Abstract
Roscovitine and flavopiridol suppress CDK7 and CDK9 activity resulting in transcription inhibition, thus providing an alternative mechanism to traditional genotoxic chemotherapy. These agents have been effective in slow or non-replicative cell types. 8-Amino-adenosine is a transcription inhibitor which has proven very effective in multiple myeloma cell lines and primary indolent leukemia cells. The objective of the current work was to define mechanisms of action that lead to transcription inhibition by 8-amino-adenosine. 8-Amino-adenosine is metabolized into the active triphosphate (8-amino-ATP) in cells. This accumulation resulted in a simultaneous decrease of intracellular ATP and RNA synthesis. When the effects of established ATP synthesis inhibitors and transcription inhibitors on intracellular ATP concentrations and RNA synthesis were studied, there was a strong correlation between ATP decline and RNA synthesis. This correlation substantiated the hypothesis that the loss of ATP in 8-amino-adenosine treated cells contributes to the decrease in transcription due to the lack of substrate needed for mRNA body and poly(A) tail synthesis. RNA polymerase II C-terminal domain phosphorylation declined sharply in 8-amino-adenosine treated cells which may have been due to the lack of an ATP phosphate donor or competitive inhibition with 8-amino-ATP at CDK7 and CDK9. Furthermore, 8-amino-ATP was incorporated into nascent RNA in a dose dependent manner at the 3′-end resulting in transcription termination. Finally, in vitro transcription assays demonstrated that 8-amino-ATP competes with ATP for incorporation into mRNA. Collectively, we have concluded that 8-amino-adenosine elicits effects on multiple mechanisms of transcription, providing a new class of transcription inhibitors.
Keywords: multiple myeloma, 8-amino-adenosine, nucleoside analog, ATP, transcription
Introduction
Cyclin dependent kinase (CDK) inhibitors are gaining success in the clinic as anticancer agents especially in hematological malignancies (1). Although originally developed as cell cycle inhibitors, several of these agents were recognized as transcription inhibitors since they inhibit the activity of CDK7 and CDK9. These kinases phosphorylate serine residues at the C-terminal domain (CTD) of RNA polymerase II which is the enzyme responsible for synthesis of mRNA (messenger RNA) (2). Roscovitine was among the first to be tested in preclinical and clinical testing (3–4). Flavopiridol is probably the most used transcription inhibitor and proof-of-concept studies have shown action against CDK7 and CDK9 as well as inhibition of transcript synthesis with an immediate effect on short-lived transcripts and proteins such as Mcl-1, cyclin D1, and cMet (5–7). Clinically, flavopiridol has been extensively used for treatment of chronic lymphocytic leukemia (8). The new agent SNS-032 (formerlyknown as BMS-387032) also belongs to the same category of CDK inhibitors with more potent inhibition of CDK2, −7, and −9 (9). In general, CDK inhibitors that suppress transcription provide a novel strategy for drug development.
The structure of roscovitine and other analogs clearly demonstrated that purine and adenine provide a scaffold which were exploited to create additional drugs and analogs (3). Our previous work determined that adenine nucleoside analogs or adenosine analogs were potent inhibitors of transcription (10–12). The two members of the carbon-8 (C8) substituted adenosine analogs that have been tested for cytotoxicity are 8-chloro-adenosine and 8-amino-adenosine. Unlike CDK inhibitors, C8-substituted analogs provide different mechanisms of transcription inhibition (11). However, similar to CDK inhibitors, these analogs are also effective in slow or non-replicating cells such as those from multiple myeloma or indolent leukemias such as chronic lymphocytic leukemia (10, 12–14). Data from replicationally quiescent primary chronic lymphocytic leukemia cells with either CDK inhibitors or C8-substituted analogs established that DNA replication was not the target of these compounds (13–14). Furthermore, the cytotoxic mechanisms were similar with these agents in that there was an inhibition of transcription and a decline in short-lived transcripts and proteins such as Mcl-1, XIAP, and cMet (5–7, 13–14). Tumors that depend on these survival proteins succumb when protein quantities fall below critical levels.
While 8-chloro-adenosine was the first to be tested in preclinical setting and is currently in a Phase I clinical study for patients with chronic lymphocytic leukemia, the congener of 8-chloro-adenosine, 8-amino-adenosine, is more potent as demonstrated by induction of poly(ADP-ribose) polymerase (PARP) cleavage by 6 hours with 8-amino-adenosine compared to 24 hours by chlorinated adenosine. The intracellular accumulation of 8-amino-adenosine triphosphate (8-amino-ATP) is 38-fold higher than 8-chloro-adenosine triphosphate (8-chloro-ATP) in myeloma cells (10, 12), and leads to a greater analog-mediated decline in ATP (adenosine triphosphate) pool and mRNA synthesis (6, 10, 12-13, 15–16).
While metabolic aspects, mechanisms of action, and cytotoxic effects of 8-amino-adenosine have been investigated, information regarding how 8-amino-adenosine inhibits transcription is not available. Previous observations of 8-amino-adenosine led to the present study focused on the multiple mechanisms of transcription inhibition by 8-amino-adenosine. The results indicate that there was first a rapid decline in the intracellular ATP pool. Second, phosphorylation at the C-terminal domain of RNA polymerase II was decreased. Third, 8-amino-ATP was incorporated at the 3′-terminus in the mRNA leading to transcription termination. Collectively, these multifaceted actions indicate that 8-amino-adenosine is a member of a new class of transcription inhibitors.
Materials and Methods
Cell Line
MM.1S cells were obtained from Drs. Steven Rosen and Nancy Krett (Robert H. Lurie Comprehensive Cancer Center, Northwestern University) and grown as a suspension culture in RPMI 1640 with L-glutamine and 10% fetal bovine serum in the presence of 5% CO2 at 37 °C. This multiple myeloma cell line was derived from the peripheral blood cells of an immunoglobin A myeloma patient (17–18). The cell line is sensititve to glucocorticoids and a detailed characterization has been published (18). The current authors have not independently tested and authenticated these cells. Routine testing for Mycoplasma infection was performed using the MycoTest kit (Invitrogen, Carlsbad, CA).
