Abstract
We developed a conditional and inducible gene knockout methodology that allows effective gene deletion in mouse cardiomyocytes. This transgenic mouse line was generated by co-injection of two transgenes, a “reverse” tetracycline-controlled transactivator (rtTA) directed by a rat cardiac troponin T (Tnnt2) promoter and a Cre recombinase driven by a tetracycline-responsive promoter (TetO). Here, Tnnt2-rtTA activated TetO-Cre expression takes place in cardiomyocytes following doxycycline treatment. Using two different mouse Cre reporter lines, we demonstrated that expression of Cre recombinase was specifically and robustly induced in the cardiomyocytes of embryonic or adult hearts following doxycycline induction, thus, allowing cardiomyocyte-specific gene disruption and lineage tracing. We also showed that rtTA expression and doxycycline treatment did not compromise cardiac function. These features make the Tnnt2-rtTA;TetO-Cre transgenic line a valuable genetic tool for analysis of spatiotemporal gene function and cardiomyocyte lineage tracing during developmental and postnatal periods.
Keywords: cardiomyocyte, Cre recombinase, doxycycline, rtTA, Tnnt2
Cardiac-specific gene disruption in the mouse has advanced our current understanding of the biological function of gene products in mammalian heart development (Robbins, 2004). Mouse cardiac gene recombination involves the use of a cardiomyocyte-specific Cre deletor and a floxed allele of a gene of interest. The efficacy and precision of cardiac recombination, therefore, depends on the expression level and tissue specificity of Cre recombinase. Since the first cardiac-specific Cre deletor line was generated (Agah et al., 1997) using a promoter of the mouse cardiac myosin heavy chain (MHC) (Gulick et al., 1991), new Cre deletors have been created to improve recombination specificity and efficiency. For example, to avoid potential transgenic insertion artifacts and to achieve ventricular myocardium-specific gene inactivation, a knock-in strategy was used in which Cre recombinase was inserted into the mouse myosin light chain 2v (MLC2v) genomic locus (Chen et al., 1998). In addition to cardiac structural genes, the genomic locus or the promoter/enhancer of Nkx2.5, which encodes a transcription factor essential for cardiogenesis (Lyons et al., 1995), was used to generate Cre lines for analysis of gene function in Nkx2.5-expressing cardiac cell lineages (Moses et al., 2001) (Stanley et al., 2002) (McFadden et al., 2005).
The major advantage of tissue-specific recombination using Cre recombinase is to avoid embryonic lethality caused by germline loss-of-function mutations that precludes analysis of gene function in postnatal hearts. However, spatial control of gene deletion alone may not be sufficient for studying cardiac function in later embryonic development or in the postnatal period when cardiac-specific gene deletion still results in severe cardiac defects and early embryonic lethality. Techniques that enable temporal control of gene deletion in the developing and postnatal hearts have been developed using a tetracycline-controlled transactivator (tTA) driven by a rat MHC promoter (Fishman et al., 1994; Yu et al., 1996), or by an attenuated mouse MHC promoter (Sanbe et al., 2003).
In the tTA or “Tet-off” system, transgene expression is repressed by a continuous long-term doxycycline (Dox) treatment. Removal of Dox activates transgene expression or initiates Cre-mediated recombination (Zhu et al., 2002). However, chronic Dox treatment may have side effects on cardiac function and gene expression. Dox may accelerate the onset of cardiac hypertrophy and the progression to congestive heart failure in mice (Vinet et al., 2008). Furthermore, Dox has a long half-life in mice (Robertson et al., 2002), therefore, removal of Dox does not allow prompt induction of gene deletion. Such disadvantages can be minimized by using a “Tet-on” system or a “reverse” tTA (rtTA) converted from the tTA by point mutations (Freundlieb et al., 1999), in which transgene expression is active in the presence of Dox. A doubly transgenic system that drives expression of rtTA from tissue-specific promoters and activates Cre recombinase expression directed by a doxycycline/rtTA-inducible promoter has emerged as a leading approach permitting tight spatiotemporal control of gene deletion in many tissues (Branda and Dymecki, 2004).
