Abstract
Nodal-signaling is required for specification of mesoderm, endoderm, establishing left-right asymmetry and craniofacial development. Wdr68 is a WD40-repeat domain-containing protein recently shown to be required for endothelin-1 (edn1) expression and subsequent lower jaw development. Previous reports detected the Wdr68 protein in multiprotein complexes containing mammalian members of the dual-specificity tyrosine-regulated kinase (dyrk) family. Here we describe the characterization of the zebrafish dyrk1b homolog. We report the detection of a physical interaction between Dyrk1b and Wdr68. We also found perturbations of Nodal signaling in dyrk1b antisense morpholino knockdown (dyrk1b-MO) animals. Specifically, we found reduced expression of lft1 and lft2 (lft1/2) during gastrulation and a near complete loss of the later asymmetric lft1/2 expression domains. Although wdr68-MO animals did not display lft1/2 expression defects during gastrulation, they displayed a near complete loss of the later asymmetric lft1/2 expression domains. While expression of ndr1 was not substantially effected during gastrulation, ndr2 expression was moderately reduced in dyrk1b-MO animals. Analysis of additional downstream components of the Nodal signaling pathway in dyrk1b-MO animals revealed modestly expanded expression of the dorsal axial mesoderm marker gsc while the pan-mesodermal marker bik was largely unaffected. The endodermal markers cas and sox17 were also moderately reduced in dyrk1b-MO animals. Notably, and similar to defects previously reported for wdr68 mutant animals, we also found reduced expression of the pharyngeal pouch marker edn1 in dyrk1b-MO animals. Taken together, these data reveal a role for dyrk1b in endoderm formation and craniofacial patterning in the zebrafish.
Keywords: Mirk, dyrk1b, wdr68, craniofacial, edn1
INTRODUCTION
Wdr68 is a WD40-repeat domain-containing protein originally identified as the vertebrate homolog of the petunia gene AN11 (de Vetten et al., 1997). AN11 is required for production of the anthocyanin pigments in flower petals. The Wdr68 protein is also 45% similar to the arabidopsis TTG1 protein that is essential for proper specification of hair cell fates in the developing shoot and root (Schiefelbein, 2003; Walker et al., 1999). More recently, the wdr68 gene was identified in the zebrafish through an insertional mutagenesis screen as essential for development of the craniofacial apparatus (Amsterdam et al., 2004; Nissen et al., 2006). The wdr68 gene is essential for both upper and lower jaw development in the zebrafish embryo. The wdr68 gene is required upstream of the endothelin-1 (edn1) ligand that is in turn essential for the expression of several transcription factors including members of the distal-less (dlx) gene family that pattern the cranial neural crest giving rise to the lower jaw (Clouthier and Schilling, 2004; Miller et al., 2000; Nissen et al., 2006). Notably, edn1 is not required for the embryonic upper jaw cartilage indicating that additional roles for wdr68 beyond a requirement for edn1 expression remain to be identified.
The mammalian Wdr68 protein has been detected in large multiprotein complexes containing two members of the dual-specificity tyrosine-regulated kinase (dyrk) family, Dyrk1a and Dyrk1b (Lim et al., 2002; Skurat and Dietrich, 2004). Initial characterizations of human DYRK1B/MIRK revealed it to encode a nuclear localized protein enriched in skeletal muscle, testes, brain, heart and spleen with low levels of expression in most other tissues (Leder et al., 2003; Leder et al., 1999; Lee et al., 2000). Several subsequent analyses showed Dyrk1b to be an important modulator of the G0-G1 cell cycle transition that is important for maintaining or achieving the differentiated state in several transformed cell lines (Deng et al., 2009; Jin et al., 2009). In the mouse myogenic C2C12 cell line, Dyrk1b inhibits association between Mef2d and the histone deacetylase MITR. This Dyrk1b-mediated differentiation switch is required for myogenin (myog) expression and subsequent myotube formation (Deng et al., 2005; Deng et al., 2003). In flies, a member of the dyrk family, minibrain (mnb), is thought to play a role in cell proliferation because mnb mutant flies have fewer neurons in specific brain regions (Tejedor et al., 1995). Similarly, a yeast homolog, yak1, is required for growth control and pseudohyphal differentiation (Garrett and Broach, 1989; Zhang et al., 2001). Collectively, these data suggest a potential role for Dyrk1b-containing protein complexes in regulating cell differentiation events in vertebrates.
The Nodal-signaling pathway is required for specification of mesoderm, endoderm, establishing left-right asymmetry, and craniofacial development (Schier, 2003; Tian and Meng, 2006). During embryogenesis, the absence of Nodal signaling yields predominantly ectoderm, low levels of Nodal signaling are needed for mesoderm specification, and high levels are needed for endoderm specification (Gritsman et al., 2000; Schier et al., 1997; Thisse et al., 2000). The Nodal-dependent transcription factors casanova (cas) and sox17 are essential for endoderm specification (Alexander and Stainier, 1999; Dickmeis et al., 2001; Kikuchi et al., 2001). Later in development, signals emanating from the pharyngeal endoderm are important for craniofacial development (David et al., 2002; Piotrowski and Nusslein-Volhard, 2000). The endothelin-1 (edn1) pathway is essential for lower jaw formation in several species (Crump et al., 2004; Depew et al., 2002; Kempf et al., 1998; Kurihara et al., 1994; Miller et al., 2000; Nissen et al., 2003). The small peptide ligand Edn1 is secreted by the pharyngeal pouch ectoderm and endoderm cells as well as by the arch core mesoderm. Edn1 signals through binding to a seven-transmembrane G protein-coupled receptor expressed on neural crest cells to regulate a downstream network of transcription factors important for lower jaw development (Clouthier et al., 1998; Clouthier et al., 2000; Ivey et al., 2003). The wdr68 gene is required for edn1 expression in the zebrafish (Nissen et al., 2006).
Although nuclear localization of both Wdr68 and a Dyrk1-family member was shown (Nissen et al., 2006), a direct physical interaction between the zebrafish Wdr68 and Dyrk1b proteins has not been demonstrated. Likewise, the potential functional relationships between Wdr68 and Dyrk1b remain uncharacterized. To facilitate studies on the Dyrk1-Wdr68 complexes, we searched the zebrafish genome for genes homologous to the mammalian dyrk1b gene and identified a single zebrafish dyrk1b gene. Here, we report the detection of a physical interaction between zebrafish Dyrk1b and Wdr68, the expression pattern for dyrk1b during early development, and an initial characterization of dyrk1b antisense morpholino (dyrk1b-MO) knockdown animals.
RESULTS
Isolation of zebrafish dyrk1b
We performed BLAST searches on the zebrafish genome (Ensembl database assembly Zv8) to identify genes encoding proteins homologous to the human Dyrk1b protein. We found the putative zebrafish gene on chromosome 16. Using Clustal W to analyze sequence similarities, we found that it encodes a predicted protein that is 60% similar to human Dyrk1b and only 56% similar to human Dyrk1a (Thompson et al., 1994). In accordance with Zebrafish nomenclature, we will from here forward refer to the chromosome 16 homolog as dyrk1b.
To characterize zebrafish dyrk1b further, we isolated cDNA fragments by performing 5′-RACE from cDNA of mixed zebrafish developmental stages using primers to the highly conserved kinase domain. Sequencing identified partial clones further confirming that the putative homolog is transcribed and spliced in embryos. Our sequencing-based assembled full-length sequence of dyrk1b was submitted to Genbank (GQ449256) and, notably, represents a 100% match to the independently predicted open reading frame contained in file XM_678964.
