exocyst complex (Farhan et al., 2004) and in tethering GAT1 to the actin cytoskeleton (Imoukhuede et al., 2009). The present data show that the primary sequence of that motif can be either the native -AYI from GAT1 or another class II motif from a different molecule (-YKV-CO2−). Both motifs have experimentally determined interaction partners (Brückner et al., 1999; Lin et al., 1999; McHugh et al., 2004; Imoukhuede et al., 2009). However, the identity and combination of the hydrophobic residues at P(0) and P(−2) is important (Harris and Lim, 2001; Sheng and Sala, 2001). The terminal -YKI-CO2− sequence of mGAT10XFP is not present at the C terminus of any known membrane protein and evidently is poorly used by PDZ domain–containing proteins in N2a cells or in vivo (Chiu et al., 2002). The sequence of the C-terminal type II PDZ–interacting motif and its interacting partners are therefore critical in the events that lead to the emergence of the third high-amplitude component of NFRET in the cell periphery.
Besides the terminal type II PDZ–interacting sequence, apparently no other motifs influence trafficking within the sequence between the P(−3) and P(−28) positions in the GAT1 C terminus. These findings corroborate previous studies of truncation mutations in neuronal SLC6 transporter trafficking (Torres et al., 2003; Farhan et al., 2004). However, a region in the C termini of SLC6 transporters corresponding to the P(−28) through P(−44) residues of mGAT1 controls constitutive export of transporter from the ER (Farhan et al., 2004, 2007, 2008) and both constitutive and regulated endocytosis (Holton et al., 2005; Boudanova et al., 2008). mGAT1XFP45 possesses two copies of this region (Fig. 2), but its uptake deficit (Fig. 4 A) is caused predominantly by poor oligomerization (Figs. 6 D and 9), rather than by the introduction of additional endocytic or exocytotic motifs. Nonetheless, mGAT1565XFP566CT showed extensive oligomerization (Fig. 10, A–D) but poor trafficking to the plasma membrane (Fig. 5 B), presumably because placing the fluorophore between residues R565 (P(−33)) and L566 (P(−32)) (Fig. 2) inhibited interaction with the COPII coat (Farhan et al., 2007; Reiterer et al., 2008). Moreover, in mGAT1565XFP566CT, the XFP is adjacent to the 562LKQR565 residues (analogous to 587FREK590 in hDAT) that slow the basal rate of transporter internalization (Boudanova et al., 2008); this may result in faster endocytosis for this construct relative to wild-type GAT1. The trafficking deficit in mGAT1565XFP566CT also supports earlier findings that mutations of this region dominate over the presence of a class II PDZ–interacting motif at the distal C terminus (Farhan et al., 2004).
For mGAT1570XFP571CT, we suggest that moving the fluorophore more distal in the C terminus partially relieved the impaired trafficking of the transporters because the 565RL566 is preserved, allowing interaction with the COPII machinery, and the XFP is further from 562LKQR565, slowing the basal internalization relative to mGAT1565XFP566CT. Nevertheless, the functional expression of mGAT1570XFP571CT at the plasma membrane is probably hampered because the fluorophore is adjacent to, and may sterically interfere with, the 568VMI570 motif, which specifies export from the ER–Golgi intermediate compartment (Farhan et al., 2008).
Almost wild-type–like function (similar Km and 78% Vmax) and strong NFRET in the periphery were achieved in the mGAT1577XFP578CT design, in which the XFP was inserted 6 residues after the 10-residue region, rich in regulatory information for transporter trafficking (Figs. 2, 5, B and C, and 10, and Table I) (Farhan et al., 2004, 2007, 2008; Holton et al., 2005; Boudanova et al., 2008). We will continue to investigate the endocytosis and exocytosis of these constructs. The functional (Fig. 5, B and C) and NFRET (Fig. 10) data suggest that both mGAT1XFP8 and mGAT1577XFP578CT assemble well as mature transporters, and the latter construct has a small deficit in the number of transporters finally inserted into the plasma membrane. mGAT1577XFP578CT and mGAT1XFP8 displayed similar proportions of low-, intermediate-, and high-amplitude components for peripheral ROI NFRET distributions, and this may imply similar oligomerization and interaction states for the constructs. Of these two pairs, the mGAT1577XFP578CT pair can indicate mGAT1 assembly more sensitively because the apparently nearby fluorophores have robust FRET.
Value of a linear, responsive cellular assay
Various cell lines and transient transfection techniques have been applied to investigate the oligomerization and trafficking of SLC6 transporters (Melikian and Buckley, 1999; Schmid et al., 2001; Whitworth and Quick, 2001; Chiu et al., 2002; Torres et al., 2003; Bjerggaard et al., 2004; Farhan et al., 2004, 2007; Just et al., 2004; Korkhov et al., 2004; Holton et al., 2005; Mazei-Robison and Blakely, 2005; Soragna et al., 2005; Bartholomäus et al., 2008). In gene transfer experiments, quantitative analysis of protein function can be vitiated by several mechanisms that cause a “ceiling,” “maximum,” “limit,” “plateau,” “leveling off,” or “saturation” of measured uptake. This study's data, as well as our laboratory's previously reported findings, show that when mGAT10GFP is transiently expressed in the widely used HEK 293T cell line, its [3H]GABA uptake appears normal despite its significant trafficking deficits in vivo (Chiu et al., 2002). Several possible mechanisms could prevent mGAT10GFP from recapitulating its in vivo behavior when expressed in HEK 293T cells. The first is overexpression caused by the presence of the simian virus 40 (SV40) large T antigen in HEK 293T cells, which allows for episomal replication of transfected plasmids, such as the pcDNA3.1(+) used in this study, that contain the SV40 origin of replication. Consistent with this hypothesis, [3H]GABA uptake was maximal in HEK 293T cells transfected with only 50–100 ng mGAT1 plasmid, yet submaximal for 500 ng of plasmid in N2a cells (which lack the SV40 T antigen) (Fig. 1). Furthermore, the total fluorescence of mGAT10GFP was almost fivefold greater when expressed in HEK 293T cells compared with N2a cells (Fig. 1, D–F). We suspect that overexpression of mGAT1 in HEK 293T cells exceeds the capacity of one or more cellular trafficking mechanisms, so that the cell cannot dynamically regulate transporter number at the cell surface.