A single transgenic mouse harboring both rtTA and Cre transgenes will provide a useful genetic tool for temporal analysis of gene function in cardiomyocytes and their lineage evolution. Therefore, we generated a new inducible cardiomyocyte-specific Cre mouse line using rtTA under the control of a rat cardiac troponin T (Tnnt2) promoter (Wang et al., 2000). The choice of this promoter over the mouse MHC promoter was based on previous observations that Tnnt2-Cre resulted in an earlier and more uniform recombination in the developing heart (Jiao et al., 2003) (Chen et al., 2006) than that induced by MHC-Cre (Agah et al., 1997).
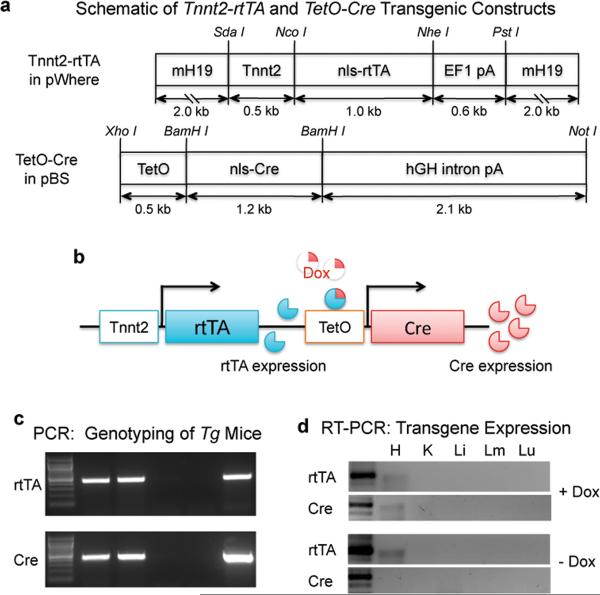
We made the Tnnt2-rtTA driver construct in the pWhere plasmid (InvivoGen, San Diego, CA), which contains two mouse H19 insulators (mH19) protecting the flanked Tnnt2-rtTA cassette from potential position effects on transgene expression (Fig. 1a). The responsive nuclear-localized Cre (nls-Cre) construct was generated using the TetO-CMV promoter (Pan et al., 2000). The two genetic constructs were simultaneously microinjected into fertilized eggs of FVB/N mice to generate founders that were then tested for Cre expression upon Dox treatment (Fig. 1b). Eight doubly transgenic founders were obtained from the microinjection. Seven of eight founders had both constructs and the transgene status of the offspring was analyzed by PCR (Fig. 1c). We found that the two transgenes consistently co-segregated and passed on to the offspring in a Mendelian fashion in all 7 lines, suggesting that the constructs inserted into the host genome at the same location. We showed by RT-PCR analysis that rtTA expression was restricted to the embryonic heart in two lines. Cre expression was only detected in the embryonic heart after Dox induction, indicating that Cre expression was tightly controlled by the `Dox-rtTA' and not leaky (Fig. 1d). Thus, rtTA expression is restricted to the heart, and its induction of cardiac-specific Cre activity requires Dox treatment.
Figure 1. Generation of Tnnt2-rtTA;TetO-Cre doubly transgenic mice.
a. Schematic of Tnnt2-rtTA and TetO-Cre transgenic constructs. b. Schematic of inducible Cre expression directed by Dox and the doubly transgenic system. c. Example of PCR genotyping shows co-segregation of rtTA and Cre transgenes in the established transgenic line. d. Example of RT-PCR reveals cardiac-specific expression of rtTA whereas the cardiac-specific expression of Cre recombinase is Dox-dependent. H, K, Li, Lm, Lu; heart, kidney, liver, limb, lung
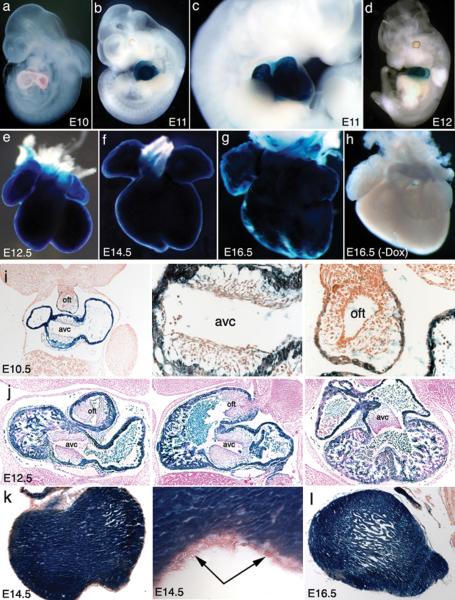
We tested the feasibility of these two lines for inducing genomic recombination by crossing them with the R26-floxstop-lacZ reporter line (R26R) (Soriano, 1999). One line (t26) gave reliable recombination after Dox treatment. This line was backcrossed with C57BL/6 for 5 generations and maintained in this background for rest of the studies. We found that one-day induction with Dox in the drinking water (1 mg/ml) at either embryonic day (E) 9, 10, 11, or 11.5 was sufficient to Cre-mediated excision of the floxed stop codon and activation of lacZ reporter expression in the whole heart at E10, 11, 11.5, or 12.5 (Fig. 2a–e). Gene recombination was not observed outside of the heart after an overnight X-gal staining, demonstrating a cardiac-specific induction of Cre expression in these embryos. Significant