Abstract
CD103+ dendritic cells (DCs) in nonlymphoid tissues are specialized in the cross-presentation of cell-associated antigens. However, little is known about the mechanisms that regulate the development of these cells. We show that two populations of CD11c+MHCII+ cells separated on the basis of CD103 and CD11b expression coexist in most nonlymphoid tissues with the exception of the lamina propria. CD103+ DCs are related to lymphoid organ CD8+ DCs in that they are derived exclusively from pre-DCs under the control of fms-like tyrosine kinase 3 (Flt3) ligand, inhibitor of DNA protein 2 (Id2), and IFN regulatory protein 8 (IRF8). In contrast, lamina propria CD103+ DCs express CD11b and develop independently of Id2 and IRF8. The other population of CD11c+MHCII+ cells in tissues, which is CD103−CD11b+, is heterogenous and depends on both Flt3 and MCSF-R. Our results reveal that nonlymphoid tissue CD103+ DCs and lymphoid organ CD8+ DCs derive from the same precursor and follow a related differentiation program.
DCs are the major antigen-presenting cell population involved in T cell priming. This is a complex group of cells that consists of several different subsets in the skin and lymphoid organs, each of which may be specialized for their role in response to different types of cellular, microbial, and protein antigens.
Consistent with this hypothesis, two functionally distinct subsets of classical DCs (cDCs) are present in the lymphoid organs of mice, including CD8+ and CD8− cDCs (Shortman and Liu, 2002). CD8+ cDCs are more potent at cross-presenting soluble and cell-associated antigens to CD8+ T cells when compared with CD8−CD11b+ cDCs (Heath et al., 2004). CD8+ cDCs preferentially prime CD8+ T cells, whereas CD8−CD11b+ cDCs are more efficient in processing and presenting antigen to CD4+ T cells (Dudziak et al., 2007).
The skin contains three DC compartments that consist of epidermal LCs and two phenotypically distinct cDC populations in the dermis (Bursch et al., 2007; Ginhoux et al., 2007; Poulin et al., 2007). These include the CD11c+CD11b+ cDC subset that is thought to be the main DC population in the dermis, as well as the recently identified CD103+langerin+CD11blocDC subset (Bursch et al., 2007; Ginhoux et al., 2007; Poulin et al., 2007; Nagao et al., 2009). CD103+ cDCs have also been identified in the lung (Sung et al., 2006) and in the intestine (Annacker et al., 2005). Dermal CD103+ cDCs resemble CD8+ cDCs in lymphoid organs in that both of these subsets excel in cross-presentation of cell-associated antigens (Bedoui et al., 2009) and also that deletion of the transcription factor Batf3 (basic leucine zipper transcription factor, ATF-like 3) eliminates dermal CD103+ cDCs and lymphoid organ CD8+ cDCs, whereas other DC subsets in these tissues remain unaffected (Hildner et al., 2008). Thus, CD8+ cDCs in lymphoid organs and dermal CD103+ cDCs appear to be related. Whether similar DC subsets exist in all nonlymphoid tissues and how they are related to cDCs and macrophages remains unclear.
Recent studies have identified the discrete cellular steps that characterize DC lineage commitment in the BM. The macrophage and DC precursor (MDP; Fogg et al., 2006) gives rise to monocytes and to the common DC precursor (CDP; Liu et al., 2009), which has lost monocyte/macrophage differential potential and gives rise exclusively to plasmacytoid DCs and lymphoid organ cDCs (Naik et al., 2007; Onai et al., 2007b). CDP also produces pre-cDC, a cDC-restricted progenitor that has lost plasmacytoid DC differentiation potential (Liu et al., 2009). Pre-cDCs circulate through the blood to localize in lymphoid organs where they differentiate into both lymphoid organ cDC subsets, including CD8+ and CD8− cDCs (Liu et al., 2009).
Thus, monocytes and DC lineages separate in the BM at the MDP stage (Liu et al., 2009). The DC-restricted lineage does not give rise to monocytes and macrophages, and monocytes are unable to give rise to lymphoid organ cDCs in the steady state (Fogg et al., 2006). Nevertheless, cultured monocytes can develop many of the phenotypic features of cDCs, including expression of CD11c and MHC class II (Banchereau et al., 2000), and monocytes can repopulate the Langerhans cell (LC) compartment in an inflamed epidermis (Ginhoux et al., 2006) and give rise to intestinal CX3CR1+ cDCs in the steady state (Bogunovic et al., 2009).
In this paper, we explore the mechanisms that control cDC homeostasis in nonlymphoid tissues in the steady state. We find that two phenotypically and developmentally distinct cDC populations coexist in most nonlymphoid tissues. Tissue CD103+ cDCs are related to lymphoid organ CD8+ cDCs and are derived from pre-cDCs, whereas tissue CD11b+ cDCs are heterogenous.
RESULTS
Two phenotypically distinct nonlymphoid tissue cDC subsets
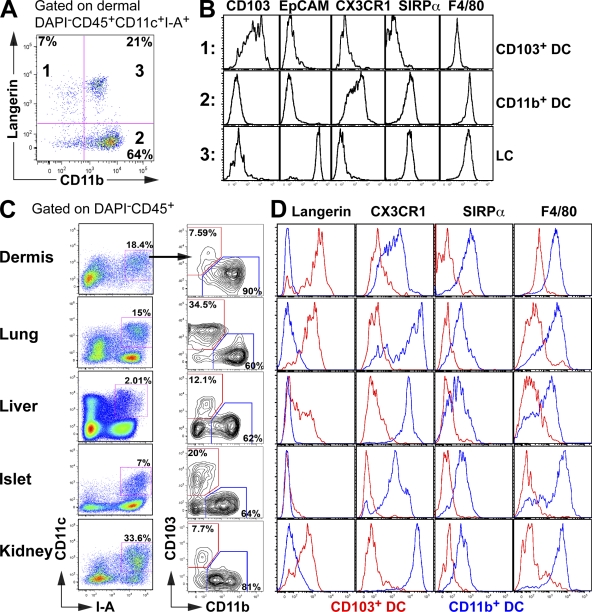
Langerin, which is highly expressed on the surface of epidermal LCs in mice and humans, was initially thought to be a specific marker for LCs in the skin (Merad et al., 2008). However, langerin is also expressed by a population of dermal cDCs, which migrates to the LN (Bursch et al., 2007; Ginhoux et al., 2007; Poulin et al., 2007), as well as by a subset of cDCs in the lungs (Sung et al., 2006). As shown in Fig. 1, dermal cDCs can be segregated into three populations that differentially express langerin and CD11b. One population expresses langerin, CD103 (Cepek et al., 1994), and low levels of CD11b but lacks several monocyte/macrophage markers including F4/80 (Austyn and Gordon, 1981), SIRP-α (van Beek et al., 2005), and CX3CR1 (Jung et al., 2000; CD103+ DC; Fig.1, A and B). The other DC subset lacks CD103 but expresses high levels of CD11b, CX3CR1, F4/80, and SIRP-α (CD11b+ DC; Fig.1, A and B). In addition, the dermis contains migratory LCs, which uniquely express epithelial cell adhesion molecule (EpCAM; Nagao et al., 2009; Fig. 1, A and B).
Figure 1.
Two major phenotypically distinct DC subsets coexist in the dermis, lung, liver, pancreatic islets, and kidney. (A and B) Dermal cell suspensions isolated from langerin/EGFP, CX3CR1/EGFP, and WT C57BL/6 mice were analyzed by flow cytometry. (A) Dot plot shows the expression of langerin and CD11b on gated DAPI−CD45+CD11c+I-A+ dermal DCs, allowing the discrimination of three DC subsets. (B) Histograms show the expression levels of CD103, EpCAM, CX3CR1, SIRP-α, and F4/80 on the gated population described in A. (C and D) Dermal, lung, liver, pancreatic islets, and kidney cell suspensions were isolated from langerin/EGFP, CX3CR1/EGFP, and WT C57BL/6 mice. (C) Dot plot shows the expression of CD103 versus CD11b (right) on gated nonlymphoid tissue DAPI−CD45+CD11c+I-A+ DCs (left). For dermal DCs analysis, DCs were also gated on EPCAM− to exclude LCs. (D) Histograms show overlays of cell surface marker expression on CD103+ (red) and CD11b+ (blue) DC subsets. These results are representative of three independent experiments (n = 2–3).
To determine whether similar cDC subsets also exist in nonlymphoid tissues other than the skin, we examined the phenotype of CD11c+MHCII+ cells in the lung, liver, pancreatic islets, and kidney. Like in the skin, cDCs in these tissues segregate into two major populations, which we refer to as CD103+ DCs (CD103+CD11bloCX3CR1−F4/80−SIRP-α−) and CD11b+ DCs (CD103−CD11bhiCX3CR1+F4/80+SIRP-α+; Fig. 1, A–D). The level of langerin expression on CD103+ cDCs varies among tissues and is absent in CD103+ cDCs in the pancreatic islets (Fig. 1, A–D). We conclude that two major subsets of cDCs exist in most mouse tissues. We have reported that two DC subsets, identified as CD103+ and CD103− cDCs, also exist in mouse lamina propria (Bogunovic et al., 2009). We recently showed that in contrast to most other nonlymphoid tissue, lamina propria CD103+ cDCs also express CD11b, F4/80, and SIRP-α (Fig. S1, A and B; Bogunovic et al., 2009). Intestinal cDCs also include a population of CD103+CD11b−CD8α+ cDCs that accumulate in Peyer's patches (Bogunovic et al., 2009).
