Abstract
A deeper understanding of stem cell-niche engagement and subsequent behaviors would be enhanced by technologies enabling the tracking of individual stem cells at the clonal level in long-term co-culture (LTC), which mimics the complexity of the bone marrow microenvironment in vivo. Here, we report the application of time-lapse imaging with intermittent fluorescence for tracking well-defined populations of GFP+ murine hematopoietic stem cells (HSCs) using LTC for more than 5 weeks. Long-term (LT) and short-term (ST) repopulating HSCs and hematopoietic progenitor cells (HPCs) were compared. The transition from cobblestone areas (CA) under the stromal cell mantle into dispersed migrating cells on top of the stroma (COS) were directly observed. The ST-HSC and LT-HSC were able to initiate multiple waves of CA formation and COS expansion beyond 2 and 4 weeks, respectively. Retrospective tracking of individual CA forming cell (CAFC) revealed a preference for residing under stroma prior to the first division and a longer interval before first division for LT-HSC. Inability to maintain quiescence in subsequent divisions was revealed. Our study represents an important starting point from which the LTC system can be augmented to provide a better in vitro model for bone marrow stem cell niches.
Keywords: hematopoietic stem cell, stem cell niche (microenvironment), long-term culture, time-lapse imaging, cobblestone area
Introduction
Physiological stem cells in vivo are able to undergo different fate choices including migration, self-renewal, differentiation, apoptosis and quiescence (1) under specific conditions in which the stem cell niche is believed to serve as one of the central elements (2). Advances in the isolation of highly purified populations of rare hematopoietic stem cells (HSC) and hematopoietic progenitor cells (HPC) from bone marrow (3–7) provide opportunities to better understand stem cell traits and to seek improvements for their use as therapeutic agents. But once these valued cells are isolated, relevant in vitro models are needed for testing features and determinants of normal stem cell function. A good in vitro model should support and permit a quantitative analysis for the different stem cell behaviors and outcomes. Such a model would be useful for evaluating variables that control these functions and for assisting the development of techniques for expansion of HSCs with specific therapeutic purposes.
Mixtures of currently known hematopoietic cytokines do not maintain the long-term in vivo repopulating ability of HSCs in liquid culture for longer than days (8). In contrast, as an early demonstration of the importance of the microenvironment for HSC preservation (9–11), co-cultures of hematopoietic cells and primary bone marrow stromal cells without added cytokines have been shown to preserve repopulating HSCs for periods of up to several weeks in vitro (11–13). Thus, the co-culture system appears to at least partially recreate the hypothesized “stem cell niche” that constitutes a specialized micro-environment involving cell-cell interactions that control stem cell numbers and function in vivo (14). Crucial cell-cell interactions are only poorly understood (15, 16), so the co-culture system can provide a useful model for investigating stem cell–stromal cell interactions, and it may also serve as a platform upon which to build a better understanding of essential components of the stem cell niche, particularly when combined with time-lapse imaging and tracking at single cell resolution (17, 18). The cobblestone areas (CAs) are regions of hematopoietic cell clusters that proliferate beneath the stromal cell mantle and have been demonstrated to be associated with the activities of both HSC and HPC, dependent upon the specific time point of the readout (19, 20). Their long-term (> 4weeks) presence of CA in co-cultures forms the basis of a classical in vitro surrogate for HSCs. CAs are often surrounded by colony cells on top of the stroma (COS), which are thought to be the differentiating progeny of CA, although direct evidence for this association is still lacking. Therefore, kinetics of CA and COS in the long-term co-culture system in part recapitulates the hematopoietic regeneration in vivo.
For high content analysis of small numbers of rare cell types, time lapse imaging has become an important tool that continues to come of age with improvements in hardware automation, software, data storage/retrieval, and computer speed and power (21–27). Multiple parameters can be monitored at high resolution over time while cells retain the capacity for further functional or molecular analysis. Although short-term fluorescent imaging has been reported(18), time-lapse analysis of HSCs in long-term co-culture has not been well documented in detail due to problems such as photo-toxicity (16) by long-term frequent fluorescence imaging, difficulty of tracking the desired input cells in the presence of overwhelming numbers of stromal cells(27), the stablity of the operating system over long time periods, and challenges of handling and analysing very large image sets.
In this study, following our previous establishment of the time lapse imaging system for cultured immortal cell lines (28), we attempted to overcome the problems associated with time-lapse analysis of HSCs in long-term co-culture by improving the imaging and tracking strategy and software using the well-defined HSC and HPC subsets from GFP-transgenic mice. We compared multiple variable of behaviors of individual GFP+ cells from the different subsets of HSCs and HPCs. We analyzed division times and tracked transmigration behavior of individual cells that gave rise to CAs, and then followed the kinetics of CA formation in early and late stages through the stage of transition from CA to COS. Moreover, we were able to monitor the CA activity for more than 5 weeks in order to correlate it with the immuno-phenotypes of HSC and HPC as defined with the in vivo model. We sought to determine whether traits such as quiescence, transmigration, division and differentiation occurred differently between hematopoietic precursors with different phenotypes and we hypothesized that such traits would be more evident using the stromal co-culture as opposed to the studies using liquid culture (24).
Methods
Isolation of hematopoietic cell subsets (29)
Bone marrow cells were obtained from 12-wk-old green fluorescent protein (GFP) transgenic C57BL/6J mice (The Jackson Laboratory, Bar Harbor, Maine). c-Kit+ cells were first enriched by immunoselection with CD117 microbeads according to manufacturer’s instructions (Miltenyi Biotec, Auburn, CA). Cells were then stained with biotinylated anti-CD34 and PE-Texas Red conjugated strepdavidin, PE-conjugated anti-Sca-1, allophycocyanin (APC)-conjugated anti–c-Kit and PE-Cy7-conjugated lineage marker cocktail consisting of anti-Gr-1, -CD11b, -B220, -CD3, -CD4, -CD8, and -Ter119 monoclonal antibodies (BD PharMingen, San Jose, CA and eBioscience, San Diego, CA). 4′,6-diamidino-2-phenylindole, dihydrochloride (DAPI, Invitrogen, Carlsbad, California) was added (2 ug/ml) for dead cell discrimination. The final hematopoietic cell subsets, CD34-negative, lineage-negative, c-Kit-positive, Sca-1-positive (LT-HSC), CD34-positive, lineage-negative, c-Kit-positive, Sca-1-positive (ST-HSC), lineage-negative, c-Kit-positive, Sca-1-negative (HPC) were sorted using a MoFlo High-Speed Cell Sorter (DakoCytomation, Denmark) based on multi-color flow cytometric analysis.