recombination was also found at later embryonic stages with one-day Dox treatment, although the recombination was not as efficient. In contrast, a 2-day treatment greatly improved induction efficiency and led to complete gene recombination in older embryos (Fig. 2f,g). Cardiac gene recombination was not found in embryos without Dox induction, indicating no or undetectable “leakiness” of Cre expression (Fig. 2h). Analysis of histological sections confirmed that cardiac-specific induction of Cre-mediated recombination was robust throughout the myocardium of the entire heart and restricted to cardiomyocytes (Fig. 2i–I). No reporter expression was found in the mesenchymal cells of the outflow tract (oft), atrioventricular cannel (avc) (Fig. 2i,j), or the epicardium (Fig. 2k). Additional analysis showed complete gene recombination in one-week old neonatal hearts when Dox was given to nursing females for 3 days starting at the day of delivery (data not shown). Taken together, these observations demonstrate that the Tnnt2-rtTA;TetO-Cre transgenic methodology allows efficient induction of Cre-mediated gene recombination in the cardiomyocytes of both embryonic and neonatal hearts.
Figure 2. Tnnt2-rtTA directs efficient recombination during heart development in the R26-floxstop-lacZ reporter line (R26R).
a–g. A panel of photos of wholemount X-gal staining shows the cardiac-specific Cre-mediated recombination of the floxstop lacZ and beta-galactosidase expression in embryonic hearts after Dox induction. Note that no sign of recombination is outside of the heart. h. A photo of wholemount X-gal staining shows no beta-galactosidase expression in the doubly transgenic heart without Dox induction. i–l Panel of histological sections shows cardiac-specific recombination (beta-galactosidase expression) in embryonic hearts. Note that no beta-galactosidase expression in the non-myocardial mesenchymal outflow tract (oft) and atrioventricular cannel (avc) at E10.5 and E12.5 as well as in the epicardium at E14.5 (arrows).
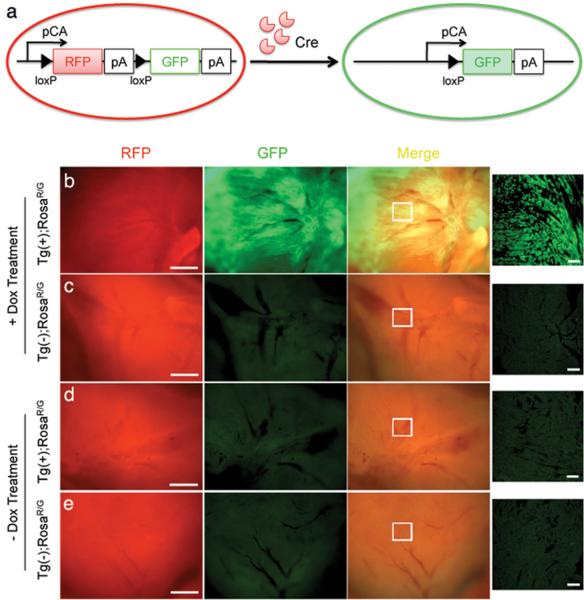
To assess whether this transgenic method is also applicable for inducible gene recombination in adult hearts, we performed a serial of analyses of Tnnt2-rtTA;TetO-Cre adult mice using a double-fluorescent Cre reporter line (RosaR/G) that contains a membrane-targeted tandem dimmer Tomato (mT), a red fluorescent protein (RFP), and a floxstop-membrane-targeted green fluorescent protein (mG) (Muzumdar et al., 2007) (Fig. 3a). The RosaR/G line also uses a strong enhancer, which improves expression of reporter genes in the postnatal heart (Muzumdar et al., 2007). We found that Dox induction (1 mg/ml in the drinking water) for 5 days resulted in significant activation of GFP expression in the hearts of triple compound heterozygous mice (Tnnt2rtTA/+;TetOCre/+;RosaR/G/+) (Fig. 3b). Attenuation of RFP intensity of the whole heart also indicated inactivation of its expression in cardiomyocytes but not in non-cardiomyocytes such as the endothelial, epithelial, smooth muscle, or fibroblast cells. In contrast, control littermates without theTnnt2-rtTA;TetO-Cre transgenes expressed only RFP, but not GFP (Fig. 3c). In the absence of Dox, there was no detectable GFP expression in either triple compound heterozygous (Fig. 3d) or control non-transgenic mice (Fig. 3e), indicating that the recombination was Dox-dependent.
