Abstract
Background
Galactokinase catalyses the first committed step of galactose catabolism in which the sugar is phosphorylated at the expense of MgATP. Recent structural studies suggest that the enzyme makes several contacts with galactose – five side chain and two main chain hydrogen bonds. Furthermore, it has been suggested that inhibition of galactokinase may help sufferers of the genetic disease classical galactosemia which is caused by defects in another enzyme of the pathway galactose-1-phosphate uridyl transferase. Galactokinases from different sources have a range of substrate specificities and a diversity of kinetic mechanisms. Therefore only studies on the human enzyme are likely to be of value in the design of therapeutically useful inhibitors.
Results
Using recombinant human galactokinase expressed in and purified from E. coli we have investigated the sugar specificity of the enzyme and the kinetic consequences of mutating residues in the sugar-binding site in order to improve our understanding of substrate recognition by this enzyme. D-galactose and 2-deoxy-D-galactose are substrates for the enzyme, but N-acetyl-D-galactosamine, L-arabinose, D-fucose and D-glucose are all not phosphorylated. Mutation of glutamate-43 (which forms a hydrogen bond to the hydroxyl group attached to carbon 6 of galactose) to alanine results in only minor changes in the kinetic parameters of the enzyme. Mutation of this residue to glycine causes a ten-fold drop in the turnover number. In contrast, mutation of histidine 44 to either alanine or isoleucine results in insoluble protein following expression in E. coli. Alteration of the residue that makes hydrogen bonds to the hydroxyl attached to carbons 3 and 4 (aspartate 46) results in an enzyme that although soluble is essentially inactive.
Conclusions
The enzyme is tolerant to small changes at position 2 of the sugar ring, but not at positions 4 and 6. The results from site directed mutagenesis could not have been predicted from the crystal structure alone and needed to be determined experimentally.
Background
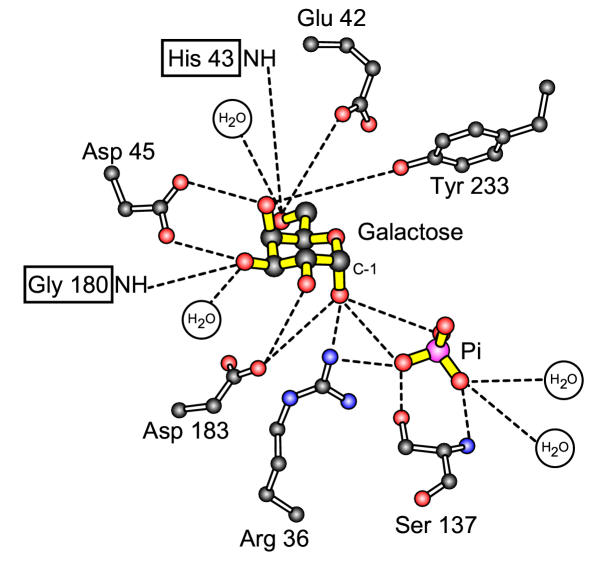
Galactose is metabolised via the Leloir pathway [1] in order to produce glucose-6-phosphate that can enter glycolysis. The first committed step of this pathway is the phosphorylation of galactose at the expense of ATP – a reaction that is catalysed by the enzyme galactokinase. Lack of functional galactokinase in humans is one cause of the inherited disease galactosemia [2-4]. The main symptom of this disease, which is treatable by the complete removal of lactose and galactose from the diet, is early onset cataracts. The enzyme has been purified and characterised from a variety of different sources including bacteria [5], yeast [6,7], plants [8,9] and mammals [10-12]. The primary sequence of these enzymes reveals only limited sequence similarity except at five highly conserved motifs [13]. The first of these motifs – the so-called galactokinase signature motif – has been implicated in galactose binding [12,13]. Recent structural data [14] on the galactokinase from the bacterium Lactococcus lactis confirms this hypothesis and shows that most of the contacts between the sugar and the protein are provided by residues in this motif (Fig. 1). The cavity in which galactose binds is, in part, defined by a histidine residue (H43 in L. lactis which is equivalent to H44 in the human enzyme). The side-chain of this residue is located close to, but not in contact with, the hydroxyl attached to carbon 6 of the sugar.