A second possible explanation for the difference in trafficking between HEK 293T cells and mouse neurons is that HEK 293T cells do not express the necessary collection of associating proteins to regulate GAT1 trafficking appropriately. For example, protein interacting with C kinase 1 (PICK1), a PDZ protein involved in GAT1 plasma membrane trafficking, is expressed in N2a cells but hardly detectable in HEK 293 cells (Bjerggaard et al., 2004). However, HEK 293T cells appear to process and traffic mGAT10GFP like wild-type mGAT1, suggesting that this cell type does not rely on PDZ interactions to deliver GAT1 to or stabilize its presence at the plasma membrane. Rather, HEK 293T cells may use distinct pathways for these processes. In fact, hNET mutants with truncated C termini function like wild-type transporter in HEK 293 cells (from which HEK 293T is derived), although in many other cell lines, truncation mutants exhibit reduced substrate uptake (Bauman and Blakely, 2002).
HEK 293T cells were valuable for pioneering heterologous expression studies of excitability proteins (Lester, 1988) and still serve well when one desires sheer quantity rather than faithful processing, assembly, sorting, regulation, and targeting. However, our studies show that N2a cells more closely resemble the native neuronal environment of mGAT1. We developed a transfection protocol and uptake assay in the N2a cell line that remained in the linear range with respect to protein expression level, substrate concentration, and assay duration. We chose to work with the N2a cell line because its provenance closely matches that of the mGAT1 protein that we were studying: mouse and neuronal. We cannot claim perfect fidelity, but several influences on expression, oligomerization, and trafficking are readily reflected by changed function under these assay conditions.
Conclusion
We have determined standardized designs for fluorescent transporter fusions, applied a sensitized emission pixel-by-pixel NFRET analysis method, and measured the radial distribution of NFRET amplitudes to investigate the basic and high-order oligomerization and peripheral interactions of the mGAT1 transporter. Each of these important events is reported as a Gaussian component of the all-pixel NFRET amplitude distribution. These techniques might also reveal the basic and high-order oligomerization of other SLC6 transporter members (Kilic and Rudnick, 2000; Hastrup et al., 2003; Just et al., 2004; Bartholomäus et al., 2008) and other multi-subunit plasma membrane proteins (Raicu et al., 2009; Son et al., 2009). Future investigations will study the dynamic changes in the interactions of transporters with regulatory and scaffolding partners in various subcellular compartments, and will examine critical events in transporter modulation, such as the recently reported substrate-mediated dissociation of transporter oligomers (Chen and Reith, 2008).
APPENDIX: FRET FOR DIMERS AND TETRAMERS
We assume that the fluorescent mGAT1 constructs with the most wild-type function most accurately report the normal oligomerization properties of mGAT1 via FRET. The lowest-order oligomer is a dimer. If we assume that CFP- and YFP-tagged fluorescent mGAT1 protomers are synthesized and fold equally efficiently, in a cell transfected by equimolar amounts of the donor and acceptor cDNAs, a binomial distribution of donor-donor–, donor-acceptor–, and acceptor-acceptor–tagged dimers will exist (Fig. 18 A). Only 50% of the total oligomer population, the donor-acceptor dimers, will contribute to the NFRET amplitude. This means that the experimentally measured NFRET amplitude for a specific pixel is not the true “pair-wise” FRET data of a single CFP-YFP pair, but is the “apparent” FRET of a specific concentration of the FRET-yielding dimers, mixed with non-FRET productive dimers, and in some organelles donor or acceptor monomers (Raicu, 2007). In oligomers larger than dimers, there is more than one pathway for nonradiative energy transfer (Fig. 18, B and C). The apparent FRET in each pixel is influenced by:
Figure 18.
Theory of FRET from three oligomer types with several fluorophore stoichiometries. (A) XFP-tagged mGAT1 protomers can assemble in three possible fluorophore stoichiometries. (B) Square tetramers can have six possible fluorophore configurations. (C) There are nine possible configurations of a rhomboid tetramer. Nomenclature for panels A–C: D, donor; A, acceptor; ExD:yA, FRET efficiency for each configuration where x and y represent the number of donor or acceptor fluorophores, respectively; R0, the Förster distance for a CFP-YFP pair; r1, the pair-wise FRET radius in the basic mGAT1 dimer; r2-r4, the three other pair-wise FRET radii present in either square tetramers or rhomboid tetramers. (D) Plot of the apparent FRET (EApp) modeled for mGAT1 dimers, square tetramers, or rhomboid tetramers with changing r1, the pair-wise FRET radius in the basic mGAT1 dimer.
(a) The number of donor-acceptor pair radii in the oligomers (e.g., two possible FRET radii exist in a square tetramer; r and .
(b) The stoichiometry of the donor and acceptor fluorophores within each oligomer.
(c) The relative concentration of each oligomer stoichiometry type.
(d) The proportions of dimers versus higher-order oligomers that exist in the pixel.