Figure 3. Tnnt2-rtTA drives effective recombination and activation of the double-fluorescent Cre reporter (RosaR/G) in the adult heart.
a. Schematic shows that Tnnt2-rtTA induces cardiac-specific recombination and expression of a membrane-targeted GFP (mG) in the RosaR/G reporter line also constitutively expresses membrane-associated Tomato RFP (mT). b. Fluorescent photos show significant GFP expression in hearts of the triple compound heterozygous mice (Tnnt2rtTA/+;TetOCre/+;RosaR/G/+) after a 5-day Dox induction. Continuous but attenuated RFP expression in the entire heart was from non-cardiomyocytes after Dox treatment. c. No GFP expression was found in non-transgenic control mice [Tg(−)] with the same treatment. d. Tnnt2-rtTA without Dox induction was unable to activate GFP expression. e. Photos show no GFP expression in control mice [Tg(−)] without Dox treatment. Bar = 200 um. The far right panels show the sectional view of the areas (indicated by the white box) on the left. Bar = 50 μm.
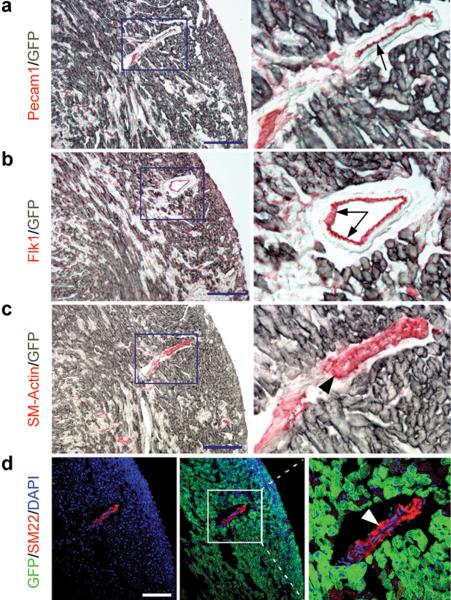
We next used co-immunohistochemistry to analyze the cell-specificity of gene recombination directed by the Tnnt2-rtTA in adult hearts in the presence of Dox. By using an antibody against GFP and antibodies that mark specifically endothelial or smooth muscle cells, we showed that gene recombination was clearly cardiomyocyte-specific, as Cre-mediated GFP expression did not overlap with expression of Pecam1 (Fig. 4a, arrow) or Flk1 (Fig. 4b, arrow) in endothelial cells. Furthermore, the recombination was not observed in smooth muscle cells, which were labeled by SM-actin (Fig. 4c, arrowhead) or SM22 antibodies (Fig. 4d, arrowhead).
Figure 4. Tnnt2-rtTA directed recombination is specific for cardiomyocytes.
a,b. Immunohistochemistry shows that the Tnnt2-rtTA-directed, Cre-mediated GFP expression (grey) is not in the endothelial cells, which expresses cell surface makers Pecam1 and Flk1 (red, indicated by arrow). c,d. The Tnnt2-rtTA directed Cre-mediated GFP expression (grey or green) is not in smooth muscle cells, which express smooth muscle cell actin (SM-Actin) and SM22 (red, indicated by arrowhead). Bar = 50 μm.
