Abstract
Mutations in the XPD subunit of the DNA repair/transcription factor TFIIH result in the rare recessive genetic disorder xeroderma pigmentosum (XP). Many XP patients are compound heterozygotes with a “causative” XPD point mutation R683W and different second mutant alleles, considered “null alleles.” However, there is marked clinical heterogeneity (including presence or absence of skin cancers or neurological degeneration) in these XPD/R683W patients, thus suggesting a contribution of the second allele. Here, we report XP patients carrying XPD/R683W and a second XPD allele either XPD/Q452X, /I455del, or /199insPP. We performed a systematic study of the effect of these XPD mutations on several enzymatic functions of TFIIH and found that each mutation exhibited unique biochemical properties. Although all the mutations inhibited the nucleotide excision repair (NER) by disturbing the XPD helicase function, each of them disrupted specific molecular steps during transcription: XPD/Q452X hindered the transactivation process, XPD/I455del disturbed RNA polymerase II phosphorylation, and XPD/199insPP inhibited kinase activity of the cdk7 subunit of TFIIH. The broad range and severity of clinical features in XP patients arise from a broad set of deficiencies in NER and transcription that result from the combination of mutations found on both XPD alleles.
The human xeroderma pigmentosum (XP) group D gene (XPD/ERCC2) is located on 19q13. XPD encodes an ATP-dependent 5′-3′ helicase of 760 amino acids, which is a subunit of the multiprotein complex, TFIIH. In addition to helicase activity, XPD is intrinsically involved in the maintenance of the TFIIH integrity by promoting the interaction between the CAK subcomplex (cdk activating kinase, containing cyclin H, MAT1, and the kinase cdk7) and the core of TFIIH (including the 3′-5′ helicase XPB and proteins p62, p52, p44, p34, and p8/TTDA). TFIIH was initially defined as a basal transcription factor for RNA polymerase II (RNA pol II). This complex is also involved in transcription mediated by RNA polymerase I (Iben et al., 2002), as well as in the nucleotide excision repair (NER) pathway. In NER, TFIIH, through the enzymatic activity of XPD and XPB, unwinds the DNA around lesions generated by UV irradiation or bulky chemical adducts. In the transcription of protein coding genes, where the preinitiation complex is assembled (including TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and RNA pol II), TFIIH opens DNA around the proximal promoter through its XPB subunit (Holstege et al., 1996) and phosphorylates the C-terminal domain of the largest subunit of RNA pol II via its kinase cdk7 (Feaver et al., 1991; O’Brien et al., 1994). This phosphorylation is a prerequisite for promoter escape (Dvir et al., 1997).
Mutations in the XPD gene result in several different rare autosomal recessive disorders, including xeroderma pigmentosum (XP), trichothiodystrophy (TTD), combined XP and Cockayne syndrome, or combined XP and TTD (Kraemer et al., 2007). Primarily defined as a DNA repair syndrome (van Steeg and Kraemer, 1999), XP is characterized by a deficiency of the NER pathway, which leads to skin sun sensitivity. XP may also be caused by defects in other genes in the NER pathway (XPA, XPB/ERCC3, XPC, XPE/DDB2, XPF/ERCC4, or XPG/ERCC5) or in the polymerase eta gene (XP variant; Masutani et al., 1999; Lehmann, 2003; Kraemer et al., 2007). XP patients have a 1,000-fold increased frequency of skin cancers, including melanomas, squamous cell carcinomas, and basal cell carcinomas (Kraemer et al., 1987, 1994). Approximately 30% of XP patients, in addition, have progressive neurological degeneration. Immature sexual development and dwarfism has been reported in a few XP patients (de Boer and Hoeijmakers, 2000), some of which may be associated with hormonal dysfunctions (Chen et al., 2002; Keriel et al., 2002; Drané et al., 2004; Compe et al., 2005, 2007).
The fact that most patients with XPD mutations are compound heterozygotes complicates the understanding of genotype/phenotype relationships. For instance, the point mutation R683W in the XPD protein, a hotspot for the XP phenotype, is found as a heterozygous mutation in >80% of XP-D patients (Taylor et al., 1997; Kobayashi et al., 2002; Boyle et al., 2008; Emmert et al., 2009). Curiously, the clinical manifestations of patients who are compound heterozygotes for XPD/R683W and a second mutation include patients with or without skin cancers and patients with or without severe neurological impairments (Taylor et al., 1997; Boyle et al., 2008; Emmert et al., 2009). This prompted us to study whether the mutation found on the second XPD allele might contribute to the heterogeneity of the clinical features.
In this study, we report XP patients in three families each carrying R683W with a different second XPD mutation and having different clinical symptoms. Two brothers with XP with cancers and neurodegeneration are compound heterozygotes for XPD R683W and an in-frame deletion of 1 aa (I455del). Another patient had >300 skin cancers and progressive neurodegeneration with R683W and a second mutation that leads to a premature stop codon (Q452X). Two siblings in the third family had neither skin cancer nor neurodegeneration and carried R683W along with mutations leading to insertion of 2 aa (199insPP). We show how each of these second XPD mutations found in patients who also have the R683W mutation specifically interfere with TFIIH repair and transcription functions in vivo and in vitro. Together, our results suggest that the mutations found on the second XPD allele participate along with R683W in the development of the XP phenotype.
RESULTS
Identification of new XP-D patients
Patients AS552 (Fig. 1, A and B) and AS553 (Fig. 1, C and D) were French brothers with XP who had clinically normal nonconsanguineous parents (AS550, mother; AS551, father; Table I, family 2). In addition to burning on minimal sun exposure, they had several skin cancers (including malignant melanoma and basal cell carcinoma). They developed progressive neurodegeneration resulting in loss of ability to walk, talk, and swallow. Computed tomography scans of the brain showed diffuse atrophy of the cerebral cortex and enlargement of the ventricles, but no calcification (unpublished data). They died in 2008 at the age of 35 and 39, respectively, of progressive cachexia associated with neurodegeneration.
Figure 1.
XP patients studied. (A and B) Patient AS552. (A) At 11 yr of age he had marked freckle-like hyperpigmentation in sun exposed portions of his face and neck. (B) By age 33 yr he used a wheelchair because of his inability to walk. He died at age 39 yr of progressive neurological degeneration. (C and D) Patient AS553, brother of AS552. (C) At 7 yr of age he had marked freckle-like hyperpigmentation in sun exposed portions of his face. (D) By 29 yr of age he used a wheelchair because of his inability to walk. He had posturing of his hands, a feature of neurodegeneration. He died at age 35 yr of progressive neurological degeneration. (E) Patient XP29BE at 32 yr of age. He has marked freckle-like hyperpigmentation in sun exposed portions of his face and neck. His face shows extensive scarring and grafting from numerous surgical procedures for removal of skin cancers. The circled lesions on his face and shoulder were being followed photographically for signs of skin cancer. His conjunctiva show telangiectasia. He died at age 37 yr of progressive neurological degeneration. (F) Patient XP34BE. He was well protected from sun exposure and at age 24 yr he had minimal freckle-like hyperpigmentation on his face and neck and no skin cancers. By age 31 yr he had no evidence of neurological degeneration. (G) Patient XP35BE, younger sister of patient XP34BE. She was well protected from sun exposure and at age 20 yr she had minimal freckle-like hyperpigmentation on her face and neck and no skin cancers. Brief sun exposure of an unprotected portion of her neck resulted in an acute sunburn followed by hyperpigmentation, scaling and peeling of the skin (arrow). By age 27 yr she had no evidence of neurological degeneration.
Table I.
Clinical features and XPD mutations of patients studied
| XPD mutations | |||||||||||
| allele1 | allele2 | allele2 | allele2 | ||||||||
| Family | Patient | Age/sex | Living/dead | Acute sun sensitivitya | Skin cancer | Progressive neurological abnormalities | R683W | Q452X | I455del | 199insPP | References |
| 1 | XP29BE | 37 yr/M | dead | +b | +cd | +b | + | + | Kraemer et al.,1988e; Wang et al., 2009; this study | ||
| 2 | AS552 | 39 yr/Mf | dead | + | +c | + | + | + | This study | ||
| 2 | AS553 | 35 yr/Mf | dead | + | +c | + | + | + | This study | ||
| 3 | XP34BE | 31 yr/Mg | alive | + | 0 | normal | + | + | Boyle et al., 2008 | ||
| 3 | XP35BE | 27 yr/Mg | alive | + | 0 | normal | + | + | Boyle et al., 2008 | ||
Skin burning on minimal sun exposure.
Present.
Including at least one melanoma.
>300 documented skin cancers, including 24 melanomas, 284 basal cell carcinomas, and 12 squamous cell carcinomas.
XP29BE was patient 7 in this study.
Siblings.
Siblings.
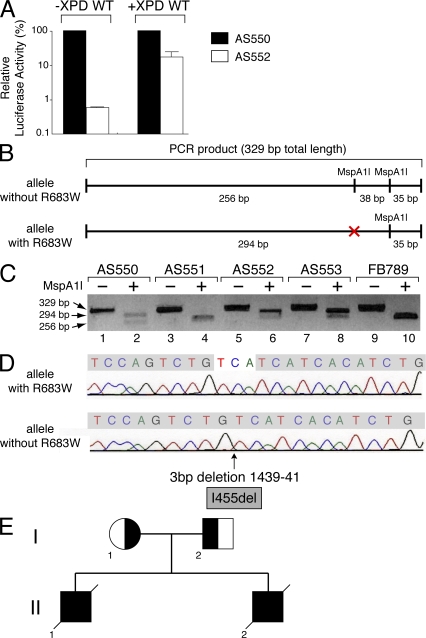
To assign the cells from these patients to an XP complementation group, we measured the DNA repair ability of fibroblasts using the host cell reactivation assay (Khan et al., 1998). A plasmid (pCMV-luc) encoding the luciferase reporter gene was exposed to 1,000 J/m2 of UVC radiation (254 nm, which generates ∼100 photolesions per 5 kb of DNA; van Hoffen et al., 1995), and then transfected into AS552 and maternal (AS550) fibroblasts (Fig. 2 A). The luciferase activity was strongly reduced in AS552 cells when compared with that observed in the maternal fibroblasts (Fig. 2 A, left two columns), demonstrating that the NER activity is deficient in these cells. Strikingly, an overexpression of the WT XPD subunit greatly increased the luciferase activity in AS552 fibroblasts (Fig. 2 A, right two columns), demonstrating that the NER defect in these patients is associated with a XPD deficiency. Similar results were obtained from AS553 cells (not depicted).