Preparation of stromal layers from primary bone marrow
Isolated murine bone marrow cells from syngeneic non-GFP+ mice were resuspended in long-term culture medium (MyeloCult™ M5300 with 10−6M hydrocortisone, StemCell Technologies, Vencouver, CA) and plated at 2×107 cells per 25 cm2 tissue cultureware flask. Flasks were incubated at 33°C, 5% CO2 in air and ≥95% humidity for two to three weeks with half-medium changes performed weekly. Prior to seeding with sorted hematopoietic cell subsets, the stromal cells were trypsinized, irradiated at 15 Gy using a cesium137 source (MDS Nordion, Ottawa, Canada), and replated into various cultureware types as indicated. For 96-well plates, 2.5 × 104 cells per 100 ul per well yielded 100% confluent stromal layers.
Single cell culture and differentiation assay
Single cells (one cell/well) were deposited into 96-well plates containing liquid culture medium with “early” cytokines (100 ng/ml of mSCF, 25 ng/ml of mTPO, 10ng/ml of mIL-3 and 2 U/ml of rhEPO) as described by Nakauchi et al (30). After culturing for 10–14 days, cell numbers were counted using a hemacytometer, and cells from each well were harvested and centrifuged onto glass slides with a Cytospin3 (Shandon, Runcorn, Cheshire, U.K). Cells were stained with Wright-Giemsa solution (Fisher Scientific, Middletown, VA) for morphological examination and identification by microscopy as neutrophils (n), monocytes/macrophages (m), erythroblasts (E), or megakaryocytes (M).
In vivo bone marrow transplantation assay
Bone marrow transplantation assay was performed using C57BL/6 (CD45.2), B6/SJL (CD45.1) congenic mice. LT-HSC (50 cells/mouse) and ST-HSC (300 cells/mouse) cells were sorted from B6/SJL (CD45.1) mice and injected through tail vein with 1 × 105 bone marrow cells from CD45.1/CD45.2 mice as competitors. C57BL/6 (CD45.2) mice as recipient were lethally irradiated with 9.5 Gy. Peripheral blood was analyzed 3 weeks, 12 weeks and 5 months after transplantation for donor contribution by flow cytometry.
Time-lapse video microscopy
The automated imaging system consisting of a Nikon 300M inverted epifluorescent microscope with automated stage positioning and focus controls and a stage-mounted incubating chamber, maintained for this experiment at 33°C, 5% CO2 in 95% humidified air, has been previously described (28). The sorted hematopoietic subsets were cultured onto the prepared primary stromal layers in 24-well plate for more than 8 weeks. Cultures were fed weekly by changing half of the medium (M5300). Multiple target cells in the plate were memorized in the system and revisited every 10 minutes. Exposure of cells and associated photo-toxicity from fluorescence excitation were minimized by automating and synchronizing lamp and camera shutters and further reduced while at the same time sufficient images produced for confirming and re-establishing tracks over extended time periods by acquiring only one fluorescent image set (at 100 minute intervals) for every 10 bright-field images taken at 10-minute intervals. A metal halide fluorescence illumination system (X-Cite, Exfo Lifesciences and Industrial Division) was used with separate excitation filters (Chroma series 86000) mounted in a filter wheel (Sutter) in combination with a Sedat-quad dichroic filter cube (Chroma 86000) containing a multiband emission filter (Chroma series 84000) for GFP fluorescence detection. Brightfield images (8bit, JPEG format) were acquired every 10 minutes using a 20x objective and a CCD camera (1392 × 1040 pixel, CoolSnap ES, Photometrics) at 2×2 binning, and corresponding fluorescent images were acquired at 1/10th this frequency.
Image processing
A fluorescent green (490 nm excitation) and red (555 nm) image set was acquired for fluorescence analysis; the red image was used to correct for autofluorescence from the primary stromal layer cells. Correction was performed using ImagePro Plus software (Media Cybernetics) by first reducing the average red intensity to match the average green image intensity, based upon control wells that contained only irradiated stromal cells, followed by subtraction of the reduced-red intensity image from the green intensity image for each matched-image set. This process isolated GFP expressing cells from autofluorescing stromal cells and fortuitously flattened the central “bowing” of the fluorescence surface. Lamp intensity fluctuations over time were compensated for by normalization of the entire green image set to the average intensity across the entire experiment, based upon background intensity of control irradiated stromal cells. For producing green color overlays for manual analysis and to better identify dim GFP+ cells, the intensity of the fluorescent green images was linearly and uniformly multiplied at two levels (2x and 10x) prior to overlaying with contrast enhanced (ImagePro “bestfit”) brightfield images. Analysis was performed using custom viewing programs that enabled interactive tracking and recording of xy coordinates and additional information (“on top” versus “under” stromal layer, division times, parent-daughter relationships, cell fates and notes) for GFP+ cells. Green color fluorescent-visible overlays were combined with bright field visible images at every tenth scan in order to establish and verify identity of the “founder” cells and their progeny. Multiple adjacent viewfields were used to follow the cells entering or emigrating the target fields.
Determination of burst radii
The original image sets were first composited into 108 viewfields for each cell type using 1/18 the original resolution. Background correction and normalization were then applied to the composites as described above. A self-developed software program was used to estimate the radii of the GFP+ bursts that were clearly evident in these image sets. Three positions were recorded throughout the relevant time sequence for each burst at 1) the approximate center of the burst, 2) a horizontal outer edge, and 3) a vertical outer edge. The simultaneous sequence of center, horizontal, and vertical points was used to estimate an average radius at each scan for the burst.