Zebrafish Dyrk1b and Wdr68 can physically interact
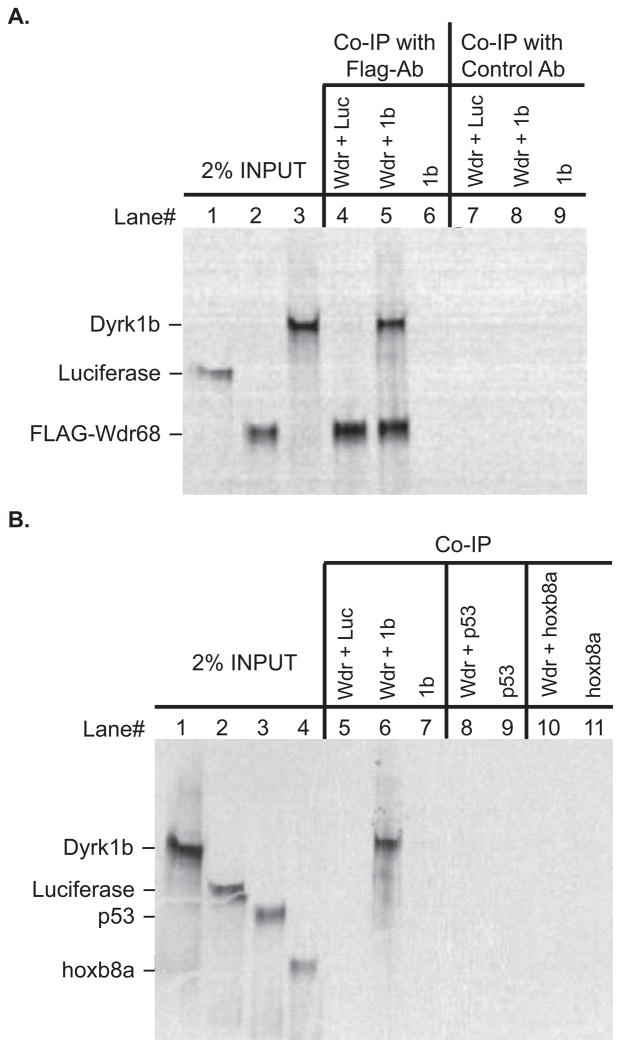
The mammalian Wdr68 and Dyrk1b proteins have been shown to physically interact in vitro (Skurat and Dietrich, 2004). To determine whether the zebrafish proteins might also form a physical complex, we tested them using an in vitro co-immunoprecipitation assay (Figure 1). We found that a FLAG-tagged zebrafish Wdr68 protein specifically co-immunoprecipitated the zebrafish Dyrk1b protein (Figure 1A, lane 5). Indicating specificity, we found that the FLAG-Wdr68 protein did not significantly co-immunoprecipitate a Luciferase control protein (Figure 1A, lane 4). Precipitated Dyrk1b protein was also not detected in the absence of the FLAG-Wdr68 protein ruling out a non-specific interaction between Dyrk1b and the FLAG antibody-bound protein-G sepharose beads (Figure 1A, lane 6). Substituting a negative control antibody for the FLAG antibody also resulted in the absence of any signal (Figure 1A, lanes 7–9). As additional negative controls, we found that neither p53 nor hoxb8a were capable of co-immunoprecipitating with FLAG-Wdr68 in our assay (Figure 1B). Since hoxb8a is similar in size to Flag-Wdr68, the experiments shown in Figure 1B were all conducted using non-radioactively labeled FLAG-Wdr68 protein that is therefore not detected in the autoradiographic image. Thus, we conclude that zebrafish Dyrk1b can physically interact with zebrafish Wdr68 in an in vitro assay.
Figure 1. The zebrafish Wdr68 and Dyrk1b proteins can physically interact.
A) Lane 1 shows 2% of the negative control Luciferase protein input. Lane 2 shows 2% of the FLAG-Wdr68 protein input. Lane 3 shows 2% of the Dyrk1b protein input. Lane 4 shows the results of a co-immunoprecipitation (co-IP) between FLAG-Wdr68 and Luciferase indicating no co-IP of the negative control Luciferase. Lane 5 shows the results of a co-IP between FLAG-Wdr68 and Dyrk1b indicating that Dyrk1b can physically interact with FLAG-Wdr68. Lane 6 shows the dependence of the Dyrk1b co-IP on the presence of FLAG-Wdr68. Lanes 7–9 are the same as lanes 4–6 but with the FLAG antibody substituted with an unrelated histone H3 control antibody. B) Lane 1 shows 2% of Dyrk1b input. Lane 2 shows 2% of negative control Luciferase input. Lane 3 shows 2% of negative control p53 input. Lane 4 shows 2% of negative control hoxb8a input. The FLAG-Wdr68 protein used in these experiments was translated with non-radioactive amino acids and is therefore not detected in the image. Lane 5 shows the results of a co-IP between FLAG-Wdr68 and Luciferase indicating no co-IP of the negative control Luciferase. Lane 6 shows the results of a co-IP between FLAG-Wdr68 and Dyrk1b indicating that Dyrk1b can physically interact with FLAG-Wdr68. Lane 7 shows the dependence of the Dyrk1b co-IP on the presence of FLAG-Wdr68. Lanes 8–9 show the lack of interaction between FLAG-Wdr68 and the negative control p53. Lanes 10–11 show the lack of interaction between FLAG-Wdr68 and the negative control hoxb8a.
dyrk1b is ubiquitously expressed during early development
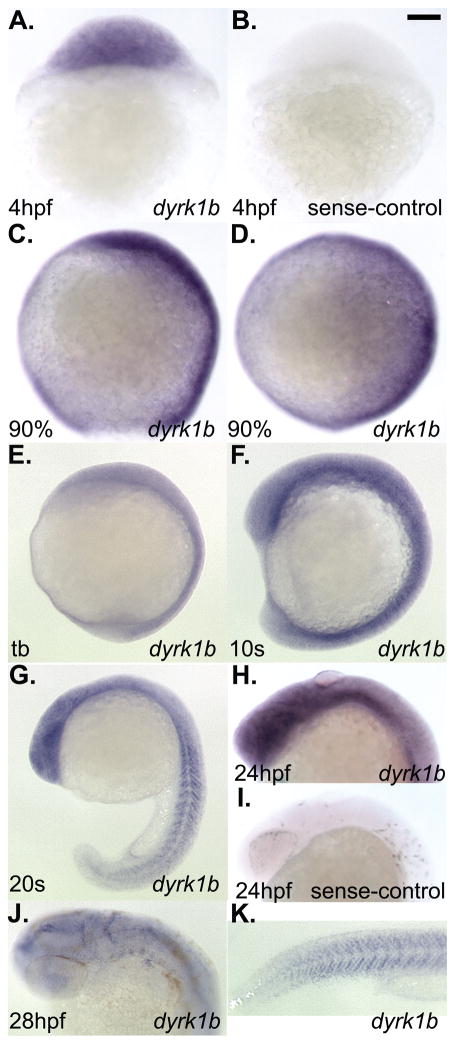
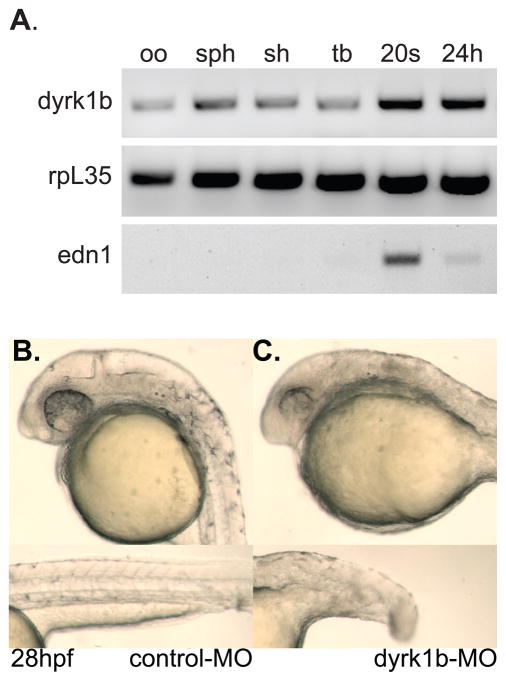
To determine the expression pattern of dyrk1b we used the partial clone isolated by 5′-RACE to generate probes for use in whole-mount in situ hybridization (ISH). Expression of dyrk1b was detected ubiquitously by an antisense-strand probe from the earliest stage examined through to approximately the 10 somites stage (Figure 2A–F). The dyrk1b sense-strand probe served as the negative control (Figure 2B). Expression of dyrk1b was readily detected in the developing somites as well (Figure 2G–K). By 28hpf, dyrk1b expression in the head and pharyngeal arches region was significantly reduced relative to the level expressed in the somites (Figure 2J, K). The sense-strand probe served as the negative control at later stages as well (Figure 2I). Consistent with the ISH experiments, we also detected expression of dyrk1b by RT-PCR (Figure 3A). Notably, we detected maternal transcripts in the unfertilized oocyte cDNA sample (Figure 3A, lane 1). Since the primers used in the RT-PCR experiments are separated by several introns and over 8kb in the genome, the detected 636bp bands cannot represent amplification from genomic DNA. As controls for a known maternally supplied gene, we also found expression of rpl35 in all samples tested (Figure 3A). As negative control, edn1 transcripts were not detected until after the tailbud stage (Figure 3A, lane 5).