We considered two possible forms of such an mGAT1 tetramer. The square tetramer would resemble the arrangement of subunits in voltage-gated potassium channels (Kerschensteiner et al., 2005; Miranda et al., 2008) (Fig. 18 B). A rhomboid organization (Fig. 18 C) was also considered because mGAT1 molecules function as monomers; therefore, an mGAT1 oligomer might not display the approximate fourfold radial symmetry that constrains voltage-gated channels (Scholze et al., 2002; Yamashita et al., 2005; Zhang and Rudnick, 2006). Fig. 18 D shows a model of the apparent FRET efficiencies that would arise from populations of mGAT1 dimers, square tetramers, or rhomboid tetramers for various distances between fluorophores. To set the initial value for the closest distance between protomers r1, we used the LeuTAa structure (PDB entry 2A65A; Yamashita et al., 2005). We assumed that the fluorophores in the mGAT1 C terminus hang directly below the substrate molecule in the protomer core. We determined the proportional values for r2–4 when we varied r1 (Fig. 18, A–C). The dimensions of the XFP barrel limit the smallest measurable distance for side-by-side chromophores in an oligomerized membrane protein to r1 = 24 Å (Ormö et al., 1996), and we assumed a largest value of r1 = 100 Å. The FRET efficiency for each tetramer configuration for each r1 was determined and multiplied by the probability of its occurrence to determine the apparent FRET, Eapp (Corry et al., 2005a). We also assumed that energy transfer always occurs in all possible FRET-yielding configurations (i.e., fluorophore bleaching or blinking was not factored into the calculations), with exactly equal representation of donor- and acceptor-tagged protomers in all compartments and equally efficient assembly of dimers and high-order oligomers.
The calculations show that, relative to Eapp from a dimer, a square tetramer and a rhomboid tetramer have 1.33- to 1.96-fold and 1.54- to 2.87-fold higher FRET, respectively, as r1 varies. For the majority of the constructs examined in this paper, the mean NFRET amplitude of the second Gaussian fit component was between 1.6- and 2.6-fold greater than for the lowest-amplitude first component.
We discuss the width of each component to the Gaussian fits to the mGAT1 NFRET distributions. Consider the simple example of a single pixel in which the only oligomer type possible is a dimer. The NFRET amplitude from that pixel is determined by the number of dimers that possess both a donor- and acceptor-tagged protomer, but the signal from these oligomers is normalized to the square root of the product of the pixel's donor and acceptor fluorophore intensities. Donor-donor dimers, acceptor-acceptor dimers, and non-oligomerized monomers can also exist in the pixel. Furthermore, immature, bleached, or misfolded XFP moieties may exist, all reducing the measured NFRET from donor-acceptor dimers (Fig. 18 A). If all pixels in an ROI contain only dimers, the fit to the NFRET amplitude distribution has a width determined by the variation in the number of FRET-yielding dimers relative to the total fluorophore concentration per pixel in the ROI. Some of these statistical considerations generate a binomial distribution; but given the additional effects, and the transformation from nF to NFRET, we assume a Gaussian distribution.
Now consider a pixel containing only mGAT1 tetramers. There are two major sources of variation in the NFRET amplitude: (1) the proportion of the total fluorophore concentration in the pixel that assembles into FRET-yielding tetramers, and (2) the relative contributions of each FRET-yielding donor and acceptor fluorophore stoichiometry in the pixel (Fig. 18, B and C). Tetramers have more sources of variation than dimers. Therefore, for an ROI whose pixels contain only tetramers, and at levels of donor and acceptor fluorophores equal to that of a pure dimer population, the NFRET distribution is fit by a single Gaussian of average amplitude greater than that for the dimer case; however, the full width at half-maximum exceeds that from the ROI containing only dimers. We observed throughout this work that the full width at half-maximum of the first low-amplitude NFRET component was less than for the second medium-amplitude component (Figs. 8 H, 10, D and H, 11 D, and 16 D). Given the presence of negative NFRET amplitudes, we think it unwise to convert the full width at half-maximum values to coefficients of variation.
Pixels are 69 nm2, but a pixel's optical signal also contains X, Y, and Z contributions from nearby regions, as appropriate to the microscope's point-spread function, therefore increasing the possible variability from a single pixel's signal. The signal from some or most pixels also arises from a mixed population of oligomerization states. Therefore, the NFRET amplitude is also determined by the numbers of the FRET-yielding dimers and tetramers and the fluorophore stoichiometries in the tetramers. For all the pixels from an ROI containing both dimers and tetramers, the NFRET distribution is fit with two overlapping Gaussian components, one for each oligomer population. The mean amplitudes of these two components represent pixels primarily but not purely containing one oligomerization state and as a result are shifted closer together than if the NFRET signal from the ROI came from pixels that contained either pure dimers or pure tetramers. Subcomponent overlap occurs because in pixels containing a heterogeneous population of dimers and tetramers, there are numerous possibilities where different mixtures of oligomer types and fluorophore stoichiometries result in the same pixel NFRET amplitude.
Acknowledgments
We thank Robert Chow, Cagdas Son, Rigo Pantoja, and Rahul Srinivasan for discussion. We thank Jo Ann Trinkle and Elisha Mackey for technical assistance. We also acknowledge Ryan Morin of Michael Smith Genome Sciences Centre, BC Cancer Agency Vancouver, BC, Canada for his assistance with the bioinformatics.
This research is supported by grants from the National Institutes of Health (grants DA-09121, DA-10509, and NS-11756). F.J. Moss received an American Heart Association Postdoctoral Fellowship. Some experiments used the facilities of the Millard and Muriel Jacobs Genetics and Genomics Laboratory at the California Institute of Technology.
Edward N. Pugh Jr. served as editor.