Figure 2.
Identification of the XPD mutations in the AS550 and AS552 patients. (A) Host cell reactivation in AS550 (filled columns) and AS552 (open columns). Fibroblasts were transfected with the pCMV-luc (500 ng) previously exposed to 1,000 J/cm2 of UVC-light (254 nm), pCH110 (100 ng, encoding the β-galactosidase which was used to normalize transfection efficiencies), and either empty pcDNA (10 ng; left pair of columns) or pcDNA XPD WT (10 ng, XPD WT; right pair of columns). Host cell reactivation was measured as the relative luciferase activity caused by the repair of the UV-damaged luciferase gene. The values of three independent experiments are presented as percentages, 100% being the level of luciferase activity obtained in AS550 cells. (B) Scheme representing the PCR products obtained from XPD allele with or without the point mutation R683W, which corresponds to a C-to-T substitution at the first base of exon 22 in the gene. This mutation abolishes the restriction site for the endonuclease MspA1l. (C) PCR products were obtained from genomic DNA of AS550, AS551, AS552, AS553, and FB789 (used as control) fibroblasts. Restriction fragment length polymorphism assay was performed from the PCR products digested by MspA1l. (D) DNA sequencing of the PCR product obtained from cDNA of AS552 cells. The XPD allele without the R683W point mutation has a deletion of three bases (TCA) at position 1439–41, giving rise to an amino acid del455 in-frame (I455del). (E) Genetic pedigree of the family of the patients AS552 and AS553. The mother AS550 (I-1) carries the XPD mutation that corresponds to an amino acid substitution at position R683W. The father AS551 (I-2) carries a deletion of three bases (TCA) at position 1439–41 that makes amino acid del455 in-frame (I455del). The deceased patients AS552 (II-1) and his brother AS553 (II-2) had compound heterozygous mutations that corresponded to the substitution at R683W (maternal) and the deletion I455del (paternal), resulting in the development of XP with skin cancer and neurological degeneration.
Many XP patients with mutations in the XPD gene have a R683W point mutation, which corresponds to a C-to-T substitution at the first base of exon 22 in the XPD gene (Taylor et al., 1997; Kobayashi et al., 2002; Boyle et al., 2008; Emmert et al., 2009). This mutation abolishes the restriction site for the endonuclease MspA1l (Fig. 2 B). Genomic DNA was extracted from AS550, AS551, AS552, and AS553 cell lines, and control normal fibroblasts (FB789). Restriction fragment length polymorphisms were assessed from a PCR product (329 bp) containing the putative R683W substitution, which was digested by MspA1l. Restriction analysis of the AS551-DNA (the father) resulted in a fragment of 256 bp that is identical to that obtained from WT FB789 fibroblasts (Fig. 2 C, lanes 4 and 10). In contrast, the two hydrolyzed fragments that were obtained from the genomic DNA of the mother AS550 (lane 2) were also found within the AS552- and the AS553-DNA (lanes 6 and 8), suggesting that the AS552/553 patients are heterozygous for the XPD point mutation R683W. We then searched for the mutation in the second XPD allele. mRNA were isolated from AS552 fibroblasts and reverse transcribed. After sequencing, we found on the second XPD allele, a deletion of three bases (TCA) at position 1,439–41 (Fig. 2 D), which makes amino acid del455 in-frame (I455del). This residue corresponds to the first amino acid of the helicase domain III of XPD (Fig. 3 A; Gorbalenya and Koonin, 1993). Altogether, these results demonstrate that patients AS552 and AS553 have the XPD point mutation R683W on one allele and the new I455del mutation on the other allele. This mutation was also found in the AS551 cells from the father (Fig. 2 E and not depicted).
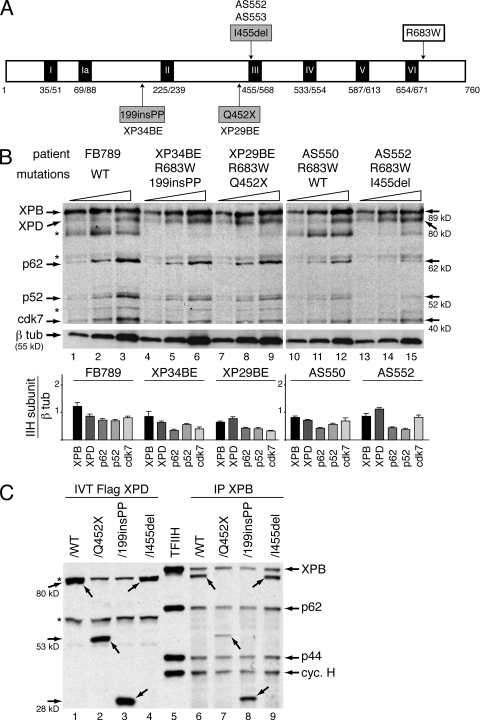
Figure 3.
TFIIH complex into XPD fibroblasts. (A) The diagram represents the 760-aa XPD protein with the 7 (I–VI) helicase motifs. Amino acid changes resulting from mutations found in the XPD/R683W patients who are compound heterozygotes are depicted. (B) Western blot analyses of TFIIH subunits (XPB, XPD, p62, p52, and cdk7) in increasing amounts of whole-cell extracts isolated from fibroblasts of FB789, XP34BE, XP29BE, AS550, and AS552 patients. * indicate nonspecific bands. β-Tubulin (β tub) was used as an internal control. The results are representative of three independent experiments. Diagram represents the ratio between each TFIIH subunit and β tub (arbitrary units). (C) Western blotting analysis of Ab-XPB ChIP (IP XPB) samples from chromatin extracts isolated from HD2 cells transfected with expression vectors encoding either Flag-XPD/WT (∼80 kD), /Q452X (∼53 kD), /199insPP (∼28 kD), or /I455del (∼80 kD). The immunoprecipitated fractions were resolved on SDS-PAGE followed by immunoblotting using antibodies raised against XPB, p62, p44, cyclin H and the Flag-tag. In vitro synthesized Flag-XPD mutated proteins (IVT Flag XPD, lanes 1 to 4) and highly purified TFIIH from HeLa cells (lane 5) were used as references. Arrows indicate the different forms of XPD. * indicate nonspecific bands. The results are representative of three independent experiments.
Clinical features differ among patients who are compound heterozygotes for the XPD R683W mutation
The clinical manifestations of the AS552 and AS553 patients were compared with those of other XP patients who are compound heterozygotes for R683W and a second mutation (Table I). Patient XP29BE (with a premature stop codon Q452X on the other XPD allele; Fig. 3 A) was a Caucasian male with severe XP phenotype (Fig. 1 E; Kraemer et al., 1988; Wang et al., 2009). In addition to severe sunburning on minimal sun exposure, and extensive freckle-like pigmentation, he had >300 skin cancers documented, including 24 cutaneous melanomas, 284 basal cell carcinomas, and 12 squamous cell carcinomas. He developed progressive neurological degeneration with sensorineural deafness and severe motor impairment (difficulty walking, talking, and swallowing). He died at age 39 yr of progressive neurological degeneration.
Patient XP34BE (bearing a mutation generating a frame shift after 2-aa PP insertion at position 199 on the other XPD allele, Fig. 3 A) is a Caucasian male with a mild XP phenotype (Fig. 1 F; Boyle et al., 2008). His sister, patient XP35BE (Fig. 1 G), had the same mutations and a similar mild XP phenotype (Table I; Boyle et al., 2008). Patient XP34BE never developed skin tumors, despite marked sun sensitivity on minimal sun exposure. He had excellent sun protection and developed only minimal freckle-like pigmentation on his face (Fig. 1 F). By age 31 yr he had no evidence of neurological abnormalities. Altogether, these observations clearly illustrate that XP patients who are compound heterozygotes and share the XPD point mutation R683W can have a wide spectrum of severity with respect to their clinical phenotypes (Table I).
XPD mutations do not affect the TFIIH cellular concentration
The aforementioned data prompted us to analyze whether the mutations found on the second XPD allele contribute to the clinical phenotype. Some XPD mutations disrupt the steady-state TFIIH levels in cells derived from TTD patients (Vermeulen et al., 2000; Botta et al., 2002; Boyle et al., 2008). We thus analyzed the TFIIH cellular concentration in fibroblasts isolated from the compound heterozygous XPD/R683W patients (Fig. 3 B). Whole-cell extracts isolated from WT FB789 and AS550 cells (used as controls), as well as from XP34BE, XP29BE, and AS552 fibroblasts were resolved by SDS-PAGE. We found that the TFIIH cellular concentration (as indicated by the Western blotting of its XPB, XPD, p62, p52 and cdk7 subunits) is similar in XPD and normal cells using β-tubulin as a reference. It is worthwhile to note that the premature truncated forms of XPD/Q452X (molecular weight [MW] = 53 kD, lanes 4–6) and XPD/199insPP (MW = 28 kD, lanes 7–9) cannot be visualized in XP34BE and XP29BE cells, respectively, because the only available antibody recognizes an epitope located within the C-terminal part of XPD. To analyze whether the XPD-mutated forms might be found in vivo in the TFIIH complex, eukaryotic expression vectors encoding either WT or the mutated forms of XPD with a Flag-tag were generated and transiently transfected into HD2 cells. HD2 cells are a cell line resulting from the fusion between human fibroblasts harboring the XPD/R683W point mutation and HeLa cells (Johnson et al., 1985). We performed chromatin immunoprecipitation (ChIP) with an antibody directed against XPB (Ab-XPB). Western blot analysis revealed that XPD/WT, /Q452X, /199insPP, and /I455del fusion proteins coimmunoprecipitated together with XPB, p62, p44, and cyclin H subunits of TFIIH (Fig. 3 C). Together, these results indicate that the XPD mutations found in XP patients who are compound heterozygotes for R683W and a second mutation do not alter the steady-state cellular concentration of components of TFIIH or the binding of this complex to chromatin.