Sister Interdivision Time Difference Analysis
To determine whether there was evidence for equivalence between sisters for early interdivision time in each pair, the image sets were analyzed and confirmed by two scientists individually to make sure all the division events were ascertainable and accurate. Since assignment of each sister as “first” or “second” was arbitrary, we included both the positive and negative difference for each pair to yield a symmetrical distribution centered about the mean difference of zero for calculating standard deviation. We graphically present only the positive half of this distribution, and reason that evidence for equivalence in interdivision time difference would be revealed by significant departure from the uni-modal normal distribution expected from symmetrical divisions.
Results
Functional validation of the immuno-phenotypically defined hematopoietic cell subsets
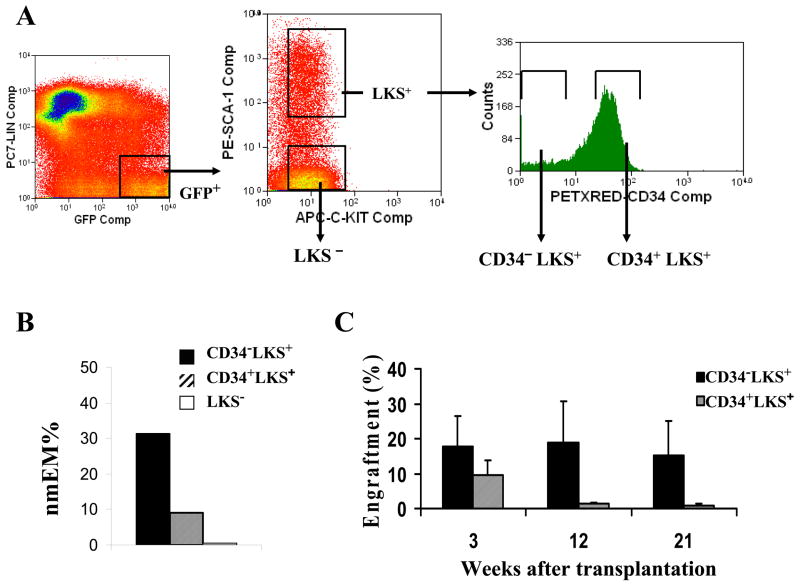
We chose to explore and compare attributes of well characterized subsets of immunophenotypically sorted HSCs based upon positive and negative CD34 expression among the lineage−, c-Kit+ and Sca-1+ (LKS+) bone marrow cells according to the method developed by others (4). The lineage−, c-Kit+ and Sca-1− (LKS−) HPC populations were also included for comparing to the two primitive HSC types (Figure 1A). Prior to performing time-lapse analysis, we validated the subsets using standard in vivo and in vitro functional assays. In the single cell culture and differentiation assay, more than 30% of CD34− LKS+ were capable of differentiation into 4 myeloid lineages compared with less than 10% of CD34+LKS+ and almost none of LKS− cells (Figure 1B). Competitive repopulation in lethally irradiated mice demonstrated sustained long-term engraftment by 50 CD34− LKS+ at over 15% peripheral blood chimerism after 21 weeks and even higher in the early stage, whereas engraftment by 300 CD34+LKS+ resulted in declining chimerism to approximately 1% after 21 weeks (Figure 1C). On the basis of these functional validations, we refer to the CD34− LKS+ subset as “long-term” repopulating HSC (LT-HSC), the CD34+LKS+ subset as “short-term” repopulating HSC (ST-HSC) and the LKS− subset as HPC throughout the remainder of this manuscript.
Figure 1. Isolation and conventional analysis of hematopoietic cell subsets.
The cell sorting strategy of Nakauchi et al (4) is illustrated for isolation of the CD34− and CD34+ subpopulations from LKS+ hematopoietic stem cells (A). The LKS− subset, referred to as “progenitor cells” (HPC), was used as a control for comparison in most assays. Multi-lineage differentiation potential was determined as the percentage of single cells (n=180 for each subset) that gave rise to all four morphological phenotypes: neutrophils, monocyte/macrophage, erythroid, and megakaryoctye after approximately two weeks in liquid culture (B). In vivo competitive repopulation was compared between CD34− LKS+ (50 cells/mouse) and CD34+LKS+ (300 cells/mouse) hematopoietic stem cells sorted from B6/SJL (CD45.1) mice and injected through tail vein with bone marrow from CD45.1/CD45.2 mice as competitors into lethally irradiated C57BL/6 (CD45.2) mice as recipients (C). Peripheral blood was analyzed as % chimerism by flow cytometry at 3 weeks, 12 weeks, and 5 months after transplantation (error bars represent mean ± SD, n=5 recipient mice).
Time-lapse imaging of hematopoietic cell subsets on stromal cells
Time-lapse imaging was performed using sorted cells from GFP transgenic mice seeded onto primary irradiated stromal cells prepared from syngeneic non-GFP+ mice. We used an incubated, fully automated imaging system that minimizes exposure of cells and associated photo-toxicity from fluorescence excitation by automating and synchronizing lamp and camera shutters (28). We further reduced fluorescence exposures by acquiring only one fluorescent image set (at 100 minute intervals) for every 10 bright-field images taken at 10-minute intervals. Both red and green fluorescent images were acquired at each 100-minute time point to allow correction for auto-fluorescence from primary stromal cells. In an effort to maximize cell numbers while observing the extent of “free-range” migration of individual cells, we performed this study in a 24-well plate and acquired adjacent view-fields that could be assembled into composite images for global viewing or that could be examined at original 20x scale for precise tracking and evaluation of individual cells moving across viewfields (data not shown).