Figure 1.
The structure of the galactose and phosphate binding sites from L. lactis galactokinase [14]. The protein was crystallised in the presence of both galactose and inorganic phosphate (both shown with yellow bonds). The following colour scheme has been used for the atoms: Carbon = black, Oxygen = red, Nitrogen = blue, Phosphate = pink. Distances of less than 0.32 nm are shown as dashed lines. Figure courtesy of Hazel Holden (University of Wisconsin).
Another, and more common, cause of galactosemia is deficiency of the next enzyme in the Leloir pathway, galactose-1-phosphate uridyl transferase (GALT) [2,3]. The symptoms of this deficiency are generally more severe and include, in addition to cataracts, damage to the brain, liver and kidneys – effects which cannot be reversed or even completely prevented by the exclusion of galactose and lactose from the diet. This increased severity is believed to result from the build up of the toxic metabolite galactose-1-phosphate. The mechanism of galactose-1-phosphate toxicity is not known. However, in brain at least it may be linked to the substantial (five-fold in conditions designed to mimic those observed in GALT-deficient patients) increases in Mg-ATPase activity and consequent depletion of ATP within the cell [15]. Recently, it has been suggested that inhibition of galactokinase in GALT-deficient patients might be used in addition to diet to prevent the build up of galactose-1-phosphate and thus the development of the more severe symptoms [16].
The yeast enzyme is reported as having high specificity for the sugar substrate with no ability to phosphorylate glucose, mannose, galactitol, arabinose, 2-deoxygalactose, fucose or lactose [6]. The rat liver enzyme can phosphorylate 2-deoxygalactose [11] and the enzyme from fenugreek seeds can use 2-deoxygalactose and fucose as substrates [8]. A detailed study of the substrate specificity of the E. coli galactokinase showed that the enzyme was moderately active (less than 10-fold reduction in kcat) with 2-deoxygalactose and 2-aminogalactose (but inactive with N-acetylgalactosamine) and weakly active (10 to 20-fold reduction in kcat) with fucose [17]. The enzymes from rat liver and yeast have the same kinetic mechanism as the human one – an ordered ternary complex mechanism in which ATP is the first substrate to bind [7,10,12]. In contrast the plant enzyme has an ordered mechanism in which galactose binds first [8] and the enzyme from E. coli has a random mechanism in which either substrate can bind first [18].
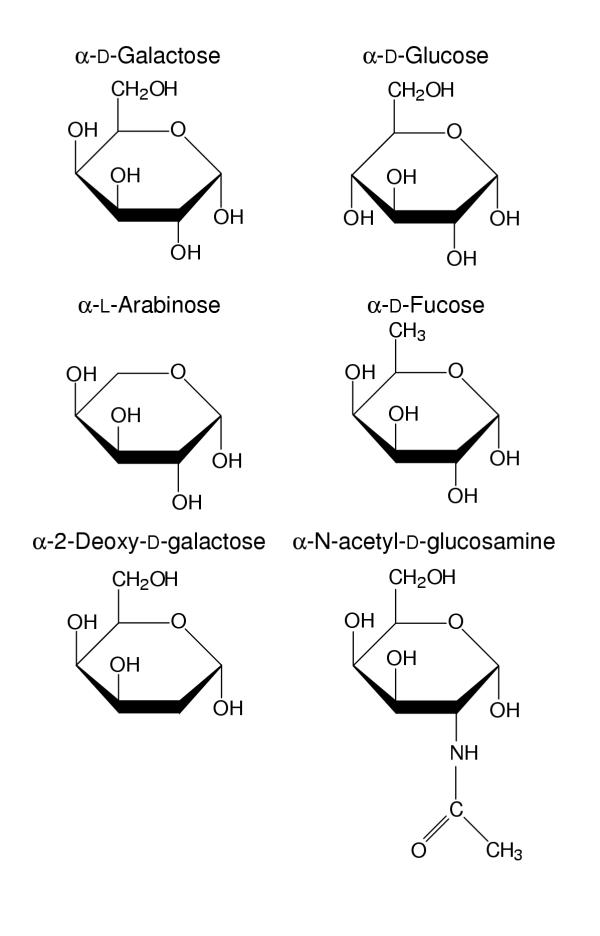
No sugar specificity study has yet been carried out with the human enzyme. The variety of substrate specificities and the diversity of reaction mechanisms mean that it is imperative that studies on the human enzyme be carried out in order to inform, accurately, any future study of therapeutically useful inhibitors of the galactokinase reaction. We were particularly interested in sugars that differ at carbons 4 and 6 – parts of the molecule which make hydrogen bonds with the protein. D-Glucose differs from D-galactose only in the configuration of the hydroxyl group at position 4 (Fig 2). D-fucose (6-deoxy-D-galactose) differs from galactose in that it lacks a hydroxyl group at position 6. L-arabinose lacks carbon 6 (and its associated hydroxyl) altogether.