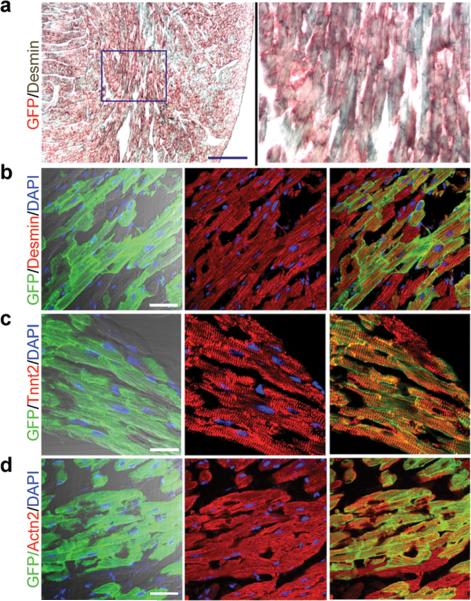
We also employed double immunostaining with antibodies against GFP or several markers of cardiomyocytes to study if they were marked by Tnnt2-rtTA;TetO-Cre-mediated gene recombination. The staining showed that recombination was indeed restricted to cardiomyocytes, as expression of the GFP reporter co-localized with desmin (Fig. 5a,b), Tnnt2 (Fig. 5c), and α2-actinin (Actn2) (Fig. 5d). Overall, these findings demonstrate that expression of the Tnnt2-rtTA transgene is restricted to cardiomyocytes, and that in the presence of Dox, it can effectively activate the TetO promoter to direct Cre-mediated recombination specifically in cardiomyocytes.
Figure 5. Tnnt2-rtTA-directed recombination marks cardiomyocyte lineages.
a,b. Immunohistochemistry shows that the Tnnt2-rtTA-directed, Cre-mediated GFP expression (red in a or green in b) in Desmin-expressing cardiomyocytes (grey in a or red in b). c. Tnnt2-rtTA directed Cre-mediated GFP expression (green) in Tnnt2-expressing cardiomyocytes (red). d. Tnnt2-rtTA directed Cre-mediated GFP expression (green) in the α2-actinin (Actn2)-expressing cardiomyocytes (red). Bar = 50 μm.
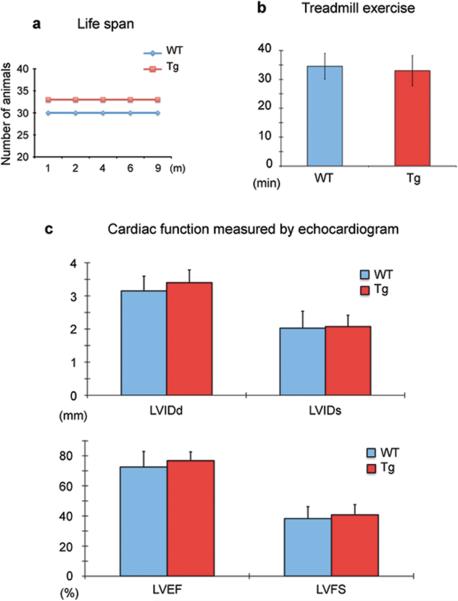
To assess the effect of transgenes on cardiac function, we analyzed the viability and cardiac physiology of Tnnt2-rtTA;TetO-Cre mice. These mice treated with Dox and with lifetime cardiac rtTA expression appeared grossly normal, healthy and fertile. Their lifespan was comparable to their non-transgene littermates up to at least 9 months of age when the breeders were retired from experimental use (Fig. 6a). To determine cardiac function in the Tnnt2-rtTA;TetO-Cre mice, we performed treadmill exercise tests and echocardiographic analysis. We showed that transgenic mice and control littermates at the age of 6 to 8 months with short-term (1 week) Dox treatment had comparable exercise duration, indicating that rtTA expression does not limit cardiac function and endurance (Fig. 6b). Furthermore, echocardiographic analysis revealed that transgenic and non-transgenic mice exhibited similar values of the left ventricular internal diameter both in diastole (LVIDd) and in systole (LVIDs) (Fig. 6c). The left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS) were also comparable (Fig. 6d). These cardiac functional measurements indicate that rtTA expression and Dox treatment have no appreciable effects on cardiac function.