Differential expression of fms-like tyrosine kinase 3 (Flt3) and the MCSF receptor (MCSF-R)
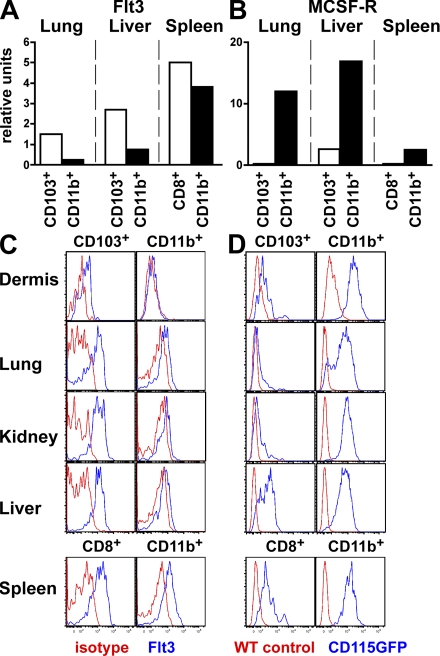
Expression of the receptor Flt3 is required for the development and maintenance of cDCs in lymphoid organs (Waskow et al., 2008). In contrast, MCSF-R is required for LCs to develop (Ginhoux et al., 2006) but is dispensable for the development of spleen cDCs (Witmer-Pack et al., 1993; Ginhoux et al., 2006). This differential cytokine requirement may in part reflect the distinct developmental origin of these two cell types. Therefore, we measured the expression of these two receptors by the different cDC subsets in nonlymphoid tissues. Flt3 messenger RNA (mRNA) and protein were expressed on both cDC subsets but at higher levels on CD103+ cDCs than CD11b+ cDCs in all tissues analyzed including the dermis, lung, kidney, and liver (Fig. 2, A and C). MCSF-R expression was measured using transgenic mice expressing GFP under the control of the MCSF-R promoter (Burnett et al., 2004) and confirmed by quantitative PCR. Although we did not detect MCSF-R protein expression by flow cytometry on cDCs and macrophages, this is likely to be a consequence of antigen degradation caused by the long tissue digestion process. In contrast to Flt3, MCSF-R was expressed at higher levels on CD11b+ cDCs compared with CD103+ cDCs in all tissues analyzed (Fig. 2, B and D). Similar differences were found among spleen CD8− and CD8+ cDCs, respectively, suggesting that these cells might be developmentally related (Fig. 2, A–D).
Figure 2.
Cytokine receptor expression profile among CD103+ and CD11b+ tissue DC subsets. (A and B) DC subsets were sorted from the lung, liver, and spleen of C57BL/6 mice and RNA expression was measured by quantitative RT-PCR. Bar graphs present the relative expression of Flt3 (A) and MCSF-R (B) in lung and liver CD103+ (white) and CD11b+ (black) DCs and spleen CD8+ (white) and CD8−CD11b+ (black) DCs. One representative experiment of three independent experiments is shown. (C and D) Dermal, lung, kidney, liver, and spleen cell suspensions were isolated from WT C57BL/6 and MCSF-R/EGFP mice and analyzed by flow cytometry. (C) Histograms show Flt3 cell surface expression (blue) or isotype antibody control (red) on gated CD103+ versus CD11b+ DC subsets in the dermis, lung, kidney, and liver and CD8+ versus CD8−CD11b+ DCs in the spleen. (D) Histograms show MCSF-R/EGFP expression (blue) or WT C57BL/6 control (red) on gated DC subsets in the same tissue. These results are representative of three independent experiments.
Flt3 is required for nonlymphoid tissue cDC homeostasis
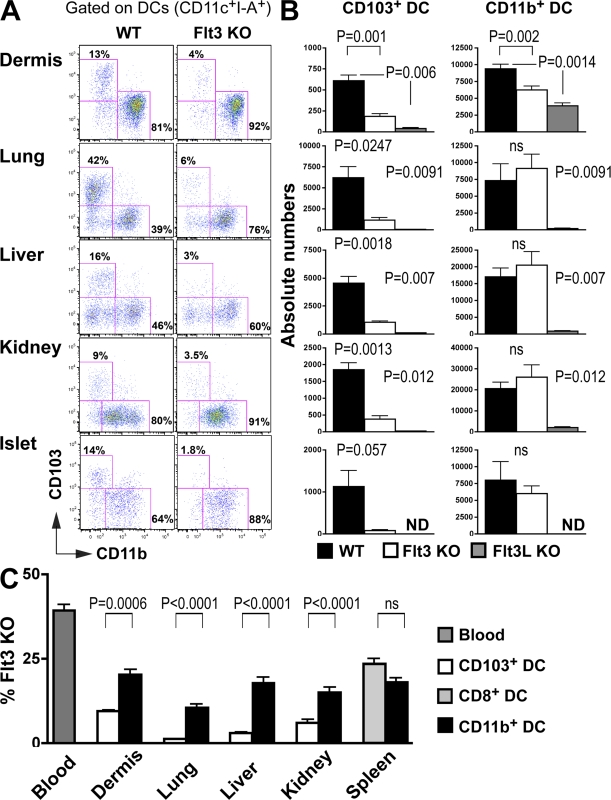
Although Flt3 and its ligand (Flt3L) are both essential for normal lymphoid organ DC development, the effects of Flt3L deletion are more pronounced, possibly as a result of the existence of a partially redundant receptor (Waskow et al., 2008). To examine the role of Flt3 in the development of nonlymphoid tissue cDCs, we measured CD103+ and CD11b+ tissue cDCs in mice that lack Flt3 or Flt3L (Fig. 3, A–C). The relative and absolute numbers of CD103+ cDCs were strongly reduced in mice that lack Flt3 or its ligand in all tissues analyzed including the dermis, lung, liver, and kidney (Fig. 3, B and C). CD11b+ cDCs were also strongly reduced in Flt3L-deficient mice (Fig. 3 C), whereas Flt3 deletion had little demonstrable effect (Fig. 3 C). In contrast, LCs do not express Flt3 and are unaffected in Flt3- or Flt3L-deficient mice (Fig. S2), whereas they were absent in mice that lack MCSF-R (Ginhoux et al., 2006), which is consistent with a distinct developmental origin for this cell type. These results are consistent with previous data showing that lung cDCs are strongly reduced in Flt3L-deficient mice, whereas tracheal cDCs are spared in these mice (Walzer et al., 2005). It seems likely that tracheal cDCs are closely related to epidermal LCs and therefore do not express Flt3.
Figure 3.
The development of tissue CD103+ DCs is dependent on Flt3 and Flt3L. (A–C) Nonlymphoid tissue DCs isolated from WT, Flt3 KO (A and B), and Flt3L KO (C) C57BL/6 mice were analyzed by flow cytometry. (A) Dot plots show CD103 and CD11b expression among gated DAPI−CD45+CD11c+I-A+ DCs in WT (left) and Flt3 KO mice (right). (B) Bar graphs show the absolute numbers of CD103+ and CD11b+ DC subsets among tissue DCs in WT (black) versus Flt3 KO (white) versus Flt3L (gray) mice. Bars represent data from six pooled experiments (n = 3–4). Error bars represent the mean ± SEM (n = 3). (C) Lethally irradiated CD45.1+ recipient mice were reconstituted with 50% CD45.1+ WT and 50% CD45.2+ WT mixed BM or with 50% CD45.1+ WT and 50% CD45.2+ Flt3 KO mixed BM and analyzed 2–3 mo after. Bar graphs show the relative contribution of Flt3 KO CD45.2+CD103+ DCs (white) among total CD103+ DCs and the relative contribution of Flt3 KO CD45.2+CD11b+ DCs (black) among total CD11b+ DCs. Spleen CD8+ (light gray) and CD11b+ DC (black) subsets are also shown. Dark gray bars represent the relative number of Flt3 KO CD45.2+ blood granulocyte chimerism. Data shown represent two pooled experiments (n = 3). Error bars represent the means ± SEM. P-values indicate the results of a Student's t test performed between the indicated groups.
To confirm the role of Flt3 in nonlymphoid tissue cDC development, we produced mixed BM chimeras in CD45.1+ recipients reconstituted with a mixture of CD45.2+ Flt3−/− and CD45.1+ WT (50:50%) or control BM cells (Fig. S3 A and Fig. 3 D). As expected, Flt3−/− BM progenitors were less efficient than their WT counterparts in giving rise to all tissue DC subsets. However, the effect was far more pronounced on CD103+ than CD11b+ cDCs (Fig. 3 D).