Prospective Analysis of Early Cobblestone Area (ECA) formation
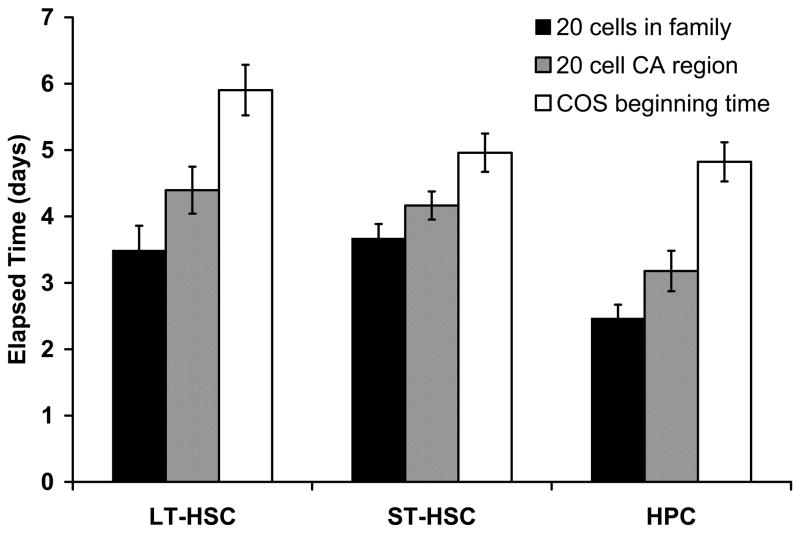
We developed a software program (CytoTracker) for manual-interactive tracking of individual cells and their progeny. To reduce selection bias, we tracked all GFP+ founder cells present in rectangular mosaics that “contained” approximately 50 total GFP+ cells for each subtype. The tracking difficulties due to the “collision” of confounded familes were around 10% in each group which suggested our imaging interval strategy was reasonable. CA formation was a highly variable process with some families forming tight clusters early in the period of expansion and others more rapidly migrating and more widely scattering before forming stable CA regions (see example in Supplementary Movie). In some cases, particularly among the HPC subset, founder cells that initially divided beneath the stroma did not achieve as many as 20 cells before they transmigrated to the top of the stromal cell mantle and dispersed. Thus in many cases, the point at which a family “officially” became a typical CA region with a certain cell number (e.g. 20-cell) was difficult to judge. Nevertheless, we attempted to determine two time points in this process: 1) where the members of the family reached 20 cells, and 2) where at least one CA region of 20 cells was evident within the family (Figure 2). HPC cells reached these stages earlier than HSC (p<0.05). Importantly, CA expansions were followed in every case by transitioning of cells from underneath to on top of the stromal cell mantle followed by phagocytosis and/or differentiation to an adherent long-lived cell type over time.
Figure 2. Times of CA formation and transition to COS.
During prospective analysis of founder cells, time-points were noted when: 1) 20 progeny cells were present in the family, whether or not these cells formed a single cobblestone area (leftmost bar for each subset), 2) 20 progeny cells were present in a single cobblestone area (center bar for each subset), and 3) colony cells first began to transition to the top of the stromal cell mantle (rightmost bar for each subset). The times of CA formation of the HSC group are significantly longer than that of the HPC group (p < 0.05).
Kinetics of expansion of CA/COS bursts
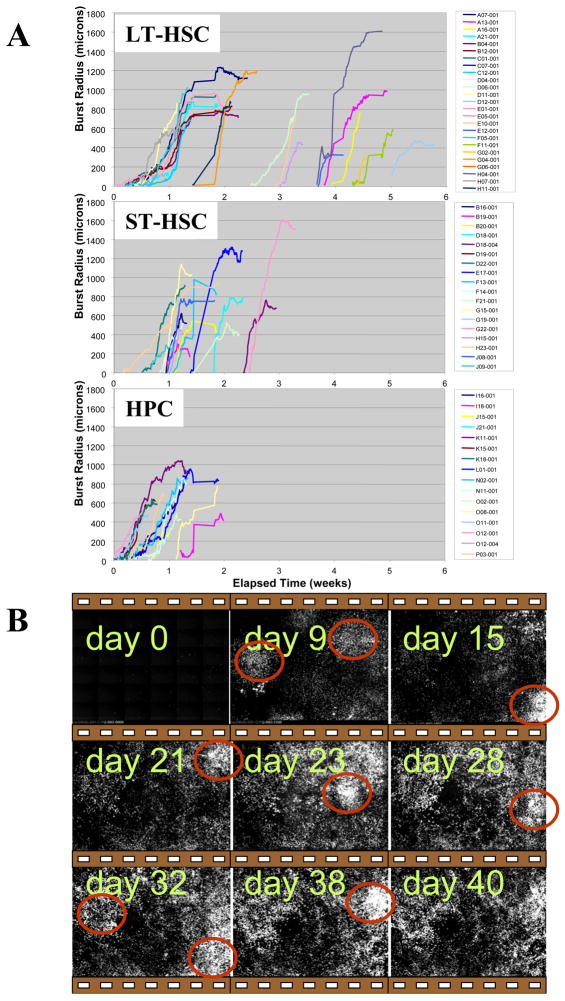
We compared the kinetics for these CA/COS expansions between hematopoietic cell subsets by estimating the radii of circular GFP+ burst regions from the time of CA formation through the phase of COS migration on top of the stromal cell mantle (Figure 3). Composites of adjacent images in rectangular mosaics were used to follow the “global” kinetics of GFP+ cell clustering into CAs and expansion of COS cells into broadly migrating bursts. The radius reflects both growth and migration activity
Figure 3. Expansion kinetics of CA/COS bursts.