Figure 2. Whole-mount in situ hybridization analysis of dyrk1b expression in wildtype zebrafish embryos.
A) ubiquitous expression of dyrk1b at 4hpf. B) sense-strand negative control for staining also at 4hpf. C) lateral view of 90% epiboly stage, D) dorsal view of 90% epiboly stage. E) lateral view of tailbud (tb) stage. F) lateral views of 10 somites (10s) stage. G) lateral view of 20 somites (20s) stage. H) lateral view of 24hpf stage. I) sense-strand negative control for staining also at 24hpf. J) dorsolateral view of 28hpf stage. K) lateral tail view of the same animal shown in panel J. Scale bar in panel B = 100 micrometers length.
Figure 3. dyrk1b is maternally supplied and essential during embryonic development in the zebrafish.
A) RT-PCR detection of dyrk1b transcripts in unfertilized oocytes (oo), sphere stage (sph), shield stage (sh), tailbud stage (tb), 20 somites stage (20s), and 24 hours post fertilization (24h) stage animals. The rpl35 gene serves as positive control. The edn1 gene serves as negative control for detection in oocytes. B) Normal phenotype of 5-mismatch control morpholino-injected (control-MO) animals at 28hpf. Upper panel shows normal head, eye, pigment and ear development. Lower panel shows normal notochord, tail and somite development. C) Phenotype of dyrk1b antisense morpholino-injected (dyrk1b-MO) animals at 28hpf. Upper panel shows small head and eyes of the morphant animals. Lower panel shows the moderately shortened length of the tail.
Zebrafish dyrk1b is important for embryonic development
To determine what role the dyrk1b gene plays in zebrafish development, we created a gene knockdown model by designing a translation-blocking antisense morpholino oligonucleotide against the dyrk1b mRNA (dyrk1b-MO) as well as a 5-base mismatch control (control-MO). While animals injected with the control-MO appeared normal (Figure 3B), dyrk1b-MO animals displayed morphological defects including a smaller head and eyes as well as a moderately shortened body (Figure 3C). Animals injected with a second antisense morpholino against a dyrk1b intron-exon junction displayed a similar but milder phenotype (not shown). While survival of the control-MO animals did not differ from that of uninjected animals, the dyrk1b-MO animals rarely survived past 48hpf precluding analysis of later developmental stages or craniofacial cartilage formation.
Zebrafish dyrk1b and wdr68 are important for lft1 and lft2 expression
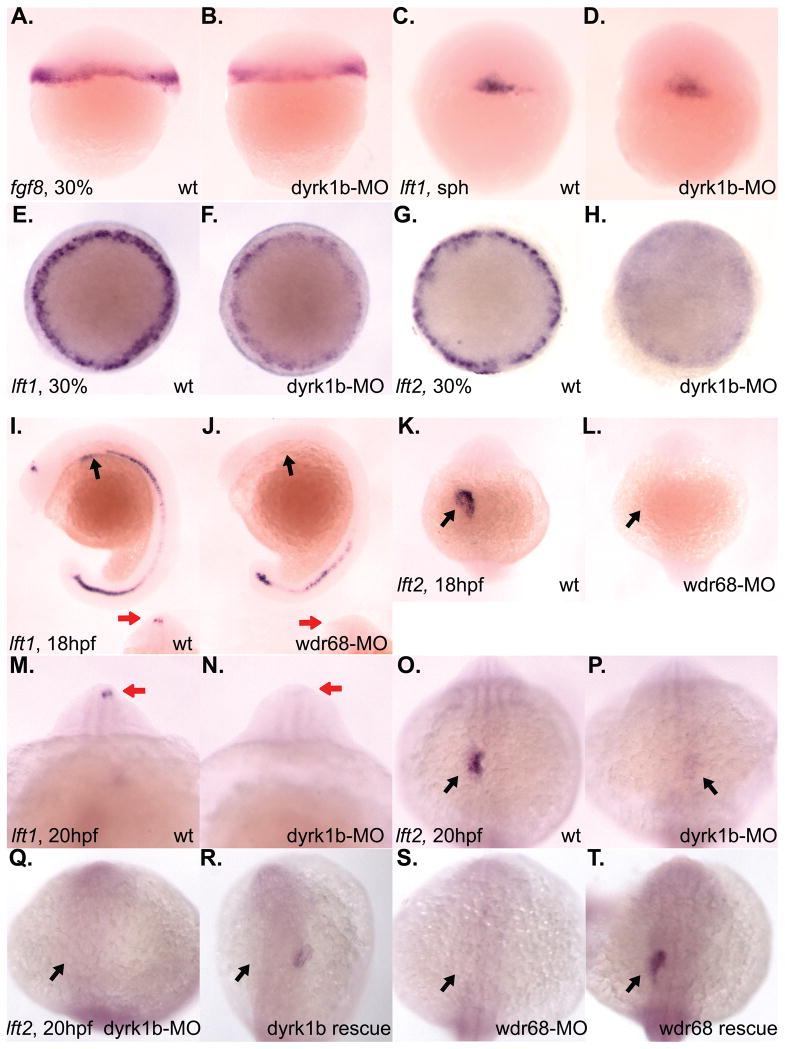
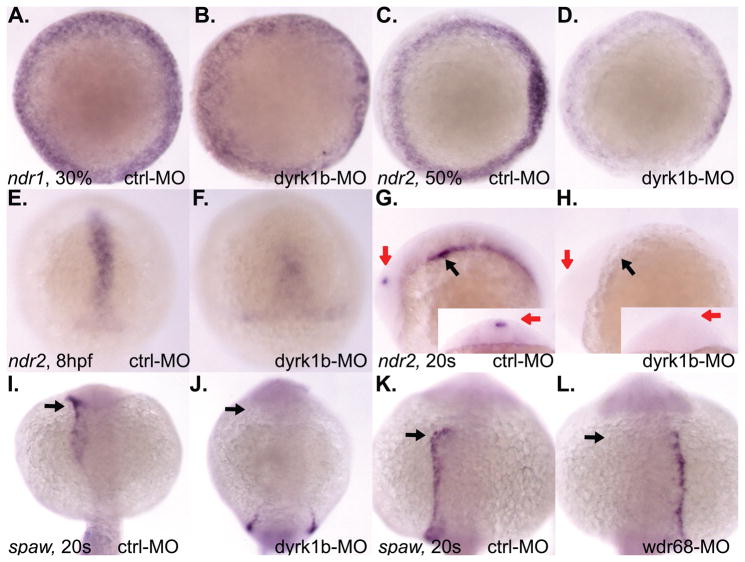
The Nodal signaling antagonists, lefty1 (lft1) and lefty2 (lft2), were originally identified as required for left-right asymmetry (Kosaki et al., 1999; Meno et al., 1997; Meno et al., 1996). By morphological inspection of both dyrk1b-MO and wdr68-MO animals, we noted that the heart developed on the right side in many of the morphant animals instead of the left side, suggesting a defect in left-right asymmetry (data not shown). To explore the possibility of a left-right patterning defect further, we performed ISH analysis on lft1 and lft2 expression in dyrk1b-MO and wdr68-MO animals (Figure 4). Although initial expression of lft1 prior to gastrulation appeared normal (Figure 4C, D), we found a subsequent failure to maintain both lft1 and lft2 expression during gastrulation in dyrk1b-MO animals (Figure 4E–H). Analyses of lft1 and lft2 expression in wdr68-MO animals did not reveal any substantial defects during gastrulation stages (data not shown). Expression of fgf8 prior to gastrulation was also not significantly affected (Figure 4A, B). We also examined the later asymmetric expression of lft1 and lft2 after gastrulation (Figure 4I–P). At 18hpf, the asymmetric expression of lft1 in the diencephalon was severely reduced in wdr68-MO animals (red arrows in Figure 4I, J, inset images). Expression of both lft1 and lft2 in the lateral plate mesoderm of the developing heart field was also severely reduced in wdr68-MO animals (black arrows in Figure 4I, J and K, L, respectively). We found similar losses in dyrk1b-MO animals (Figure 4M–P). At 20hpf in dyrk1b-MO animals, we found severely reduced expression of lft1 in the diencephalon (red arrows in Figure 4M, N) and reduced expression of lft2 in the lateral plate mesoderm of the developing heart field (black arrows in Figure 4O, P).