MCSF-R is not required for the development of CD103+ cDCs
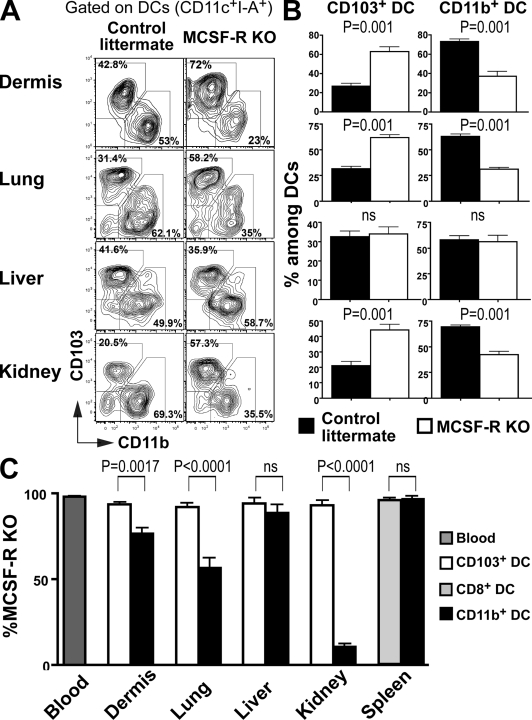
To determine whether MCSF-R mRNA expression correlates with a requirement for this cytokine for cDC development, we examined whether the absence of MCSF-R differentially affects the development of nonlymphoid tissue cDC populations. MCSF-R−/− mice have several defects, including severe osteopetrosis and tissue trophic defects, that may indirectly affect tissue cDC development (Dai et al., 2002). Nevertheless, we were able to measure CD103+ and CD11b+ cDCs in mice that lack MCSF-R. Consistent with the higher MCSF-R expression on CD11b+ cDCs (Fig. 2, B and D), lack of MCSF-R led to a developmental bias against CD11b+ cDCs. CD11b+ cDCs were partially reduced in the dermis, lung, and kidney but not in the liver, which is consistent with the moderate role of MCSF-R on liver macrophages (Dai et al., 2002; Fig. 4, A and B). In contrast, tissue CD103+ cDCs developed normally and remained unaffected in MCSF-R−/− mice (Fig. 4, A and B).
Figure 4.
MCSF-R is required for normal development of some nonlymphoid tissue CD11b+ DCs. (A) Dot plots show CD103 and CD11b expression among tissue CD45+CD11c+I-A+ DCs in MCSF-R KO (right) or control littermate (left) mice. (B) Bar graphs show the relative numbers of CD103+ and CD11b+ DC subsets among tissue DCs in control littermates (black) versus MCSF-R KO mice (white). Bars represent data from three pooled experiments (n = 2–3). Error bars represent the mean ± SEM. (C) Lethally irradiated CD45.1+ recipient mice were reconstituted with 5% CD45.1+ WT and 95% CD45.2+ WT fetal liver or 5% CD45.1+ WT and 95% CD45.2+ MCSF-R KO fetal liver. Bar graphs show the relative contribution of MCSF-R KO CD45.2+CD103+ DCs (white) among total CD103+ DCs and the relative contribution of MCSF-R KO CD45.2+CD11b+ DCs (black) among total CD11b+ DCs. Spleen CD8+ (light gray) and CD11b+ DC (black) subsets are also shown. Dark gray bars represent the percentage of MCSF-R KO CD45.2+ blood granulocytes chimerism. Data shown represent three pooled experiments (n = 3). Error bars represent the means ± SEM. P-values indicate the results of a Student's t test performed between the indicated groups.
To determine whether the role of MCSF-R in CD11b+ cDC development is cell intrinsic, we produced mixed BM chimeras by reconstituting CD45.1+ mice with CD45.2+ MCSF-R−/− fetal liver cells and CD45.1+ WT BM cells (95:5%) or control littermate fetal liver cells to favor the engraftment of CD45.2+ progenitors, which is compromised in the absence of MCSF-R (Fig. 4 C). Consistent with the observations in MCSF-R KO mice, MCSF-R−/− progenitors were less efficient than controls in producing CD11b+ cDCs in the dermis, lung, and kidney, whereas CD11b+ cDCs developed normally in the liver in the same mice (Fig. S3 B and Fig. 4 C). In contrast, MCSF-R−/− progenitors were as efficient as controls in giving rise to tissue CD103+ cDCs (Fig. 4 C). We conclude that MCSF-R plays a role in the development of CD11b+ cDCs in the dermis, lung, and kidney but not the liver. Furthermore, it is entirely dispensable for the differentiation of CD103+ DCs in these same tissues.
Nonlymphoid tissue CD103+ DCs share a similar developmental program than CD8+ lymphoid organ DCs
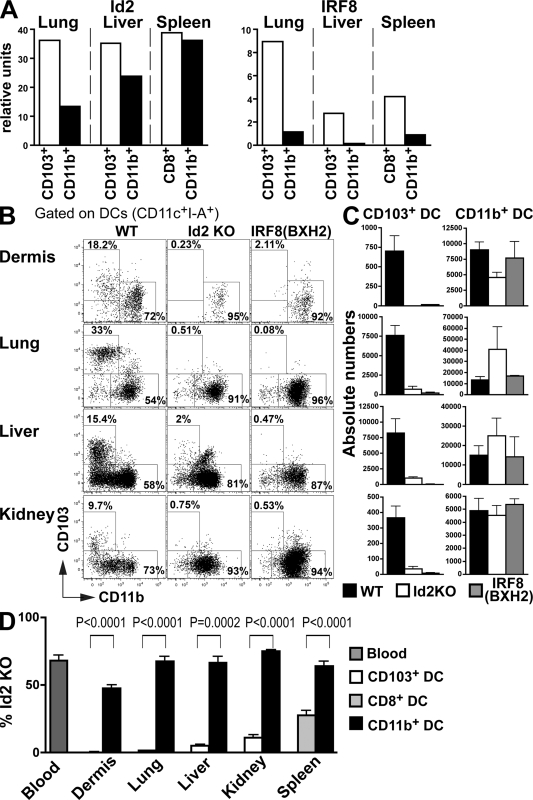
The cell surface phenotype of nonlymphoid tissue CD103+ cDCs closely resembles that of lymphoid tissue CD8+ cDCs. The transcription factor Batf3 is required for CD8+ cDCs in the spleen and dermal CD103+ cDCs but not for other cDC populations in those organs (Hildner et al., 2008). Both CD103+ and CD8+ cDCs excel in cross-presentation of cell-associated antigens (Bedoui et al., 2009), which suggest that these two subsets are closely related. Two other molecules have also been shown to control the development of spleen CD8+ cDCs, the inhibitor of DNA protein 2 (Id2; Hacker et al., 2003) and the IFN regulatory protein 8 (IRF8; Aliberti et al., 2003; Tailor et al., 2008).
Both Id2 and IRF8 are expressed at higher levels on CD103+ cDCs compared with CD11b+ cDCs in nonlymphoid tissues (Fig. 5 A). Consistent with differential expression of Id2, its absence blocked the development of CD103+ but only had a minor effect on CD11b+ cDCs in the dermis, lung, liver, and kidney (Fig. 5, B and C). To determine whether the effect of Id2 was cell autonomous, we produced mixed BM chimeras. In chimeric mice, Id2−/− BM failed to differentiate into CD103+ cDCs but produced CD11b+ cDCs (Fig. S3C and Fig. 5 D). Similar to Id2, absence of IRF8 is critical for the development of CD103+ cDCs but is dispensable for the development of CD11b+ cDCs (Fig. 5, B and C). Id2 and IRF8 were also required for the development of Peyer's patch CD103+CD11b−CD8+ cDCs but were dispensable for the development of lamina propria CD103+CD11b+ cDC (Fig. S1, C–F). Thus, in addition to controlling the development of lymphoid organ CD8+ cDCs, Id2 and IRF8 are also required for the differentiation of nonlymphoid CD103+ cDCs but not lamina propria CD103+ cDCs or tissue CD11b+ cDCs.
Figure 5.