The approximate radii of expanding regions of GFP+ cells were determined using composited adjacent image sets and a manual interactive viewer that output a horizontal and a vertical radius for each burst at 100-minute intervals (i.e. for each fluorescent image) (A). The mean radius at each time point is shown here. The time axis (x) represents actual elapsed time. Representative fluorescent images from one 6×6 mosiac show multiple waves of LT-HSCs in long-term culture (B)
An initial “wave” of burst activity began 2 to 3 days after seeding for HPC, whereas LT and ST-HSC burst expansions began toward the end of the first and into the second week. Following this initial wave, hematopoiesis within the active regions declined. For HPC cells, no further bursts were observed after 2 weeks. But for ST-HSC and LT-HSC cell types, new bursts continued to form up to 4 and 5 weeks after seeding, respectively. Late appearing CAs were not a continuation of early ones, but instead, CAs appeared in multiple waves in time and space, rather than a longlasting unimodal curve, as thought previously, in the LT-HSC and ST-HSC subsets. The CA underwent a phase of expansion, highly variable in cell number, prior to appearance of COS cells. Expanding CA formations frequently “flowed” into adjacent regions of the stroma as a result of coordinated migration, sometimes splitting apart and sometimes merging together in a fluid-like manner. In all cases, CA cells eventually gave rise to COS cells, which continued to divide to some extent while on top of the stromal cell mantle. COS activity was maintained as long as a CA region was associated with it, and the eventual cessation of COS activity was always preceded by decline and disappearance of an associated CA. The majority of COS cells dispersed from their CA region of formation to face two options: 1) differentiation to an adherent cell type, or 2) phagocytosis by stromal cells.
Retrospective Analysis of Cobblestone Area Forming Cells
To look into the early behaviors of the hematopoietic precursor cells in depth, a total of 30 founder cells (CAFCs) were analyzed retrospectively in the early stages (to 5th generation) using full resolution bright-field image sets that included green-fluorescent overlays at every tenth scan.
Embedding of CAFC beneath the stromal cell mantle
The majority of CAFCs (28/30) were on top of the stromal cell mantle at the beginning of imaging, approximately three hours after seeding. There was little difference between cell types for initial embedding times, but on average the proportion of time spent beneath the stroma was greatest for the LT-HSC cell type (p<0.05) and intermediate between LT-HSC and HPC for the ST-HSC cell type (Table 1). The majority of CAFCs remained under the stromal cell mantle at the time of first division (23/30).
Table 1.
Features of Embedding Beneath the Stromal Mantle
| LT -HSC | ST -HSC | HPC | |
|---|---|---|---|
| Time of initial embedding (hr) | 6.9 ± 3.0 | 9.6 ± 3.3 | 8.9 ± 2.1 |
| Time beneath stroma (hr) | 26.6 ± 1.9 | 19.1 ± 3.6 | 12.8 ± 2.0 |
| Time to 1st division (hr) | 37.8 ± 3.0 | 31.9 ± 2.0 | 24.0 ± 2.9 |
| Time beneath/Time to 1st division (%) | 74 ± 6 | 59 ± 10 | 51 ± 8 |
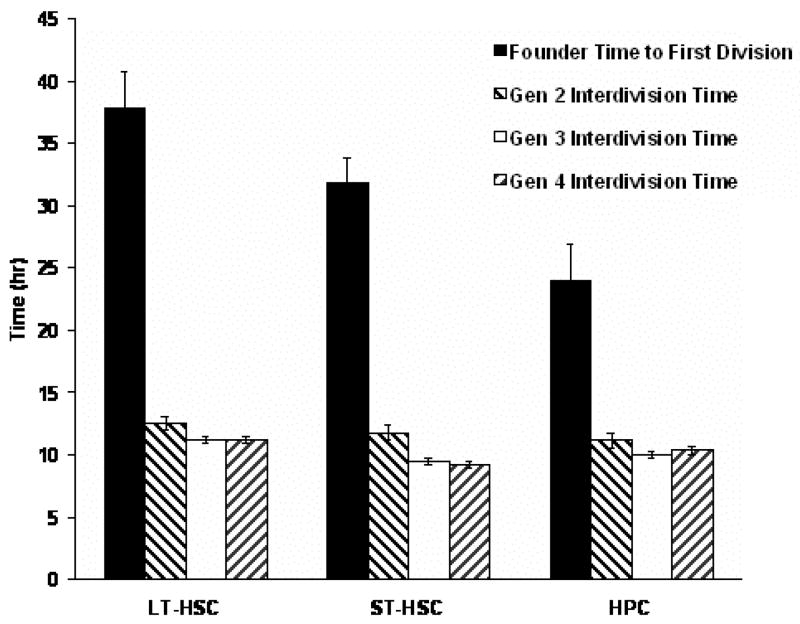
Division time analysis of CAFC
Division time analysis revealed that the average time to first division for CAFCs (1st generation) was 2 to 3-fold longer than the average inter-division time of the daughters (p<0.05), and the 2nd generation daughter inter-division time was in turn only slightly longer than that of the 3rd and 4th generations (Figure 4). These data support the notion that founder cells were not actively cycling at the time of seeding, but that cycling rates ramped up immediately thereafter. The LT-HSC population exhibited a longer time to first division in comparison to the other cell types (p<0.05), but by the 2nd generation, there were no significant differences in inter-division times between cell types. This confirmed a more quiescent state of LT-HSC over the other two cell populations that were isolated from bone marrow.
Figure 4. Early Division Times of Cobblestone Area Founder Cells and Progeny.
Division times were analysed from the 1st to 4th generation in a total of 30 CAFC families. The duration of 1st generation is significantly longer (2 to 3-fold) than that of other generations in all three cell types (p < 0.05). The LT-HSC population exhibited a longer time to first division in comparison to the other cell types (p<0.05), but by the 2nd generation, there were no significant differences in inter-division times between cell types.