Figure 4. Reduced expression of lft1 and lft2 in dyrk1b and wdr68 knockdown animals.
A) lateral view of normal fgf8 expression in control animals at 30% epiboly stage. B) normal fgf8 expression in dyrk1b-MO animals at 30% epiboly stage. C) lateral view of normal lft1 expression in control animals at sphere (sph) stage. D) normal lft1 expression in dyrk1b-MO animals at sphere stage. E) dorsal view of normal lft1 expression in control animals at 30% epiboly stage. F) reduced lft1 expression in dyrk1b-MO animals at 30% epiboly stage. G) dorsal view of normal lft2 expression in control animals at 30% epiboly stage. H) reduced lft2 expression in dyrk1b-MO animals at 30% epiboly stage. I) lateral view of normal lft1 expression in control animals at 18hfp. Black arrows indicate asymmetric expression in the lateral plate mesoderm of the developing heart field. Red arrows in inset images are on the left side of the animals and indicate asymmetric expression in the diencephalon. J) severely reduced lft1 expression in the asymmetric heart and diencephalon territories of wdr68-MO animals at 18hpf. K) dorsal view of normal lft2 expression in control animals at 18hpf. Black arrows indicate asymmetric expression in the lateral plate mesoderm of the developing heart field. L) severely reduced lft2 expression in the asymmetric heart and diencephalon territories of wdr68-MO animals at 18hpf. M) anterior view of normal lft1 expression in the asymmetric diencephalon territory of control animals at 20hpf. Red arrows are on the left side of the animals and indicate asymmetric expression in the diencephalon. N) severely reduced lft1 expression in the asymmetric diencephalon territory of dyrk1b-MO animals at 20hpf. O) dorsal view of normal lft2 expression in the asymmetric heart territory of control animals at 20hpf. Black arrows indicate asymmetric expression in the lateral plate mesoderm of the developing heart field. P) severely reduced lft2 expression in the asymmetric heart territory of dyrk1b-MO animals at 20hpf. Q) dorsal view of severely reduced lft2 expression in dyrk1b-MO animals co-injected with EF1alpha transcripts. R) partial rescue of lft2 expression in dyrk1b-MO animals co-injected with Dyrk1b transcripts. S) dorsal view of severely reduced lft2 expression in wdr68-MO animals co-injected with EF1alpha transcripts. T) partial rescue of lft2 expression in wdr68-MO animals co-injected with FLAG-Wdr68 transcripts.
To test the specificity of the observed phenotypes, we injected control, dyrk1b-MO and wdr68-MO animals with in vitro transcribed RNA encoding EF1-alpha, Dyrk1b or FLAG-Wdr68 (Figure 4Q–T). We found that transcripts encoding Dyrk1b partially rescued the lft2 expression defect in 40% of dyrk1b-MO animals (Figure 4R and Table 1). We also found that transcripts encoding FLAG-Wdr68 partially rescued the lft2 expression defect in 43% of wdr68-MO animals (Figure 4T and Table 1). Notably, over-expression of Dyrk1b in wildtype animals disrupted lft2 expression in 19% of animals injected while EF1-alpha over-expression had no substantial effect (Table 1).
Table 1.
Partial RNA rescue of lft2 expression defects
| MO and transcript injected | lft2 detected | lft2 not detected |
|---|---|---|
| Control-MO + EF1alpha | 107 (98%) | 2 (2*%) |
| dyrk1b-MO + EF1alpha | 20 (19%) | 83 (81%) |
| dyrk1b-MO + Dyrk1b | 25 (40%) | 38 (60%) |
| wdr68-MO + EF1alpha | 7 (16%) | 37 (84%) |
| wdr68-MO + FLAG-Wdr68 | 24 (43%) | 32 (57%) |
| EF1alpha | 31 (100%) | 0 (0%) |
| Dyrk1b | 62 (81%) | 15 (19%) |
dyrk1b is important for ndr2 expression in zebrafish
While in mammals there is only one Nodal gene, in zebrafish there are three, ndr1/sqt, ndr2/cyc, and spaw (Long et al., 2003; Rebagliati et al., 1998a). Since the ndr2 gene is predominantly responsible for mediating lft1 and lft2 expression (Feldman et al., 2002), we examined the expression patterns of the zebrafish nodal genes (Figure 5). Expression of ndr1 was not significantly reduced in dyrk1b-MO animals (Figure 5A, B). In contrast, expression of ndr2 was significantly reduced at mid-gastrulation (Figure 5C, D) and later stages of gastrulation (Figure 5E, F). We also found severe reductions in the later asymmetric ndr2 expression in the lateral plate mesoderm (black arrows in Figure 5G, H) and diencephalon of 20 somites stage dyrk1b-MO animals (red arrows in Figure 5G, H).
Figure 5. dyrk1b and wdr68 are important for ndr2 and spaw expression in the zebrafish.
A) dorsal view of normal ndr1 expression in control animals at 30% epiboly stage. B) normal ndr1 expression in dyrk1b-MO animals at 30% epiboly stage. C) A) normal ndr2 expression in control animals at 50% epiboly stage. D) reduced ndr2 expression in dyrk1b-MO animals at 50% epiboly stage. E) shield view of normal ndr2 expression in control animals at 8hpf. F) reduced ndr2 expression in dyrk1b-MO animals at 8hpf. G) normal asymmetric ndr2 expression in control animals at 20 somites stage. Black arrow indicates asymmetric expression in lateral plate mesoderm. Red arrows indicate asymmetric expression in diencephalon. Red arrow in inset image is on left side of animal. H) reduced ndr2 expression in dyrk1b-MO animals at 20 somites stage. I) dorso-posterior view of normal asymmetric spaw expression in control animals at 20 somites stage. J) reduced expression of spaw in dyrk1b-MO animals at 20 somites stage. K) dorsal view of normal asymmetric spaw expression in control animals at 20 somites stage. L) loss of asymmetry in the expression of spaw in wdr68-MO animals at 20 somites stage.