Mutations in XPD alter its helicase activity and the composition of TFIIH
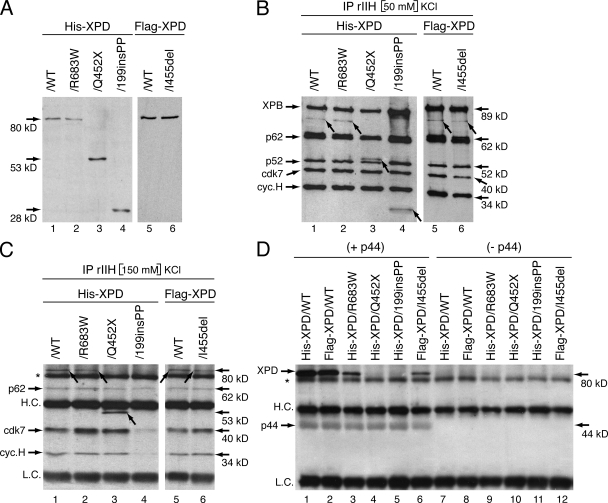
We next analyzed whether the mutated XPD had an effect on the TFIIH architecture, as some XPD mutations weaken the interaction between XPD and the p44 subunit, which anchors the CAK to the core of TFIIH (Coin et al., 1998). The mutated forms of XPD were tagged with either a Histidine-tag (His-XPD/WT, /R683W, /Q452X, and /199insPP) or a Flag-tag (Flag-XPD/WT and /I455del) and then expressed in Sf9 cells by using the baculovirus expression system. The size and extent of expression of each mutated form of XPD was verified by Western blots by using antibody raised against either the His- or the Flag-tag (Fig. 4 A).
Figure 4.
XPD mutations disrupt TFIIH integrity. (A) Production in SF9 insect cells of the recombinant mutated forms of XPD tagged with either the His-tag (His-XPD/WT, /R683W, Q452X, and 199insPP) or the Flag-tag (Flag-XPD/WT and /I455del). Whole-cell extracts were resolved by SDS-PAGE and blotted with antibodies raised against either the His- or the Flag-tag. Arrows indicate the theoretical molecular weight of each XPD mutated form. The results are representative of four independent experiments. (B) Purification of the recombinant TFIIH (rIIH). Insect cells were infected with baculoviruses overexpressing the subunits of TFIIH including either WT or mutated XPD, and complexes were immunoprecipitated by using antibody (Ab) directed toward the p44 subunit of the core TFIIH in low salt conditions (50mM KCl). After elution with a synthetic peptide recognized by Ab-p44, equal amounts of purified rIIHs were then resolved by SDS-PAGE and blotted with antibodies against XPB, p62, p52, cdk7 and cyclin H subunits of TFIIH. His-XPD/WT, /R683W, /Q452X, /199insPP, and Flag-XPD/WT, /I455del were visualized with antibodies raised against either the His- or the Flag-tag, respectively. Arrows indicate the different forms of XPD. The results are representative of five independent experiments. (C) Immunoprecipitation with Ab-p44 of the various rIIH in a higher salt condition (150 mM KCl). rIIHs immunoprecipitated with Ab-p44 cross-linked on agarose beads were boiled, resolved by SDS-PAGE, and blotted with antibodies against p62, cdk7, cyclin H, and either the His- or the Flag-tag. Arrows show the different forms of XPD. HC., heavy chain of Ab-p44; LC, light chain of Ab-p44; * indicates a nonspecific band. The results are representative of three independent experiments. (D) Infected Sf9 cell lysates containing either WT or mutated XPD as indicated at the top of the panel, were incubated with (+ p44) or without (− p44) WT p44, immunoprecipitated to agarose beads and further washed with 350 mM KCl. The immunoprecipitated fractions were then resolved on SDS-PAGE, followed by immunoblotting using Ab-p44 and either anti-His or anti-Flag antibodies. * indicates a nonspecific band. The results are representative of three independent experiments.
Recombinant TFIIH complexes (rIIH), resulting from coinfection by baculoviruses containing the XPD mutated forms found in patients together with baculoviruses expressing each of the other subunits of TFIIH, were produced in Sf9 cells (Dubaele et al., 2003) and then immunoprecipitated by using antibody directed toward the p44 subunit. Under low-salt conditions (50 mM KCl), the XPD/R683W, /Q452X, /199insPP, and /I455del coimmunoprecipitated with all the other subunits of TFIIH (Fig. 4 B). Strikingly, at higher salt concentration (150 mM) the composition of the immunoprecipitated rIIH containing the XPD/WT, /R683W, /Q452X, and /I455del was maintained (Fig. 4 C, lanes 1–3 and 5–6). However, we failed to obtain an entire XPD/199insPP rIIH (lane 4). Indeed, neither the CAK subcomplex (illustrated by cdk7 and cyclin H) nor the truncated XPD/199insPP coimmunoprecipitated at 150 mM KCl. Collectively, the above data indicate that XPD mutations affect the stability of TFIIH in different ways.
Knowing that XPD contributes to the anchoring of the CAK subcomplex to the core of TFIIH, we analyzed whether XPD mutations impede its interaction with p44 (Dubaele et al., 2003). Equal amounts of either WT or mutated XPD subunits were incubated with or without p44 (when indicated) in the presence of Ab-p44 cross-linked to agarose beads (Fig. 4 D). After extensive washing at high-salt concentration (350 mM KCl), we observed that p44 is able to retain much less XPD/R683W and XPD/I455del than XPD/WT (Fig. 4 D, compare lanes 1 and 3 and compare lanes 2 and 6), whereas no XPD binding was found in absence of p44 (Fig. 4 D, lanes 7–12). Strikingly, the truncated XPD/Q452X and XPD/199insPP did not coimmunoprecipitate with p44 (Fig. 4 D, lanes 4 and 5), which is not surprising because p44 interacts with the C-terminal end of XPD (Coin et al., 1998). Our data show how each of the mutations of the XPD helicase specifically affect its interaction with the p44 subunit of TFIIH.
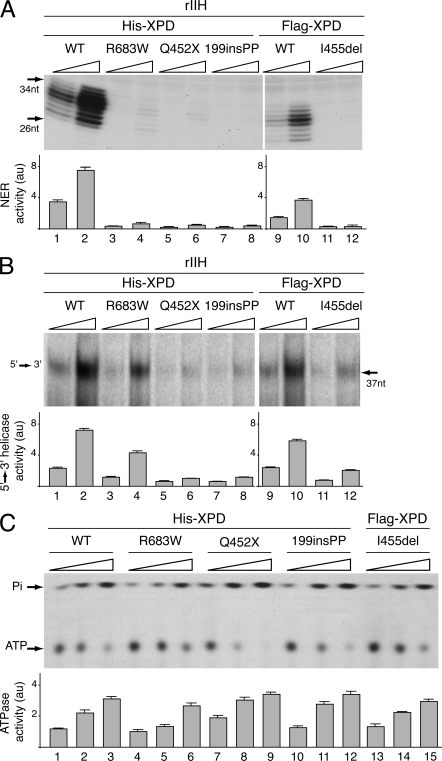
XPD mutations inhibit NER activity
XPD is crucial for the functioning of TFIIH in DNA repair (Schaeffer et al., 1993; Drapkin et al., 1994). We thus investigated the effect of the XPD mutations in in vitro NER assays. The rIIHs (immunoprecipitated at 50 mM KCl; Fig. 4 B) were tested in a dual-incision assay using the purified NER factors and damaged plasmid containing a single cisplatin adduct (Araújo et al., 2000). Whereas the rIIH XPD/WT potentiates the removal of the damaged oligonucleotides when added to the reaction (Fig. 5 A, lanes 1–2 and 9–10; Fig. S1 A shows the complete gel), rIIH-XPD/R683W, /Q452X, /199insPP, and /I455del were inactive (Fig. 5 A, lanes 3–8 and 11–12).
Figure 5.
DNA Repair Activities of the rIIHs. (A) Equivalent increasing amounts of rIIHs (adjusted according to Western blotting) were added to an incision/excision assay using recombinant NER factors. The area of the gel containing the excision products is shown (the complete gel is shown in Fig. S1 A). The excised oligonucleotides signals were quantified and plotted in arbitrary units (au). The results are representative of three independent experiments. (B) Helicase activity of the WT and mutated forms of XPD. Equivalent increasing amounts of the various immunoprecipitated rIIH were tested for their 5′–3′ XPD helicase activity. The density of radiolabeled oligo displaced by the XPD helicase activity was measured and plotted in arbitrary units (au). The results are representative of three independent experiments. The complete gel is shown in Fig. S1 B. (C) Equal amounts of immunoprecipitated XPD were tested in an ATPase assay for 120 min. Nonhydrolyzed ATP as well as Pi are indicated. The ATP hydrolysis was quantified and plotted in arbitrary units (au). The results are representative of three independent experiments.
This prompted us to analyze whether the XPD mutations would affect either the helicase and/or the ATPase activity of XPD. The 5′-3′ XPD helicase activity was measured with an in vitro assay that detects the displacement of a 32P-labeled DNA fragment previously annealed to a single-stranded circular DNA (Schaeffer et al., 1993). When added to the in vitro assay, rIIH (immunoprecipitated at 50 mM salt concentration) containing XPD/Q452X, /199insPP, or /I455del did not displace the labeled oligonucleotide from the single-stranded plasmid (Fig. 5 B, lanes 5–8 and 11–12; Fig. S1 B shows the complete gel). In this case, we also observed that rIIH-XPD/R683W exhibits a lower helicase activity than rIIH-XPD/WT (reduction of 40 ± 2%; compare Fig. 5 B, lanes 3–4 to 1–2). We next investigated the ATPase activity of the XPD-mutated forms (Fig. 5 C). The WT and mutated XPD proteins were immunoprecipitated, extensively washed at 500 mM NaCl, and incubated in the presence of [γ-32P]ATP and double-stranded DNA. We observed that the ATPase activity of the mutated forms of XPD was not affected. This result was anticipated because the ATP binding site of XPD is located at position K48 and should not be altered by these mutations (Sung et al., 1988).
Taken together, the above results clearly demonstrate that mutations found in XP patients who are compound heterozygotes for R683W and a second mutation affect the TFIIH activity during NER by disturbing the helicase function of XPD without affecting its ATPase activity.