The development of CD103+ DCs in tissues is also dependent on Id2 and IRF8. (A) Lung and liver CD103+ and CD11b+ DC subsets and spleen CD8+ and CD8−CD11b+ DCs were isolated from C57BL/6 mice and mRNA expression of Id2 and IRF8 was measured by quantitative RT-PCR. Bar graphs present the relative expression of Id2 (left) and IRF8 (right). One representative experiment of two independent experiments is shown. (B and C) Nonlymphoid tissue DCs isolated from WT, Id2 KO, and IRF8 (BXH2) C57BL/6 mice were analyzed by flow cytometry. (B) Dot plots show CD103 and CD11b expression among gated DAPI−CD45+CD11c+I-A+ DCs in WT (left), Id2 KO mice (middle), and IRF8 (BXH2; right). (C) Bar graphs show the absolute numbers of CD103+ and CD11b+ DC subsets among tissue DCs in WT (black), Id2 KO (white), and IRF8 (BXH2; gray) mice. Bars represent data from two pooled experiments (n = 2–3). Error bars represent the mean ± SEM. (D) Lethally irradiated CD45.1+ recipient mice were reconstituted with 50% CD45.1+ WT and 50% CD45.2+ WT mixed BM or with 50% CD45.1+ WT and 50% CD45.2+ Id2 KO mixed BM and analyzed 2–3 mo after. Bar graphs show the relative contribution of Id2 KO CD45.2+CD103+ DCs (white) among total CD103+ DCs and the relative contribution of Id2 KO CD45.2+CD11b+ DCs (black) among total CD11b+ DCs. Spleen CD8+ (light gray) and CD11b+ DC (black) subsets are also shown. Dark gray bars represent the percentage of Id2 KO CD45.2+ blood granulocytes chimerism. Data shown represent two pooled experiments (n = 3). Error bars represent means ± SEM. P-values indicate the results of a Student's t test performed between the indicated groups.
Turnover of nonlymphoid tissue DCs
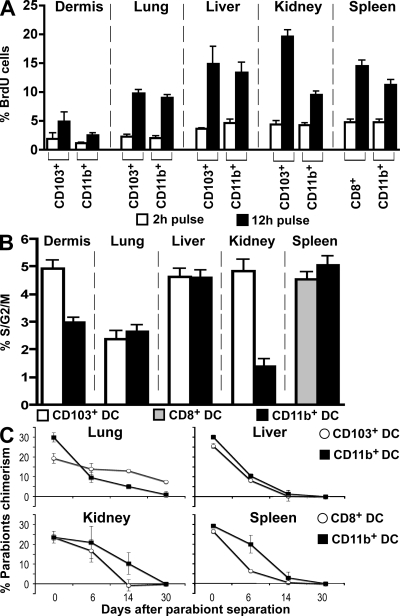
In the spleen and LNs, cDCs are dividing cells that live for 10–14 d (Liu et al., 2007). To determine whether nonlymphoid tissue cDCs are also dividing, we measured the cells’ BrdU uptake and cell cycle distribution in the dermis, lung, liver, and kidney. 12 h after BrdU injection, 5–10% of the cDCs in the dermis and lung, and 10–20% of the cDCs in the liver and kidney were labeled with BrdU (Fig. 6 A). In all tissues analyzed, the relative numbers of BrdU-labeled CD103+ cDCs and CD11b+ cDCs were similar, except in the kidney where the CD11b+ cDC subset was labeled to a lower level (Fig. 6 A). These results were confirmed by cell cycle analysis (Fig. 6 B). We conclude that both major cDC subsets in tissues are dividing in the steady state.
Figure 6.
Turnover of nonlymphoid tissue DC subsets. (A and B) BrdU incorporation and proliferation index of tissue DC subsets were measured by flow cytometry. Dermal, lung, liver, kidney, and spleen cell suspensions were isolated from WT C57BL/6 mice. (A) Bar graphs show the relative numbers of BrdU+ cells among CD103+ and CD11b+ DC subsets in nonlymphoid tissues and among CD8+ and CD8−CD11b+ spleen DCs after a 2-h (white) or a 12-h (black) BrdU pulse. Bars represent data from two pooled experiments (n = 3). Error bars represent the mean ± SEM. (B) Bar graph shows the percentage of proliferating cells (% S/G2/M) measured by DNA content among CD103+ (white) and CD11b+ (black) DC subsets in nonlymphoid tissues and among CD8+ (grey) and CD8−CD11b+ (black) spleen DCs. Bars represent data from three pooled experiments (n = 3). Error bars represent the mean ± SEM. (C) Tissue DC mixing in parabiotic mice. CD45.2 and CD45.1 C57BL/6 mice were surgically joined for 55 d before being separated. Graphs show the percentage of parabiont-derived DCs among each DC subset in the lung, liver, kidney, and spleen at different times after parabiont separation. Bars represent two separate parabionts at each time point. Error bars represent the mean ± SEM.
To examine the kinetics of cDC turnover in nonlymphoid tissues, we established parabiosis between mice expressing different CD45 alleles (Fig. 6 C). After 2 mo, the level of chimerism was 20% for CD103+ cDCs and 30% for CD11b+ cDCs, which is similar to the levels for CD8+ and CD8− cDCs in the spleen, respectively (Fig. 6 C; Liu et al., 2009). Chimerism in parabionts is a function of DC turnover in tissue, their rate of replacement by blood precursors, and the half-life of the precursors in the blood (Liu et al., 2007). To determine the rate of replacement of lymphoid tissue cDCs by blood precursors, we separated the parabionts after 2 mo and measured the decay of parabiont-derived cDCs in the lung, liver, kidney, and spleen. Parabiont-derived CD11b+ and CD103+ cDCs were lost in the 14 d after separation in all tissues analyzed (half-life 7 d; Fig. 6 C), except in the lung. Lung cDCs had a much longer half-life than cDCs in other tissues (Fig. 6 C). In addition, lung CD103+ cDCs had a much longer half-life than CD11b+ cDCs in the same tissue. 100% of the lung CD11b+ DCs was lost in 30 d after separation (half-life 15 d), whereas only 50% of parabiont-derived lung CD103+ cDCs was lost at that time (half-life 30 d; Fig. 6 C). We conclude that cDCs in nonlymphoid tissues undergo a limited number of divisions and must be continually replaced by blood-borne precursors.
Pre-DCs give rise to nonlymphoid tissue CD103+ cDCs
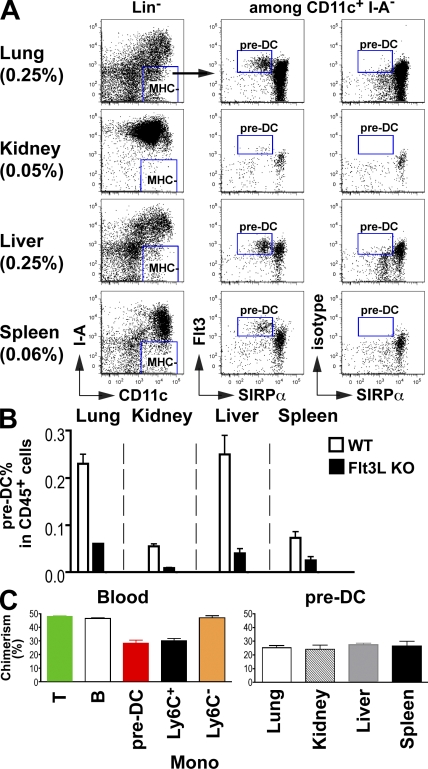
Despite their phenotypic and functional differences, both major subsets of cDCs in the spleen and LNs arise from a common progenitor, the pre-cDC, which is found in the BM, blood, and lymphoid organs (Liu et al., 2009). Little is known about the origin of cDCs in nonlymphoid tissues. However, several studies suggests that monocytes give rise to cDCs in nonlymphoid tissues (Geissmann et al., 2003; Yrlid et al., 2006; Varol et al., 2007; Jakubzick et al., 2008). To determine whether pre-cDCs might also give rise to nonlymphoid tissue cDCs, we looked for these cells in the lung, liver, and kidney (Fig. 7 A). We were unable to assess the presence of pre-cDCs in the dermis because of the low recovery yield of dermal cells. Pre-cDCs were present in each of these tissues and their accumulation in nonlymphoid tissues was strongly dependent on Flt3L (Fig. 7, A and B). In the absence of Flt3L, we found fewer pre-cDCs in all nonlymphoid organs, and this decrease in pre-cDC was also directly proportional to the decrease in both CD103+ and CD11b+ cDC populations (Fig. 7 C and Fig. 3 C). Parabiosis experiments established that pre-cDC chimerism was similar in the blood, liver, lung, kidney, and spleen and proportional to both CD103+ and CD11b+ cDC chimerism in these organs (Fig. 7 C). Therefore, in addition to migrating to lymphoid organs, pre-cDCs also migrate from the BM through the blood to nonlymphoid tissues.
Figure 7.
Pre-DCs are present in nonlymphoid tissues. (A) Dot plots show the relative numbers of CD45+MHCII+CD11c−Flt3hiSIRP-α− pre-DCs among hematopoietic cells in the lung, kidney, liver, and spleen. (B) Bar graph shows the relative numbers of pre-DCs in indicated organs of Flt3L+/+ (WT, white) and Flt3L−/− (black) mice. One representative experiment of three (A) and two (B) independent experiments is shown (n = 2). (C) CD45.2 and CD45.1 C57BL/6 mice were surgically joined for 55 d. Graphs show the percentage of parabiont-derived T cells, B cells, and pre-DCs in the blood and the percentage of parabiont-derived pre-DCs in the lung, kidney, liver, and spleen 55 d after parabiosis was established. The bar graphs summarize two independent experiments with more than three mice in total. Error bars represent the mean ± SEM.