Footnotes
Abbreviations used in this paper:
- ACU
- arbitrary calibration unit
- CFP
- cyan fluorescent protein
- DMEM
- Dulbecco's modified Eagle's medium
- FRET
- Förster resonance energy transfer
- GABA
- γ-aminobutyric acid
- GFP
- green fluorescent protein
- HEK 293T
- human embryonic kidney T/17 cell line
- hGAT1
- human GAT1
- IQR
- interquartile range
- mGAT1
- mouse GAT1
- N2a
- neuroblastoma 2a
- nAChR
- nicotinic acetylcholine receptor
- NFRET
- normalized FRET
- PDZ
- postsynaptic density 95/discs large/zona occludens 1
- ROI
- region of interest
- SLC6
- solute carrier 6
- SV40
- simian virus 40
- TM
- transmembrane domain
- XFP
- cyan or yellow fluorescent proteins, collectively
- YFP
- yellow fluorescent protein
References
- Abramoff M.D., Magelhaes P.J., Ram S.J. 2004. Image processing with ImageJ. Biophotonics International. 11:36–42 [Google Scholar]
- Bal M., Zaika O., Martin P., Shapiro M.S. 2008a. Calmodulin binding to M-type K+ channels assayed by TIRF/FRET in living cells. J. Physiol. 586:2307–2320 10.1113/jphysiol.2008.152777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bal M., Zhang J., Zaika O., Hernandez C.C., Shapiro M.S. 2008b. Homomeric and heteromeric assembly of KCNQ (Kv7) K+ channels assayed by total internal reflection fluorescence/fluorescence resonance energy transfer and patch clamp analysis. J. Biol. Chem. 283:30668–30676 10.1074/jbc.M805216200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartholomäus I., Milan-Lobo L., Nicke A., Dutertre S., Hastrup H., Jha A., Gether U., Sitte H.H., Betz H., Eulenburg V. 2008. Glycine transporter dimers: evidence for occurrence in the plasma membrane. J. Biol. Chem. 283:10978–10991 10.1074/jbc.M800622200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman P.A., Blakely R.D. 2002. Determinants within the C-terminus of the human norepinephrine transporter dictate transporter trafficking, stability, and activity. Arch. Biochem. Biophys. 404:80–91 10.1016/S0003-9861(02)00232-1 [DOI] [PubMed] [Google Scholar]
- Bjerggaard C., Fog J.U., Hastrup H., Madsen K., Loland C.J., Javitch J.A., Gether U. 2004. Surface targeting of the dopamine transporter involves discrete epitopes in the distal C terminus but does not require canonical PDZ domain interactions. J. Neurosci. 24:7024–7036 10.1523/JNEUROSCI.1863-04.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackman S.M., Piston D.W., Beth A.H. 1998. Oligomeric state of human erythrocyte band 3 measured by fluorescence resonance energy homotransfer. Biophys. J. 75:1117–1130 10.1016/S0006-3495(98)77601-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudanova E., Navaroli D.M., Stevens Z., Melikian H.E. 2008. Dopamine transporter endocytic determinants: carboxy terminal residues critical for basal and PKC-stimulated internalization. Mol. Cell. Neurosci. 39:211–217 10.1016/j.mcn.2008.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd C., Hughes T., Pypaert M., Novick P. 2004. Vesicles carry most exocyst subunits to exocytic sites marked by the remaining two subunits, Sec3p and Exo70p. J. Cell Biol. 167:889–901 10.1083/jcb.200408124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brückner K., Pablo Labrador J., Scheiffele P., Herb A., Seeburg P.H., Klein R. 1999. EphrinB ligands recruit GRIP family PDZ adaptor proteins into raft membrane microdomains. Neuron. 22:511–524 10.1016/S0896-6273(00)80706-0 [DOI] [PubMed] [Google Scholar]
- Chen N., Reith M.E. 2008. Substrates dissociate dopamine transporter oligomers. J. Neurochem. 105:910–920 10.1111/j.1471-4159.2007.05195.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C.S., Kartalov E., Unger M., Quake S., Lester H.A. 2001. Single-molecule measurements calibrate green fluorescent protein surface densities on transparent beads for use with ‘knock-in' animals and other expression systems. J. Neurosci. Methods. 105:55–63 10.1016/S0165-0270(00)00354-X [DOI] [PubMed] [Google Scholar]
- Chiu C.S., Jensen K., Sokolova I., Wang D., Li M., Deshpande P., Davidson N., Mody I., Quick M.W., Quake S.R., Lester H.A. 2002. Number, density, and surface/cytoplasmic distribution of GABA transporters at presynaptic structures of knock-in mice carrying GABA transporter subtype 1-green fluorescent protein fusions. J. Neurosci. 22:10251–10266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole N.B., Smith C.L., Sciaky N., Terasaki M., Edidin M., Lippincott-Schwartz J. 1996. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 273:797–801 10.1126/science.273.5276.797 [DOI] [PubMed] [Google Scholar]
- Corry B., Jayatilaka D., Rigby P. 2005a. A flexible approach to the calculation of resonance energy transfer efficiency between multiple donors and acceptors in complex geometries. Biophys. J. 89:3822–3836 10.1529/biophysj.105.069351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corry B., Rigby P., Liu Z.W., Martinac B. 2005b. Conformational changes involved in MscL channel gating measured using FRET spectroscopy. Biophys. J. 89:L49–L51 10.1529/biophysj.105.072009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deken S.L., Wang D., Quick M.W. 2003. Plasma membrane GABA transporters reside on distinct vesicles and undergo rapid regulated recycling. J. Neurosci. 23:1563–1568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty G.J., McMahon H.T. 2009. Mechanisms of endocytosis. Annu. Rev. Biochem. 78:857–902 [DOI] [PubMed]