Since the asymmetric expression of both ndr2 and the lft1 and lft2 genes are known to be dependent on spaw (Long et al., 2003), we also examined the expression of spaw. We found loss of anterior spaw expression accompanied by symmetric posterior spaw expression in 20 somites stage dyrk1b-MO animals (black arrows in Figure 5I, J, are on left side of animals). In wdr68-MO animals, the milder phenotype of incorrect spaw expression on the right side of the animals was observed in 42% (10/24) of the animals examined compared to only 6% (3/46) of the control animals (black arrows in Figure 5K, L, are on the left side of animals).
dyrk1b is important for endoderm formation and edn1 expression
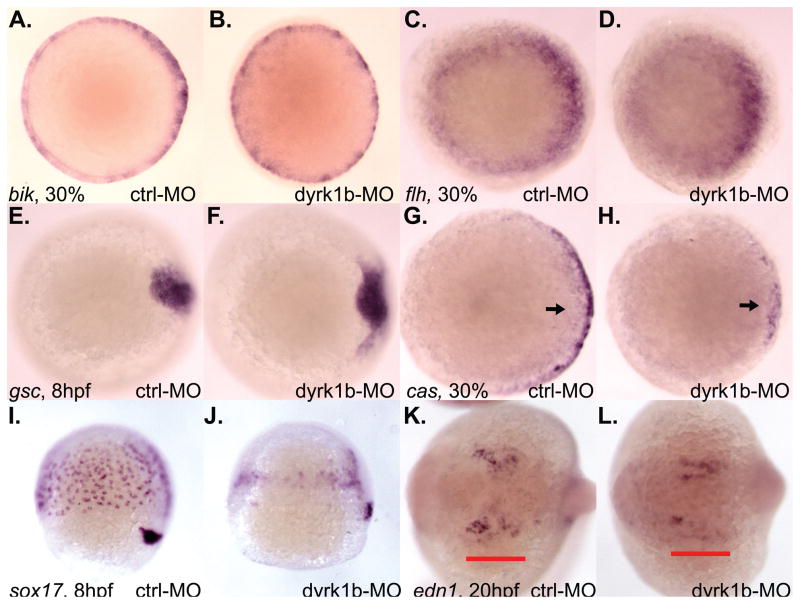
While only moderate levels of Nodal signaling are needed for mesoderm specification, high levels are needed for endoderm specification (Gritsman et al., 2000; Schier et al., 1997; Thisse et al., 2000). Therefore, we examined the expression of both mesoderm and endoderm markers in dyrk1b-MO animals (Figure 6). Expression of the pan-mesodermal marker bik was unaffected in dyrk1b-MO animals (Figure 6A, B). Likewise, expression of flh was not substantially affected (Figure 6C, D). Expression of the dorsal axial mesoderm marker gsc was modestly expanded in dyrk1b-MO animals (Figure 6E, F). In contrast, expression of the endoderm marker cas was significantly reduced during gastrulation in dyrk1b-MO animals (Figure 6G, H). Later expression of the cas-dependent endoderm marker sox17 revealed reduced numbers of sox17-positive cells in dyrk1b-MO animals (Figure 6I, J).
Figure 6. dyrk1b is important for endoderm induction and edn1 expression in the zebrafish.
A) normal bik expression in control animals at 30% epiboly stage. B) normal bik expression in dyrk1b-MO animals at 30% epiboly stage. C) normal flh expression in control animals at 30% epiboly stage. D) normal flh expression in dyrk1b-MO animals at 30% epiboly stage. E) normal gsc expression in control animals at 8hpf. F) modest expansion of gsc expression in dyrk1b-MO animals at 8hpf. G) normal cas expression in control animals at 30% epiboly stage. H) reduced cas expression in dyrk1b-MO animals at 30% epiboly stage. I) normal sox17 expression in control animals at 8hpf. J) reduced sox17 expression in dyrk1b-MO animals at 8hpf. K) normal edn1 expression in control animals at 20hpf. Red bar underlines pharyngeal expression domain of edn1. L) reduced edn1 expression in dyrk1b-MO animals at 20hpf.
The wdr68 gene is required for expression of the edn1 gene that, in turn, is required for lower jaw development in the zebrafish (Clouthier and Schilling, 2004; Miller et al., 2000; Nissen et al., 2006). Analysis of edn1 expression revealed a significant reduction in the number of edn1-positive cells in dyrk1b-MO animals (compare regions underlined in red, Figure 6K, L).
DISCUSSION
Using 5′RACE and RT-PCR, we isolated transcripts representing a single zebrafish dyrk1b gene containing an open reading frame exactly matching an ORF independently proposed by an ORF prediction algorithm. In mammals, dyrk1b is subject to alternative splicing and an alternative exon1 has been identified in mice (Leder et al., 2003). Nonetheless, our 5′-RACE experiments using primers in the dyrk1b kinase domain failed to detect an alternative 5′-exon variant in the zebrafish. Since the experiments presented here focused on early developmental stages, further analyses that extend into adult stages might reveal the existence of an alternative exon1 in zebrafish.
Consistent with previous results using the mammalian proteins (Skurat and Dietrich, 2004), we found that the zebrafish Dyrk1b protein was capable of engaging in a specific physical interaction with Wdr68 (Figure 1). In tissue culture cells, Wdr68 localizes to the nucleus (Nissen et al., 2006). Several reports indicate that Dyrk1b can also localize to the nucleus (Deng et al., 2004; Leder et al., 1999; Lee et al., 2000). Taken together, these data suggest that a nuclear Wdr68-Dyrk1b protein complex may exist.
Our analysis revealed the near ubiquitous expression of dyrk1b during early development in the zebrafish with enrichment in the developing somites at later stages (Figure 2). Expression of dyrk1b in the developing somites is consistent with the reported role for dyrk1b in muscle differentiation (Deng et al., 2003). In C2C12 cells, dyrk1b is required for myogenin expression and inhibits association between Mef2d and the histone deacetylase MITR revealing a role as a transcriptional co-regulator (Deng et al., 2005; Deng et al., 2003). Coupled with the Wdr68 interaction assay results, we speculate that a Wdr68-Dyrk1b transcriptional co-regulator complex may exist in the zebrafish.
Several of our results reveal that dyrk1b and wdr68 share common regulatory targets (Figure 4, 5, 6). We found that both dyrk1b and wdr68 are required for the asymmetric expression of lft1 and lft2 (Figure 4). Similarly, both genes are important for normal asymmetric expression of spaw (Figure 5). Likewise, both dyrk1b and wdr68 are important for edn1 expression (Figure 6)(Nissen et al., 2006). We also found that dyrk1b is important for ndr2, cas, sox17, lft1 and lft2 expression during gastrulation stages. However, we did not find significant defects in ndr2, cas, sox17, lft1 or lft2 expression during gastrulation stages in wdr68-MO animals (data not shown). While it is possible that dyrk1b and wdr68 function independently in the regulation of these target genes, we speculate that the loss of the catalytic Dyrk1b subunit of the Wdr68-Dyrk1b complex might be more detrimental to overall complex function than the loss of the Wdr68 subunit, for which a biochemical activity remains unknown. Consistently, the spaw expression defect was less severe in wdr68-MO animals than in dyrk1b-MO animals (Figure 5).