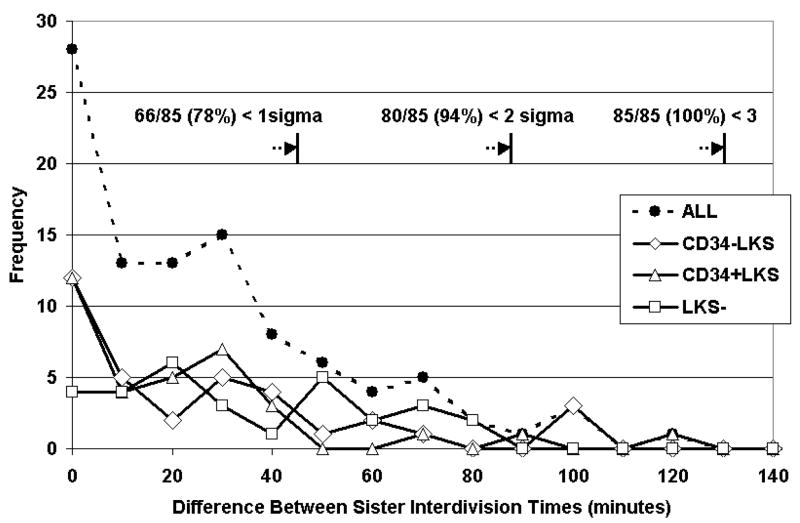
Equivalence in sister inter-division times
To determine whether there was time equivalence between pairs of sisters, the 2nd and 3rd generation sister-differences were analyzed. Interdivision times were measured as the elapsed time from the first image in which the parent cell divided into two daughter cells to the last image before each sister divided. Histograms show uni-modal normal distributions for sister-differences that would be expected from synchronized division only (Figure 5), and we did not find evidence for greater heterogeneity between sisters among the LT-HSC cells in comparison with ST-HSC or HPC subsets (standard deviations for sister pairs were 0.76 hours, 0.65 hours, and 0.75 hours, respectively).
Figure 5. Sister inter-division time difference frequency polygons.
The absolute values of differences between inter-division times of sisters illustrate constancy of all data points with expectations of a normal distribution, indicating the time equivalence of sister-pairs from generations 2 and 3. Symmetry around the midpoint at zero is assumed, so absolute values were used to remove any artificial bias. For calculation of standard deviation, the distribution was interpreted as the right half of a normal distribution centered on zero (see Methods).
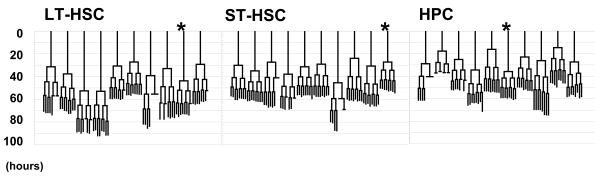
Family trees
Family trees illustrate that synchronization of division times within generations was a prominent feature for progeny of all three subtypes (Figure 6), such that sisters and cousins (i.e. family members within a generation) tend to have more similar inter-division times to each other than they do to their parents or their progeny. This feature was supported by significant interaction between family and generation in analysis of variance (Supplementary Table 1), and may suggest that determinants of interdivision time were equally distributed between sisters at the time of division.
Figure 6. Family trees.
Graphical presentations of CAFC family trees were prepared from a total of 30 families where both the beginning and ending time points were ascertainable. The patterns illustrate synchronization of sister inter-division times, particularly where “cousin” inter-division times differ. The statistical interaction factor between family and generation is apparent in families where short inter-division times precede longer ones, indicated by astericks (*).
Discussion
Our study demonstrates for the first time that the long-term hematopoietic co-culture system can be monitored for more than 5 weeks in high spatial and temporal resolution with the time-lapse imaging system. The kinetics of late CA expansion, which correlates with in vivo self-renewal potential of HSC, as well as multiple variables of early behaviors such as transmigration, mitotic quiescence, and differentiation (1) can be in part quantitatively measured with our in vitro system. Differences between hematopoietic subsets were evident even with limited numbers analyzed in this study. This approach allows a direct and high-resolution analysis of cobblestone formation and differentiated cell production from single purified cells, and allows direct assessment of some of the long-standing assumptions on the stem cell and progenitor behavior in the CAFC assay.
We have established the feasibility of using combined brightfield and fluorescence time-lapse imaging to distinguish and track individual GFP+ CAFCs in co-cultures as a method for measuring early division times, tracing family lineages, monitoring aspects of early stromal cell-hematopoietic cell interaction. Subsequently with composites of fluorescent images from adjacent viewfields we followed the kinetics of CA formation and COS transition out to more than 5 weeks elapsed time. This is a helpful starting point to guide the fluorescent protein application in the long-term tracking system with a high-frequency imaging technique.
Direct evidence was provided for the derivation of COS from CA, which could serve as qualitative measure for hematopoietic differentiation. We have shown the predominant cell behaviors and outcomes, including initial localization, CA formation and consequent COS emergence as common features of all three subsets of sorted hematopoietic cells on the stroma. However, the CA as defined by cell morphology was not uniquely associated with either HSC or HPC. We did not observe significant morphological differences of CAs generated from LT-HSC, ST-HSC and HPC. Rather, the appearance of CAs at different time points is correlated with the hierarchy of the hematopoietic cells as also indicated in previous studies by others (20). We found that late appearing CAs were not a continuation of early ones, but instead, CAs appeared in multiple waves in time and space, rather than a longlasting single wave as thought previously, in the LT-HSC and ST-HSC subsets. In addition CAs can merge, split and flow into adjacent areas in a fluid-like manner, exhibiting an intercellular attraction that keeps them clustered despite their migratory activity. Short-interval imaging even in the late stage as well as in the early stage helps better tracking of the dynamic traits revealed above.
Our retrospective analysis demonstrated that, on average, CAFC from the more primitive LT-HSC population showed greater preference for residing beneath the stromal cell mantle than CAFC from the less primitive ST-HSC and HPC populations. As expected, LT-HSC also exhibited a longer elapsed time before first division (quiescence), which has been well documented previously in other culture systems. It is known that stem cells exit the quiescent state when cultured in vitro. How long they maintain quiescence in LTC is as yet undetermined, but our study suggests that rapid cycling is uniformly initiated after first division. The longer average delay of LT-HSC before first division was associated with the known later CA formation (31, 32) in comparison to ST-HSC and HPC, In a 4-day study in liquid culture (i.e. without accessory stromal cells but with added cytokines), Dykstra and colleagues reported longer elapsed time before first division for HSCs that were subsequently demonstrated to be functionally competent in comparison to non-competent cells by competitive repopulation in vivo (24). In our study, stem cell-associated functionality, as set forth by Ploemacher et al (33), was reflected by continued CA activity (multiple waves of CA/COS expansion) for the LT-HSC population out to 5 weeks, in contrast with the HPC and ST-HSC populations, which did not support CA activity beyond 2 and 4 weeks, respectively, thereby also indicating a greater self-renewal potential of LT-HSC. Moreover, these important findings are highly consistent with our functional validations for these different hematopoietic cell subsets with both in vitro and in vivo state-of-the-art assays (Fig. 1). Therefore, this work with a real time imaging system provides insights into the classical long-term culture system in which physiological features of HSCs and HPCs in the context of stroma-based microenvironment can be at least partially recapitulated.