Figure 2.
The structures of the sugars used in this investigation.
The importance of the aspartate and histidine residues in the recognition of the sugar is underlined by the observation that the related sugar kinase, arabinose kinase, has an almost identical sequence in motif I except that these two residues are altered to glycine and isoleucine, respectively [19]. We complemented our sugar specificity studies by mutating three key, conserved residues (Glu-43, His-44 and Asp-46) in the sugar binding site of the human galactokinase and assessing the kinetic consequences of these changes. Interestingly although the abolition of the carbon 6 hydroxyl in arabinose and fucose results in no activity, deleting the side chain which makes a hydrogen bond with this part of the sugar causes little change in the steady state kinetics of the galactokinase reaction.
Results
Human galactokinase shows high specificity at carbons 4 and 6 of the sugar
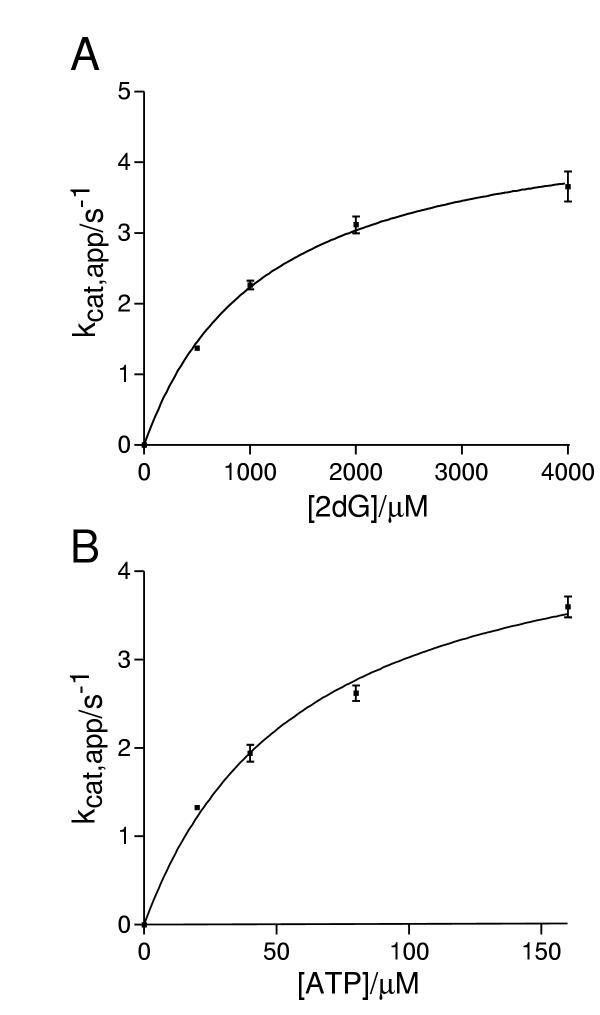
The ability of recombinant human galactokinase to catalyse the phosphorylation of sugars with structures similar to the natural substrate galactose was tested. No activity was observed with D-glucose, D-fucose, L-arabinose or N-acetyl-D-galactosamine, even when these sugars were present at high (100 mM) concentrations. Nor did any of these sugars act as inhibitors of the galactokinase reaction (data not shown). In contrast, 2-deoxy-D-galactose was a substrate for the enzyme (Fig. 3). The kinetic parameters were not much changed from those with galactose as substrate with no change greater than four-fold.