XPD mutations inhibit basal transcription activity
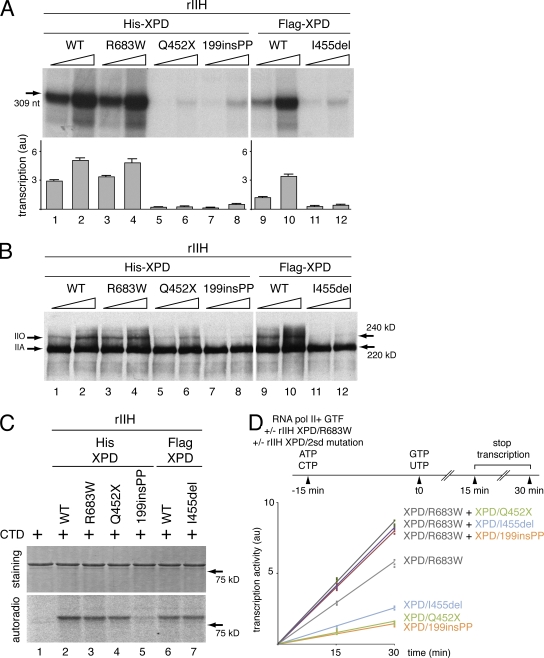
The rIIHs were next tested for their transcription activity. When added to an incubation mixture containing the AdMLP template, RNA pol II, and the basal transcription factors (Dubaele et al., 2003), rIIH-XPD/R683W exhibit a transcription activity as strong as rIIH-XPD/WT (Fig. 6 A, lanes 3–4 and 1–2, respectively; see Fig. S2 A for the complete gel). On the contrary, rIIH-XPD/Q452X, /199insPP, and /I455del result in barely detectible in vitro RNA synthesis (Fig. 6 A, lanes 5–8 and 11–12).
Figure 6.
Transcription activity of the rIIHs. (A) Basal transcription activity of the rIIHs. The purified rIIHs were added to an in vitro reconstituted transcription system lacking TFIIH. The length of the corresponding transcript is indicated on the left side. The transcription activity of all variants was assessed using increasing amounts of rIIHs for 30 min. The signals were quantified and plotted in arbitrary units (au). The results are representative of three independent experiments. The complete gel is shown in Fig. S2 A. (B) Phosphorylation of the RNA polymerase II during in vitro reconstituted transcription assays. The RNA pol II kinase activity of low salt immunopurifed rIIHs was analyzed in an in vitro assay containing all the basal transcription factors and the AdMLP. Arrows indicate hypo (IIA) and hyper (IIO) phosphorylated forms of RNA pol II. The results are representative of two independent experiments. The complete gel is shown in Fig. S2 B. (C) In vitro phosphorylation of the GST-CTD fusion protein was performed with equal amounts of rIIHs in the presence of 0.14 µM [γ-32P] ATP. Coomassie blue–stained gel (staining; top) and autoradiography (autoradio; bottom) of the incubated fractions are shown. The results are representative of three independent experiments. The complete gels are shown in Fig. S2 C. (D) In vitro reconstituted transcription assays were performed following the protocol scheme. rIIH XPD/R683W, rIIH XPD/Q452X, /199insPP, or /I455del were preincubated either alone or in combination, in the presence of RNA pol II, the general transcription factors (GTF), the AdMLP template, ATP, and CTP. 15 min later, the transcription process was initiated by addition of GTP and UTP. The reactions were performed for 15 and 30 min. The signals of three independent experiments were then quantified and plotted in arbitrary units (au).
The phosphorylation of the C-terminal domain (CTD) of the largest subunit of RNA pol II by the cdk7 subunit of TFIIH is a prerequisite to allow the transition between initiation and elongation and further promoter escape (Akoulitchev et al., 1995; Dvir et al., 1997). We thus checked the phosphorylation status of RNA pol II after the in vitro transcription. Antibody that recognizes the hypo- (IIA) and hyperphosphorylated (IIO) forms of the largest subunit of RNA pol II showed that the phosphorylation status of the RNA pol II was similar when rIIH-XPD/WT and /R683W were present in the assay (Fig. 6 B, compare lanes 1–2 and 3–4, respectively; Fig. S2 B shows the complete gel). However, we observed the lack of RNA pol IIO in the presence of either rIIH-XPD/199insPP or rIIH-XPD/I455del (Fig. 6 B, lanes 7–8 and 11–12). Moreover, the phosphorylation of RNA pol II was not totally abrogated by rIIH-XPD/Q452X (Fig. 6 B, lanes 5–6; see Discussion). Having observed that some of the mutated rIIHs prevent the phosphorylation of the CTD of RNA pol II in the context of a transcription assay, we next investigated whether the XPD mutations disturb the enzymatic activity of the cdk7 kinase itself. In vitro kinase assays show that rIIH-XPD/R683W, /Q452X, and /I455del, as well as rIIH-XPD/WT, phosphorylated the CTD fused to the GST protein (Fig. 6 C, compare lanes 3–4 to lane 2 and lane 7 to lane 6; Fig. S2 C shows the complete gel). On the contrary, rIIH-XPD/199insPP did not phosphorylate CTD (Fig. 6 C, lane 5).
We thus investigated how transcription occurs in the presence of the combinations of TFIIH found in the XP-D patients. rIIH XPD/R683W, rIIH XPD/Q452X, /199insPP, or /I455del were preincubated either alone or in combination, in the presence of RNA pol II, the general transcription factors (GTF), the AdMLP template, ATP, and CTP (Fig. 6 D). 15 min later, the transcription process was initiated by addition of GTP and UTP (Fig. 6 D, see protocol scheme). rIIH XPD/R683W had greater transcription activity than rIIH XPD/Q452X, /199insPP, or /I455del separately. Furthermore, rIIH XPD/Q452X, /199insPP, and /I455del did not counteract the transcription activity when combined with rIIH XPD/R683W. In contrast, we observed an additive effect. Similar results were obtained when we performed transcription assays with preincubation of rIIH XPD/Q452X, /199insPP and /I455del before addition of rIIH XPD/R683W (unpublished data).
Impaired transactivation of nuclear receptors in XP-D cells
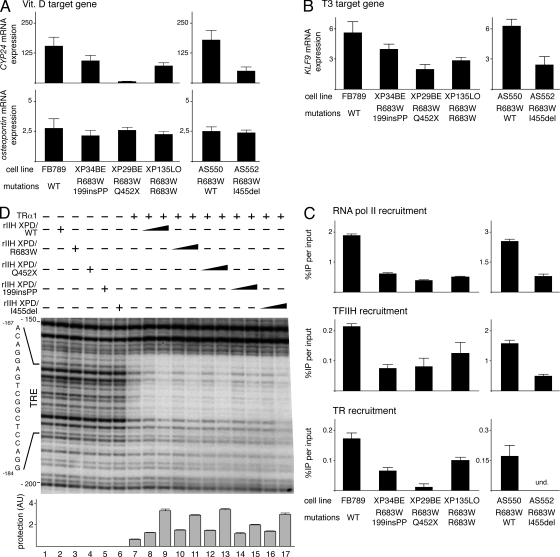
TFIIH is involved in the transactivation of nuclear receptors by phosphorylating (via its cdk7 kinase subunit) either their A/B domain (as shown for ER, PPAR, or RAR; Rochette-Egly et al., 1997; Chen et al., 2002; Compe et al., 2007) or one of their partners (such as Ets1 for the vitamin D receptor; Drané et al., 2004). We thus analyzed by real-time quantitative RT-PCR the expression of genes known to be under the control of nuclear receptors in fibroblasts isolated from the compound heterozygote XPD/R683W patients (XP34BE, XP29BE, and AS552) and from the homozygous XPD/R683W patient (XP135LO; Taylor et al., 1997), as well as from normal fibroblasts (FB789) and from an XPD/R683W heterozygote (AS550; Fig. 2 C). We first examined the expression of a vitamin D nuclear receptor (VDR) target gene, vitamin D-24-hydroxylase (CYP24; Akeno et al., 1997), and the osteopontin gene whose expression is known to be unaltered in XPD-deficient cells (Drané et al., 2004). Upon induction by vitamin D (100 nM), the expression of CYP24 (Akeno et al., 1997) was much lower in the XPD-deficient fibroblasts than in WT cells (Fig. 7 A, upper panel). Moreover, we particularly noticed that the CYP24 expression shows a greater reduction in XP29BE than in XP34BE, XP135LO, or AS552 fibroblasts. In contrast, the expression of the osteopontin gene was unaltered in any of the XPD-deficient cells (Fig. 7 A, bottom), suggesting that the expression of some genes can normally occur in these XPD-deficient cells.
Figure 7.
Incidence of XPD mutations during the transactivation mediated by nuclear receptors. (A) Expression of the VDR target genes CYP24 and osteopontin in WT (FB789, AS550) and XPD mutated (XP34BE, XP29BE, XP135LO, AS552) fibroblasts after treatment during 8h with vitamin D (vit. D, 100 nM). The values were normalized relative to 18S RNA expression. The results of three independent experiments are presented as n-fold induction relative to nontreated cells. (B) Expression of the TR target gene KLF9 in FB789, XP34BE, XP29BE, XP135LO, AS550, and AS552 fibroblasts after treatment during 24 h with T3 (10 nM). The values were normalized relative to 18S RNA expression. The results of three independent experiments are presented as n-fold induction relative to nontreated cells. (C) Recruitment of RNA pol II, TFIIH, and TR on the KLF9 promoter. After T3 treatment, the recruitment of RNA pol II and TFIIH (via its p44 subunit) was analyzed by ChIP assays on the KLF9 proximal promoter. The TR recruitment was studied on a TRE located at −2.9 kb in the KLF9 promoter. The results of three independent experiments are presented as percentage of DNA immunoprecipitated relative to the input; und, undetectable. (D) DNase1 footprint analysis on the TRE located into the TR target gene mbp. The MBP fragment (−256/+21) was labeled at the 5′ end and incubated with TRα1 and increasing amounts of rIIH XPD/WT (lanes 8–9), /R683W (lanes 10 and 11), /Q452X (lanes 12 and 13), /199insPP (lanes 14 and 15), and /I455del (lanes 16 and 17). Cis-element for the TRs (from nt-184 to nt-167) is positioned. Autoradiography of the gel is shown from nt-200 to nt-150. Graph depicts the protection level on the MBP-TRE (see Materials and methods). The complete gel is shown in Fig. S3 A.