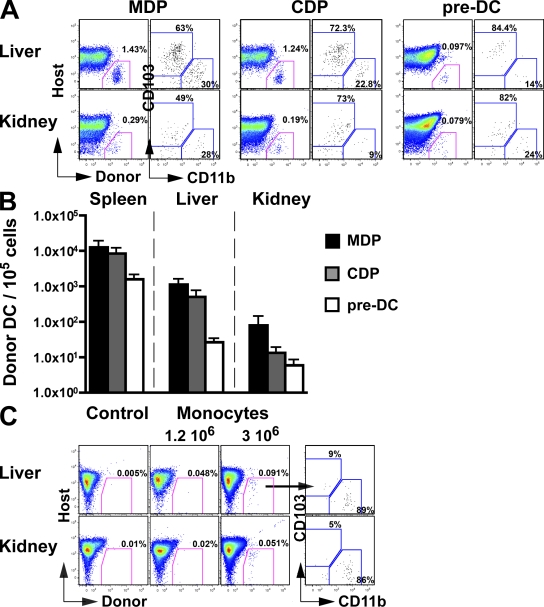
To determine the relative contribution of DC progenitors and monocytes to the development of nonlymphoid tissue cDCs, we injected pre-cDCs, monocytes, CDPs, and MDPs from C57BL/6 mice into untreated congenic recipient mice. We then measured the presence of donor-derived cDCs in nonlymphoid tissues 1 wk after adoptive transfer. Donor-derived cDCs were nearly undetectable in the lung and dermis, which limited our analysis to the liver and kidney (Fig. 8 and not depicted). As expected, MDPs, which produce monocytes and DCs, gave rise to CD103+ and CD11b+ cDCs (Fig. 8, A and B). CDPs, which produce DCs but not monocytes (Naik et al., 2007; Onai et al., 2007b), also gave rise to both CD103+ and CD11b+ cDCs (Fig. 8, A and B). The ratio of CD103+ to CD11b+ cDCs derived from CDPs was comparable to that obtained from MDPs (Fig. 8, A and B), which suggests that despite losing the ability to produce monocytes, CDP remained able to give rise to CD103+ and CD11b+ cDCs. Consistent with this idea, pre-DCs, which are restricted to producing cDCs, gave rise to both CD103+ and CD11b+ cDCs in the liver and kidney (Fig. 8, A and B). Monocytes were much less efficient in giving rise to cDCs in this assay. We were unable to detect monocyte-derived cDCs in four experiments when 106 monocytes were transferred but found small numbers of CD11b+ DCs and no CD103+ cDCs when we transferred 3 × 106 monocytes (Fig. 8 C). We conclude that nonlymphoid tissue CD103+ cDCs derive from pre-cDCs, whereas CD11b+ cDCs are heterogenous.
Figure 8.
Tissue DCs arise from pre-DCs in vivo. (A and B) MDPs, CDPs, and pre-DCs were purified from the BM of CD45.1+ mice, as described in the Material and methods, and adoptively transferred into unconditioned CD45.2+ congenic host. (A) Dot plots show the percentage of CD45.1+ donor-derived DCs among total kidney and liver DAPI−CD11c+MHCII+ DCs in mice injected with MDP, CDP, and pre-DCs. Host corresponds to the endogenous (CD45.2+) DC population. (B) Bar graph shows the mean absolute numbers of CD45.1+ donor-derived MHCII+CD11c+ DCs normalized to a 105 cells input of MDPs (black), CDPs (gray), and pre-DCs (white). Bars represent data from four pooled experiments (n = 1–2). Error bars represent the means ± SEM. (C) Monocytes were purified from the BM of CD45.2+ mice, as described in the Material and methods, and adoptively transferred into unconditioned CD45.1+ congenic host. Dot plot show the percentage of donor CD45.2+ monocyte-derived DCs among total kidney and liver DAPI−CD11c+MHCII+ DCs in mice injected with 1.2 × 106 or 3 × 106 monocytes. Host corresponds to the endogenous (CD45.1+) DC population. Control represents noninjected mice.
DISCUSSION
Our results show that two types of cDCs are present in most nonlymphoid tissues, with the exception of the lamina propria, and that different developmental programs regulate these cells. CD103+ cDCs resemble lymphoid tissue CD8+ cDCs in that they are critically dependent on Flt3L, Id2, and IRF8 and they are derived from pre-cDCs. In contrast, CD11b+ cDCs are dependent on both Flt3L and MCSF-R but are independent of Id2 and IRF8.
Flt3 and MCSF-R are differentially expressed by DCs and monocyte/macrophages (Pixley and Stanley, 2004; Onai et al., 2007a). As could be expected from this phenotypic difference, cDC development in lymphoid tissues is dependent on Flt3 and Flt3L, whereas monocyte/macrophage development depends on MCSF-R (Pixley and Stanley, 2004; Onai et al., 2007a). CD103+ cDCs express low levels of MCSF-R, as shown in MCSF-R reporter mice, they do not express macrophages markers, and they do not require MCSF-R for their development. Like CD8+ cDCs in lymphoid organs, CD103+ cDCs in tissues originate from pre-cDCs, not from monocytes, and are critically dependent on Flt3L.
Nonlymphoid tissue CD11b+ cDCs share several cell surface markers with macrophages and appear to be more closely related to monocyte/macrophages than to cDCs. They express F4/80, SIRP-α, CD11b, and CX3CR1. Consistent with these features, CD11b+ cDCs in the skin, lung, and kidney, including epidermal LCs (Ginhoux et al., 2006), are partially MCSF-R dependent. MCSF-R expression is gradually lost by cDCs as they develop from MDPs, CDPs, and pre-cDCs, but MCSF-R continues to be expressed in nonlymphoid tissue CD11b+ cDCs to a greater extent than in CD103+ cDCs. Despite these phenotypic differences, and their dependence on MCSF-R, some nonlymphoid tissue CD11b+ cDCs can be reconstituted by pre-DCs. Finally, both subsets of nonlymphoid tissue cDCs are also depend on Flt3L for their development.
Flt3 is expressed throughout the DC lineage (Naik et al., 2007; Onai et al., 2007b; Waskow et al., 2008; Liu et al., 2009) but it is strongly reduced in nonlymphoid tissue CD11b+ cDCs compared with CD103+ cDCs. Consistent with this difference in gene expression, the development of CD103+ cDCs is more strongly affected in Flt3- and Flt3L-deficient mice compared with CD11b+ cDCs, as was recently suggested by Kingston et al. (2009). The defect in both nonlymphoid tissue cDCs in Flt3L-deficient mice likely reflects the role of this cytokine in the commitment of early hematopoietic progenitors to the DC lineage (Christensen and Weissman, 2001; Karsunky et al., 2003; Naik et al., 2007; Onai et al., 2007b; Liu et al., 2009), whereas the more pronounced reduction of CD103+ cDCs in these mice may be a result of the additional role of Flt3L in the survival or proliferation of CD103+ cDCs in nonlymphoid tissues (Waskow et al., 2008).
Id2 is a dominant-negative inhibitor of E2A, a basic helix-loop-helix transcription factor that plays a key role in hematopoietic cell differentiation (Engel et al., 2001). The balance of Id2 and E2A is thought to influence lineage fate decision in hematopoiesis. For example, targeted mutation of Id2 inhibits NK cell development (Yokota et al., 1999), whereas targeted mutation of E2A inhibits B and T cell development (Engel et al., 2001). Id2−/− mice show normal tissue macrophages in all tissues analyzed (unpublished data), whereas they specifically lack CD8+, but not CD8−, cDCs in the spleen (Hacker et al., 2003) and CD103+ cDCs in nonlymphoid tissues. IRF-8/IFN consensus sequence-binding protein (ICSBP) is an IFN-γ–inducible transcription factor of the IRF family that controls myeloid cell differentiation (Gabriele and Ozato, 2007). BXH2 mice that carry a spontaneous point mutation in ICSBP1/IRF-8 (IRF8C294; Turcotte et al., 2005) lack CD8+ cDCs but not CD8− cDCs or plasmacytoid DCs in lymphoid organs (Tailor et al., 2008). In addition to Id2, IRF8 also plays a key role in the development of nonlymphoid tissue CD103+ but not CD11b+ cDCs. Importantly, Id2 and IRF8 also controlled the differentiation of CD103+CD11b−CD8+ Peyer's patch cDCs but failed to control the differentiation of CD103+CD11b+ lamina propria cDCs. These results further emphasize the common requirements for development and homeostasis of lymphoid CD8+ cDCs and most nonlymphoid organ CD103+ cDCs with the exception of lamina propria CD103+ cDCs.
Pre-cDCs develop in the BM and migrate though blood to seed lymphoid organs, where they give rise to CD8+ and CD8− cDCs (Liu et al., 2009). Our results revealed that pre-cDCs also migrate to nonlymphoid tissues, where they differentiate into CD103+ and CD11b+ cDCs with a development bias toward CD103+ cDCs. Both DC types divide in situ, but they have a limited number of divisions and their half-life in the tissues is variable, ranging from 7 d in the kidney and liver to 30 d in the lung.