The cycling rates ramped up quickly suggesting the lack of critical components for maintaining the quiescence of HSC with the current co-culture system. This might be due to manipulations such as radiation exposure of the stromal cells (34). A hypoxic environment similar to that in bone marrow (35) may be required. But with the measurable parameters (such as transmigration, cell division time and CA wave), our system, can guide future strategies of building a more physiologically-relevant in vitro microenvironment with other components such as the osteoblastic and endothelial niche cells subsets or non-cellular factors that may include unique combinations involving extracellular matrix, membrane bound ligands, short lived and short range secreted molecules (2). Nevertheless, our current study provides an important starting point for refinement of the method and further investigation of possible factors involved in supporting and maintaining the stem cell phenotype over a long-term of culture. An in vitro imaging system as demonstrated in our study should be complementary to in vivo imaging analyses (36–38) though its physiological relevance needs to be further developed. Moreover, given our current demonstration, we envision that the interaction between the microenvironment and malignant cell types such as leukemia stem cells should be also testable in vitro with the time-lapse imaging.
Supplementary Material
Acknowledgments
We thank Dr. Sallie Boggs for helpful discussion and stimulating suggestions, and Julie Goff for help collecting data. We thank Charalambos Athanassiou and Lei Qian for maintaining the automatic system and for statistical analysis. This investigation was supported by the NIH grants RO1 EB 001051 (to R.H.), R01 AI080424 and RO1 HL 075601 (to T.C.). T.C was a recipient of the Scholar Award from the Leukemia & Lymphoma Society (2008).
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com/leu)
References
- 1.Cheng T. Toward ‘SMART’ stem cells. Gene Ther. 2008 Jan;15(2):67–73. doi: 10.1038/sj.gt.3303066. [DOI] [PubMed] [Google Scholar]
- 2.Scadden DT. The stem-cell niche as an entity of action. Nature. 2006 Jun 29;441(7097):1075–1079. doi: 10.1038/nature04957. [DOI] [PubMed] [Google Scholar]
- 3.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183(4):1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34− low/negative hematopoietic stem cell. Science. 1996;273(5272):242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 5.Ema H, Takano H, Sudo K, Nakauchi H. In vitro self-renewal division of hematopoietic stem cells. J Exp Med. 2000 Nov 6;192(9):1281–1288. doi: 10.1084/jem.192.9.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morrison SJ, Wandycz AM, Akashi K, Globerson A, Weissman IL. The aging of hematopoietic stem cells [see comments] Nat Med. 1996;2(9):1011–1016. doi: 10.1038/nm0996-1011. [DOI] [PubMed] [Google Scholar]
- 7.Wagers AJ, Weissman IL. Differential expression of alpha2 integrin separates long-term and short-term reconstituting Lin-/loThy1.1(lo)c-kit+ Sca-1+ hematopoietic stem cells. Stem Cells. 2006 Apr;24(4):1087–1094. doi: 10.1634/stemcells.2005-0396. [DOI] [PubMed] [Google Scholar]
- 8.Krosl J, Austin P, Beslu N, Kroon E, Humphries RK, Sauvageau G. In vitro expansion of hematopoietic stem cells by recombinant TAT-HOXB4 protein. Nat Med. 2003 Oct 26;9(11):1428–1432. doi: 10.1038/nm951. [DOI] [PubMed] [Google Scholar]
- 9.Bianco P, Robey PG, Simmons PJ. Mesenchymal stem cells: revisiting history, concepts, and assays. Cell Stem Cell. 2008 Apr 10;2(4):313–319. doi: 10.1016/j.stem.2008.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dexter TM, Allen TD, Lajtha LG. Conditions controlling the proliferation of haemopoietic stem cells in vitro. J Cell Physiol. 1977;91(3):335–344. doi: 10.1002/jcp.1040910303. [DOI] [PubMed] [Google Scholar]
- 11.Dexter TM, Moore MA, Sheridan AP. Maintenance of hemopoietic stem cells and production of differentiated progeny in allogeneic and semiallogeneic bone marrow chimeras in vitro. J Exp Med. 1977 Jun 1;145(6):1612–1616. doi: 10.1084/jem.145.6.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4(1–2):7–25. [PubMed] [Google Scholar]
- 13.Frimberger AE, Stering AI, Quesenberry PJ. Characterization of engraftable hematopoietic stem cells in murine long-term bone marrow cultures. Exp Hematol. 2001 May;29(5):643–652. doi: 10.1016/s0301-472x(01)00629-4. [DOI] [PubMed] [Google Scholar]
- 14.Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006 Mar 31;311(5769):1880–1885. doi: 10.1126/science.1110542. [DOI] [PubMed] [Google Scholar]
- 15.Frimberger AE, McAuliffe CI, Werme KA, Tuft RA, Fogarty KE, Benoit BO, et al. The fleet feet of haematopoietic stem cells: rapid motility, interaction and proteopodia. Br J Haematol. 2001 Mar;112(3):644–654. doi: 10.1046/j.1365-2141.2001.02542.x. [DOI] [PubMed] [Google Scholar]
- 16.Wagner W, Saffrich R, Wirkner U, Eckstein V, Blake J, Ansorge A, et al. Hematopoietic progenitor cells and cellular microenvironment: behavioral and molecular changes upon interaction. Stem Cells. 2005 Sep;23(8):1180–1191. doi: 10.1634/stemcells.2004-0361. [DOI] [PubMed] [Google Scholar]
- 17.Rieger MA, Schroeder T. Exploring hematopoiesis at single cell resolution. Cells Tissues Organs. 2008;188(1–2):139–149. doi: 10.1159/000114540. [DOI] [PubMed] [Google Scholar]