We also examined the expression of Krüppel-like factor 9 (KLF9; Fig. 7 B), a thyroid hormone receptor (TR) responsive gene in human fibroblasts (Moeller et al., 2005). After 24 h of treatment with triiodothyronine T3 (10 nM), the expression of KLF9 was clearly reduced in the XPD-mutated fibroblasts when compared with WT. Moreover, similar to that observed for the vitamin D target gene CYP24 (Fig. 7 A), the expression of KLF9 showed a much greater reduction in XP29BE than in XP34BE, AS552, or XP135LO fibroblasts. It should be noted that expression of these genes in cells from the compound heterozygote XPD/R683W patients (XP34BE, XP29BE, and AS552) was different from that in the homozygous XPD/R683W patient (XP135LO).
Chromatin immunoprecipitation (ChIP) assays were then undertaken to investigate whether the expression pattern of the KLF9 gene in XPD-deficient cells could be correlated with transcriptional defects (Fig. 7 C). The genomic DNA fragments bound to RNA pol II and TFIIH were immunoprecipitated with corresponding antibodies and were further analyzed by quantitative (q)PCR with primers designed to amplify the transcription initiation start site of the KLF9 promoter (from − 41 to +39). After 24 h of treatment with triiodothyronine T3 (10 nM), the recruitment of RNA pol II as well as of TFIIH was clearly reduced in the XPD mutated fibroblasts when compared with that observed in the corresponding WT cells. Antibodies directed against TR were also used to immunoprecipitate DNA fragments encompassing a thyroid hormone response element (TRE) located in the distal KLF9 promoter (from −2,891 to −2,875; Denver and Williamson, 2009). After T3 treatment, we clearly observed that the TR recruitment on the TRE was reduced in the XPD-mutated fibroblasts when compared with WT (Fig. 7 C, bottom), suggesting that the lower amounts of mRNA observed in the mutated cells (Fig. 7 B) are caused by defects at a transcriptional level.
TFIIH contributes to the binding stability of the nuclear receptors on their response elements (Compe et al., 2007), thus we analyzed whether the XPD mutations might affect such coactivator function of TFIIH. The proximal promoter of the myelin basic protein (mbp) gene, which is known to contain a response element for the TRs (from nt-184 to nt-167; Farsetti et al., 1991), was used as a radiolabeled probe during DNase1 in vitro footprinting assays (see Materials and methods). The radiolabeled-mbp promoter was incubated with highly purified TRα1 in the presence of either rIIH-XPD/WT, /R683W, /Q452X, or /I455del (Fig. 7 D; Fig. S3 A shows the complete gel). Whereas the rIIH complexes did not modify the pattern of digestion by the DNase1 of the mbp promoter in absence of TRα1 (Fig. 7 D, compare lanes 2–6 to lane 1), we observed that increasing amounts of rIIH XPD/WT, /R683W, /Q452X, and /I455del similarly promote the recruitment of TRα1 on its response element TRE (Fig. 7 D, lanes 7–13). Strikingly, we found that rIIH XPD/199insPP, which affects the TFIIH architecture (Fig. 4 C), does not efficiently stabilize TRα1 on its TRE (Fig. 7 D, compare lanes 14–15 with lanes 8–9).
Collectively, our results demonstrate that the transactivation mediated by nuclear receptors is differentially affected in cells isolated from several patients who are compound heterozygotes for R683W/XPD and a second mutation. Strikingly, such defects result from deficiencies in the recruitment of the basal transcription machinery and the nuclear receptors, in which the product of the second XPD allele might be implicated.
DISCUSSION
Marked heterogeneity of clinical features is observed in XP patients who are compound heterozygotes for XPD/R683W and a second mutation (Table I). Because some putative biallelic effects might be difficult to distinguish from the influence of environment and genetic background, we engaged a systematic study of the incidence of XPD mutations on the numerous TFIIH functions during NER and transcription (Table II).
Table II.
Summary of the effects of the different XPD mutations
| XPD mutation | ||||||
| R683W | Q452X | I455del | 199insPP | |||
| DNA repair | NER | |||||
| helicase activity | ± | |||||
| ATPase | + | + | + | + | ||
| TFIIH structure | 50 mM KCl | + | + | + | + | |
| 150 mM KCl | + | + | + | |||
| p44–XPD interaction | 350 mM KCl | + | + | |||
| Transcription | RNA pol II phosphorylation | + | ± | |||
| cdk7 kinase activity | + | + | + | |||
| basal transcription | + | |||||
| transactivation by nuclear receptors | ||||||
| nuclear receptor stabilization | + | + | + | ± | ||
We designed recombinant rIIH XPD/R683W, /Q452X, /I455del, and /199insPP in which the XPD subunit carries different amino acid changes found in XP patients who are compound heterozygotes for XPD/R683W and a second mutation, and have different clinical symptoms. Each of the recombinant TFIIH mutants exhibited unique biochemical properties (Table II). None of them displayed in vitro DNA repair (NER) activity (Fig. 5 A). All the mutations inhibited the 5′–3′ helicase activity of XPD, except the point mutation R683W. The XPD/R683W mutant, however, exhibited a much lower helicase activity when compared with XPD/WT. It seems that XPD mutations affect the helicase activity either by altering the helicase domain (the mutation I455del is located in the helicase motif III, whereas Q452X and 199insPP generate truncated forms) or by disrupting the interaction between the XPD CTD and the regulatory p44 subunit within TFIIH (such as the point mutation R683W, Fig. 4 D). We also noticed that all the mutations retained the ATPase activity of XPD, which was not surprising because these mutations do not affect the ATP binding site (Sung et al., 1988). However, our results for human XPD/R683W differ from those reported for an archeobacteria homologue of XPD bearing the same mutation (Fan et al., 2008). Besides the fact that these proteins come from different organisms, this apparent discrepancy might also be explained by the nature of the DNA substrate used in the ATPase assays (double-stranded versus single-stranded DNA; Singleton et al., 2007). We next observed that rIIH XPD/Q452X, /I455del, and /199insPP were highly defective during in vitro transcription assays that only contained the minimal set of basal transcription factors in addition to the RNA pol II. Under our in vitro experimental conditions, the RNA synthesis was similar in the presence of rIIH XPD/R683W to that observed with rIIH XPD/WT (Fig. 6 A), underlying the fact that the XPD unwinding activity is not essential for transcription (Dubaele et al., 2003; Coin et al., 2007).
In dissecting which molecular processes were affected during RNA synthesis, we found that each XPD mutation differently weakens XPD/p44 interactions (Table II). Knowing that such interactions direct the anchoring of the CAK kinase complex, and consequently the positioning of TFIIH within the transcription complex, it is not surprising to observe that beside defect in basal transcription, each mutation will also differently affect the transactivation process. We thus found that rIIH XPD/199insPP and rIIH XPD/I455del failed to phosphorylate RNA pol II (Fig. 6 B), although the cdk7 kinase activity rIIH XPD/I455del was not directly disrupted by the mutation (Fig. 6 C and Table II). Interestingly, the CTD of the RNA pol II was correctly phosphorylated in the presence of rIIH XPD/Q452X during in vitro transcription as well as in vitro kinase assays (Fig. 6 B and 6C). These observations show that a given XPD mutation affects specific molecular processes during transcription, resulting in a different biochemical phenotype for each. These differences were more clearly illustrated in the in vivo transactivation assays. Indeed, we observed that the transactivation mediated by nuclear receptors, such as VDR, TR, and the retinoic acid receptors RAR (Fig. 7 A and 7B, and unpublished data) was differently affected in the fibroblasts from different compound heterozygous XPD/R683W patients. Whereas the expression of some genes, such as the osteopontin, is unaltered in these cells (Fig. 7 A, lower panel; Drané et al., 2004), the severity of the transactivation defect depends on the nature of the mutation found on the second XPD allele. While dissecting the transactivation mechanism, by either ChIP or DNase1 in vitro footprinting assays (Fig. 7 C and 7D), we observed that TR were differently recruited on the promoter of target genes and consequently were unable to transactivate properly their responsive genes. More precisely we demonstrated that TFIIH-XPD/199insPP does not fully stabilize TR on its response element (Fig. 7 D).
Although compound heterozygosity is common in human recessive disorders, its incidence in the development of disease such as XP has not been well established. XPD mutations exclusively related to XP (such as XPD/R683W) were considered as causative mutations, and other alleles that were associated with more than one disorder were regarded as null alleles (Taylor et al., 1997; Cleaver et al., 1999). In support of this hypothesis, some analogous XPD mutations in Schizosaccharomyces pombe rad15 failed to support viability in the haploid state and were thus considered devoid of signficant biological activity. However, XP, XP/Cockayne syndrome, TTD, or XP/TTD patients who are compound heterozygous for the same XPD causative mutation develop different clinical features (Table I; Broughton et al., 2001; Fujimoto et al., 2005; Rao et al., 2007; Botta et al., 2009), which is difficult to explain from a monoallelic assumption. Moreover, if the phenotype depended only on one XPD causative mutation, there should not be striking differences between XPD/R683W heterozygous and homo-/hemizygous patients. We clearly observed that each XPD mutation affected different functions of TFIIH (Table II). Gene expression was differently affected depending on the mutation found on the second allele, indicating that, in absence of a WT allele, genetic interactions between recessive XPD alleles might result in different transcriptional outcomes. Thus, the induction of the KLF9 gene by T3 was much less reduced in the XPD R683W/Q452X compound heterozygous cells than in the XPD R683W homozygous, suggesting that the transactivation defect directed by XPD/R683W might be partially counteracted by the XPD/Q452X. Conversely, the expression of the vit-D–induced CYP24 was similarly affected in the XPD R683W/199insPP cells when compared with that observed in the XPD/R683W homozygous cells, suggesting that the deficiency associated with XPD/199insPP might be complemented by XPD/R683W. This could be correlated with recent analyses of compound heterozygous mouse models showing a variety of biallelic effects, including interallelic complementation, a phenotype attributable to combinations of recessive XPD alleles (Andressoo et al., 2006). It is worthwhile to note that patients with XP or TTD with XPD splice mutations (as in XP34BE and XP35BE) may have mild symptoms in conjunction with a small amount of WT XPD mRNA, as previously described (Boyle et al., 2006; Botta et al., 2009). In addition, we cannot exclude that the XPD/199insPP mutated protein might be more unstable than other XPD mutated forms, such as XPD/Q452X and XPD/I455del, allowing the XPD/R683W mutated protein to counteract the defects associated with XPD/199insPP.