Materials
The drug 8-amino-adenosine and its phosphorylated metabolite 8-amino-ATP were obtained from R.I. Chemical (Brea, CA) and ChemCyte (San Diego, CA), respectively. [2-3H]-8-Amino-adenosine (3.8 Ci/mmol) and [5, 6-3H]-uridine (41.2 Ci/mmol) were purchased from Moravek Biochemicals (Brea, CA). [α-32P]-UTP (3000 Ci/mmol) was purchased from Perkin Elmer (Waltham, MA). Flavopiridol (Drug Synthesis and Chemistry Branch, Division of Cancer Treatment) and deoxycoformycin (Dr. V. N. Narayanan) were obtained from the National Cancer Institute.
Actinomycin D, α-amanitin, antimycin A, 2-deoxy-D-glucose, and 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB) were purchased from Sigma-Aldrich (St. Louis, MO).
RNA Synthesis
Exponentially growing MM.1S cells were treated with various concentrations of 8-amino-adenosine or other inhibitors. Thirty minutes prior to the end of the incubation time, 2 μCi [5, 6-3H]-uridine was added to the cells. Samples were collected and washed before applying to glass fiber filters (Whatman, Piscataway, NJ) on a Millipore vacuum manifold (Bedford, MD) and isolating acid-insoluble material by perchloric acid extraction. Radioactivity on the filters was quantified by liquid scintillation counting (Packard Bioscience, Perkin Elmer Life and Analytical Sciences Inc., Waltham, MA). Data were expressed as percent of untreated (control) cells.
Measurement of Intracellular Nucleotides
Ten mL of MM.1S cells were plated in suspension at 3 ×105 cells/mL and treated with 8-amino-adenosine or other inhibitors as indicated. Cells were collected and nucleotides were extracted using perchloric acid followed by potassium hydroxide neutralization as previously described (19–20). To determine ATP and 8-amino-ATP concentrations, nucleotide extracts were applied to an anion-exchange Partisil-10 SAX column at a flow rate of 1.5 mL/min using a Waters 2697 Separations Module (Waters Corp, Milford, MA). The nucleotides were separated with a 60 minute concave gradient and quantitated as previously described (12).
Immunoblot Analysis
MM.1S cells were grown to a concentration of 3 × 105 cells/mL and treated with 8-amino-adenosine, DRB, or flavopiridol as indicated. Cells were harvested and lysed in fresh working radioimmunoprecipitation buffer (RIPA buffer) with sodium orthovanadate and one Complete, Mini, EDTA-free protease inhibitor cocktail tablet (Roche Applied Science, Mannheim, Germany). Protein concentration was determined by DC Protein Assay (Bio-Rad Laboratories, Hercules, CA). Colorimetric detection and quantitation were performed using the PowerWave XS Microplate Spectrometer and KC4 Data Analysis software (BioTek Instruments Inc., Winooski, VT). Protein separated on a 4-12% Criterion XT gel in XT MOPS buffer (Bio-Rad Laboratories, Hercules, CA) and transferred onto a polyvinylidene difluoride membrane (Immobilon-P, Millipore, Bedford, MA) in transfer buffer containing 20% methanol, 25 mM tris base, and 190 mM glycine. Membranes were blocked with blocking buffer (LI-COR Biosciences, Lincoln, NE) followed by incubations with a primary antibody and then secondary antibody. The membranes were washed before visualizing and quantifying proteins using the Odyssey Infrared Imaging System and associated software (LI-COR Biosciences, Lincoln, NE). Antibodies were purchased from various sources: β-actin (AC-15) (Sigma Aldrich, St. Louis, MO); CDK7 (c-4), and CDK9 (c-20), (Santa Cruz Biotechnology, Santa Cruz, CA); RNA Polymerase II 8WG16, H5 and H14, (Covance, Berkeley, CA); Alexa-Fluor 680 goat anti-mouse immunoglobulin G, Alexa-Fluor 680 goat anti-mouse immunoglobulin M, and Alexa-Fluor 800 goat anti-rabbit immunoglobulin G (Molecular Probes, Eugene, OR).
[3H]-8-Amino-adenosine Incorporation into RNA Assay
Exponentially growing MM.1S cells were treated for 1 hour with various concentrations of [2-3H]-8-amino-adenosine. Samples were collected, extracted, and quantified as described for the RNA synthesis method (11).
Site of Nucleoside Incorporation Assay
MM.1S cells were treated with [2-3H]-8-amino-adenosine or [5, 6-3H]-uridine for two hours. RNA was isolated with Trizol LS (Invitrogen, Carlsbad, CA) and resuspended in diethylpyrocarbonate-treated water. RNA (20,000 dpm) was digested into internal nucleotides and 3′ terminal nucleosides as previously described (11, 21). Nucleosides and nucleotides were extracted by perchloric acid and neutralized with potassium hydroxide. High pressure liquid chromatography (HPLC) analysis was performed using a XTerra MS C18 Column (3 mm X 100 mm 3.5 μm) at a flow rate of 0.3 mL/min and monitored at an absorption of 265 nm. [5, 6-3H]-Uridine treated samples were eluted in 100% 20 mM triethylammonium acetate buffer for 15 minutes. The retention times were: UMP, 5.5 minutes; uridine, 7.5 minutes. 8-Amino-adenosine treated samples were eluted using 95% buffer A (98% 80 mM ammonium acetate, 0.04% acetic acid) and 5% buffer B (8% 80 mM ammonium acetate, 0.04% acetic acid, 35% acetonitrile) for 30 minutes. The retention times were: AMP, 5.2 minutes; adenine, 6.9 minutes; adenosine 19.5 minutes; and 8-amino-adenosine 23.5 minutes. An 8-amino-adenosine monophosphate (8-amino-AMP) standard has not been obtained, however based on the known retention time of 8-chloro-adenosine monophosphate (8-chloro-AMP), 8-amino-AMP is predicted to elute at the same retention time as AMP. Radioactive standards were detected by an online scintillation counting. Sample fractions were collected every 20 seconds and quantitated by liquid scintillation counting.