- Drenan R.M., Nashmi R., Imoukhuede P., Just H., McKinney S., Lester H.A. 2008. Subcellular trafficking, pentameric assembly, and subunit stoichiometry of neuronal nicotinic acetylcholine receptors containing fluorescently labeled α6 and β3 subunits. Mol. Pharmacol. 73:27–41 10.1124/mol.107.039180 [DOI] [PubMed] [Google Scholar]
- Elangovan M., Wallrabe H., Chen Y., Day R.N., Barroso M., Periasamy A. 2003. Characterization of one- and two-photon excitation fluorescence resonance energy transfer microscopy. Methods. 29:58–73 10.1016/S1046-2023(02)00283-9 [DOI] [PubMed] [Google Scholar]
- Farhan H., Korkhov V.M., Paulitschke V., Dorostkar M.M., Scholze P., Kudlacek O., Freissmuth M., Sitte H.H. 2004. Two discontinuous segments in the carboxyl terminus are required for membrane targeting of the rat γ-aminobutyric acid transporter-1 (GAT1). J. Biol. Chem. 279:28553–28563 10.1074/jbc.M307325200 [DOI] [PubMed] [Google Scholar]
- Farhan H., Freissmuth M., Sitte H.H. 2006. Oligomerization of neurotransmitter transporters: a ticket from the endoplasmic reticulum to the plasma membrane. Neurotransmitter Transporters. Sitte H.H., Freissmuth M., Springer, Berlin: 233–249 [DOI] [PubMed] [Google Scholar]
- Farhan H., Reiterer V., Korkhov V.M., Schmid J.A., Freissmuth M., Sitte H.H. 2007. Concentrative export from the endoplasmic reticulum of the γ-aminobutyric acid transporter 1 requires binding to SEC24D. J. Biol. Chem. 282:7679–7689 10.1074/jbc.M609720200 [DOI] [PubMed] [Google Scholar]
- Farhan H., Reiterer V., Kriz A., Hauri H.P., Pavelka M., Sitte H.H., Freissmuth M. 2008. Signal-dependent export of GABA transporter 1 from the ER-Golgi intermediate compartment is specified by a C-terminal motif. J. Cell Sci. 121:753–761 10.1242/jcs.017681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige J.N., Sage D., Wahli W., Desvergne B., Gelman L. 2005. PixFRET, an ImageJ plug-in for FRET calculation that can accommodate variations in spectral bleed-throughs. Microsc. Res. Tech. 68:51–58 10.1002/jemt.20215 [DOI] [PubMed] [Google Scholar]
- Gauthier N.C., Monzo P., Gonzalez T., Doye A., Oldani A., Gounon P., Ricci V., Cormont M., Boquet P. 2007. Early endosomes associated with dynamic F-actin structures are required for late trafficking of H. pylori VacA toxin. J. Cell Biol. 177:343–354 10.1083/jcb.200609061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser M., Cèbe R., Drewello D., Schmitz R. 2001. Integration of PCR fragments at any specific site within cloning vectors without the use of restriction enzymes and DNA ligase. Biotechniques. 31:88–90, 92 [DOI] [PubMed] [Google Scholar]
- Gordon G.W., Berry G., Liang X.H., Levine B., Herman B. 1998. Quantitative fluorescence resonance energy transfer measurements using fluorescence microscopy. Biophys. J. 74:2702–2713 10.1016/S0006-3495(98)77976-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guastella J., Nelson N., Nelson H., Czyzyk L., Keynan S., Miedel M.C., Davidson N., Lester H.A., Kanner B.I. 1990. Cloning and expression of a rat brain GABA transporter. Science. 249:1303–1306 10.1126/science.1975955 [DOI] [PubMed] [Google Scholar]
- Harris B.Z., Lim W.A. 2001. Mechanism and role of PDZ domains in signaling complex assembly. J. Cell Sci. 114:3219–3231 [DOI] [PubMed] [Google Scholar]
- Hastrup H., Sen N., Javitch J.A. 2003. The human dopamine transporter forms a tetramer in the plasma membrane: cross-linking of a cysteine in the fourth transmembrane segment is sensitive to cocaine analogs. J. Biol. Chem. 278:45045–45048 10.1074/jbc.C300349200 [DOI] [PubMed] [Google Scholar]
- Hess S.T., Gould T.J., Gudheti M.V., Maas S.A., Mills K.D., Zimmerberg J. 2007. Dynamic clustered distribution of hemagglutinin resolved at 40 nm in living cell membranes discriminates between raft theories. Proc. Natl. Acad. Sci. USA. 104:17370–17375 10.1073/pnas.0708066104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton K.L., Loder M.K., Melikian H.E. 2005. Nonclassical, distinct endocytic signals dictate constitutive and PKC-regulated neurotransmitter transporter internalization. Nat. Neurosci. 8:881–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J., Quick M.W. 2008. Substrate-mediated regulation of γ-aminobutyric acid transporter 1 in rat brain. Neuropharmacology. 54:309–318 10.1016/j.neuropharm.2007.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung A.Y., Sheng M. 2002. PDZ domains: structural modules for protein complex assembly. J. Biol. Chem. 277:5699–5702 10.1074/jbc.R100065200 [DOI] [PubMed] [Google Scholar]
- Imoukhuede P.I., Moss F.J., Michael D.J., Chow R.H., Lester H.A. 2009. Ezrin mediates tethering of the γ-aminobutyric acid transporter GAT1 to actin filaments via a C-terminal PDZ-interacting domain. Biophys. J. 96:2949–2960 10.1016/j.bpj.2008.11.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman E.A., Jovin T.M. 2006. Imaging molecular interactions in living cells by FRET microscopy. Curr. Opin. Chem. Biol. 10:409–416 10.1016/j.cbpa.2006.08.021 [DOI] [PubMed] [Google Scholar]