Although differences in the genetic background may also account for the phenotypic variability (the identification of such modifier genes could also be of clinical significance), our results suggest that some presumed null XPD alleles might have an impact on the disease development in compound heterozygous humans. Strikingly, as summarized in Table I, the two brothers in family two had the same XPD mutations and similar severe XP symptoms of skin cancer and progressive neurological degeneration leading to death in the fourth decade. In contrast, the affected brother and sister in family three had the same XPD mutations and similar mild symptoms of sun sensitivity without skin cancer or neurological abnormalities (Table I). Because all four XP patients had the XPD/R683W mutation, this suggests that the second XPD mutation in each family had a major role in determining the different clinical symptoms.
Collectively, these observations cast doubt the monoallelic assumption to explain the clinical heterogeneity in XPD compound heterozygous, and instead support a biallelic hypothesis in which both alleles contribute to the phenotype (Andressoo et al., 2006). Previous studies hypothesized that XP is primarily a defect of NER and TTD a defect of transcription (van Steeg and Kraemer, 1999; Lehmann, 2001). However, the present study indicates that XPD mutations in XP patients affect both NER and transcription (Table II). Interestingly, it has been previously shown that the patient XP29BE had a poor response to isotretinoin, an oral retinoid used for chemoprevention of skin cancers (Kraemer et al., 1988). Our data suggest that this lower response might be caused by a weaker transactivation mediated by RAR. Moreover, depending on the nature of the combination of XPD mutations, this type of pharmacological treatment might have to be specifically adapted to efficiently prevent skin cancers in compound heterozygotes for XPD/R683W. Further studies should thus be undertaken to reveal phenotypic effects attributable to combinations of differentially mutated XPD alleles and to examine the effect of concomitant mutations on the DNA repair and/or transcription processes.
MATERIALS AND METHODS
Experiments.
The French Ministry of Higher Education and Research (Statement of Collections of Human Biological Samples for Scientific Purposes n°DC-2008-785), the French Ethical Research Committee EST IV, and the National Cancer Institute Institutional Review Board approved the human experiments.
Host cell reactivation assay.
The pCMV-luc vector was UV irradiated (254 nm, 1,000 J/m2) at a concentration of 1 mg/ml in 10 mM Tris-HCl (pH 8.0) and 1 mM EDTA. AS550 and AS552 cells were transfected with 500 ng of pCMV (UV±) and 10 ng of pcDNA XPD WT in a 6-well plate at a confluence of 95% using Lipofectamine Plus (Invitrogen). To normalize transfection efficiencies, 100 ng of nonirradiated pCH110 vector expressing the β-galactosidase (Invitrogen) was co-transfected. Cells were lysed after 24 h to measure the luciferase and the β-galactosidase activities. The results are the mean of at least three measurements.
ChIP Western blot assays.
HD2 cells were grown in DME/HamF10 (vol/vol) containing 10% FCS and 40 µg/ml gentamycin and were plated to ∼50% confluency before transfection. The cells were then transiently transfected using the transfection reagent JetPei (Polyplus-Transfection). In all experiments, 12 µg of each expression vector were used per 15-cm dish (6–8 dishes were used for each expression vector). At 48 h after transfection, soluble chromatin was prepared as previously described (Coin et al., 2008). In each assay, 750 µg of protein from cross-linked chromatin was immunoprecipitated with polyclonal antibodies raised against the p89 subunit of TFIIH (sc-293; Santa Cruz Biotechnology, Inc.). The immunocomplexes were collected overnight at 4°C by adsorption to protein A/G–Sepharose beads. The beads were next washed and resuspended in 1× Laemmli SDS buffer. Samples were incubated at 95°C for 90 min for de–cross-linking prior electrophoresis.
Construction of baculoviruses.
Baculoviruses overexpressing XPB, p62, p52, p44, p34, cdk7, cyclin H, MAT1, and p8 were as previously described (Dubaele et al., 2003; Coin et al., 2007). cDNAs encoding the various mutated XPD proteins were obtained by PCR site-directed mutagenesis and inserted into either the PVL1392 or the pSK278 vector. The resulting vectors were recombined with baculovirus DNA (BaculoGold; PharMingen). The recombinant viruses were purified from isolated plaques and viral stocks were prepared by three-step growth amplification.
Purification of complexes.
Sf9 insect cells were infected with Flag-XPB, WT or mutated XPD, p62, p52, p44, p34, Flag-cdk7, cyclin H, and Mat1 baculoviruses. The whole-cell extracts were initially incubated 4 h, with agarose beads bound to anti–M2-Flag antibody at 4°C. After washing with a buffer containing 50 mM Tris/HCl (pH 7.9), 20% glycerol, and 50 mM KCl, rIIH9 complexes were eluted for 8–12 h with the same buffer containing 0.2 mg/ml of the epitope peptide (Jawhari et al., 2002). The eluted fraction was then incubated with anti-p44 (1H5) antibody in a buffer containing 50 mM Tris/HCl (pH 7.9), 20% glycerol, and either 50 or 150 mM KCl. The immunoprecipitated fraction was then either boiled or eluted with a synthetic peptide recognized by Ab-p44.
Interaction assays.
WT or mutated XPDs were coexpressed with p44 by infecting Sf9 insect cells with the corresponding baculoviruses. Cell extracts were prepared by homogenizing in 50 mM Tris/HCl (pH 7.9), 150 mM KCl 20% glycerol, 0.1% NP-40, 5 mM β-mercaptoethanol, 1× protease cocktail inhibitor, and clarification by centrifugation (15,000 rpm, 1 h) at 4°C. Immunoprecipitations were performed using Ab-p44 (1H5) cross-linked to protein A–Sepharose beads. After extensive washings (350 mM KCl), Western blots were performed using antibodies raised against p44, the His, or the Flag-tag.
NER assays.
The single-lesion (Pt-GTG) plasmid was prepared as described previously (Araújo et al., 2000). Dual-incision assay was performed in 10 µl of a buffer containing 45 mM HepeS-KOH (pH 7.8), 5 mM MgCl2, 1 mM DTT, 0.3 mM EDTA, 10% glycerol, 2.5 µg BSA, 50 mM KCl, and 2 mM ATP. Each reaction contained XPG (5 ng), XPF/ERCC1 (15 ng), XPC/hHR23B (10 ng), RPA (50 ng), XPA (25 ng), and 30 ng of rIIH complexes, and the reaction was conducted as previously described (Dubaele et al., 2003).
Helicase assays.
Helicase probe was prepared by mixing an oligonucleotide (5 ng) corresponding to fragment 6219–6253 of the single-stranded M13mp18(+) DNA with single-stranded M13mp18(−) phage (1 µg) in the presence of NaCl (25 mM) and MgCl2 (2.5 mM). The mixture was heated for 2 min at 100°C and cooled slowly to room temperature to allow annealing of the DNA heteroduplex. Probe labeling was performed using Klenow fragment in the presence of dTTP (50 mM) and [α-32P]dATP (70 µCi; 3000 Ci/mmol; GE Healthcare). After phenol/chloroform extraction, the labeled probe was purified on a gel filtration column. Helicase assays were then performed as previously described (Coin et al., 1998).
ATPase assays.
The WT and mutated XPD proteins were immunoprecipitated to protein A–Sepharose beads and then incubated 30 min with 1 µCi [γ-32P]ATP (7,000 Ci/mmol; GE Healthcare), 2 nM of cold ATP, 4 µg/ml of pUC309 plasmid in 20 mM Tris/HCl (pH 7.9), 4 mM MgCl2, 50 µg/ml BSA, and 1 mM DTT. The reaction was stopped by adding EDTA (50 mM final). The ATPase hydrolysis was then analyzed as previously described (Marinoni et al., 1997).
In vitro kinase assays.
The highly purified rIIH complexes were incubated 30 min at 30°C with 1 µg of GST-CTD recombinant protein (Drané et al., 2004) in the presence of [γ-32P]ATP (0.14 µM).
Transcription reactions.
Run-off transcription assays were performed using recombinant TFIIB, TFIIE, TFIIF, TBP, endogenous RNA pol II, and the different rIIHs, as previously described (Gerard et al., 1991).
Cell culture.
XP patient (AS552 and AS553) and parental (AS550 and AS551) fibroblasts were a generous gift from A. Sarasin (Institut Gustave Roussy, Villejuif, France). Fibroblasts isolated from the XP135LO patient were a generous gift from A. Lehman (University of Sussex, Brighton, England, UK). XP29BE (GM11613), XP34BE (GM16955) and XP35BE (GM16957) fibroblasts were obtained from the NIGMS Human Genetic Cell Repository, Coriell Institute for Medical Research, Camden, NJ. Normal and XPD-mutated fibroblasts were grown in DME (1 g glucose/liter; Invitrogen) supplemented with 10% FCS and 40 µg/ml gentamicin. Treatments with either triiodothyronine T3 (10 nM) or vitamin D (100 nM) were performed in the presence of red phenol–free medium containing 10% charcoal-treated FCS and 40 µg/ml gentamicin.
Retrotranscription and real-time qPCR.
Total RNAs (1 µg) were reverse transcribed with Moloney murine leukemia virus RT (Invitrogen) using random hexanucleotides. Real-time qPCR was done using the FastStart DNA Master SYBR Green kit and the Lightcycler apparatus (Roche). The primer sequences and the PCR conditions for each target gene are available upon request.
ChIP assays.
15-cm dishes of subconfluent fibroblasts were treated for 24 h with T3 (10 nM) and ChIP experiments were next performed as previously described (Compe et al., 2005). Nonspecific controls have been performed with samples incubated with Sepharose beads in the absence of antibodies. Primers were designed to amplify either the region containing TRE (−2,898 to −2,814) or the proximal promoter (−41 to +39) of KLF9. The nomenclature of the amplicon is according to the transcription initiation site being +1. The primers sequences and the PCR conditions for each target gene are available upon request.