In Vitro Transcription Assay
AFP7 plasmid (alpha-fetoprotein promoter driving β-galactosidase gene) and HeLa nuclear extracts were provided by Drs. Michelle Barton and Meghan Minard at the University of Texas M.D. Anderson Cancer Center (22-23). The plasmid was digested with Age1 and calf alkaline phosphatase to limit transcription to 250 nucleotides. In vitro transcription was performed in a 50 μL reaction containing 3 mg digested DNA (5 μL), 95 μg HeLa nuclear extract in nuclear dialysis buffer (15 μL), 10 μL nuclear dialysis buffer, 71 mM potassium chloride, 20 mM HEPES, pH 7.9, 5 mM magnesium chloride, 5 mM creatine phosphate, 0.6 mM ATP, 0.6 mM GTP, 0.6 mM CTP, 2 mM dithiothreitol, 0.2 mg/mL bovine serum albumin, 10 U/mL creatine kinase, 50 μM UTP, and 10 μCi α-[32P]-UTP for 1 hour at 30 °C followed by an incubation with 10 μg tRNA and 2.5 U RQ-1 DNAse (Promega) and another incubation with 10 μg Proteinase K and 2.5 μL 5% SDS/0.125 M EDTA. The product was purified and resuspended in denaturing loading dye. The samples were separated in a prerun 8 M Urea/ 8% polyacrylamide gel at 600 V in 1X tris-borate-EDTA buffer. The gel was exposed to Amersham Hyperfilm MP (GE Healthcare Life Sciences, Piscataway, NJ) overnight and developed. The film was scanned and quantitations were determined using the Odyssey Infrared Imaging System software.
Statistical Analysis
Relationships between different parameters were obtained using GraphPad Prism Software (La Jolla, CA). Student t-test was used for analyses.
Results
8-Amino-adenosine affects RNA synthesis and intracellular ATP
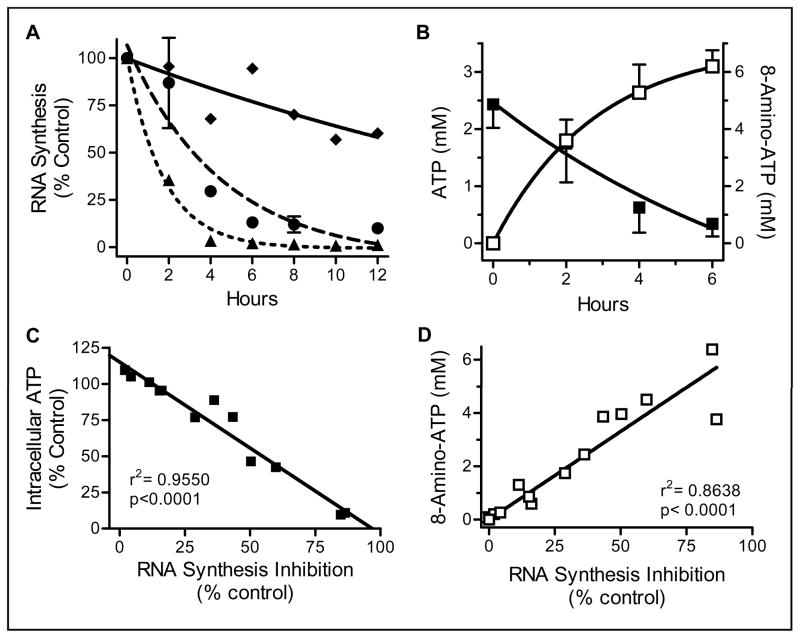
To determine the actions of 8-amino-adenosine on global RNA synthesis, both time- and dose-dependent experiments were performed (Fig. 1A). While 1 μM 8-amino-adenosine treatment resulted in approximately 50% inhibition of RNA synthesis after 12 hours, cells treated with 10 μM and 30 μM 8-amino-adenosine had a 50% RNA synthesis inhibition by 2 and 4 hours, respectively. RNA synthesis inhibition was in parallel to the decline in the intracellular ATP pool over time as well as the accumulation of the metabolite 8-amino-ATP in cells (Fig. 1B). Reduction in the endogenous ATP pool was time dependent and by 6 hours of 10 μM 8-amino-adenosine treatment less than 20% of the initial intracellular ATP level remained. Accumulation of 8-amino-ATP was efficient; at 2 and 6 hours more than 3 and 6 mM 8-amino-ATP was formed. Additional analyses (Fig. 1C and Fig. 1D) demonstrated that there were highly significant and linear correlations between RNA synthesis inhibition and the decline in ATP pool and the accumulation of 8-amino-ATP in treated cells.
Figure 1. Actions of 8-amino-adenosine on RNA synthesis (A) and intracellular ATP pool (B).
A. Cells were incubated with 1 (diamond), 10 (circle), and 30 μM (triangle) 8-amino-adenosine and RNA synthesis was measured using uridine incorporation assay. B. Cells were incubated with 10 μM 8-amino-adenosine for the indicated times and the ATP (solid) and 8-amino-ATP (open) intracellular concentrations were measured by HPLC. C. Relationship between ATP decline and inhibition of RNA synthesis. Data from A and B were plotted. D. Relationship between 8-amino-ATP accumulation and inhibition of RNA Synthesis. Data from A and B were plotted.