- Just H., Sitte H.H., Schmid J.A., Freissmuth M., Kudlacek O. 2004. Identification of an additional interaction domain in transmembrane domains 11 and 12 that supports oligomer formation in the human serotonin transporter. J. Biol. Chem. 279:6650–6657 10.1074/jbc.M306092200 [DOI] [PubMed] [Google Scholar]
- Kanner B.I. 2003. Transmembrane domain I of the γ-aminobutyric acid transporter GAT-1 plays a crucial role in the transition between cation leak and transport modes. J. Biol. Chem. 278:3705–3712 10.1074/jbc.M210525200 [DOI] [PubMed] [Google Scholar]
- Kanner B.I. 2006. Structure and function of sodium-coupled GABA and glutamate transporters. J. Membr. Biol. 213:89–100 10.1007/s00232-006-0877-5 [DOI] [PubMed] [Google Scholar]
- Kerschensteiner D., Soto F., Stocker M. 2005. Fluorescence measurements reveal stoichiometry of K+ channels formed by modulatory and delayed rectifier α-subunits. Proc. Natl. Acad. Sci. USA. 102:6160–6165 10.1073/pnas.0500468102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khakh B.S., Fisher J.A., Nashmi R., Bowser D.N., Lester H.A. 2005. An angstrom scale interaction between plasma membrane ATP-gated P2X2 and α4β2 nicotinic channels measured with fluorescence resonance energy transfer and total internal reflection fluorescence microscopy. J. Neurosci. 25:6911–6920 10.1523/JNEUROSCI.0561-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilic F., Rudnick G. 2000. Oligomerization of serotonin transporter and its functional consequences. Proc. Natl. Acad. Sci. USA. 97:3106–3111 10.1073/pnas.060408997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korkhov V.M., Farhan H., Freissmuth M., Sitte H.H. 2004. Oligomerization of the γ-aminobutyric acid transporter-1 is driven by an interplay of polar and hydrophobic interactions in transmembrane helix II. J. Biol. Chem. 279:55728–55736 10.1074/jbc.M409449200 [DOI] [PubMed] [Google Scholar]
- Kuryatov A., Luo J., Cooper J., Lindstrom J. 2005. Nicotine acts as a pharmacological chaperone to up-regulate human α4β2 acetylcholine receptors. Mol. Pharmacol. 68:1839–1851 [DOI] [PubMed] [Google Scholar]
- Lakowicz J.R. 2006. a. Energy transfer. Principles of Fluorescence Spectroscopy. Lakowicz J.R., editor Springer Science, New York: 443–476 [Google Scholar]
- Lakowicz J.R. 2006. b. Time-resolved energy transfer and conformational distributions of biopolymers. Principles of Fluorescence Spectroscopy. Lakowicz J.R., editor Springer Science, New York: 477–506 [Google Scholar]
- Lester H.A. 1988. Heterologous expression of excitability proteins: route to more specific drugs? Science. 241:1057–1063 10.1126/science.2457947 [DOI] [PubMed] [Google Scholar]
- Lester H.A., Cao Y., Mager S. 1996. Listening to neurotransmitter transporters. Neuron. 17:807–810 10.1016/S0896-6273(00)80213-5 [DOI] [PubMed] [Google Scholar]
- Li M., Farley R.A., Lester H.A. 2000. An intermediate state of the γ-aminobutyric acid transporter GAT1 revealed by simultaneous voltage clamp and fluorescence. J. Gen. Physiol. 115:491–508 10.1085/jgp.115.4.491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Gish G.D., Songyang Z., Pawson T. 1999. The carboxyl terminus of B class ephrins constitutes a PDZ domain binding motif. J. Biol. Chem. 274:3726–3733 10.1074/jbc.274.6.3726 [DOI] [PubMed] [Google Scholar]
- Loder M.K., Melikian H.E. 2003. The dopamine transporter constitutively internalizes and recycles in a protein kinase C-regulated manner in stably transfected PC12 cell lines. J. Biol. Chem. 278:22168–22174 10.1074/jbc.M301845200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo D.D., Eskandari S., Boorer K.J., Sarkar H.K., Wright E.M. 2000. Role of Cl− in electrogenic Na+-coupled cotransporters GAT1 and SGLT1. J. Biol. Chem. 275:37414–37422 10.1074/jbc.M007241200 [DOI] [PubMed] [Google Scholar]
- Madsen K.L., Beuming T., Niv M.Y., Chang C.W., Dev K.K., Weinstein H., Gether U. 2005. Molecular determinants for the complex binding specificity of the PDZ domain in PICK1. J. Biol. Chem. 280:20539–20548 10.1074/jbc.M500577200 [DOI] [PubMed] [Google Scholar]
- Mager S., Min C., Henry D.J., Chavkin C., Hoffman B.J., Davidson N., Lester H.A. 1994. Conducting states of a mammalian serotonin transporter. Neuron. 12:845–859 10.1016/0896-6273(94)90337-9 [DOI] [PubMed] [Google Scholar]
- Manley S., Gillette J.M., Patterson G.H., Shroff H., Hess H.F., Betzig E., Lippincott-Schwartz J. 2008. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat. Methods. 5:155–157 10.1038/nmeth.1176 [DOI] [PubMed] [Google Scholar]
- Mazei-Robison M.S., Blakely R.D. 2005. Expression studies of naturally occurring human dopamine transporter variants identifies a novel state of transporter inactivation associated with Val382Ala. Neuropharmacology. 49:737–749 10.1016/j.neuropharm.2005.08.012 [DOI] [PubMed] [Google Scholar]
- McHugh E.M., Zhu W., Milgram S., Mager S. 2004. The GABA transporter GAT1 and the MAGUK protein Pals1: interaction, uptake modulation, and coexpression in the brain. Mol. Cell. Neurosci. 26:406–417 10.1016/j.mcn.2004.03.006 [DOI] [PubMed] [Google Scholar]