Monocytes were unable to differentiate into nonlymphoid tissue CD103+ cDCs. However, they can produce lamina propria CD103−CD11b+ cDCs without contribution from CDPs or pre-cDCs (Bogunovic et al., 2009). In contrast, in the kidney and liver, CDPs and pre-cDCs can contribute to the CD11b+ cDC pool. Monocytes only gave rise to CD11b+ cDCs at low efficiency in the steady state, but we cannot rule out the possibility that poor monocyte survival in adoptive transfer is, in part, responsible for this effect. The development of genetic lineage tracing tools should resolve this issue. These results are consistent with previous studies showing that monocytes fail to give rise to lung CD11c+CD11b− cells (Landsman et al., 2007; Varol et al., 2007) but can produce lung CD11c+CD11b+ cells in the steady state. Although in these studies lung CD11c+CD11b− cells were originally called macrophages, it is likely that lung CD103+ cDCs were also included in this population. In addition, in these studies (Landsman et al., 2007; Varol et al., 2007) monocyte contribution to lung cDCs was revealed in mice depleted of cDCs using diphtheria toxin. In contrast, we have adoptively transferred monocytes only in naive untreated animals and it is possible that the poor monocyte contribution to the tissue DC pool observed in our study could be enhanced in mice depleted of endogenous cDCs.
Although less is known about the physiological function of DC subsets in nonlymphoid than lymphoid tissues, CD103+ and CD11b+ cDCs appear to be functionally specialized. CD103+ cDCs cross-present extracellular antigens to CD8+ T cells in skin (Bedoui et al., 2009), and in the lung they are required for optimal influenza virus responses (GeurtsvanKessel et al., 2008). These results are quite remarkable because CD103+ cDCs are only a minor fraction of the DCs in the dermis and in the lung, suggesting that CD103+ cDCs play a unique role in tissue immunity, which is consistent with their common origin with CD8+ lymphoid tissue cDCs.
In conclusion, our results identify two distinct populations of DCs in nonlymphoid tissues that originate from pre-cDCs. Like CD8+ cDCs in lymphoid tissues, nonlymphoid tissue CD103+ DCs, with the exception of lamina propria CD103+ cDCs, require Flt3L, Id2, and IRF8 for their differentiation and play a nonredundant and essential role in initiating CD8+ T cell responses.
MATERIALS AND METHODS
Mice.
C57BL/6 (CD45.2+) mice, congenic C57BL/6 CD45.1+ mice, MCSF-R/GFP C57BL/6 mice (macrophage fas-induced apoptosis; Burnett et al., 2004) and BXH2 C57BL/6J C3H/HeJ mice that carry a spontaneous point mutation (R294C) in the transcriptional regulator Icsbp1/IRF8 (Icsbp1C294; Turcotte et al., 2005) were purchased from The Jackson Laboratory. Flt3L−/− mice (McKenna et al., 2000) were purchased from Taconic. CX3CR1/GFP C57BL/6 mice (Jung et al., 2000) were provided by D. Littman (Skirball Institute, New York, NY), Langerin/GFP C57BL/6 and Langerin/DTR-GFP C57BL/6 mice (Kissenpfennig et al., 2005) were provided by B. Malissen (Centre d'Immunologie Marseille-Luminy, Marseille, France), and Flt3−/− C57BL/6 mice (Mackarehtschian et al., 1995) were provided by I. Lemischka (Mount Sinai School of Medicine, New York, NY). Id2−/− C57BL/6 (Yokota et al., 1999) were provided by Y. Yokota (University of Fukui, Fukui, Japan). MCSF-R−/− FVB/NJ mice (Dai et al., 2002) were produced as previously described and bred in our animal facility. All mice were analyzed between 8 and 12 wk of age, with the exception of MCSF-R−/− mice which were analyzed at 3 wk of age. All animal protocols were approved by the Institutional Committee on Animal Welfare of the Mount Sinai Medical School (New York, NY).
In vivo reconstitution assays.
In Flt3−/− and Id2−/− mixed BM chimeras, CD45.1+ C57BL/6 mice were lethally irradiated (2× 600 rad, 3 h apart using a Cesium source) and reconstituted with a 1:1 mixture of CD45.1+ WT BM and CD45.2+ BM isolated from WT or mutant mice. In MCSF-R−/− chimera, fetal liver cells were used as a source of MCSF-R−/− hematopoietic precursor cells because C57BL/6 MCSF-R−/− mice are embryonically lethal (Dai et al., 2002). Lethally irradiated CD45.1+ C57BL/6 mice received a mixture of MCSF-R−/− CD45.2+ fetal liver cells and syngeneic WT CD45.1+ BM (95: 5%) or MCSF-R+/+ CD45.2+ control littermates fetal liver cells and syngeneic WT CD45.1+ BM (95:5%). In all transplanted mice, engraftment was assessed by measuring the percentage of donor cells among blood Ly6C/G+ MCSF-R−/− granulocytes 3–4 wk after transplantation.
Cell suspensions preparation.
Skin cell suspensions were isolated as previously described (Ginhoux et al., 2007) and analyzed by flow cytometry. In brief, mouse ears were split in two (dorsal and ventral) parts and incubated for 60 min in HBSS containing 2.4 mg/ml Dispase (working activity of 1.7 U/mg; Invitrogen) to allow for separation of dermal and epidermal sheets. Epidermal and dermal sheets were then cut into small pieces, incubated for 2 h in HBSS containing 10% FBS and 0.2 mg/ml collagenase type IV (working activity of 770 U/mg; Sigma-Aldrich), and then passed trough a 19G syringe to obtain homogeneous cell suspension. Lung, liver, kidney, and pancreatic islets (isolated as previously described [Zhang et al., 2003]) were isolated, homogenized, incubated for 1 h in 10% FBS HBSS containing 0.2 mg/ml collagenase type IV (working activity of 770 U/mg), and passed trough a 19G syringe to obtain homogeneous cell suspension. Cell suspensions were then run either trough a room temperature NycoPrep gradient 1.077 (Axis-Shield; for lung and kidney) or a room temperature isotonic Percoll density (GE Healthcare; for liver) centrifugation to enrich the percentage of hematopoietic cells, as per the manufacturer's instructions. Lamina propria cell suspensions were prepared as described by Bogunovic et al. (2009). The analysis of hematopoietic cell populations present in tissue cell suspensions was assessed by flow cytometry by gating on singlets DAPI− (Invitrogen) CD45+ cells for extracellular staining or singlets CD45+ cells for intracellular langerin staining.
Flow cytometry and cell sorting.
Multiparameter analyses of stained cell suspensions were performed on an LSR II (BD) and analyzed with FlowJo software (Tree Star, Inc.). Fluorochrome- or biotin-conjugated mAbs specific to mouse B220 (clone RA3-6B2), CD8a (clone 53–6.7), IA/IE (clone M5/114.15.2), CD103 (clone 2E7), SIRP-α (clone P84), CD117 (clone 2B8), CD135 (clone A2F10), CD11b (clone M1/70), CD11c (clone N418), CD45 (clone 30F11), CD45.1 (clone A20), CD45.2 (clone 104), MCSF-R (also called CD115; clone AFS98), Gr-1Ly6C/G (clone RB6-8C5), CD3 (clone 17A2), and CD4 (clone L3T4), the corresponding isotype controls, and the secondary reagents (allophycocyanin, peridinin chlorophyll protein and phycoerythrin–indotricarbocyanine-conjugated streptavidin) were purchased either from BD or eBioscience. Anti-F4/80 (A3-1) mAb was purchased from AbD Serotec. Anti-EpCAM mAb (G8.8) was provided by M. Udey (National Institutes of Health, Bethesda, MD). Anti-langerin antibody (E-17) was purchased from Santa Cruz Biotechnology, Inc. Intracellular staining for langerin was performed with the Cytofix/Cytoperm kit (BD) according to the manufacturer's protocol. For quantitative RT-PCR assays, nonlymphoid tissue DC subsets were sorted using an inFlux cell sorter (BD) to achieve 98% purity.
BrdU incorporation and cell cycle assays.
C57BL/6 mice were injected intraperitoneally with 1 mg BrdU (Sigma-Aldrich) and euthanized after a 2- or 12-h BrdU pulse. Tissues were prepared for flow cytometry analysis as described in the previous section and intracellular staining for BrdU was performed with the BrdU Flow kit (BD) according to the manufacturer's protocol. For cell cycle assays, tissues were prepared from flow cytometry analysis as described in the previous section. Stained cell suspensions were fixed with the Cytofix/Cytoperm solution (BD), washed with Perm/Wash buffer solution (BD), and resuspended in Perm/Wash buffer solution containing 10 µg/ml RNase A and 20 µg/ml DAPI (Invitrogen) for overnight incubation.
Quantitative RT-PCR.