Figure 3.
2-deoxy-D-galactose (2dG) is a substrate for human galactokinase. Steady state kinetic analysis was carried out as described in Materials and Methods. Each kcat,app value is derived, using non-linear curve-fitting [25], from a set of rates data at a single sub-saturating substrate concentration. Error bars represent the standard error in kcat,app as derived by from the fitting process. The lines are non-linear curve fits of the kcat,app data to the equation kcat,app = kcat[substrate]/(Km + [substrate]). The following parameters were derived from this fitting: kcat = 4.8 ± 0.3 s-1; Km,ATP = 59 ± 9 μM; Km,2dG = 1100 ± 110 μM; kcat/Km,ATP = (8.1 ± 0.8) × 104 l.mol-1.s-1; kcat/Km,2dG = 4200 ± 270 l.mol-1.s-1.
Mutation of glutamate 43 to alanine has little effect on the steady state kinetics
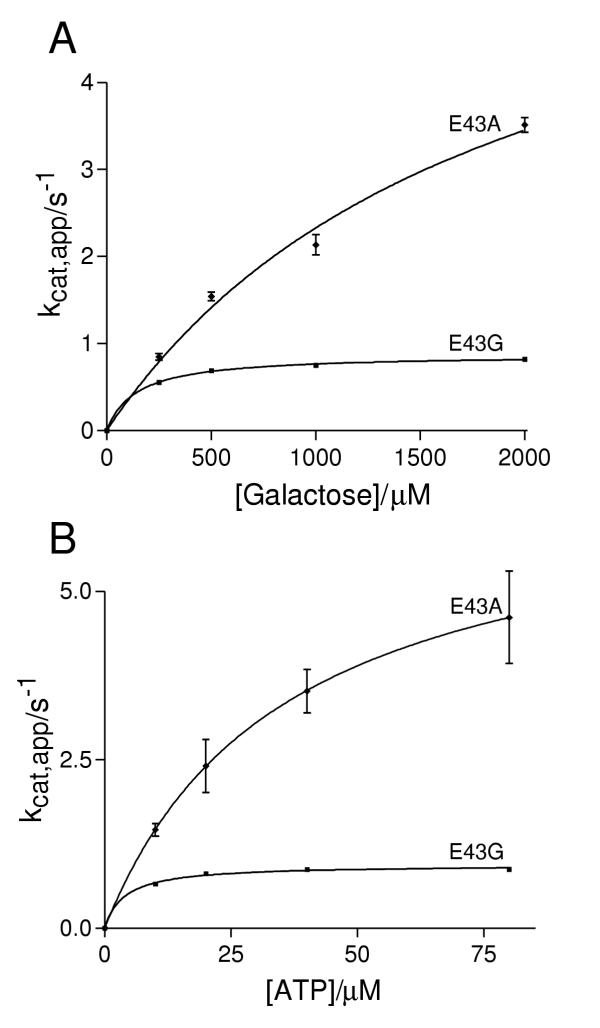
Since the elimination of the carbon-6 hydroxyl (in L-arabinose and D-fucose) results in a major reduction in the sugar-enzyme affinity we mutated glutamate 43 to alanine (the equivalent residue to Glu-42 in Fig. 1) in order to eliminate the side chain which contacts this part of the sugar. This mutant protein was soluble following expression in E. coli and could be purified. However, the yield obtained was approximately one third that of the wild type protein. Steady state kinetic analysis of the E43A mutant (Fig. 4) revealed few changes in the kinetic parameters (Table 1). This mutant, in common with the wild-type, has no detectable activity with D-fucose, L-arabinose and D-glucose as substrates.
Figure 4.
Kinetic analysis of the soluble mutants, E43A and E43G. Error bars represent the standard error as derived from the non-linear curve fitting (see materials and methods). The kinetic parameters derived from these curve fits are detailed in table 1.
Table 1.