- Melikian H.E., Buckley K.M. 1999. Membrane trafficking regulates the activity of the human dopamine transporter. J. Neurosci. 19:7699–7710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda M., Sorkina T., Grammatopoulos T.N., Zawada W.M., Sorkin A. 2004. Multiple molecular determinants in the carboxyl terminus regulate dopamine transporter export from endoplasmic reticulum. J. Biol. Chem. 279:30760–30770 10.1074/jbc.M312774200 [DOI] [PubMed] [Google Scholar]
- Miranda P., Manso D.G., Barros F., Carretero L., Hughes T.E., Alonso-Ron C., Domínguez P., de la Peña P. 2008. FRET with multiply labeled HERG K(+) channels as a reporter of the in vivo coarse architecture of the cytoplasmic domains. Biochim. Biophys. Acta. 1783:1681–1699 10.1016/j.bbamcr.2008.06.009 [DOI] [PubMed] [Google Scholar]
- Morel E., Parton R.G., Gruenberg J. 2009. Annexin A2-dependent polymerization of actin mediates endosome biogenesis. Dev. Cell. 16:445–457 10.1016/j.devcel.2009.01.007 [DOI] [PubMed] [Google Scholar]
- Munro S. 2004. Organelle identity and the organization of membrane traffic. Nat. Cell Biol. 6:469–472 10.1038/ncb0604-469 [DOI] [PubMed] [Google Scholar]
- Nashmi R., Dickinson M.E., McKinney S., Jareb M., Labarca C., Fraser S.E., Lester H.A. 2003. Assembly of α4β2 nicotinic acetylcholine receptors assessed with functional fluorescently labeled subunits: effects of localization, trafficking, and nicotine-induced upregulation in clonal mammalian cells and in cultured midbrain neurons. J. Neurosci. 23:11554–11567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashmi R., Xiao C., Deshpande P., McKinney S., Grady S.R., Whiteaker P., Huang Q., McClure-Begley T., Lindstrom J.M., Labarca C., et al. 2007. Chronic nicotine cell specifically upregulates functional α 4* nicotinic receptors: basis for both tolerance in midbrain and enhanced long-term potentiation in perforant path. J. Neurosci. 27:8202–8218 10.1523/JNEUROSCI.2199-07.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ormö M., Cubitt A.B., Kallio K., Gross L.A., Tsien R.Y., Remington S.J. 1996. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 273:1392–1395 10.1126/science.273.5280.1392 [DOI] [PubMed] [Google Scholar]
- Quick M.W. 2006. The role of SNARE proteins in trafficking and function of neurotransmitter transporters. Neurotransmitter Transporters. Sitte H.H., Freissmuth M., Springer, Berlin: 181–196 [DOI] [PubMed] [Google Scholar]
- Raicu V. 2007. Efficiency of resonance energy transfer in homo-oligomeric complexes of proteins. J. Biol. Phys. 33:109–127 10.1007/s10867-007-9046-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raicu V., Stoneman M.R., Fung R., Melnichuk M., Jansma D.B., Pisterzi L.F., Rath S., Fox M., Wells J.W., Saldin D.K. 2009. Determination of supramolecular structure and spatial distribution of protein complexes in living cells. Nat. Photonics. 3:107–113 10.1038/nphoton.2008.291 [DOI] [Google Scholar]
- Ramamoorthy S., Giovanetti E., Qian Y., Blakely R.D. 1998. Phosphorylation and regulation of antidepressant-sensitive serotonin transporters. J. Biol. Chem. 273:2458–2466 10.1074/jbc.273.4.2458 [DOI] [PubMed] [Google Scholar]
- Reiterer V., Maier S., Sitte H.H., Kriz A., Rüegg M.A., Hauri H.P., Freissmuth M., Farhan H. 2008. Sec24- and ARFGAP1-dependent trafficking of GABA transporter-1 is a prerequisite for correct axonal targeting. J. Neurosci. 28:12453–12464 10.1523/JNEUROSCI.3451-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith M.E., Zhen J., Chen N. 2006. The importance of company: Na+ and Cl− influence substrate interaction with SLC6 transporters and other proteins. Neurotransmitter Transporters. Sitte H.H., Freissmuth M., Springer, Berlin: 75–93 [DOI] [PubMed] [Google Scholar]
- Riven I., Iwanir S., Reuveny E. 2006. GIRK channel activation involves a local rearrangement of a preformed G protein channel complex. Neuron. 51:561–573 10.1016/j.neuron.2006.08.017 [DOI] [PubMed] [Google Scholar]
- Rizzo M.A., Springer G., Segawa K., Zipfel W.R., Piston D.W. 2006. Optimization of pairings and detection conditions for measurement of FRET between cyan and yellow fluorescent proteins. Microsc. Microanal. 12:238–254 10.1017/S1431927606060235 [DOI] [PubMed] [Google Scholar]
- Roux A., Uyhazi K., Frost A., De Camilli P. 2006. GTP-dependent twisting of dynamin implicates constriction and tension in membrane fission. Nature. 441:528–531 10.1038/nature04718 [DOI] [PubMed] [Google Scholar]
- Rudnick G. 2002. Mechanisms of biogenic amine neurotransmitter transporters. Neurotransmitter Transporters: Structure, Function, and Regulation. Reith M.E.A., editor Humana Press, Totowa, NJ: 25–52 [Google Scholar]
- Rudnick G. 2006. Structure/function relationships in serotonin transporter: new insights from the structure of a bacterial transporter. Neurotransmitter Transporters. Sitte H.H., Freissmuth M., Springer, Berlin: 59–73 [DOI] [PubMed] [Google Scholar]
- Sallette J., Pons S., Devillers-Thiery A., Soudant M., Prado de Carvalho L., Changeux J.P., Corringer P.J. 2005. Nicotine upregulates its own receptors through enhanced intracellular maturation. Neuron. 46:595–607 10.1016/j.neuron.2005.03.029 [DOI] [PubMed] [Google Scholar]