Total cellular RNA was extracted from the indicated DC subsets (CD11c+MHCII+CD103+ for CD103+ DCs and CD11c+MHCII+CD11b+ for CD11b+ DCs for liver and lung, CD11c+MHCII+CD8+ and CD11c+MHCII+CD4+CD11b+ for spleen) using the RNeasy Mini kit (QIAGEN) and reverse transcribed into complementary DNA using the SuperScript II RT kit (Invitrogen) according to the manufacturer's instructions. The samples were treated with DNase I (Roche), and mRNA was quantified by quantitative RT-PCR performed using the Express SYBR GreenER kit (Invitrogen) on an ABI PRISM 7900HT (Applied Biosystems). Relative expression was calculated as 2cycle threshold [Ct] control − Ct gene, using β-actin RNA as endogenous control. The following primer pairs were used: ID2 forward, 5′-TCCTGTCCTTGCAGGCATCTGAAT-3′; ID2 reverse, 5′-AACGTGTTCTCCTGGTGAAATGGC-3′; MCSFR forward, 5′-GCGCAAGCTTGAGTTTATCACCCA-3′; MCSFR reverse, 5′-ATATCGCAGGGTGAGCTCAAAGGT-3′; Flt3 forward, 5′-CGTGGATCTTCTCTCAAGCC-3′; Flt3 reverse, 5′-GAACTGGGCGTCATCATTTT-3′; IRF8 forward, 5′-GAGCGAAGTTCCTGAGATGG-3′; IRF8 reverse, 5′-TGGGCTCCTCTTGGTCATAC-3′; β-actin forward, 5′-TGACAGGATGCAGAAGGAGA-3′; and β-actin reverse, 5′-CGCTCAGGAGGAGCAATG-3′.
Adoptive transfer of DC precursors and monocytes.
For adoptive transfer experiment, MDPs, CDPs, and pre-DCs were preenriched by biotin anti-Flt3 antibody, followed by anti-biotin and anti-CD11c microbeads (Miltenyi Biotec), and positive fraction from columns (Miltenyi Biotec) was eluted, stained, and sorted on a FACs Aria (BD) to >95% purity as previously described (Liu et al., 2009). Gr1hi BM monocytes were preenriched by biotin anti-CD115 antibody, followed by anti-biotin microbeads (Miltenyi Biotec), and positive fraction was eluted, stained, and sorted based on the Gr1hiCD11b+CD115+ phenotype. Purified precursor cells (from 1 × 105 to 4 × 105) were injected intravenously and their contribution to nonlymphoid tissue DCs was analyzed 7–8 d after adoptive transfer.
Parabiosis.
Parabiosis and separation were done, as previously reported (Liu et al., 2007), with 5–6-wk-old male B6 and B6 CD45.1+ mice that were matched for body weight. For each pair of joined mice, mean chimerism was calculated as follows: (%CD45.1+ cells in CD45.2 mouse + %CD45.2+ cells in CD45.1 mouse)/2. For each separated parabiont, net DC chimerism was calculated as follows: %donor-derived DCs − %donor-derived hematopoietic stem cells.
Statistical analysis.
Mann-Whitney and unpaired Student's t tests (with a 95% confidence) were performed using Prism 4.0 (GraphPad Software, Inc.). All p-values are two-tailed.
Online Supplemental material.
Fig. S1 shows that lamina propria CD103+ DCs are distinct from most nonlymphoid tissue CD103+ DCs. Fig. S2 shows that epidermal LCs develop independently of Flt3 and Flt3L. Fig. S3 shows WT control chimerism for mixed chimera experiments. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091756/DC1.
Acknowledgments
We would like to thank Dr. Yoshifumi Yokota (University of Fukui, Fukui, Japan) for providing the Id2−/− mice. We thank Dr. Xiao-Hua Zong, Ranu Basu, and Marylene Leboeuf for technical assistance.
M. Merad was supported by National Institutes of Health grants CA112100 and CA086899. E.R. Stanley was supported by National Institutes of Health grants CA32551 and CA26504. F. Ginhoux is supported by a grant from the Leukemia and Lymphoma Society (LLS 3220-08). K. Liu was supported by a C.H. Li Memorial Scholarship Award from The Rockefeller University. This work was supported in part by grants from the National Institutes of Health to M. Nussenzweig. M. Nussenzweig is a Howard Hughes Medical Institute investigator.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- CDC
- classical DC
- CDP
- common DC precursor
- EpCAM
- epithelial cell adhesion molecule
- Flt3
- fms-like tyrosine kinase 3
- ICSBP
- IRF-8/IFN consensus sequence-binding protein
- Id2
- inhibitor of DNA protein 2
- IRF8
- IFN regulatory protein 8
- LC
- Langerhans cell
- MDP
- macrophage and DC precursor
- mRNA
- messenger RNA
References
- Aliberti J., Schulz O., Pennington D.J., Tsujimura H., Reis e Sousa C., Ozato K., Sher A. 2003. Essential role for ICSBP in the in vivo development of murine CD8alpha + dendritic cells. Blood. 101:305–310 10.1182/blood-2002-04-1088 [DOI] [PubMed] [Google Scholar]
- Annacker O., Coombes J.L., Malmstrom V., Uhlig H.H., Bourne T., Johansson-Lindbom B., Agace W.W., Parker C.M., Powrie F. 2005. Essential role for CD103 in the T cell–mediated regulation of experimental colitis. J. Exp. Med. 202:1051–1061 10.1084/jem.20040662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austyn J.M., Gordon S. 1981. F4/80, a monoclonal antibody directed specifically against the mouse macrophage. Eur. J. Immunol. 11:805–815 10.1002/eji.1830111013 [DOI] [PubMed] [Google Scholar]
- Banchereau J., Briere F., Caux C., Davoust J., Lebecque S., Liu Y.J., Pulendran B., Palucka K. 2000. Immunobiology of dendritic cells. Annu. Rev. Immunol. 18:767–811 10.1146/annurev.immunol.18.1.767 [DOI] [PubMed] [Google Scholar]
- Bedoui S., Whitney P.G., Waithman J., Eidsmo L., Wakim L., Caminschi I., Allan R.S., Wojtasiak M., Shortman K., Carbone F.R., et al. 2009. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 10:488–495 10.1038/ni.1724 [DOI] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnett S.H., Kershen E.J., Zhang J., Zeng L., Straley S.C., Kaplan A.M., Cohen D.A. 2004. Conditional macrophage ablation in transgenic mice expressing a Fas-based suicide gene. J. Leukoc. Biol. 75:612–623 10.1189/jlb.0903442 [DOI] [PubMed] [Google Scholar]
- Bursch L.S., Wang L., Igyarto B., Kissenpfennig A., Malissen B., Kaplan D.H., Hogquist K.A. 2007. Identification of a novel population of Langerin+ dendritic cells. J. Exp. Med. 204:3147–3156 10.1084/jem.20071966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cepek K.L., Shaw S.K., Parker C.M., Russell G.J., Morrow J.S., Rimm D.L., Brenner M.B. 1994. Adhesion between epithelial cells and T lymphocytes mediated by E-cadherin and the alpha E beta 7 integrin. Nature. 372:190–193 10.1038/372190a0 [DOI] [PubMed] [Google Scholar]
- Christensen J.L., Weissman I.L. 2001. Flk-2 is a marker in hematopoietic stem cell differentiation: a simple method to isolate long-term stem cells. Proc. Natl. Acad. Sci. USA. 98:14541–14546 10.1073/pnas.261562798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X.M., Ryan G.R., Hapel A.J., Dominguez M.G., Russell R.G., Kapp S., Sylvestre V., Stanley E.R. 2002. Targeted disruption of the mouse colony-stimulating factor 1 receptor gene results in osteopetrosis, mononuclear phagocyte deficiency, increased primitive progenitor cell frequencies, and reproductive defects. Blood. 99:111–120 10.1182/blood.V99.1.111 [DOI] [PubMed] [Google Scholar]
- Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.W., Park C.G., et al. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science. 315:107–111 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- Engel I., Johns C., Bain G., Rivera R.R., Murre C. 2001. Early thymocyte development is regulated by modulation of E2A protein activity. J. Exp. Med. 194:733–745 10.1084/jem.194.6.733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., Geissmann F. 2006. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 311:83–87 10.1126/science.1117729 [DOI] [PubMed] [Google Scholar]
- Gabriele L., Ozato K. 2007. The role of the interferon regulatory factor (IRF) family in dendritic cell development and function. Cytokine Growth Factor Rev. 18:503–510 10.1016/j.cytogfr.2007.06.008 [DOI] [PubMed] [Google Scholar]
- Geissmann F., Jung S., Littman D.R. 2003. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 19:71–82 10.1016/S1074-7613(03)00174-2 [DOI] [PubMed] [Google Scholar]