DNaseI footprinting.
The MBP promoter fragment (from −256 to +21) was amplified by using forward primer previously radiolabeled by the T4 Polynucleotide kinase (Biolabs). The probe (20,000 cpm) was incubated with rIIHs (purified at low salt concentration) and TRα1 as previously described (Compe et al., 2007). The MBP-TRE protection was quantified using Genetool software (Syngene). Ratio between the signal intensity in the nonprotected region and the MBP-TRE–protected region were quantified and substracted from the ratio obtained from the digested probe alone.
Online supplemental material.
Fig. S1 shows the complete gels for the excision products obtained during in vitro NER assays and the 5′-3′ helicase activity of the WT and mutated forms of XPD shown in Fig. 5. Fig. S2 shows the complete gels for the basal transcription activity of the rIIHs, the phosphorylation of the RNA polymerase II during in vitro reconstituted transcription assays, and the in vitro phosphorylation by rIIHs of the GST-CTD fusion protein shown in Fig. 6. Fig. S3 A shows the complete gel for the DNase1 footprint analysis shown in Fig. 7 D. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091892/DC1.
Acknowledgments
We wish to dedicate this work to Mathieu, François Guy, and Lydie D. We are grateful to C. Braun for her technical expertise; Ph. Thevenet (Centre Hospitalier de Saint-Malo), H. Dollfus, and V. Laugel (Centre Hospitalier Universitaire de Strasbourg) for their clinical expertise; A. Sarasin and A. Lehmann for the AS550-AS553 and XP135LO fibroblasts, respectively; I. Kolb-Cheynel for the design and production of recombinant baculoviruses; S. Lyonnet and A. Sarasin for fruitful discussions; R. Velez-Cruz for critical reading of the manuscript; and Dr. J.J. DiGiovanna and D. Tamura, R.N. for assisting in the care of the National Institutes of Health patients.
Work in J.M. Egly's laboratory is supported by a European Research Council Advanced Grant (2009). Funds from the French League Against Cancer (Equipe Labellisée) and the French National Research Agency (ANR-08GENOPAT042) supported this study. This research was supported in part by the Intramural Research Program of the U.S. National Institutes of Health, National Cancer Institute, Center for Cancer Research, Bethesda, MD.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- CTD
- C-terminal domain
- NER
- nucleotide excision repair
- TR
- thyroid hormone receptor
- TRE
- thyroid hormone response element
- TTD
- trichothiodystrophy
- XP
- xeroderma pigmentosum
- XPD
- xeroderma pigmentosum group D gene
References
- Akeno N., Saikatsu S., Kawane T., Horiuchi N. 1997. Mouse vitamin D-24-hydroxylase: molecular cloning, tissue distribution, and transcriptional regulation by 1alpha,25-dihydroxyvitamin D3. Endocrinology. 138:2233–2240 10.1210/en.138.6.2233 [DOI] [PubMed] [Google Scholar]
- Akoulitchev S., Mäkelä T.P., Weinberg R.A., Reinberg D. 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature. 377:557–560 10.1038/377557a0 [DOI] [PubMed] [Google Scholar]
- Andressoo J.O., Jans J., de Wit J., Coin F., Hoogstraten D., de Ven M.V., Toussaint W., Huijmans J., Thio H.B., van Leeuwen W.J., et al. 2006. Rescue of progeria in trichothiodystrophy by homozygous lethal Xpd alleles. PLoS Biol. 4:e322 10.1371/journal.pbio.0040322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo S.J., Tirode F., Coin F., Pospiech H., Syväoja J.E., Stucki M., Hübscher U., Egly J.M., Wood R.D. 2000. Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes Dev. 14:349–359 [PMC free article] [PubMed] [Google Scholar]
- Botta E., Nardo T., Lehmann A.R., Egly J.M., Pedrini A.M., Stefanini M. 2002. Reduced level of the repair/transcription factor TFIIH in trichothiodystrophy. Hum. Mol. Genet. 11:2919–2928 10.1093/hmg/11.23.2919 [DOI] [PubMed] [Google Scholar]
- Botta E., Nardo T., Orioli D., Guglielmino R., Ricotti R., Bondanza S., Benedicenti F., Zambruno G., Stefanini M. 2009. Genotype-phenotype relationships in trichothiodystrophy patients with novel splicing mutations in the XPD gene. Hum. Mutat. 30:438–445 10.1002/humu.20912 [DOI] [PubMed] [Google Scholar]
- Boyle J., Ueda T., Gonzalez V., Oh K.S., Imoto K., Inui H., Busch D.B., Khan S.G., Tamura D., DiGiovanna J.J., Kraemer K.H. 2006. Splice mutations in the XPD gene and absence of neurological symptoms. J. Invest. Dermatol. 126:S79 [Google Scholar]
- Boyle J., Ueda T., Oh K.S., Imoto K., Tamura D., Jagdeo J., Khan S.G., Nadem C., Digiovanna J.J., Kraemer K.H. 2008. Persistence of repair proteins at unrepaired DNA damage distinguishes diseases with ERCC2 (XPD) mutations: cancer-prone xeroderma pigmentosum vs. non-cancer-prone trichothiodystrophy. Hum. Mutat. 29:1194–1208 10.1002/humu.20768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broughton B.C., Berneburg M., Fawcett H., Taylor E.M., Arlett C.F., Nardo T., Stefanini M., Menefee E., Price V.H., Queille S., et al. 2001. Two individuals with features of both xeroderma pigmentosum and trichothiodystrophy highlight the complexity of the clinical outcomes of mutations in the XPD gene. Hum. Mol. Genet. 10:2539–2547 10.1093/hmg/10.22.2539 [DOI] [PubMed] [Google Scholar]
- Chen D., Washbrook E., Sarwar N., Bates G.J., Pace P.E., Thirunuvakkarasu V., Taylor J., Epstein R.J., Fuller-Pace F.V., Egly J.M., et al. 2002. Phosphorylation of human estrogen receptor alpha at serine 118 by two distinct signal transduction pathways revealed by phosphorylation-specific antisera. Oncogene. 21:4921–4931 10.1038/sj.onc.1205420 [DOI] [PubMed] [Google Scholar]
- Cleaver J.E., Thompson L.H., Richardson A.S., States J.C. 1999. A summary of mutations in the UV-sensitive disorders: xeroderma pigmentosum, Cockayne syndrome, and trichothiodystrophy. Hum. Mutat. 14:9–22 [DOI] [PubMed] [Google Scholar]
- Coin F., Marinoni J.C., Rodolfo C., Fribourg S., Pedrini A.M., Egly J.M. 1998. Mutations in the XPD helicase gene result in XP and TTD phenotypes, preventing interaction between XPD and the p44 subunit of TFIIH. Nat. Genet. 20:184–188 10.1038/2491 [DOI] [PubMed] [Google Scholar]
- Coin F., Oksenych V., Egly J.M. 2007. Distinct roles for the XPB/p52 and XPD/p44 subcomplexes of TFIIH in damaged DNA opening during nucleotide excision repair. Mol. Cell. 26:245–256 10.1016/j.molcel.2007.03.009 [DOI] [PubMed] [Google Scholar]
- Coin F., Oksenych V., Mocquet V., Groh S., Blattner C., Egly J.M. 2008. Nucleotide excision repair driven by the dissociation of CAK from TFIIH. Mol. Cell. 31:9–20 10.1016/j.molcel.2008.04.024 [DOI] [PubMed] [Google Scholar]
- Compe E., Drané P., Laurent C., Diderich K., Braun C., Hoeijmakers J.H., Egly J.M. 2005. Dysregulation of the peroxisome proliferator-activated receptor target genes by XPD mutations. Mol. Cell. Biol. 25:6065–6076 10.1128/MCB.25.14.6065-6076.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compe E., Malerba M., Soler L., Marescaux J., Borrelli E., Egly J.M. 2007. Neurological defects in trichothiodystrophy reveal a coactivator function of TFIIH. Nat. Neurosci. 10:1414–1422 10.1038/nn1990 [DOI] [PubMed] [Google Scholar]
- de Boer J., Hoeijmakers J.H. 2000. Nucleotide excision repair and human syndromes. Carcinogenesis. 21:453–460 10.1093/carcin/21.3.453 [DOI] [PubMed] [Google Scholar]
- Denver R.J., Williamson K.E. 2009. Identification of a thyroid hormone response element in the mouse Kruppel-like factor 9 gene to explain its postnatal expression in the brain. Endocrinology. 150:3935–3943 10.1210/en.2009-0050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drané P., Compe E., Catez P., Chymkowitch P., Egly J.M. 2004. Selective regulation of vitamin D receptor-responsive genes by TFIIH. Mol. Cell. 16:187–197 10.1016/j.molcel.2004.10.007 [DOI] [PubMed] [Google Scholar]
- Drapkin R., Reardon J.T., Ansari A., Huang J.C., Zawel L., Ahn K., Sancar A., Reinberg D. 1994. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 368:769–772 10.1038/368769a0 [DOI] [PubMed] [Google Scholar]
- Dubaele S., Proietti De Santis L., Bienstock R.J., Keriel A., Stefanini M., Van Houten B., Egly J.M. 2003. Basal transcription defect discriminates between xeroderma pigmentosum and trichothiodystrophy in XPD patients. Mol. Cell. 11:1635–1646 10.1016/S1097-2765(03)00182-5 [DOI] [PubMed] [Google Scholar]
- Dvir A., Tan S., Conaway J.W., Conaway R.C. 1997. Promoter escape by RNA polymerase II. Formation of an escape-competent transcriptional intermediate is a prerequisite for exit of polymerase from the promoter. J. Biol. Chem. 272:28175–28178 10.1074/jbc.272.45.28175 [DOI] [PubMed] [Google Scholar]