During embryogenesis, low levels of Nodal signaling specify mesoderm while high levels specify endoderm (Gritsman et al., 2000; Schier et al., 1997; Thisse et al., 2000). We found that ndr2 expression was reduced, but not lost, in dyrk1b-MO animals (Figure 5C–F). We also found a modest expansion of the expression of the dorsal axial mesoderm marker gsc in dyrk1b-MO animals (Figure 6E–F). At similar developmental stages we also found reduced expression of the endoderm markers cas and sox17 in dyrk1b-MO animals (Figure 6G–J). These results are consistent with the current model for Nodal signaling in which high levels of signal are needed for endoderm induction while only low levels are needed for mesoderm induction. Notably, the zebrafish phenotype for complete loss of ndr2 activity is cyclopia (Rebagliati et al., 1998b), a phenotype we never observed in dyrk1b-MO animals. The absence of cyclopia in dyrk1b-MO animals is consistent with our observed moderate reduction, but not loss, of ndr2 expression.
The wdr68 gene is important for craniofacial development in the zebrafish (Nissen et al., 2006). We previously found that wdr68 is essential for expression of the lower jaw patterning gene edn1 (Clouthier and Schilling, 2004; Miller et al., 2000; Nissen et al., 2006). Here, we found that dyrk1b is also important for edn1 expression suggesting a role for dyrk1b in craniofacial development in the zebrafish (Figure 6K–L).
A mouse knockout of dyrk1b was made but unfortunately lost before a detailed characterization of the mutant animals could be completed (Leder et al., 2003). Although it is unlikely that defects fully comparable to those described here for the zebrafish were present but overlooked in the mutant mice, the unavailability of the mutant mouse line precludes a definitive assessment. Regardless, a possible explanation for any phenotypic discrepancies suspected between the mouse and fish, based on the preliminary characterization of the now lost mouse mutant, may lie in the evolutionary differences in the Nodal signaling pathway between the two organisms. Namely, zebrafish have 3 nodal-related genes while mice appear to only have one. The evolutionary split of the single mammalian nodal gene functions amongst multiple teleost nodal-related genes might also have been accompanied by neomorphic changes in gene regulatory roles for various other loci such as dyrk1b and/or wdr68. Recreation of a dyrk1b mutant mouse line will be needed to fully explore these possibilities.
Taken together, these results suggest that a Wdr68-Dyrk1b complex may mediate some of the roles previously observed for wdr68 in the zebrafish. Future studies will be needed to further explore the roles of this complex.
MATERIALS AND METHODS
Animal husbandry
Wildtype TAB5 and TAB14 zebrafish strains were maintained essentially as described (Nissen et al., 2006).
5′-RACE, PCR subcloning and RT-PCR
RACE was performed using the SMART RACE kit (Clontech) as previously described (Golling et al., 2002). 5′-RACE fragments were subcloned into pCR-bluntII vectors (Invitrogen) for sequencing. The 5′-RACE primers were dyrk1b-r1 5′-TTGTAGGAGAGAAGCTCAAACACCAG-3′ and dyrk1b-r2 5′-AGGCACAGGTGGTTACGGAACAT-3′. The 5′-RACE fragment was subcloned into a pCRbluntII vector yielding the plasmid pCRbluntII-dyrk1b-5′R. To clone full-length dyrk1b, the following additional primers were used: dyrk1b-start 5′-ATGTCGAGCCAGCACAGCCAC-3′, dyrk1b-stop 5′-TCAAGAGTTGGCTGCACTCTGACC-3′, dyrk1b-sub1 5′-ttcttcggatccgatatcgcgaattccaccATGTCGAGCCAGCACAGC-3′, dyrk1b-sub2 5′-ttcttcctcgagTCAAGAGTTGGCTGCACTCTG-3′. A full-length ORF clone was inserted into a pCRbluntII vector yielding the plasmid pCRbluntII-dyrk1b-2256. The plasmid pCS2+dyrk1b-2256 was generated by subcloning the EcoRI fragment from plasmid pCRbluntII-dyrk1b-2256 into the vector pCS2+. For stage-specific RT-PCR analysis of dyrk1b expression, total RNA was isolated from specific developmental stages and used for 1st strand synthesis as previously described (Nissen et al., 2006). The primers used to detect dyrk1b transcripts are dyrk1b-kin-f1 5′-GTGGCCATCAAGATCATCAAGAACAAG-3′ and dyrk1b-kin-r1 5′-GGTCGAAGTATTTGCGTGCTTTAGG-3′. The primers for rpL35 were previously described (Nissen et al., 2006).
Co-immunoprecipitation assay
The FLAG-tagged zebrafish Wdr68 construct was previously described (Nissen et al., 2006). Translation-grade 35S-methionine was purchased from GE Healthcare. Co-immunoprecipitation assays were performed using a TNT Quick-Coupled Transcription-Translation Kit as per the manufacturer recommendations (Promega, catalog # L2080). The pCS2+Dyrk1b-2256 plasmid was used to generate full-length zebrafish Dyrk1b and the pCS2+FLAG-Wdr68 plasmid was used to generate FLAG-Wdr68 protein. The plasmid for generating the luciferase control protein was provided in the TNT kit. The plasmid for generating the zebrafish p53 control protein was kindly provided by Dr. Kirsten Edepli. The plasmid for generating the zebrafish hoxb8a control protein was previous described (Mansfield et al., 2004). Briefly, after pre-blocking protein-G sepharose beads (Invitrogen, catalog # 10-1242) with 1uL reticulocyte lysate per 100uL packed beads, we washed the beads with Wash buffer (50mM Tris pH 7.8; 250mM NaCl; 0.1% Triton X-100; 1% BSA; 1mM β-mercaptoethanol) three times. To create immunocomplexes, 4uL of each translated 35S-labeled protein to be tested was mixed in a 25uL total volume of Wash buffer containing 1uL of FLAG-Antibody (Rockland catalog # NC9120608) and incubated at 4°C for 1 hour. The reaction was then centrifuged for 5 minutes at 13,000 rpm and the supernatant transferred to 30μl of a 50% slurry of the blocked protein-G sepharose beads and mixed at 4°C for 1 hour with mild agitation. After capturing the immunocomplexes, the beads were washed 3 times with 1.5mL Wash buffer to remove unbound material followed by two additional washes with Wash buffer lacking the BSA. Bound complexes were eluted from the beads in SDS-PAGE Buffer and ran on an SDS-PAGE gel. The gel was dried, exposed to autoradiographic film overnight, developed and scanned into a digital image. All co-immunoprecipitation experiments were performed at least twice.
Antisense morpholinos used in this study
The translation blocking morpholino sequence used in this study for the dyrk1b gene was dyrk1b-3 5′-gtgctggctcgacatggttggctca-3′ and the sequence complementary to the start codon is underlined. A second morpholino sequence dyrk1b-4 5′-CCTCTTTAAGTGGActgaggagaca-3′ designed against a dyrk1b splicing junction was also used and the upper case sequence corresponds to the exon sequence while lowercase corresponds to the intron sequence. The 5-base mismatch control morpholino sequence for the translation blocking morpholino used in this study is dyrk1b-5mis 5′-gtgctggctcCaGatgCttgCctGa-3′ and the capital letters indicate the mismatch changes. The morpholino against wdr68 was previously described (Nissen et al., 2006). Wildtype embryos were free-hand injected at the 1–4 cell stages with approximately 1–4nL of a solution containing 200uM morpholino using pulled glass needles and a picospritzer as previously described (Nissen et al., 2003). A hemacytometer was used to calibrate needles and quantify volumes injected into animals.