Decline in ATP pool leads to RNA synthesis inhibition
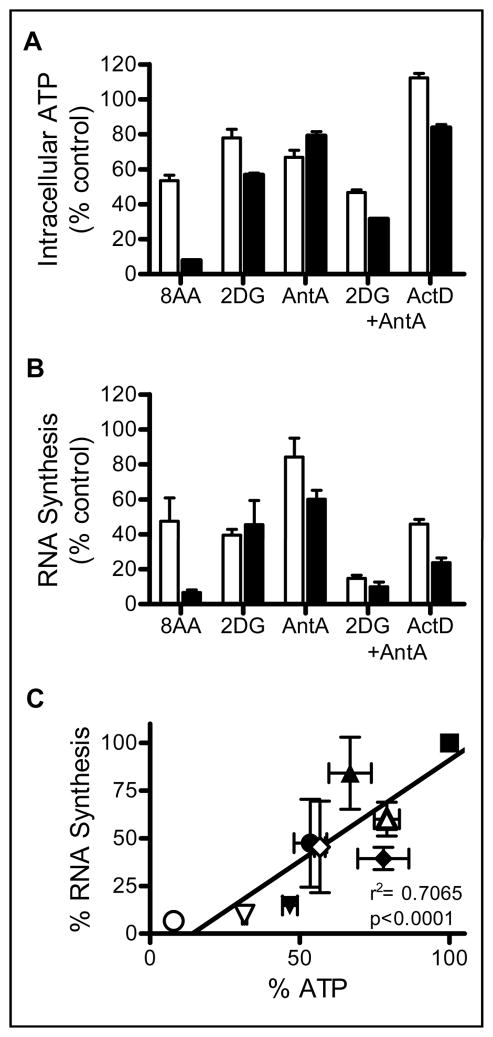
Because a relationship was established between the loss of RNA synthesis and the decline in the ATP pool (Fig. 1C), previously known ATP synthesis inhibitors (2-deoxy-D-glucose and antimycin A) and a RNA synthesis inhibitor (actinomycin D) were used to determine the effect of each on the ATP pool and RNA synthesis (Fig. 2A and 2B). 2-Deoxy-D-glucose inhibits ATP production via glycolysis in cells and cells treated with 5 mM 2-deoxy-D-glucose exhibited a 45% decline in the ATP pool and 55% decrease in RNA synthesis within 4 hours. ATP generated by the electron transport pathway is inhibited at complex III by antimycin A. ATP pool and RNA synthesis were decreased by 22% and 40% respectively in cells treated with 2 μM antimycin A. Combined treatment with 2-deoxy-D-glucose and antimycin A resulted in a 69% decrease in intracellular ATP and 90% inhibition of RNA synthesis. Actinomycin D treatment contributed to a 17% decrease in the ATP pool and 76% inhibition of RNA synthesis. 8-Amino-adenosine at 10 μM had the greatest effect on the ATP pool and RNA synthesis which were inhibited by 92% and 94% respectively at 4 hours under these conditions. To determine a relationship between ATP pool decline and RNA synthesis inhibition, data obtained with bonafide ATP inhibitors (Fig. 2A and 2B) were plotted with the associated decrease in RNA synthesis. These analyses (Fig. 2C) demonstrated a statistically significant (p<0.0001) strong linear relationship (r2= 0.7065) between the decline of the ATP pool by ATP inhibitors and a decrease in RNA synthesis indicating that the effect of 8-amino-adenosine on the decline of the ATP pool is one mechanism of 8-amino-adenosine mediated transcription inhibition.
Figure 2. Effect of RNA and ATP synthesis inhibitors on intracellular ATP pool and RNA synthesis.
A, B. Cells were incubated with 10 μM 8-amino-adenosine (8AA), 5 mM 2-deoxy-D-glucose (2DG), 2 μM antimycin A (AntA), a combination of 2 μM antimycin A and 5 mM 2-deoxy-glucose (2DG + AntA), and 0.1 μg/mL actinomycin D (ActD) for 2 (white bars) and 4 (black bars) hours. Intracellular ATP pool (A) and RNA synthesis (B) were measured as described in materials and methods. C. Relationship between the ATP pool and RNA synthesis inhibition by ATP synthesis inhibitors, untreated (square), 8-amino-adenosine (circle), antimycin A (triangle), 2-deoxy-D-glucose (diamond), antimycin A + 2-deoxy-D-glucose (upside down triangle) at 2 (open) and 4 (solid) hours. Data obtained in A and B were plotted.
8-Amino-adenosine inhibits RNA polymerase II CTD phosphorylation
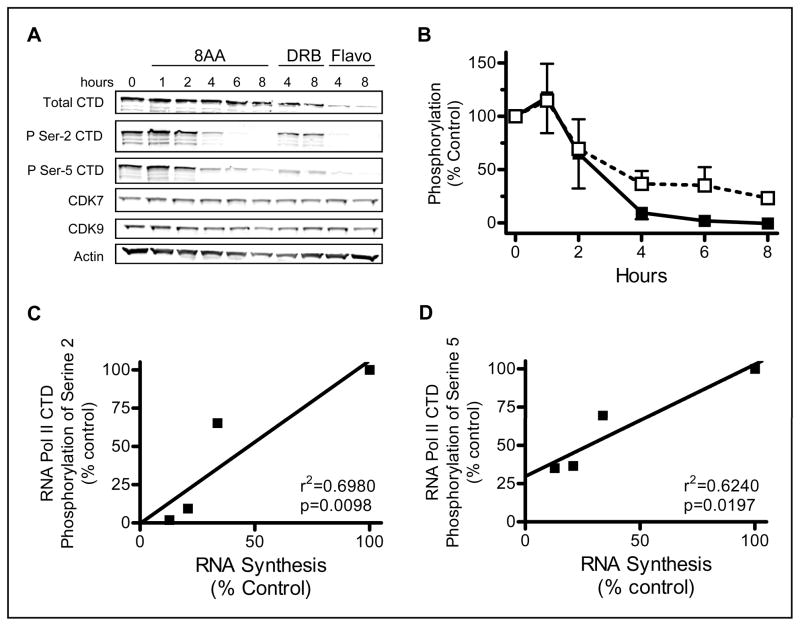
In order to understand the RNA transcription inhibition mechanism by 8-amino-adenosine in MM.1S cells, immunoblots were employed to evaluate total and phosphorylated RNA polymerase II. The RNA polymerase II C-terminal domain is phosphorylated at Serine-5 and Serine-2 by CDK7 and CDK9, respectively, and this is necessary for transcription initiation and elongation. Phosphorylation of the RNA polymerase II CTD at Serine-5 and Serine-2 was decreased with 8-amino-adenosine treatment indicating that transcription initiation and elongation activity could not proceed (Fig. 3A and 3B). DRB and flavopiridol were used as positive controls for decreased RNA polymerase II phosphorylation. DRB is a pharmacological inhibitor of CDK9 and thus leads to a decrease Serine-2 phosphorylation (24–25). Flavopiridol was developed as a general CDK inhibitor and has been shown to be a potent inhibitor of CDK9 (2, 5). To rule out the possibility of a decrease in kinase levels, the total protein expression of CDK7 and CDK9 was also examined and determined to be unaffected in 8-amino-adenosine treated cells (Fig. 3A). Thus, decline in CTD phosphorylation may be attributed to either the lack of available ATP for these kinases or 8-amino-ATP may competitively inhibit the activity of CDK7 and CDK9. There was a linear relationship between the decline in RNA synthesis and the decrease in CTD phosphorylation at Serine-5 and Serine-2 (Fig. 3C and 3D). These data indicate that the decline in CTD phosphorylation upon 8-amino-adenosine treatment is an additional mechanism of transcription inhibition.