- 18.Wu M, Kwon HY, Rattis F, Blum J, Zhao C, Ashkenazi R, et al. Imaging hematopoietic precursor division in real time. Cell Stem Cell. 2007 Nov;1(5):541–554. doi: 10.1016/j.stem.2007.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ploemacher RE, van der Sluijs JP, van Beurden CA, Baert MR, Chan PL. Use of limiting-dilution type long-term marrow cultures in frequency analysis of marrow-repopulating and spleen colony-forming hematopoietic stem cells in the mouse. Blood. 1991;78(10):2527–2533. [PubMed] [Google Scholar]
- 20.de Haan G, Van Zant G. Intrinsic and extrinsic control of hemopoietic stem cell numbers: mapping of a stem cell gene. J Exp Med. 1997;186(4):529–536. doi: 10.1084/jem.186.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Absher PM, Absher RG. Clonal variation and aging of diploid fibroblasts. Cinematographic studies of cell pedigrees. Exp Cell Res. 1976 Dec;103(2):247–255. doi: 10.1016/0014-4827(76)90261-5. [DOI] [PubMed] [Google Scholar]
- 22.Nadarajah B, Alifragis P, Wong RO, Parnavelas JG. Neuronal migration in the developing cerebral cortex: observations based on real-time imaging. Cereb Cortex. 2003 Jun;13(6):607–611. doi: 10.1093/cercor/13.6.607. [DOI] [PubMed] [Google Scholar]
- 23.DiMilla PA, Stone JA, Quinn JA, Albelda SM, Lauffenburger DA. Maximal migration of human smooth muscle cells on fibronectin and type IV collagen occurs at an intermediate attachment strength. J Cell Biol. 1993 Aug;122(3):729–737. doi: 10.1083/jcb.122.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dykstra B, Ramunas J, Kent D, McCaffrey L, Szumsky E, Kelly L, et al. High-resolution video monitoring of hematopoietic stem cells cultured in single-cell arrays identifies new features of self-renewal. Proc Natl Acad Sci U S A. 2006 May 23;103(21):8185–8190. doi: 10.1073/pnas.0602548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Frimberger AE, Stering AI, Quesenberry PJ. An in vitro model of hematopoietic stem cell homing demonstrates rapid homing and maintenance of engraftable stem cells. Blood. 2001 Aug 15;98(4):1012–1018. doi: 10.1182/blood.v98.4.1012. [DOI] [PubMed] [Google Scholar]
- 26.Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, et al. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods. 2006 May;3(5):385–390. doi: 10.1038/nmeth876. [DOI] [PubMed] [Google Scholar]
- 27.Schroeder T. Tracking hematopoiesis at the single cell level. Ann N Y Acad Sci. 2005;1044:201–209. doi: 10.1196/annals.1349.025. [DOI] [PubMed] [Google Scholar]
- 28.Bahnson A, Athanassiou C, Koebler D, Qian L, Shun T, Shields D, et al. Automated measurement of cell motility and proliferation. BMC Cell Biol. 2005 Apr 14;6(1):19. doi: 10.1186/1471-2121-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yuan Y, Shen H, Franklin DS, Scadden DT, Cheng T. In vivo self-renewing divisions of haematopoietic stem cells are increased in the absence of the early G1-phase inhibitor, p18INK4C. Nat Cell Biol. 2004 May;6(5):436–442. doi: 10.1038/ncb1126. [DOI] [PubMed] [Google Scholar]
- 30.Takano H, Ema H, Sudo K, Nakauchi H. Asymmetric division and lineage commitment at the level of hematopoietic stem cells: inference from differentiation in daughter cell and granddaughter cell pairs. J Exp Med. 2004 Feb 2;199(3):295–302. doi: 10.1084/jem.20030929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.van der Sluijs JP, de Jong JP, Brons NH, Ploemacher RE. Marrow repopulating cells, but not CFU-S, establish long-term in vitro hemopoiesis on a marrow-derived stromal layer. Exp Hematol. 1990 Sep;18(8):893–896. [PubMed] [Google Scholar]
- 32.Breems DA, Blokland EA, Siebel KE, Mayen AE, Engels LJ, Ploemacher RE. Stroma-contact prevents loss of hematopoietic stem cell quality during ex vivo expansion of CD34+ mobilized peripheral blood stem cells. Blood. 1998 Jan 1;91( 1):111–117. [PubMed] [Google Scholar]
- 33.Ploemacher RE, van der Sluijs JP, Voerman JS, Brons NH. An in vitro limiting-dilution assay of long-term repopulating hematopoietic stem cells in the mouse. Blood. 1989;74(8):2755–2763. [PubMed] [Google Scholar]
- 34.Gorbunov NV, Pogue-Geile KL, Epperly MW, Bigbee WL, Draviam R, Day BW, et al. Activation of the nitric oxide synthase 2 pathway in the response of bone marrow stromal cells to high doses of ionizing radiation. Radiat Res. 2000 Jul;154(1):73–86. doi: 10.1667/0033-7587(2000)154[0073:aotnos]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 35.Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci U S A. 2007 Mar 27;104(13):5431–5436. doi: 10.1073/pnas.0701152104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008 Dec 12;135(6):1118–1129. doi: 10.1016/j.cell.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 37.Kohler A, Schmithorst V, Filippi MD, Ryan MA, Daria D, Gunzer M, et al. Altered cellular dynamics and endosteal location of aged early hematopoietic progenitor cells revealed by time-lapse intravital imaging in long bones. Blood. 2009 Jul 9;114(2):290–298. doi: 10.1182/blood-2008-12-195644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xie Y, Yin T, Wiegraebe W, He XC, Miller D, Stark D, et al. Detection of functional haematopoietic stem cell niche using real-time imaging. Nature. 2009 Jan 1;457(7225):97–101. doi: 10.1038/nature07639. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.