- Schmid J.A., Scholze P., Kudlacek O., Freissmuth M., Singer E.A., Sitte H.H. 2001. Oligomerization of the human serotonin transporter and of the rat GABA transporter 1 visualized by fluorescence resonance energy transfer microscopy in living cells. J. Biol. Chem. 276:3805–3810 10.1074/jbc.M007357200 [DOI] [PubMed] [Google Scholar]
- Scholze P., Freissmuth M., Sitte H.H. 2002. Mutations within an intramembrane leucine heptad repeat disrupt oligomer formation of the rat GABA transporter 1. J. Biol. Chem. 277:43682–43690 10.1074/jbc.M205602200 [DOI] [PubMed] [Google Scholar]
- Sekar R.B., Periasamy A. 2003. Fluorescence resonance energy transfer (FRET) microscopy imaging of live cell protein localizations. J. Cell Biol. 160:629–633 10.1083/jcb.200210140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M., Sala C. 2001. PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24:1–29 10.1146/annurev.neuro.24.1.1 [DOI] [PubMed] [Google Scholar]
- Son C.D., Moss F.J., Cohen B.N., Lester H.A. 2009. Nicotine normalizes intracellular subunit stoichiometry of nicotinic receptors carrying mutations linked to autosomal dominant nocturnal frontal lobe epilepsy. Mol. Pharmacol. 75:1137–1148 10.1124/mol.108.054494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soragna A., Bossi E., Giovannardi S., Pisani R., Peres A. 2005. Functionally independent subunits in the oligomeric structure of the GABA cotransporter rGAT1. Cell. Mol. Life Sci. 62:2877–2885 10.1007/s00018-005-5322-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorkina T., Doolen S., Galperin E., Zahniser N.R., Sorkin A. 2003. Oligomerization of dopamine transporters visualized in living cells by fluorescence resonance energy transfer microscopy. J. Biol. Chem. 278:28274–28283 10.1074/jbc.M210652200 [DOI] [PubMed] [Google Scholar]
- Spudich G., Fernández-Suárez X.M., Birney E. 2007. Genome browsing with Ensembl: a practical overview. Brief. Funct. Genomics Proteomics. 6:202–219 10.1093/bfgp/elm025 [DOI] [PubMed] [Google Scholar]
- Tadross M.R., Park S.A., Veeramani B., Yue D.T. 2009. Robust approaches to quantitative ratiometric FRET imaging of CFP/YFP fluorophores under confocal microscopy. J. Microsc. 233:192–204 10.1111/j.1365-2818.2008.03109.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J., Rowning B.A., Coughlin M.L., Wu M., Moon R.T., Mitchison T.J., Larabell C.A. 2000. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J. Cell Biol. 148:519–530 10.1083/jcb.148.3.519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Imamoto N., Sakata-Sogawa K. 2008. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 5:159–161 10.1038/nmeth1171 [DOI] [PubMed] [Google Scholar]
- Torres G.E., Carneiro A., Seamans K., Fiorentini C., Sweeney A., Yao W.D., Caron M.G. 2003. Oligomerization and trafficking of the human dopamine transporter. Mutational analysis identifies critical domains important for the functional expression of the transporter. J. Biol. Chem. 278:2731–2739 10.1074/jbc.M201926200 [DOI] [PubMed] [Google Scholar]
- Torres R., Firestein B.L., Dong H., Staudinger J., Olson E.N., Huganir R.L., Bredt D.S., Gale N.W., Yancopoulos G.D. 1998. PDZ proteins bind, cluster, and synaptically colocalize with Eph receptors and their ephrin ligands. Neuron. 21:1453–1463 10.1016/S0896-6273(00)80663-7 [DOI] [PubMed] [Google Scholar]
- Wallrabe H., Periasamy A. 2005. Imaging protein molecules using FRET and FLIM microscopy. Curr. Opin. Biotechnol. 16:19–27 10.1016/j.copbio.2004.12.002 [DOI] [PubMed] [Google Scholar]
- Wang D., Quick M.W. 2005. Trafficking of the plasma membrane γ-aminobutyric acid transporter GAT1. Size and rates of an acutely recycling pool. J. Biol. Chem. 280:18703–18709 10.1074/jbc.M500381200 [DOI] [PubMed] [Google Scholar]
- Whitworth T.L., Quick M.W. 2001. Substrate-induced regulation of γ-aminobutyric acid transporter trafficking requires tyrosine phosphorylation. J. Biol. Chem. 276:42932–42937 10.1074/jbc.M107638200 [DOI] [PubMed] [Google Scholar]
- Xia Z., Liu Y. 2001. Reliable and global measurement of fluorescence resonance energy transfer using fluorescence microscopes. Biophys. J. 81:2395–2402 10.1016/S0006-3495(01)75886-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita A., Singh S.K., Kawate T., Jin Y., Gouaux E. 2005. Crystal structure of a bacterial homologue of Na+/Cl−-dependent neurotransmitter transporters. Nature. 437:215–223 10.1038/nature03978 [DOI] [PubMed] [Google Scholar]
- Zacharias D.A., Violin J.D., Newton A.C., Tsien R.Y. 2002. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 296:913–916 10.1126/science.1068539 [DOI] [PubMed] [Google Scholar]
- Zahniser N.R., Doolen S. 2001. Chronic and acute regulation of Na+/Cl−-dependent neurotransmitter transporters: drugs, substrates, presynaptic receptors, and signaling systems. Pharmacol. Ther. 92:21–55 10.1016/S0163-7258(01)00158-9 [DOI] [PubMed] [Google Scholar]
- Zhang Y.W., Rudnick G. 2006. The cytoplasmic substrate permeation pathway of serotonin transporter. J. Biol. Chem. 281:36213–36220 10.1074/jbc.M605468200 [DOI] [PubMed] [Google Scholar]