Kinetic parameters of the mutant galactokinases. All reactions were carried out at 37°C as described in the materials and methods. Values were derived by non-linear curve fitting as implemented in the program GraphPad Prism and are shown ± standard error as derived from this procedure.
| Mutant | kcat/s-1 | Km,gal/μM | Km,ATP/μM | kcat/Km,gal/l.mol-1.s-1 | kcat/Km,ATP/105 × l.mol-1.s-1 |
| Wild Type(1) | 8.7 ± 0.5 | 970 ± 220 | 34 ± 4 | 8900 ± 2900 | 2.6 ± 0.4 |
| E43A | 6.7 ± 0.02 | 1900 ± 500 | 35 ± 0.3 | 3400 ± 430 | 1.9 ± 0.009 |
| E43G | 0.9 ± 0.02 | 140 ± 10 | 3.9 ± 0.6 | 6100 ± 330 | 2.4 ± 0.3 |
| H44A | Not soluble | ||||
| H44I | Not soluble | ||||
| E43G/H44I | Not soluble | ||||
| D46A | No detectable activity |
Notes (1) Previously reported in [12]
Changing the sequence to match that of arabinose kinase results in insoluble protein
A double mutant E43G/H44I was constructed so as to alter the galactokinase sequence to that of arabinose kinase in the region which interacts with carbon 6. Although this protein could be expressed in E. coli, the majority was not in the soluble fraction following sonication of the bacterial cells. The small amount of protein that was soluble had minimal activity with D-galactose, D-glucose, L-arabinose or D-fucose (data not shown). Similar results were observed with the single mutants H44I and H44A. The single amino acid change, E43G does result in soluble, active protein (Fig. 4) although like E43A the yield is reduced on purification. This mutation causes an approximate ten-fold reduction in the turnover number (Table 1). However the specificity constants for both substrates are essentially unchanged compared to the wild-type.
Alteration of aspartate 46 results in an inactive enzyme
Mutation of the residue that contacts C3-OH and C4-OH on galactose results in a protein which can be readily expressed and purified from E. coli (the yield after purification was comparable to wild-type, data not shown). However, we were unable to detect any activity towards D-galactose, L-arabinose, D-fucose and D-glucose with this mutant, even at high concentrations of substrate (50 mM) and enzyme (0.48 μM enzyme compared to 0.067 μM with the E43A and E43G mutants).
Discussion
The availability of both the structure of the galactokinase from L. lactis [14] and the readily expressible human form of the enzyme [12] mean that, for the first time, it is possible to interrogate the structure and function of the enzyme by site-directed mutagenesis. Here we report the results of some mutations to residues in motif I, which contributes most of the residues involved in galactose binding and recognition.
Elimination of the Glu-43 to C6-OH hydrogen bond contact by mutating the side chain to alanine resulted in no major changes in the steady state kinetic parameters. In contrast altering this residue to glycine caused a substantial drop in the turnover number with no corresponding drop in the specificity constants. A reduction in kcat with no change in the specificity constant can only occur if Km drops by a similar factor to the turnover number. It is often (erroneously) assumed that Km is a measure of the enzyme-substrate affinity. In fact it is a summed measure of the interaction between the substrate and the enzyme across all stages of the catalytic cycle (not just the initial enzyme-substrate encounter). In contrast, the specificity constant is a rate constant which does report on this initial encounter [20]. Thus, in the case of E43G the kinetic constants tell us that the mutation causes little or no change in the rate of interaction between either ATP or galactose and the enzyme, that in at least one stage in the catalytic cycle the enzyme binds each substrate more tightly than the wild-type but that the overall rate of catalysis is reduced. A similar situation was observed in our studies of disease-causing mutants in human galactokinase – the mutant G346S which had a severely impaired kcat, but a reduction in the Km for ATP means that the specificity constant for this substrate is not much changed compared to wild-type [12].