Whole-mount in situ hybridization (WISH) analysis
WISH was performed as previously described (Nissen et al., 2003). The plasmid pCRbluntII-dyrk1b-5′R was digested with SpeI and transcribed with T7 to generate an approximately 1.5kb antisense probe against dyrk1b. The plasmid pCRbluntII-dyrk1b-5′R was digested with EcoRV and transcribed with SP6 to generate an approximately 1.5kb control sense probe for the same region of dyrk1b as that targeted by the antisense probe. The lft1 and lft2 probes were synthesized from DNA fragments generated by RT-PCR with the following primer pairs: Lft1-F1 5′-ctcctgcaccttgaaaagATGACT-3′, Lft1-R1 5′-CTTTCTGAGGTTCCACCCAGTAGA-3′, Lft1-t7R1 5′-TAATACGACTCACTATAGGGCTTTCTGAGGTTCCACCCAGTAGA-3′, Lft2-F1 5′-CTGTTCATCCAGCTGTTCATTTTG-3′, Lft2-R1 5′-ACTTCGGTGTTTTCCTGAAGTCTG-3′, Lft2-t7R1 5′-TAATACGACTCACTATAGGGACTTCGGTGTTTTCCTGAAGTCTG-3′. The cas probe was synthesized from DNA fragments generated by RT-PCR with the following primer pairs: cas-f1 5′-ATGTATCTCGACCGGATGCTCC-3′ and cas-t7r1 5′-TAATACGACTCACTATAGGgcagcaatctggatggaagc-3′. The sox17 probe was synthesized from DNA fragments generated by RT-PCR with the following primer pairs: sox17-f1 5′-gatgagcgcaagaggcttgc-3′ and sox17-t7r1 5′-TAATACGACTCACTATAGGgctcaaactccacctcgtcc-3′. The flh probe was synthesized from DNA fragments generated by RT-PCR with the following primer pairs: flh-f1 5′-ctaaacagacgccatgcagattcc-3′ and flh-t7r1 5′-TAATACGACTCACTATAGGgactcttctgtgaaatccctctcc-3′. The plasmids for generating probes to detect the ndr1, ndr2 (Rebagliati et al., 1998b), spaw (Long et al., 2003), fgf8 (Reifers et al., 1998), gsc (Schulte-Merker et al., 1994), bik (Chen and Schier, 2002) and edn1 (Miller et al., 2000) transcripts were previously described. All in situ hybridizations described were repeated at least twice and representative images were selected for inclusion in all figures.
In vitro transcription and mRNA injections
The plasmids pCS2+dyrk1b-2256 and pCS2+FLAG-Wdr68 were digested with NotI and used for in vitro transcription using the SP6 mMessage mMachine kit as per the manufacturers recommendations (Ambion, catalog # AM1340). The EF1-alpha control plasmid was supplied in the kit. After spin column purification and quantification, approximately 1–4nL of a 200ng/uL solution was injected into embryos at the 1–4 cell stage either alone or in combinations with antisense morpholinos described earlier. Embryos were harvested at the 20 somites stage and processed for in situ hybridization. The data presented in Table 1 represent the combined results from two independent experiments.
Acknowledgments
Contract grant sponsor: National Science Foundation IOS-0744454
We thank Jenny Arvizu for providing excellent management of the Cal State LA zebrafish colony. This work was supported by a grant from CSUPERB and grant #IOS-0744454 from the National Science Foundation.
References
- Alexander J, Stainier DY. A molecular pathway leading to endoderm formation in zebrafish. Curr Biol. 1999;9:1147–1157. doi: 10.1016/S0960-9822(00)80016-0. [DOI] [PubMed] [Google Scholar]
- Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Schier AF. Lefty proteins are long-range inhibitors of squint-mediated nodal signaling. Curr Biol. 2002;12:2124–2128. doi: 10.1016/s0960-9822(02)01362-3. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Schilling TF. Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res C Embryo Today. 2004;72:190–199. doi: 10.1002/bdrc.20007. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Crump JG, Swartz ME, Kimmel CB. An integrin-dependent role of pouch endoderm in hyoid cartilage development. PLoS Biol. 2004;2:E244. doi: 10.1371/journal.pbio.0020244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David NB, Saint-Etienne L, Tsang M, Schilling TF, Rosa FM. Requirement for endoderm and FGF3 in ventral head skeleton formation. Development. 2002;129:4457–4468. doi: 10.1242/dev.129.19.4457. [DOI] [PubMed] [Google Scholar]
- de Vetten N, Quattrocchio F, Mol J, Koes R. The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 1997;11:1422–1434. doi: 10.1101/gad.11.11.1422. [DOI] [PubMed] [Google Scholar]
- Deng X, Ewton DZ, Friedman E. Mirk/Dyrk1B maintains the viability of quiescent pancreatic cancer cells by reducing levels of reactive oxygen species. Cancer Res. 2009;69:3317–3324. doi: 10.1158/0008-5472.CAN-08-2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Ewton DZ, Mercer SE, Friedman E. Mirk/dyrk1B decreases the nuclear accumulation of class II histone deacetylases during skeletal muscle differentiation. J Biol Chem. 2005;280:4894–4905. doi: 10.1074/jbc.M411894200. [DOI] [PubMed] [Google Scholar]
- Deng X, Ewton DZ, Pawlikowski B, Maimone M, Friedman E. Mirk/dyrk1B is a Rho-induced kinase active in skeletal muscle differentiation. J Biol Chem. 2003;278:41347–41354. doi: 10.1074/jbc.M306780200. [DOI] [PubMed] [Google Scholar]
- Deng X, Mercer SE, Shah S, Ewton DZ, Friedman E. The cyclin-dependent kinase inhibitor p27Kip1 is stabilized in G(0) by Mirk/dyrk1B kinase. J Biol Chem. 2004;279:22498–22504. doi: 10.1074/jbc.M400479200. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Lufkin T, Rubenstein JL. Specification of jaw subdivisions by Dlx genes. Science. 2002;298:381–385. doi: 10.1126/science.1075703. [DOI] [PubMed] [Google Scholar]
- Dickmeis T, Mourrain P, Saint-Etienne L, Fischer N, Aanstad P, Clark M, Strahle U, Rosa F. A crucial component of the endoderm formation pathway, CASANOVA, is encoded by a novel sox-related gene. Genes Dev. 2001;15:1487–1492. doi: 10.1101/gad.196901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman B, Concha ML, Saude L, Parsons MJ, Adams RJ, Wilson SW, Stemple DL. Lefty antagonism of Squint is essential for normal gastrulation. Curr Biol. 2002;12:2129–2135. doi: 10.1016/s0960-9822(02)01361-1. [DOI] [PubMed] [Google Scholar]
- Garrett S, Broach J. Loss of Ras activity in Saccharomyces cerevisiae is suppressed by disruptions of a new kinase gene, YAKI, whose product may act downstream of the cAMP-dependent protein kinase. Genes Dev. 1989;3:1336–1348. doi: 10.1101/gad.3.9.1336. [DOI] [PubMed] [Google Scholar]
- Golling G, Amsterdam A, Sun Z, Antonelli M, Maldonado E, Chen W, Burgess S, Haldi M, Artzt K, Farrington S, Lin SY, Nissen RM, Hopkins N. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat Genet. 2002;31:135–140. doi: 10.1038/ng896. [DOI] [PubMed] [Google Scholar]
- Gritsman K, Talbot WS, Schier AF. Nodal signaling patterns the organizer. Development. 2000;127:921–932. doi: 10.1242/dev.127.5.921. [DOI] [PubMed] [Google Scholar]
- Ivey K, Tyson B, Ukidwe P, McFadden DG, Levi G, Olson EN, Srivastava D, Wilkie TM. Galphaq and Galpha11 proteins mediate endothelin–1 signaling in neural crest-derived pharyngeal arch mesenchyme. Dev Biol. 2003;255:230–237. doi: 10.1016/s0012-1606(02)00097-0. [DOI] [PubMed] [Google Scholar]