Figure 6. Cardiac expression of rtTA and Dox treatment has no adverse effect on cardiac function.
a. Schematic shows comparable viability between transgenic mice (Tg) and wild-type (WT) littermates within the first 9 months. b. A graph shows comparable exercise endurance between transgenic mice and wild-type littermates in treadmill tests. c. Graphs of echocardiographic measurements show normal cardiac function in transgenic mice and wild-type littermates. LVIDd, left ventricular internal diameter in diastole; LVIDs, left ventricular internal diameter in systole; LVEF, left ventricular ejection fraction; LVFS, left ventricular fractional shortening
In summary, we have generated and characterized a new Tnnt2-rtTA;TetO-Cre transgenic mouse line that allows temporally controlled and tissue-specific gene recombination in the embryonic, perinatal, and adult hearts. At desirable time points, short-term Dox treatment efficiently activates gene recombination in cardiomyocytes for investigating gene functions in the heart. This transgenic method is also useful for labeling cardiomyocyte lineages. These features make this transgenic line a useful genetic tool for spatiotemporal studies of gene functions in cardiomyocytes during embryonic heart development, perinatal heart maturation, and post-natal cardiac physiology and pathology.
MATERIALS AND METHODS
Animal Maintenance and Treatment
Maintenance of mice and animal experiments was performed according to protocols approved by the Institutional Animal Care and Use Committee of Albert Einstein College of Medicine, Vanderbilt University, Boston Children's Hospital, and Stanford University. R26R and RosaR/G/+ mice were purchased from Jackson Laboratory (Bar Harbor, MN). Noontime on the day of detecting vaginal plugs was designated as E0.5. Dox (Sigma-Aldrich, St. Louis, MO) treatment was carried out in the drinking water at a concentration of 1 mg/ml and provided to the pregnant female mice at different gestation time points, nursing females or adult mice. For adult mice, Dox water was administrated for 5–7 days, and changed every other day.
Generation of the Tnnt2-rtTA;TetO-Cre Transgenic Line
The Tnnt2-rtTA driver construct was made by inserting a 0.5-kb Tnnt2 promoter into the pWhere in vivo reporter plasmid between Sda I and Nco I sites, and a 1.0-kb nuclear localized (nls) rtTA cDNA into Nco I and Nhe I sites (Fig. 1a). The pWhere plasmid contains two H19 insulators, which we have previously shown are able to effectively prevent ectopic transcription influences from random insertion sites (Zhou et al., 2005). The responsive Cre construct was generated by cloning an nls-Cre into a TetO-CMV promoter plasmid between BamH I sites in (Pan et al., 2000). The constructs were confirmed by enzymatic digestion and sequencing. Transgenic mice were generated by co-injection of the two constructs into fertilized eggs from FVB/N mice.
Transgenic founder mice were genotyped by PCR analysis on tail tip genomic DNA using primers for rtTA and Cre. The PCR-positive founder mice were crossed with C57BL/6 mice, and transgenic offsprings were identified by PCR. Primer pairs used for genotyping included 5'-rtTA: 5'-TCGACGCCTTAGCCATTGAGAT-3', 3'-rtTA: 5'GGCTGTACGCGGACCCACTTTC-3', yielding 476-bp PCR product; and 5'-Cre: 5'-GGCGCGGCAACACCATTTTT-3 ', 3 '-C r e : 5 '-TCCGGGCTGCCACGACCAA-3', producing 446-bp PCR product.
X-gal Staining
Wholemount X-gal staining was carried out as described before (Zhou et al. 2005). Embryos from E9.5 to E12.5, or the hearts isolated at later stages, were fixed in 4% paraformaldehyde/PBS on ice for 30 minutes to 2 hours according to the stages, washed in PBS, and then stained with fresh prepared X-gal solution for 2 hours at 37°C or overnight at 30°C. After staining, the samples were washed with PBS, post-fixed, and processed for histological analysis or cleared in a glycerol gradient before being photographed under a stereoscope (SMZ800, Nikon Inc. Melville, NY).
Immunofluorescence and Immunohistochemistry
Mouse hearts (3 to 4-month old) were excised in half and fixed in 4% PFA for 4 hours at 4°C, followed by a PBS wash at 4°C. Hearts were soaked in a sucrose gradient (10% – 30%) in PBS before embedding in OCT. Sections (8 μm) were obtained using a cryostat, collected on poly-L-lysine-coated slides and fixed in 1:1 methanol/acetone for 10 minutes at −20°C. Staining was performed according to protocols as described (Zhou et al., 2006). Primary antibodies used in these experiments were GFP (A21311, Invitrogen, Carlsbad, CA), Desmin (213M, Biomeda, Foster City, CA), Actn2 (A7811, Sigma, St. Louis, MO), Tnnt2 (MS-295, Lab Vision, Fremont, CA), Pecam 1 (553370, BD Pharmingen, San Jose, CA), Flk1 (NB600–1433, Novus, Littleton, CO), (SM-Actin (C6198, Sigma, St. Louis, MO), and SM-22 (Ab14106, Abcam, Cambridge, MA). Secondary antibodies used were conjugated with Alexa Fluor fluorescent dyes (Invitrogen, Carlsbad, California) or HRP and developed with a chromagenic system according to the manufacturer's protocol (Vector Lab, Burlingame, CA). The immunostained tissues were imaged using confocal microscopy (FV1000, Olympus, Center Valley, PA).