It is interesting to note that although elimination of the Glu-43 side chain causes only modest changes to the kinetic parameters of galactokinase, whereas the elimination of the group it hydrogen bonds to in the sugar (C6-OH) in either D-fucose or L-arabinose, results in the complete loss of detectable activity. Furthermore, these compounds are not inhibitors of the reaction, suggesting that they have essentially no affinity for the enzyme. This apparently paradoxical result can be explained by the different thermodynamic consequences of eliminating the charged and uncharged group in a charged hydrogen bond. Deletion of the uncharged group (i.e., in this case, the hydroxyl group on the sugar) leaves the charged carboxyl group in the protein unpaired. This is considerably energetically destabilised compared to the hydrogen bond (approximately 15 to 20 kJ.mol-1 [21,22]). In contrast, elimination of the charged group leaving the uncharged group is only slightly destabilised compared to the hydrogen bond (2 to 6 kJ.mol-1 [21]). Mutating Glu-43 to glycine not only eliminates the charged component of the hydrogen bond, but also increases the flexibility of the peptide backbone. This increased mobility is the most likely explanation for the dramatic effects seen with this mutant on kcat. A less rigid active site structure may well have a lower affinity for the transition state and, consequently, result in reduced activity.
Mutation of His-44 to either alanine or isoleucine resulted in insoluble protein following expression in E. coli. In our previous studies of the disease-causing mutations in human galactokinase, mutation of this residue to tyrosine resulted in an enzyme which was soluble but which was deficient in its interaction with galactose [12]. Taken together this suggests that, in addition to any catalytic roles that this residue might play, it also has an important role in maintaining the structural integrity of the protein as a whole.
Although it might be expected that the D46A mutation would have similar consequences to E43A since both eliminated the charged group involved in two charged hydrogen bonds, it does not. Indeed this mutant has no detectable galactokinase activity. It is possible that this residue has important roles to play in forming the active site prior to galactose binding. Detailed speculation may therefore have to wait until the availability of a structure in the absence of a bound sugar molecule.
The combination of a readily expressible form of the human galactokinase [12] and the first crystal structure of the enzyme [14] mean that it is possible to design mutations in the enzyme to address specific questions. These results extend our studies on the enzyme which previously concentrated on point mutations which had been implicated in causing the genetic disease galactosemia [12]. In that study we looked at several mutations either in motif I (G36R, H44Y), close to it in sequence (P28T, V32M) or close in space (G346S, G347S, G349S). Of these, three (P28T, V32M and G36R) proved to be insoluble on induction in E. coli suggesting that, like H44A, H44I and E43G/H44I in the current study these mutations affect the overall folding and stability of the protein. The soluble mutants (H44Y, G346S, G347S and G349S) all had effects on the constants which report on the interaction between the enzyme and galactose (Km,gal and kcat/Km,gal) suggesting that these residues formed part of, or were close to, the sugar binding site – a conclusion confirmed by the recent crystal structure [14].
Conclusions
Recently, there has been a resurgence of interest in galactokinase as an enzyme and in the type galactosemia caused by its deficiency. If it does prove clinically viable to treat GALT-deficient galactosemia sufferers with galactokinase inhibiting drugs [16], the design of these drugs will require a thorough understanding of both the structure of the sugar binding site and how this site interacts with the substrate. Here we show that the enzyme will tolerate minor changes in the substrate at position 2 of the sugar ring, but not at positions 4 and 6. Alteration of residues by site directed mutagenesis which are close to, or in contact, with the sugar result in variety of effects ranging from insoluble protein through inactive enzyme to little observable change. These results could not have been predicted directly from the crystal structure and had to be determined experimentally. The combined approach of structural analysis and mutagenesis is likely to yield further insights in the coming months and years.
Methods
Protein expression and purification
Human galactokinase was expressed in E. coli as a His6-fusion protein and purified from this source by nickel agarose chromatography as described previously [12]. Point mutations were introduced in to the coding sequence using the QuikChange method [23] and the mutant proteins expressed and purified using the same protocol. The full DNA sequence of each mutant galactokinase was verified (DNA Sequencing Service, Department of Medicine, University of Manchester). Protein concentrations were estimated by the method of Bradford [24].
Galactokinase kinetics
The kinetics of the galactokinase reaction was measured by coupling the production of ADP to the reactions of pyruvate kinase and lactate dehydrogenase at 37°C in a total reaction volume of 150 μl [12,13]. The decrease in A340nm, which results from the oxidation of NADH, was measured in a Multiskan Ascent micro-titre plate reader. The reaction mix contained 20 mM HEPES-OH pH 8.0, 150 mM NaCl, 5 mM MgCl2, 1 mM KCl, 10%(v/v) glycerol, 1 mM NADH, 1 mM DTT, 400 μM PEP, 7.5 U pyruvate kinase (Sigma) and 10 U lactate dehydrogenase (Sigma). All sugars were purchased from Sigma-Aldrich. Reactions were initiated by the addition of enzyme.