- Jin K, Ewton DZ, Park S, Hu J, Friedman E. Mirk regulates the exit of colon cancer cells from quiescence. J Biol Chem. 2009 doi: 10.1074/jbc.M109.035519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempf H, Linares C, Corvol P, Gasc JM. Pharmacological inactivation of the endothelin type A receptor in the early chick embryo: a model of mispatterning of the branchial arch derivatives. Development. 1998;125:4931–4941. doi: 10.1242/dev.125.24.4931. [DOI] [PubMed] [Google Scholar]
- Kikuchi Y, Agathon A, Alexander J, Thisse C, Waldron S, Yelon D, Thisse B, Stainier DY. casanova encodes a novel Sox-related protein necessary and sufficient for early endoderm formation in zebrafish. Genes Dev. 2001;15:1493–1505. doi: 10.1101/gad.892301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki K, Bassi MT, Kosaki R, Lewin M, Belmont J, Schauer G, Casey B. Characterization and mutation analysis of human LEFTY A and LEFTY B, homologues of murine genes implicated in left-right axis development. Am J Hum Genet. 1999;64:712–721. doi: 10.1086/302289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao WH, Kamada N, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Leder S, Czajkowska H, Maenz B, De Graaf K, Barthel A, Joost HG, Becker W. Alternative splicing variants of dual specificity tyrosine phosphorylated and regulated kinase 1B exhibit distinct patterns of expression and functional properties. Biochem J. 2003;372:881–888. doi: 10.1042/BJ20030182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder S, Weber Y, Altafaj X, Estivill X, Joost HG, Becker W. Cloning and characterization of DYRK1B, a novel member of the DYRK family of protein kinases. Biochem Biophys Res Commun. 1999;254:474–479. doi: 10.1006/bbrc.1998.9967. [DOI] [PubMed] [Google Scholar]
- Lee K, Deng X, Friedman E. Mirk protein kinase is a mitogen-activated protein kinase substrate that mediates survival of colon cancer cells. Cancer Res. 2000;60:3631–3637. [PubMed] [Google Scholar]
- Lim S, Zou Y, Friedman E. The transcriptional activator Mirk/Dyrk1B is sequestered by p38alpha/beta MAP kinase. J Biol Chem. 2002;277:49438–49445. doi: 10.1074/jbc.M206840200. [DOI] [PubMed] [Google Scholar]
- Long S, Ahmad N, Rebagliati M. The zebrafish nodal-related gene southpaw is required for visceral and diencephalic left-right asymmetry. Development. 2003;130:2303–2316. doi: 10.1242/dev.00436. [DOI] [PubMed] [Google Scholar]
- Mansfield JH, Harfe BD, Nissen R, Obenauer J, Srineel J, Chaudhuri A, Farzan-Kashani R, Zuker M, Pasquinelli AE, Ruvkun G, Sharp PA, Tabin CJ, McManus MT. MicroRNA-responsive ‘sensor’ transgenes uncover Hox-like and other developmentally regulated patterns of vertebrate microRNA expression. Nat Genet. 2004;36:1079–1083. doi: 10.1038/ng1421. [DOI] [PubMed] [Google Scholar]
- Meno C, Ito Y, Saijoh Y, Matsuda Y, Tashiro K, Kuhara S, Hamada H. Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: their distinct expression domains, chromosomal linkage and direct neuralizing activity in Xenopus embryos. Genes Cells. 1997;2:513–524. doi: 10.1046/j.1365-2443.1997.1400338.x. [DOI] [PubMed] [Google Scholar]
- Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Left-right asymmetric expression of the TGF beta-family member lefty in mouse embryos. Nature. 1996;381:151–155. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K, Parker J, Kimmel CB. sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3828. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Nissen RM, Amsterdam A, Hopkins N. A zebrafish screen for craniofacial mutants identifies wdr68 as a highly conserved gene required for Endothelin-1 expression. BMC Dev Biol. 2006;6:28. doi: 10.1186/1471-213X-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen RM, Yan J, Amsterdam A, Hopkins N, Burgess SM. Zebrafish foxi one modulates cellular responses to Fgf signaling required for the integrity of ear and jaw patterning. Development. 2003;130:2543–2554. doi: 10.1242/dev.00455. [DOI] [PubMed] [Google Scholar]
- Piotrowski T, Nusslein-Volhard C. The endoderm plays an important role in patterning the segmented pharyngeal region in zebrafish (Danio rerio) Dev Biol. 2000;225:339–56. doi: 10.1006/dbio.2000.9842. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Fricke C, Haffter P, Dawid IB. Zebrafish nodal-related genes are implicated in axial patterning and establishing left-right asymmetry. Dev Biol. 1998a;199:261–272. doi: 10.1006/dbio.1998.8935. [DOI] [PubMed] [Google Scholar]
- Rebagliati MR, Toyama R, Haffter P, Dawid IB. cyclops encodes a nodal-related factor involved in midline signaling. Proc Natl Acad Sci U S A. 1998b;95:9932–9937. doi: 10.1073/pnas.95.17.9932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reifers F, Bohli H, Walsh EC, Crossley PH, Stainier DY, Brand M. Fgf8 is mutated in zebrafish acerebellar (ace) mutants and is required for maintenance of midbrain-hindbrain boundary development and somitogenesis. Development. 1998;125:2381–2395. doi: 10.1242/dev.125.13.2381. [DOI] [PubMed] [Google Scholar]
- Schiefelbein J. Cell-fate specification in the epidermis: a common patterning mechanism in the root and shoot. Curr Opin Plant Biol. 2003;6:74–78. doi: 10.1016/s136952660200002x. [DOI] [PubMed] [Google Scholar]
- Schier AF. Nodal signaling in vertebrate development. Annu Rev Cell Dev Biol. 2003;19:589–621. doi: 10.1146/annurev.cellbio.19.041603.094522. [DOI] [PubMed] [Google Scholar]
- Schier AF, Neuhauss SC, Helde KA, Talbot WS, Driever W. The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development. 1997;124:327–342. doi: 10.1242/dev.124.2.327. [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S, Hammerschmidt M, Beuchle D, Cho KW, De Robertis EM, Nusslein-Volhard C. Expression of zebrafish goosecoid and no tail gene products in wild- type and mutant no tail embryos. Development. 1994;120:843–852. doi: 10.1242/dev.120.4.843. [DOI] [PubMed] [Google Scholar]
- Skurat AV, Dietrich AD. Phosphorylation of Ser640 in muscle glycogen synthase by DYRK family protein kinases. J Biol Chem. 2004;279:2490–2498. doi: 10.1074/jbc.M301769200. [DOI] [PubMed] [Google Scholar]
- Tejedor F, Zhu XR, Kaltenbach E, Ackermann A, Baumann A, Canal I, Heisenberg M, Fischbach KF, Pongs O. minibrain: a new protein kinase family involved in postembryonic neurogenesis in Drosophila. Neuron. 1995;14:287–301. doi: 10.1016/0896-6273(95)90286-4. [DOI] [PubMed] [Google Scholar]
- Thisse B, Wright CV, Thisse C. Activin- and Nodal-related factors control antero-posterior patterning of the zebrafish embryo. Nature. 2000;403:425–428. doi: 10.1038/35000200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian T, Meng AM. Nodal signals pattern vertebrate embryos. Cell Mol Life Sci. 2006;63:672–685. doi: 10.1007/s00018-005-5503-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AR, Davison PA, Bolognesi-Winfield AC, James CM, Srinivasan N, Blundell TL, Esch JJ, Marks MD, Gray JC. The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell. 1999;11:1337–1350. doi: 10.1105/tpc.11.7.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Smith MM, Mymryk JS. Interaction of the E1A oncoprotein with Yak1p, a novel regulator of yeast pseudohyphal differentiation, and related mammalian kinases. Mol Biol Cell. 2001;12:699–710. doi: 10.1091/mbc.12.3.699. [DOI] [PMC free article] [PubMed] [Google Scholar]