Analysis of Cardiac Function
Treadmill exercise was carried out as follows. Transgenic mice at 6 to 8 months of age were placed into treadmill chambers 10 minutes before starting to run to allow acclimation. Starting speed was 7.5 m/min and starting incline was 4°. Every three minutes, speed was increased by 2.5m/min, and incline was increased by 2°. When speed reached 20 m/min, only the incline was increased. The workout was terminated when mice were lagging and stayed on the electric grid for 10 seconds. Cardiac function and structure of 6 to 8-month old transgenic and control wild-type mice was also examined by echocardiogram after one-week of Dox treatment using the Vevo 770 machine (VisualSonics Inc. Toronto, Ontario, Canada). Statistic analysis was performed using the Student t-Test (unpaired). The value p < 0.05 was designated as having significant difference between the testing groups.
ACKNOWLEDGEMENTS
The authors thank Kai Jiao and Chi-Wing Chow for providing plasmids for these studies; Kevin Tompkins for technical support in pronuclear injection; and Bernice Morrow and Laina Freyer for comments on the manuscript. C.T.H. was supported in part by a Predoctoral Fellowship from American Heart Association. C.P.C. was supported by an R01 from the NIH and a grant from the Tobacco-Related Disease Research Program. B.Z. was supported by an R01 from the NIH and the Turner-Hazinski Award from Vanderbilt Children's Hospital, Vanderbilt University. This work was initiated at Vanderbilt University, continued and finished at Albert Einstein College of Medicine.
LITERATURE CITED
- Agah R, Frenkel PA, French BA, Michael LH, Overbeek PA, Schneider MD. Gene recombination in postmitotic cells. Targeted expression of Cre recombinase provokes cardiac-restricted, site-specific rearrangement in adult ventricular muscle in vivo. J Clin Invest. 1997;100:169–179. doi: 10.1172/JCI119509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Chen J, Kubalak SW, Chien KR. Ventricular muscle-restricted targeting of the RXRalpha gene reveals a non-cell-autonomous requirement in cardiac chamber morphogenesis. Development. 1998;125:1943–1949. doi: 10.1242/dev.125.10.1943. [DOI] [PubMed] [Google Scholar]
- Chen JW, Zhou B, Yu QC, Shin SJ, Jiao K, Schneider MD, Baldwin HS, Bergelson JM. Cardiomyocyte-specific deletion of the coxsackievirus and adenovirus receptor results in hyperplasia of the embryonic left ventricle and abnormalities of sinuatrial valves. Circ Res. 2006;98:923–930. doi: 10.1161/01.RES.0000218041.41932.e3. [DOI] [PubMed] [Google Scholar]
- Fishman GI, Kaplan ML, Buttrick PM. Tetracycline-regulated cardiac gene expression in vivo. J Clin Invest. 1994;93:1864–1868. doi: 10.1172/JCI117174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freundlieb S, Schirra-Muller C, Bujard H. A tetracycline controlled activation/repression system with increased potential for gene transfer into mammalian cells. J Gene Med. 1999;1:4–12. doi: 10.1002/(SICI)1521-2254(199901/02)1:1<4::AID-JGM4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Gulick J, Subramaniam A, Neumann J, Robbins J. Isolation and characterization of the mouse cardiac myosin heavy chain genes. J Biol Chem. 1991;266:9180–9185. [PubMed] [Google Scholar]
- Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BL. An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev. 2003;17:2362–2367. doi: 10.1101/gad.1124803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons I, Parsons LM, Hartley L, Li R, Andrews JE, Robb L, Harvey RP. Myogenic and morphogenetic defects in the heart tubes of murine embryos lacking the homeo box gene Nkx2–5. Genes Dev. 1995;9:1654–1666. doi: 10.1101/gad.9.13.1654. [DOI] [PubMed] [Google Scholar]