It has previously been established that human galactokinase follows an ordered, ternary complex mechanism [12] for which the rate equation is given by v = (kcat. [E]0. [gal]. [ATP])/(KI,ATP.Km,gal + Km,gal. [ATP] + Km,ATP. [gal] + [ATP]. [gal]) where [gal] and [ATP] are the concentrations of galactose and ATP respectively, KI,ATP is a constant relating to the dissociation of the enzyme-ATP complex and Km,gal and Km,ATP are the Michaelis constants for galactose and ATP respectively. At any constant value of [gal] this simplifies to v = kcat,app. [E]0. [ATP]/(Km,ATP,app + [ATP]) where kcat,app = kcat. [gal]/(Km,gal + [gal]). A similar situation holds when [ATP] is held constant [12]. A 5 × 5 concentration grid with a range of sub-saturating constant concentrations of ATP and galactose was set up. Each row or column gave a set of rates at a constant concentration of one substrate. Using these data, kcat,app for each value of [galactose] and [ATP] and the associated standard error were calculated using non-linear curve fitting [25]. Secondary plots were then constructed in which these kcat,app values were plotted against the relevant substrate concentration. Non-linear curve fitting [25] was then used to derive the absolute kcat, Km and kcat/Km values and their associated standard errors. This method has the advantage of avoiding using "saturating" concentrations of substrate which may not be truly saturating, may not be easy to obtain experimentally and may result in unwanted substrate inhibition [26].
Authors' Contributions
RJR conceived of and directed the project. DJT constructed (or supervised the construction of) the mutants, purified the proteins and carried out the kinetic assays and analysis. Both authors contributed to the interpretation of the data and have read and approved the contents of this manuscript.
Acknowledgments
Acknowledgements
We thank David Rice (University of Sheffield, UK) and Hazel Holden (University of Wisconsin, USA) for sharing data prior to publication. We also thank Alison Swinnerton who constructed the E43G mutant as part of her undergraduate project. This work was funded by the BBSRC and the Leverhulme Trust.
Contributor Information
David J Timson, Email: d.timson@qub.ac.uk.
Richard J Reece, Email: Richard.Reece@man.ac.uk.
References
- Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. FASEB J. 1996;10:461–470. [PubMed] [Google Scholar]
- Novelli G, Reichardt JK. Molecular basis of disorders of human galactose metabolism: past, present, and future. Mol Genet Metab. 2000;71:62–65. doi: 10.1006/mgme.2000.3073. [DOI] [PubMed] [Google Scholar]
- Petry KG, Reichardt JK. The fundamental importance of human galactose metabolism: lessons from genetics and biochemistry. Trends Genet. 1998;14:98–102. doi: 10.1016/S0168-9525(97)01379-6. [DOI] [PubMed] [Google Scholar]
- Stambolian D, Ai Y, Sidjanin D, Nesburn K, Sathe G, Rosenberg M, Bergsma DJ. Cloning of the galactokinase cDNA and identification of mutations in two families with cataracts. Nat Genet. 1995;10:307–312. doi: 10.1038/ng0795-307. [DOI] [PubMed] [Google Scholar]
- Verhees CH, Koot DG, Ettema TJ, Dijkema C, de Vos WM, van der Oost J. Biochemical adaptations of two sugar kinases from the hyperthermophilic archaeon Pyrococcus furiosus. Biochem J. 2002;366:121–127. doi: 10.1042/BJ20011597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Wilson DB. Purification and properties of galactokinase from Saccharomyces cerevisiase. J Biol Chem. 1977;252:1162–1166. [PubMed] [Google Scholar]