- Emmert S., Ueda T., Zumsteg U., Weber P., Khan S.G., Oh K.S., Boyle J., Laspe P., Zachmann K., Boeckmann L., et al. 2009. Strict sun protection results in minimal skin changes in a patient with xeroderma pigmentosum and a novel c.2009delG mutation in XPD (ERCC2). Exp. Dermatol. 18:64–68 10.1111/j.1600-0625.2008.00763.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L., Fuss J.O., Cheng Q.J., Arvai A.S., Hammel M., Roberts V.A., Cooper P.K., Tainer J.A. 2008. XPD helicase structures and activities: insights into the cancer and aging phenotypes from XPD mutations. Cell. 133:789–800 10.1016/j.cell.2008.04.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farsetti A., Mitsuhashi T., Desvergne B., Robbins J., Nikodem V.M. 1991. Molecular basis of thyroid hormone regulation of myelin basic protein gene expression in rodent brain. J. Biol. Chem. 266:23226–23232 [PubMed] [Google Scholar]
- Feaver W.J., Gileadi O., Li Y., Kornberg R.D. 1991. CTD kinase associated with yeast RNA polymerase II initiation factor b. Cell. 67:1223–1230 10.1016/0092-8674(91)90298-D [DOI] [PubMed] [Google Scholar]
- Fujimoto M., Leech S.N., Theron T., Mori M., Fawcett H., Botta E., Nozaki Y., Yamagata T., Moriwaki S., Stefanini M., et al. 2005. Two new XPD patients compound heterozygous for the same mutation demonstrate diverse clinical features. J. Invest. Dermatol. 125:86–92 10.1111/j.0022-202X.2005.23745.x [DOI] [PubMed] [Google Scholar]
- Gerard M., Fischer L., Moncollin V., Chipoulet J.M., Chambon P., Egly J.M. 1991. Purification and interaction properties of the human RNA polymerase B(II) general transcription factor BTF2. J. Biol. Chem. 266:20940–20945 [PubMed] [Google Scholar]
- Gorbalenya A.E., Koonin U.V. 1993. Helicases: amino-acid sequence comparisons and structure-function relationships. Curr. Opin. Struct. Biol. 3:419–429 10.1016/S0959-440X(05)80116-2 [DOI] [Google Scholar]
- Holstege F.C., van der Vliet P.C., Timmers H.T. 1996. Opening of an RNA polymerase II promoter occurs in two distinct steps and requires the basal transcription factors IIE and IIH. EMBO J. 15:1666–1677 [PMC free article] [PubMed] [Google Scholar]
- Iben S., Tschochner H., Bier M., Hoogstraten D., Hozák P., Egly J.M., Grummt I. 2002. TFIIH plays an essential role in RNA polymerase I transcription. Cell. 109:297–306 10.1016/S0092-8674(02)00729-8 [DOI] [PubMed] [Google Scholar]
- Jawhari A., Uhring M., Crucifix C., Fribourg S., Schultz P., Poterszman A., Egly J.M., Moras D. 2002. Expression of FLAG fusion proteins in insect cells: application to the multi-subunit transcription/DNA repair factor TFIIH. Protein Expr. Purif. 24:513–523 10.1006/prep.2001.1597 [DOI] [PubMed] [Google Scholar]
- Johnson R.T., Squires S., Elliott G.C., Koch G.L.E., Rainbow A.J. 1985. Xeroderma pigmentosum D-HeLa hybrids with low and high ultraviolet sensitivity associated with normal and diminished DNA repair ability, respectively. J. Cell Sci. 76:115–133 [DOI] [PubMed] [Google Scholar]
- Keriel A., Stary A., Sarasin A., Rochette-Egly C., Egly J.M. 2002. XPD mutations prevent TFIIH-dependent transactivation by nuclear receptors and phosphorylation of RARalpha. Cell. 109:125–135 10.1016/S0092-8674(02)00692-X [DOI] [PubMed] [Google Scholar]
- Khan S.G., Levy H.L., Legerski R., Quackenbush E., Reardon J.T., Emmert S., Sancar A., Li L., Schneider T.D., Cleaver J.E., Kraemer K.H. 1998. Xeroderma pigmentosum group C splice mutation associated with autism and hypoglycinemia. J. Invest. Dermatol. 111:791–796 10.1046/j.1523-1747.1998.00391.x [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Uchiyama M., Fukuro S., Tanaka K. 2002. Mutations in the XPD gene in xeroderma pigmentosum group D cell strains: confirmation of genotype-phenotype correlation. Am. J. Med. Genet. 110:248–252 10.1002/ajmg.10465 [DOI] [PubMed] [Google Scholar]
- Kraemer K.H., Lee M.M., Scotto J. 1987. Xeroderma pigmentosum. Cutaneous, ocular, and neurologic abnormalities in 830 published cases. Arch. Dermatol. 123:241–250 10.1001/archderm.123.2.241 [DOI] [PubMed] [Google Scholar]
- Kraemer K.H., DiGiovanna J.J., Moshell A.N., Tarone R.E., Peck G.L. 1988. Prevention of skin cancer in xeroderma pigmentosum with the use of oral isotretinoin. N. Engl. J. Med. 318:1633–1637 [DOI] [PubMed] [Google Scholar]
- Kraemer K.H., Lee M.M., Andrews A.D., Lambert W.C. 1994. The role of sunlight and DNA repair in melanoma and nonmelanoma skin cancer. The xeroderma pigmentosum paradigm. Arch. Dermatol. 130:1018–1021 10.1001/archderm.130.8.1018 [DOI] [PubMed] [Google Scholar]
- Kraemer K.H., Patronas N.J., Schiffmann R., Brooks B.P., Tamura D., DiGiovanna J.J. 2007. Xeroderma pigmentosum, trichothiodystrophy and Cockayne syndrome: a complex genotype-phenotype relationship. Neuroscience. 145:1388–1396 10.1016/j.neuroscience.2006.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A.R. 2001. The xeroderma pigmentosum group D (XPD) gene: one gene, two functions, three diseases. Genes Dev. 15:15–23 10.1101/gad.859501 [DOI] [PubMed] [Google Scholar]
- Lehmann A.R. 2003. DNA repair-deficient diseases, xeroderma pigmentosum, Cockayne syndrome and trichothiodystrophy. Biochimie. 85:1101–1111 10.1016/j.biochi.2003.09.010 [DOI] [PubMed] [Google Scholar]
- Marinoni J.C., Rossignol M., Egly J.M. 1997. Purification of the transcription/repair factor TFIIH and evaluation of its associated activities in vitro. Methods. 12:235–253 10.1006/meth.1997.0476 [DOI] [PubMed] [Google Scholar]
- Masutani C., Araki M., Yamada A., Kusumoto R., Nogimori T., Maekawa T., Iwai S., Hanaoka F. 1999. Xeroderma pigmentosum variant (XP-V) correcting protein from HeLa cells has a thymine dimer bypass DNA polymerase activity. EMBO J. 18:3491–3501 10.1093/emboj/18.12.3491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller L.C., Dumitrescu A.M., Walker R.L., Meltzer P.S., Refetoff S. 2005. Thyroid hormone responsive genes in cultured human fibroblasts. J. Clin. Endocrinol. Metab. 90:936–943 10.1210/jc.2004-1768 [DOI] [PubMed] [Google Scholar]
- O’Brien T., Hardin S.E., Greenleaf A., Lis J.T. 1994. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 370:75–77 10.1038/370075a0 [DOI] [PubMed] [Google Scholar]
- Rao T., DiGiovanna J., Tamura D., Ueda T., Boyle J., Nadem C., Oh K.-S., Khan S., Patronas N., Schiffmann R., et al. 2007. Features of both xeroderma pigmentosum and trichothiodystrophy presenting in patients with mutations in the XPD gene. J. Invest. Dermatol. 127:S630 [Google Scholar]
- Rochette-Egly C., Adam S., Rossignol M., Egly J.M., Chambon P. 1997. Stimulation of RAR alpha activation function AF-1 through binding to the general transcription factor TFIIH and phosphorylation by CDK7. Cell. 90:97–107 10.1016/S0092-8674(00)80317-7 [DOI] [PubMed] [Google Scholar]
- Schaeffer L., Roy R., Humbert S., Moncollin V., Vermeulen W., Hoeijmakers J.H., Chambon P., Egly J.M. 1993. DNA repair helicase: a component of BTF2 (TFIIH) basic transcription factor. Science. 260:58–63 10.1126/science.8465201 [DOI] [PubMed] [Google Scholar]
- Singleton M.R., Dillingham M.S., Wigley D.B. 2007. Structure and mechanism of helicases and nucleic acid translocases. Annu. Rev. Biochem. 76:23–50 10.1146/annurev.biochem.76.052305.115300 [DOI] [PubMed] [Google Scholar]
- Sung P., Higgins D., Prakash L., Prakash S. 1988. Mutation of lysine-48 to arginine in the yeast RAD3 protein abolishes its ATPase and DNA helicase activities but not the ability to bind ATP. EMBO J. 7:3263–3269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor E.M., Broughton B.C., Botta E., Stefanini M., Sarasin A., Jaspers N.G., Fawcett H., Harcourt S.A., Arlett C.F., Lehmann A.R. 1997. Xeroderma pigmentosum and trichothiodystrophy are associated with different mutations in the XPD (ERCC2) repair/transcription gene. Proc. Natl. Acad. Sci. USA. 94:8658–8663 10.1073/pnas.94.16.8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Hoffen A., Venema J., Meschini R., van Zeeland A.A., Mullenders L.H. 1995. Transcription-coupled repair removes both cyclobutane pyrimidine dimers and 6-4 photoproducts with equal efficiency and in a sequential way from transcribed DNA in xeroderma pigmentosum group C fibroblasts. EMBO J. 14:360–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steeg H., Kraemer K.H. 1999. Xeroderma pigmentosum and the role of UV-induced DNA damage in skin cancer. Mol. Med. Today. 5:86–94 10.1016/S1357-4310(98)01394-X [DOI] [PubMed] [Google Scholar]
- Vermeulen W., Bergmann E., Auriol J., Rademakers S., Frit P., Appeldoorn E., Hoeijmakers J.H., Egly J.M. 2000. Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat. Genet. 26:307–313 10.1038/81603 [DOI] [PubMed] [Google Scholar]
- Wang Y., Digiovanna J.J., Stern J.B., Hornyak T.J., Raffeld M., Khan S.G., Oh K.S., Hollander M.C., Dennis P.A., Kraemer K.H. 2009. Evidence of ultraviolet type mutations in xeroderma pigmentosum melanomas. Proc. Natl. Acad. Sci. USA. 106:6279–6284 10.1073/pnas.0812401106 [DOI] [PMC free article] [PubMed] [Google Scholar]