Figure 3. Effect of 8-amino-adenosine on RNA Polymerase II.
A. Immunoblot of MM.1S cells treated with 10 μM 8-amino-adenosine (8AA), 50 μM 5,6-dichloro-1-β-D-ribofuranosylbenzimidazole (DRB), and 3 μM flavopiridol (Flavo) for the times indicated. B. Quantitation of the effect of 8-amino-adenosine on the phosphorylation at Serine-2 (black square) and Serine-5 (white square). Phosphorylation was normalized to the total RNA polymerase II CTD protein and actin protein. C, D. Relationship between RNA synthesis decline and RNA Polymerase II phosphorylation at Serine-2 and Serine-5. Data obtained from figure 3B and 1A were plotted to determine the correlation.
8-Amino-adenosine is incorporated into nascent RNA transcripts
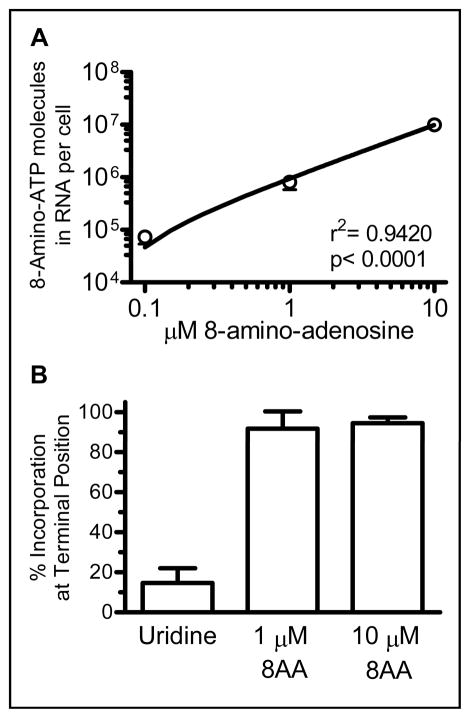
High accumulation of 8-amino-ATP in cells and its similarity to ATP suggested possible incorporation into RNA transcripts which may then prevent further elongation of the transcript by RNA polymerase II (11). Therefore, it was important to determine if 8-amino-adenosine incorporates into RNA. As shown in Fig. 4A, titrated 8-amino-adenosine incorporates into RNA in a dose dependent manner. Additionally, more than 90% of the nucleoside analog was incorporated into RNA at a 3′-terminal position suggesting that mRNA synthesis stops upon addition of 8-amino-adenosine into transcript by RNA polymerase II (Fig. 4B). Taken together, these data demonstrate that the metabolized 8-amino-adenosine is incorporated into nascent RNA transcripts resulting in transcription termination.
Figure 4. RNA incorporation of 8-amino-adenosine.
A. Dose-dependent incorporation of 8-amino-adenosine in RNA of whole cells. Cells were treated with 0.1, 1, and 10 μM 8-amino-adenosine for one hour. Incorporation was measured as described in materials and methods. B. Percent of [3H]-uridine or [3H]-8-amino-adenosine incorporation into RNA at a terminal position. Cells were incubated with 1 and 10 μM 8-amino-adenosine for 2 hours. RNA was isolated and digested to determine incorporation position of uridine or 8-amino-adenosine molecules as described in materials and methods.
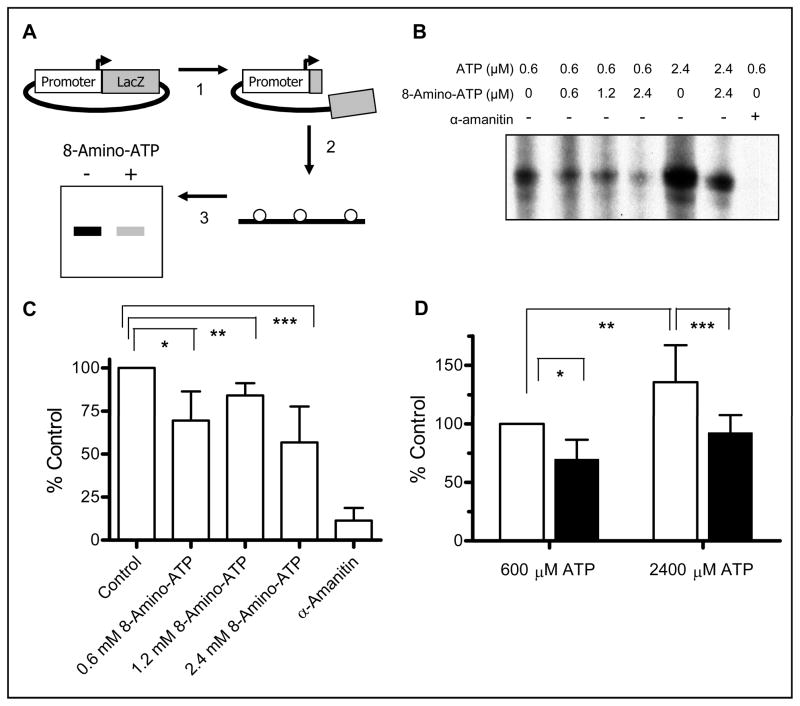
In vitro transcription competition of ATP and 8-amino-ATP
To further demonstrate incorporation of 8-amino-ATP into mRNA, inhibition of further transcript elongation, and competition between 8-amino-ATP and ATP for incorporation, an in vitro transcription assay was employed. As illustrated in Fig. 5A, the AFP7 DNA plasmid construct was cleaved so that the only 250 nucleotides from the Lac Z gene could be transcribed upon activation of the alpha-fetoprotein promoter. The in vitro transcription reaction was a cocktail of the DNA template, HeLa nuclear extract, and nucleotides. In order to specifically label only new RNA transcripts, α-[32P]-UTP was included in the reaction mixture. The mRNA products were purified, separated by gel electrophoresis, and then exposed to film in order to image the results (Fig. 5B). For each experiment, the transcription inhibitor α-amanitin was added to one reaction as a negative control. The full length 250 nucleotide RNA product was detected and quantitated. When the data from three experiments were analyzed, there appeared to be a trend to decrease mRNA product when 8-amino-ATP was used at 0.6, 1.2, and 2.4 mM levels in the presence of 0.6 mM ATP (Fig. 5C). Increases in ATP concentration in the transcription mixture augmented product formation. However, competition with 8-amino-ATP at a 1:1 ratio affected the product generation. Again, there was a decline when 8-amino-ATP was added to the transcription mixture (Fig. 5D).