- McFadden DG, Barbosa AC, Richardson JA, Schneider MD, Srivastava D, Olson EN. The Hand1 and Hand2 transcription factors regulate expansion of the embryonic cardiac ventricles in a gene dosage-dependent manner. Development. 2005;132:189–201. doi: 10.1242/dev.01562. [DOI] [PubMed] [Google Scholar]
- Moses KA, DeMayo F, Braun RM, Reecy JL, Schwartz RJ. Embryonic expression of an Nkx2–5/Cre gene using ROSA26 reporter mice. Genesis. 2001;31:176–180. doi: 10.1002/gene.10022. [DOI] [PubMed] [Google Scholar]
- Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45:593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- Pan ZZ, Parkyn L, Ray A, Ray P. Inducible lung-specific expression of RANTES: preferential recruitment of neutrophils. Am J Physiol Lung Cell Mol Physiol. 2000;279:L658–666. doi: 10.1152/ajplung.2000.279.4.L658. [DOI] [PubMed] [Google Scholar]
- Robbins J. Genetic modification of the heart: exploring necessity and sufficiency in the past 10 years. J Mol Cell Cardiol. 2004;36:643–652. doi: 10.1016/j.yjmcc.2004.02.012. [DOI] [PubMed] [Google Scholar]
- Robertson A, Perea J, Tolmachova T, Thomas PK, Huxley C. Effects of mouse strain, position of integration and tetracycline analogue on the tetracycline conditional system in transgenic mice. Gene. 2002;282:65–74. doi: 10.1016/s0378-1119(01)00793-4. [DOI] [PubMed] [Google Scholar]
- Sanbe A, Gulick J, Hanks MC, Liang Q, Osinska H, Robbins J. Reengineering inducible cardiac-specific transgenesis with an attenuated myosin heavy chain promoter. Circ Res. 2003;92:609–616. doi: 10.1161/01.RES.0000065442.64694.9F. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nature Genetics. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stanley EG, Biben C, Elefanty A, Barnett L, Koentgen F, Robb L, Harvey RP. Efficient Cre-mediated deletion in cardiac progenitor cells conferred by a 3'UTR-ires-Cre allele of the homeobox gene Nkx2–5. Int J Dev Biol. 2002;46:431–439. [PubMed] [Google Scholar]
- Vinet L, Rouet-Benzineb P, Marniquet X, Pellegrin N, Mangin L, Louedec L, Samuel JL, Mercadier JJ. Chronic doxycycline exposure accelerates left ventricular hypertrophy and progression to heart failure in mice after thoracic aorta constriction. Am J Physiol Heart Circ Physiol. 2008;295:H352–360. doi: 10.1152/ajpheart.01101.2007. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sigmund CD, Lin JJ. Identification of cis elements in the cardiac troponin T gene conferring specific expression in cardiac muscle of transgenic mice. Circ Res. 2000;86:478–484. doi: 10.1161/01.res.86.4.478. [DOI] [PubMed] [Google Scholar]
- Yu Z, Redfern CS, Fishman GI. Conditional transgene expression in the heart. Circ Res. 1996;79:691–697. doi: 10.1161/01.res.79.4.691. [DOI] [PubMed] [Google Scholar]
- Zhou B, Bi YY, Han ZB, Ren H, Fang ZH, Yu XF, Poon MC, Han ZC. G-CSF-mobilized peripheral blood mononuclear cells from diabetic patients augment neovascularization in ischemic limbs but with impaired capability. J Thromb Haemost. 2006;4:993–1002. doi: 10.1111/j.1538-7836.2006.01906.x. [DOI] [PubMed] [Google Scholar]
- Zhou B, Wu B, Tompkins KL, Boyer KL, Grindley JC, Baldwin HS. Characterization of Nfatc1 regulation identifies an enhancer required for gene expression that is specific to pro-valve endocardial cells in the developing heart. Development. 2005;132:1137–1146. doi: 10.1242/dev.01640. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Zheng T, Lee CG, Homer RJ, Elias JA. Tetracycline-controlled transcriptional regulation systems: advances and application in transgenic animal modeling. Semin Cell Dev Biol. 2002;13:121–128. doi: 10.1016/s1084-9521(02)00018-6. [DOI] [PubMed] [Google Scholar]