- GeurtsvanKessel C.H., Willart M.A., van Rijt L.S., Muskens F., Kool M., Baas C., Thielemans K., Bennett C., Clausen B.E., Hoogsteden H.C., et al. 2008. Clearance of influenza virus from the lung depends on migratory langerin+CD11b− but not plasmacytoid dendritic cells. J. Exp. Med. 205:1621–1634 10.1084/jem.20071365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Tacke F., Angeli V., Bogunovic M., Loubeau M., Dai X.M., Stanley E.R., Randolph G.J., Merad M. 2006. Langerhans cells arise from monocytes in vivo. Nat. Immunol. 7:265–273 10.1038/ni1307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F., Collin M.P., Bogunovic M., Abel M., Leboeuf M., Helft J., Ochando J., Kissenpfennig A., Malissen B., Grisotto M., et al. 2007. Blood-derived dermal langerin+ dendritic cells survey the skin in the steady state. J. Exp. Med. 204:3133–3146 10.1084/jem.20071733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker C., Kirsch R.D., Ju X.S., Hieronymus T., Gust T.C., Kuhl C., Jorgas T., Kurz S.M., Rose-John S., Yokota Y., Zenke M. 2003. Transcriptional profiling identifies Id2 function in dendritic cell development. Nat. Immunol. 4:380–386 10.1038/ni903 [DOI] [PubMed] [Google Scholar]
- Heath W.R., Belz G.T., Behrens G.M., Smith C.M., Forehan S.P., Parish I.A., Davey G.M., Wilson N.S., Carbone F.R., Villadangos J.A. 2004. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol. Rev. 199:9–26 10.1111/j.0105-2896.2004.00142.x [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C., Tacke F., Ginhoux F., Wagers A.J., van Rooijen N., Mack M., Merad M., Randolph G.J. 2008. Blood monocyte subsets differentially give rise to CD103+ and CD103- pulmonary dendritic cell populations. J. Immunol. 180:3019–3027 [DOI] [PubMed] [Google Scholar]
- Jung S., Aliberti J., Graemmel P., Sunshine M.J., Kreutzberg G.W., Sher A., Littman D.R. 2000. Analysis of fractalkine receptor CX(3)CR1 function by targeted deletion and green fluorescent protein reporter gene insertion. Mol. Cell. Biol. 20:4106–4114 10.1128/MCB.20.11.4106-4114.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsunky H., Merad M., Cozzio A., Weissman I.L., Manz M.G. 2003. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198:305–313 10.1084/jem.20030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston D., Schmid M.A., Onai N., Obata-Onai A., Baumjohann D., Manz M.G. 2009. The concerted action of GM-CSF and Flt3-ligand on in vivo dendritic cell homeostasis. Blood. 114:835–843 10.1182/blood-2009-02-206318 [DOI] [PubMed] [Google Scholar]
- Kissenpfennig A., Henri S., Dubois B., Laplace-Builhé C., Perrin P., Romani N., Tripp C.H., Douillard P., Leserman L., Kaiserlian D., et al. 2005. Dynamics and function of Langerhans cells in vivo: dermal dendritic cells colonize lymph node areas distinct from slower migrating Langerhans cells. Immunity. 22:643–654 10.1016/j.immuni.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Landsman L., Varol C., Jung S. 2007. Distinct differentiation potential of blood monocyte subsets in the lung. J. Immunol. 178:2000–2007 [DOI] [PubMed] [Google Scholar]
- Liu K., Waskow C., Liu X., Yao K., Hoh J., Nussenzweig M. 2007. Origin of dendritic cells in peripheral lymphoid organs of mice. Nat. Immunol. 8:578–583 10.1038/ni1462 [DOI] [PubMed] [Google Scholar]
- Liu K., Victora G.D., Schwickert T.A., Guermonprez P., Meredith M.M., Yao K., Chu F.F., Randolph G.J., Rudensky A.Y., Nussenzweig M. 2009. In vivo analysis of dendritic cell development and homeostasis. Science. 324:392–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackarehtschian K., Hardin J.D., Moore K.A., Boast S., Goff S.P., Lemischka I.R. 1995. Targeted disruption of the flk2/flt3 gene leads to deficiencies in primitive hematopoietic progenitors. Immunity. 3:147–161 10.1016/1074-7613(95)90167-1 [DOI] [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B., et al. 2000. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 95:3489–3497 [PubMed] [Google Scholar]
- Merad M., Ginhoux F., Collin M. 2008. Origin, homeostasis and function of Langerhans cells and other langerin-expressing dendritic cells. Nat. Rev. Immunol. 8:935–947 10.1038/nri2455 [DOI] [PubMed] [Google Scholar]
- Nagao K., Ginhoux F., Leitner W.W., Motegi S., Bennett C.L., Clausen B.E., Merad M., Udey M.C. 2009. Murine epidermal Langerhans cells and langerin-expressing dermal dendritic cells are unrelated and exhibit distinct functions. Proc. Natl. Acad. Sci. USA. 106:3312–3317 10.1073/pnas.0807126106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O'Keeffe M., Bahlo M., Papenfuss A., et al. 2007. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 8:1217–1226 10.1038/ni1522 [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Manz M.G. 2007a. Flt3 in regulation of type I interferon-producing cell and dendritic cell development. Ann. N. Y. Acad. Sci. 1106:253–261 10.1196/annals.1392.015 [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Ohteki T., Jarrossay D., Manz M.G. 2007b. Identification of clonogenic common Flt3+M-CSFR+ plasmacytoid and conventional dendritic cell progenitors in mouse bone marrow. Nat. Immunol. 8:1207–1216 10.1038/ni1518 [DOI] [PubMed] [Google Scholar]
- Pixley F.J., Stanley E.R. 2004. CSF-1 regulation of the wandering macrophage: complexity in action. Trends Cell Biol. 14:628–638 10.1016/j.tcb.2004.09.016 [DOI] [PubMed] [Google Scholar]
- Poulin L.F., Henri S., de Bovis B., Devilard E., Kissenpfennig A., Malissen B. 2007. The dermis contains langerin+ dendritic cells that develop and function independently of epidermal Langerhans cells. J. Exp. Med. 204:3119–3131 10.1084/jem.20071724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortman K., Liu Y.J. 2002. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2:151–161 10.1038/nri746 [DOI] [PubMed] [Google Scholar]
- Sung S.S., Fu S.M., Rose C.E., Jr., Gaskin F., Ju S.T., Beaty S.R. 2006. A major lung CD103 (alphaE)-beta7 integrin-positive epithelial dendritic cell population expressing Langerin and tight junction proteins. J. Immunol. 176:2161–2172 [DOI] [PubMed] [Google Scholar]
- Tailor P., Tamura T., Morse H.C., III, Ozato K. 2008. The BXH2 mutation in IRF8 differentially impairs dendritic cell subset development in the mouse. Blood. 111:1942–1945 10.1182/blood-2007-07-100750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte K., Gauthier S., Tuite A., Mullick A., Malo D., Gros P. 2005. A mutation in the Icsbp1 gene causes susceptibility to infection and a chronic myeloid leukemia–like syndrome in BXH-2 mice. J. Exp. Med. 201:881–890 10.1084/jem.20042170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beek E.M., Cochrane F., Barclay A.N., van den Berg T.K. 2005. Signal regulatory proteins in the immune system. J. Immunol. 175:7781–7787 [DOI] [PubMed] [Google Scholar]
- Varol C., Landsman L., Fogg D.K., Greenshtein L., Gildor B., Margalit R., Kalchenko V., Geissmann F., Jung S. 2007. Monocytes give rise to mucosal, but not splenic, conventional dendritic cells. J. Exp. Med. 204:171–180 10.1084/jem.20061011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walzer T., Brawand P., Swart D., Tocker J., De Smedt T. 2005. No defect in T-cell priming, secondary response, or tolerance induction in response to inhaled antigens in Fms-like tyrosine kinase 3 ligand-deficient mice. J. Allergy Clin. Immunol. 115:192–199 10.1016/j.jaci.2004.08.046 [DOI] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., Nussenzweig M. 2008. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 9:676–683 10.1038/ni.1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witmer-Pack M.D., Hughes D.A., Schuler G., Lawson L., McWilliam A., Inaba K., Steinman R.M., Gordon S. 1993. Identification of macrophages and dendritic cells in the osteopetrotic (op/op) mouse. J. Cell Sci. 104:1021–1029 [DOI] [PubMed] [Google Scholar]
- Yokota Y., Mansouri A., Mori S., Sugawara S., Adachi S., Nishikawa S., Gruss P. 1999. Development of peripheral lymphoid organs and natural killer cells depends on the helix-loop-helix inhibitor Id2. Nature. 397:702–706 10.1038/17812 [DOI] [PubMed] [Google Scholar]
- Yrlid U., Jenkins C.D., MacPherson G.G. 2006. Relationships between distinct blood monocyte subsets and migrating intestinal lymph dendritic cells in vivo under steady-state conditions. J. Immunol. 176:4155–4162 [DOI] [PubMed] [Google Scholar]
- Zhang N., Schröppel B., Chen D., Fu S., Hudkins K.L., Zhang H., Murphy B.M., Sung R.S., Bromberg J.S. 2003. Adenovirus transduction induces expression of multiple chemokines and chemokine receptors in murine beta cells and pancreatic islets. Am. J. Transplant. 3:1230–1241 10.1046/j.1600-6143.2003.00215.x [DOI] [PubMed] [Google Scholar]