Figure 5. Effect of 8-amino-ATP on in vitro transcription.
A. Schema of Assay. Step 1: The AFP7 plasmid was linearized by restriction enzyme digest and dephosphorylated. Step 2: In vitro transcription reaction was performed using α-32P-UTP to label nascent RNA. Step 3: Nascent RNA was purified and separated by gel electrophoresis. B. Effect of various concentrations of 8-amino-ATP and ATP on in vitro transcription of 250 nucleotide RNA product. Experiments were performed as described in schema. C. Quantitation and normalization of the in vitro transcription reactions with 0.6 μM ATP and various concentrations of 8-amino-ATP or the known RNA synthesis inhibitor α-amanitin. D. Quantitation and normalization of the reactions with 0.6 or 2.4 μM ATP with or without an equal concentration of 8-amino-ATP.
Discussion
The focus of this study was to identify the multiple mechanism of transcription inhibition by 8-amino-adenosine.
Decline in ATP pool and effect on transcription
Data presented in the present work have shown that lowering intracellular ATP by previously established ATP synthesis inhibitors as well as 8-amino-adenosine is strongly related to a decline in RNA synthesis (Fig. 1A and 2B). 2-Deoxy-glucose inhibits ATP synthesis by competing with hexokinase in the glycolysis pathway (26). Antimycin A acts by inhibiting the complex II of the pentose phosphate pathway (27). Since, ATP is the primary energy source in cells and is used as a phosphate donor for numerous enzymes, the decline in ATP likely results in transcription inhibition. More important than its role in the energetics of the cell, ATP is a substrate for mRNA synthesis including the body of the transcript and the required poly(A) tail. All adenosine derived nucleoside analogs do not inhibit RNA synthesis and the intracellular ATP pool. Cordycepin (3′-deoxyadenosine) can inhibit RNA synthesis, however, there is no effect on the intracellular ATP pool as the metabolite, 3′-deoxyadenosine triphosphate accumulates in these MM.1S cells (28). Another congener, 8-chloro-adenosine, causes an ATP pool decline as well as an inhibition of RNA synthesis (11, 13, 29–32). The precise mechanism by which 8-amino-adenosine causes a decline in the ATP pool has not been determined although it is feasible that, similar to the chlorinated adenosine analog, 8-amino-ADP is a substrate for ATP synthase and/or 8-amino-ATP may act as an inhibitor of the ATP synthase (33).
8-Amino-adenosine and RNA polymerase II activity
Another mechanism of transcription inhibition by 8-amino-adenosine is the decrease in serine phosphorylation on RNA polymerase II (Fig. 3A and 3B). RNA polymerase II phosphorylation is a two step process which requires a sequential phosphorylation of the CTD at Serine-5 and Serine-2 (34–38). An array of agents affects this process. DRB acts by inhibiting transcription elongation mediated by positive transcription elongation factor b (PTEF-b) (39–41). Flavopiridol, a cyclin dependent kinase inhibitor, also suppresses transcription by inhibiting PTEF-b (5, 7, 38, 42–43). SNS-032 is currently being developed as a chemotherapeutic agent which inhibits CDK2, CDK7, and CDK9 and consequently decreases the transcription of short-lived oncoproteins (9, 44). Roscovitine is another cyclin dependent kinase inhibitor which has diminutive effects on transcription (45–47).
RNA polymerase II phosphorylation decline is evident in cells within two hours of treatment with 8-amino-adenosine (Fig. 3A). Published work has shown that the decline in ATP pool does not lead to a global decrease in protein phosphorylation (15). Therefore, a lack of intracellular ATP substrate for the CDK7 and CDK9 proteins that mediate the phosphorylation of the RNA polymerase II C-terminal domain may not provide an appropriate rationale for this observation. While there was no effect of 8-amino-adenosine incubation on total kinase level, it is possible that 8-amino-ATP can inactivate CDK7 and CDK9 which warrants future investigation.
8-Amino-ATP incorporation and inhibition of mRNA synthesis
ATP is a natural substrate for RNA polymerase II activation as well as incorporation into nascent RNA chains. Evidence has been provided here that demonstrates 8-amino-ATP is a substrate for RNA incorporation and furthermore that this incorporation at the 3′ terminus causes chain termination (Fig. 4). These effects have also been observed in 8-chloro-adenosine treated cells (11). The chain termination is likely a consequence of the inability of the cellular machinery to proceed past the site of analog incorporation as well as an inability to excise the analog from the transcript. It has been generally accepted that errors in RNA transcription result in the decay of the new transcript and the RNA transcription machinery would restart the synthesis since the rate of transcription is so great in cells that repair mechanisms would not be useful (48). However, RNA polymerases do have a 3′–5′ nuclease activity that is enhanced by cleavage stimulatory factors which facilitate the backtracking and hydrolyzing of nascent transcripts when elongation is stalled. The polymerase is then able to reinitiate elongation (49). It is possible that the amino group may interfere with the sterics of the RNA polymerase and either cause the RNA polymerase to pause at the site, unsuccessfully attempt to repair the incorporation or perhaps cause the RNA polymerase to dissociate from the DNA.