- Timson DJ, Reece RJ. Kinetic analysis of yeast galactokinase: Implications for transcriptional activation of the GAL genes. Biochimie. 2002;84:265–272. doi: 10.1016/S0300-9084(02)01399-8. [DOI] [PubMed] [Google Scholar]
- Foglietti MJ, Percheron F. Purification et mécanisme d'action d'une galactokinase végétale. Biochimie. 1976;58:499–504. doi: 10.1016/s0300-9084(76)80218-0. [DOI] [PubMed] [Google Scholar]
- Dey PM. Galactokinase of Vicia faba seeds. Eur J Biochem. 1983;136:155–159. doi: 10.1111/j.1432-1033.1983.tb07720.x. [DOI] [PubMed] [Google Scholar]
- Ballard FJ. Kinetic studies with liver galactokinase. Biochem J. 1966;101:70–75. doi: 10.1042/bj1010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DG, Khan HH. Some properties of galactokinase in developing rat liver. Biochem J. 1968;108:169–175. doi: 10.1042/bj1080169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timson DJ, Reece RJ. Functional analysis of disease-causing mutations in human galactokinase. Eur J Biochem. 2003;270:1767–1774. doi: 10.1046/j.1432-1033.2003.03538.x. [DOI] [PubMed] [Google Scholar]
- Platt A, Ross HC, Hankin S, Reece RJ. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc Natl Acad Sci USA. 2000;97:3154–3159. doi: 10.1073/pnas.97.7.3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoden JB, Holden HM. Molecular structure of galactokinase. J Biol Chem. 2003;278:33305–33311. doi: 10.1074/jbc.M304789200. [DOI] [PubMed] [Google Scholar]
- Tsakiris S, Marinou K, Schulpis KH. The in vitro effects of galactose and its derivatives on rat brain Mg2+-ATPase activity. Pharmacol Toxicol. 2002;91:254–257. doi: 10.1034/j.1600-0773.2002.910506.x. [DOI] [PubMed] [Google Scholar]
- Bosch AM, Bakker HD, van Gennip AH, van Kempen JV, Wanders RJ, Wijburg FA. Clinical features of galactokinase deficiency: a review of the literature. J Inherit Metab Dis. 2002;25:629–634. doi: 10.1023/A:1022875629436. [DOI] [PubMed] [Google Scholar]
- Yang J, Fu X, Jia Q, Shen J, Biggins JB, Jiang J, Zhao J, Schmidt JJ, Wang PG, Thorson JS. Studies on the substrate specificity of Escherichia coli galactokinase. Org Lett. 2003;5:2223–2226. doi: 10.1021/ol034642d. [DOI] [PubMed] [Google Scholar]
- Gulbinsky JS, Cleland WW. Kinetic studies of Escherichia coli galactokinase. Biochemistry. 1968;7:566–575. doi: 10.1021/bi00842a009. [DOI] [PubMed] [Google Scholar]
- Sherson S, Gy I, Medd J, Schmidt R, Dean C, Kreis M, Lecharny A, Cobbett C. The arabinose kinase, ARA1, gene of Arabidopsis is a novel member of the galactose kinase gene family. Plant Mol Biol. 1999;39:1003–1012. doi: 10.1023/A:1006181908753. [DOI] [PubMed] [Google Scholar]
- Fersht A. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. New York: WH Freeman. 1999.
- Fersht AR, Shi JP, Knill-Jones J, Lowe DM, Wilkinson AJ, Blow DM, Brick P, Carter P, Waye MM, Winter G. Hydrogen bonding and biological specificity analysed by protein engineering. Nature. 1985;314:235–238. doi: 10.1038/314235a0. [DOI] [PubMed] [Google Scholar]
- Fersht AR. Relationships between apparent binding energies measured in site-directed mutagenesis experiments and energetics of binding and catalysis. Biochemistry. 1988;27:1577–1580. doi: 10.1021/bi00405a027. [DOI] [PubMed] [Google Scholar]
- Wang W, Malcolm BA. Two-stage PCR protocol allowing introduction of multiple mutations, deletions and insertions using QuikChange site-directed mutagenesis. Biotechniques. 1999;26:680–682. doi: 10.2144/99264st03. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- Marquardt D. An algorithm for least squares estimation of nonlinear parameters. SIAM J Appl Math. 1963;11:431–441. [Google Scholar]
- Cornish-Bowden A. Fundamentals of enzyme kinetics. London, UK: Portland Press. 1995.