It may be inferred that as 8-amino-ATP accumulates in the cell, it competes with ATP for incorporation into mRNA. Evidence is provided that illustrates that as the ratio of 8-amino-ATP to ATP increases, there is a decline of in vitro transcription generated mRNA products (Fig. 5). Based on these experiments, it appears that an in vivo concentration of 8-amino-ATP must greatly exceed the concentration of ATP in order to fully inhibit transcription. This excess may be enhanced in 8-amino-adenosine treated cells by the rapid decline in the ATP pool.
8-Amino-adenosine and polyadenylation
The stability of nascent RNA products is dependant upon posttranscriptional modifications. Polyadenylation at the 3′-end of the transcript is one of these modifications critical to the stability of newly transcribed mRNA. Previous work has investigated 8-amino-ATP in the context of polyadenylation. In vitro polyadenylation assays have shown that 8-amino-ATP is a substrate for yeast poly(A) polymerase. However, once the analog is incorporated into the poly(A) tail, further polyadenylation extension is inhibited (16). In the context of the cell, 8-amino-ATP is likely to compete with ATP as a substrate for poly(A) polymerase as shown in the dose-dependent decrease of the poly(A) tail length (16). Similar data were obtained using the mammalian poly(A) polymerase enzyme (50). The inhibition of polyadenylation by 8-amino-ATP provides another mechanism by which 8-amino-adenosine regulates and inhibits transcription.
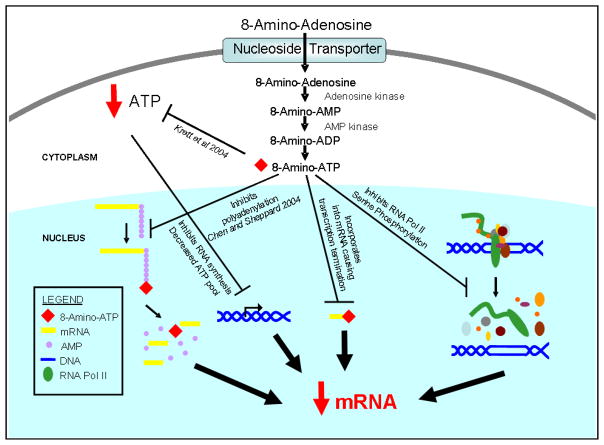
8-Amino-adenosine has proven to be cytotoxic in several cell lines. The cytotoxicity of this nucleoside analog is dependent upon its metabolism to the nucleoside triphosphate by adenosine kinase and other enzymes. The active metabolite has consequences in the cell that lead to RNA synthesis inhibition, decline in the ATP pool, and cell death. Present data and previous work have demonstrated that 8-amino-adenosine can inhibit transcription by four distinct mechanisms. The decline in the ATP pool, decrease in RNA polymerase II activity due to a decline in phosphorylation, incorporation of the 8-amino-adenosine metabolite in nascent mRNA with subsequent RNA chain termination, and inhibition of polyadenylation of transcripts are all mechanisms by which 8-amino-adenosine functions to inhibit transcription (Fig. 6).
Figure 6. Model for 8-amino-adenosine-mediated multiple effects on transcription.
8-Amino-adenosine inhibits transcription by inducing a rapid decline in the ATP pool, decreasing the phosphorylation at the C-terminal domain of RNA polymerase II, causing mRNA transcript termination and inhibiting polyA tail elongation upon the incorporation of 8-amino-ATP at the 3′-terminus.
Premature chain termination may have significant consequences as it affects transcripts with short half lives. Survival proteins such as MCL1 have short-lived transcripts which results in a loss of protein and survival protection when transcription is impeded (31). Because of the loss of essential transcripts and their protein products, the incorporation and subsequent chain termination in the cell can have far reaching effects that lead to cytotoxicity. Further examination of this process of chain termination may provide insight into the utility of RNA chain terminators as therapeutic agents.
In conclusion, the present work provides rationales and suggestions for future studies that will lead to the development of 8-amino-adenosine as a RNA-directed nucleoside analog for cancer therapeutics.
Acknowledgments
The authors would like to especially thank Dr. Michelle Barton (University of Texas M.D. Anderson Cancer Center) for her insightful and valuable discussions as well as Dr. Meghan Minard (University of Texas M.D. Anderson Cancer Center) for her assistance with the in vitro transcription assays.
Abbreviations
- 8-amino-AMP
8-amino-adenosine monophosphate
- 8-amino-ATP
8-amino-adenosine triphosphate
- 8-chloro-AMP
8-chloro-adenosine monophosphate
- 8-chloro-ATP
8-chloro-adenosine triphosphate
- CDK
cyclin dependent kinase
- CTD
C-terminal domain
- DRB
5,6-dichloro-1-β-D-ribofuranosylbenzimidazole
- HPLC
high pressure liquid chromatography
- PARP
poly(ADP-ribose) polymerase
- poly(A)
polyadenylated
- PTEF-b
positive transcription elongation factor b
- RIPA buffer
radioimmunoprecipitation buffer
References
- 1.Christian BA, Grever MR, Byrd JC, Lin TS. Flavopiridol in the treatment of chronic lymphocytic leukemia. Curr Opin Oncol. 2007;19:573–8. doi: 10.1097/CCO.0b013e3282efb9da. [DOI] [PubMed] [Google Scholar]
- 2.Chao SH, Price DH. Flavopiridol inactivates P-TEFb and blocks most RNA polymerase II transcription in vivo. J Biol Chem. 2001;276:31793–9. doi: 10.1074/jbc.M102306200. [DOI] [PubMed] [Google Scholar]
- 3.Meijer L, Raymond E. Roscovitine and other purines as kinase inhibitors. From starfish oocytes to clinical trials. Acc Chem Res. 2003;36:417–25. doi: 10.1021/ar0201198. [DOI] [PubMed] [Google Scholar]
- 4.Legraverend M, Ludwig O, Bisagni E, et al. Synthesis and in vitro evaluation of novel 2,6,9-trisubstituted purines acting as cyclin-dependent kinase inhibitors. Bioorg Med Chem. 1999;7:1281–93. doi: 10.1016/s0968-0896(99)00064-4. [DOI] [PubMed] [Google Scholar]
- 5.Chen R, Keating MJ, Gandhi V, Plunkett W. Transcription inhibition by flavopiridol: mechanism of chronic lymphocytic leukemia cell death. Blood. 2005;106:2513–9. doi: 10.1182/blood-2005-04-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stellrecht CM, Phillip CJ, Cervantes-Gomez F, Gandhi V. Multiple myeloma cell killing by depletion of the MET receptor tyrosine kinase. Cancer Res. 2007;67:9913–20. doi: 10.1158/0008-5472.CAN-07-0770. [DOI] [PubMed] [Google Scholar]
- 7.Phillip CJ, Stellrecht CM, Nimmanapalli R, Gandhi V. Targeting MET transcription as a therapeutic strategy in multiple myeloma. Cancer Chemother Pharmacol. 2009;63:587–97. doi: 10.1007/s00280-008-0770-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Byrd JC, Lin TS, Dalton JT, et al. Flavopiridol administered using a pharmacologically derived schedule is associated with marked clinical efficacy in refractory, genetically high-risk chronic lymphocytic leukemia. Blood. 2007;109:399–404. doi: 10.1182/blood-2006-05-020735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R, Wierda WG, Chubb S, et al. Mechanism of action of SNS-032, a novel cyclin dependent kinase inhibitor, in chronic lymphocytic leukemia. Blood. 2009;113:4637–45. doi: 10.1182/blood-2008-12-190256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gandhi V, Ayres M, Halgren RG, Krett NL, Newman RA, Rosen ST. 8-chloro-cAMP and 8-chloro-adenosine act by the same mechanism in multiple myeloma cells. Cancer Res. 2001;61:5474–9. [PubMed] [Google Scholar]
- 11.Stellrecht CM, Rodriguez CO, Jr, Ayres M, Gandhi V. RNA-directed actions of 8-chloro-adenosine in multiple myeloma cells. Cancer Res. 2003;63:7968–74. [PubMed] [Google Scholar]
- 12.Krett NL, Davies KM, Ayres M, et al. 8-amino-adenosine is a potential therapeutic agent for multiple myeloma. Mol Cancer Ther. 2004;3:1411–20. [PubMed] [Google Scholar]
- 13.Balakrishnan K, Wierda WG, Keating MJ, Gandhi V. Mechanisms of cell death of chronic lymphocytic leukemia lymphocytes by RNA-directed agent, 8-NH2-adenosine. Clin Cancer Res. 2005;11:6745–52. doi: 10.1158/1078-0432.CCR-05-0553. [DOI] [PubMed] [Google Scholar]
- 14.Balakrishnan K, Nimmanapalli R, Ravandi F, Keating MJ, Gandhi V. Forodesine, an inhibitor of purine nucleoside phosphorylase, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2006;108:2392–8. doi: 10.1182/blood-2006-03-007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghias K, Ma C, Gandhi V, Platanias LC, Krett NL, Rosen ST. 8-Amino-adenosine induces loss of phosphorylation of p38 mitogen-activated protein kinase, extracellular signal-regulated kinase 1/2, and Akt kinase: role in induction of apoptosis in multiple myeloma. Mol Cancer Ther. 2005;4:569–77. doi: 10.1158/1535-7163.MCT-04-0303. [DOI] [PubMed] [Google Scholar]
- 16.Chen LS, Sheppard TL. Chain termination and inhibition of Saccharomyces cerevisiae poly(A) polymerase by C-8-modified ATP analogs. J Biol Chem. 2004;279:40405–11. doi: 10.1074/jbc.M401752200. [DOI] [PubMed] [Google Scholar]
- 17.Krett NL, Zell JL, Halgren RG, Pillay S, Traynor AE, Rosen ST. Cyclic adenosine-3′,5′-monophosphate-mediated cytotoxicity in steroid sensitive and resistant myeloma. Clin Cancer Res. 1997;3:1781–7. [PubMed] [Google Scholar]
- 18.Goldman-Leikin RE, Salwen HR, Herst CV, et al. Characterization of a novel myeloma cell line, MM.1. J Lab Clin Med. 1989;113:335–45. [PubMed] [Google Scholar]
- 19.Gandhi V, Danhauser L, Plunkett W. Separation of 1-beta-D-arabinofuranosylcytosine 5′-triphosphate and 9-beta-D-arabinofuranosyl-2-fluoroadenine 5′-triphosphate in human leukemia cells by high-performance liquid chromatography. J Chromatogr. 1987;413:293–9. doi: 10.1016/0378-4347(87)80242-6. [DOI] [PubMed] [Google Scholar]
- 20.Xie C, Plunkett W. Metabolism and actions of 2-chloro-9-(2-deoxy-2-fluoro-beta-D-arabinofuranosyl)-adenine in human lymphoblastoid cells. Cancer Res. 1995;55:2847–52. [PubMed] [Google Scholar]
- 21.Cohen SS, Plunkett W. The utilization of nucleotides by animal cells. Ann N Y Acad Sci. 1975;255:269–86. doi: 10.1111/j.1749-6632.1975.tb29235.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee KC, Crowe AJ, Barton MC. p53-mediated repression of alpha-fetoprotein gene expression by specific DNA binding. Mol Cell Biol. 1999;19:1279–88. doi: 10.1128/mcb.19.2.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–89. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng J, Marshall NF, Price DH. Identification of a cyclin subunit required for the function of Drosophila P-TEFb. J Biol Chem. 1998;273:13855–60. doi: 10.1074/jbc.273.22.13855. [DOI] [PubMed] [Google Scholar]
- 25.Peng J, Zhu Y, Milton JT, Price DH. Identification of multiple cyclin subunits of human P-TEFb. Genes Dev. 1998;12:755–62. doi: 10.1101/gad.12.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Letnansky K. The influence of 2-deoxy-D-glucose on the nucleotide content of ehrlich ascites carcinoma cells. Biochim Biophys Acta. 1964;87:1–8. doi: 10.1016/0926-6550(64)90040-4. [DOI] [PubMed] [Google Scholar]
- 27.Vandemark PJ, Smith PF. Respiratory pathways in the mycoplasma. Pathway of electron transport during oxidation of reduced nicotinamide adenine dinucleotide by mycoplasma hominis. J Bacteriol. 1964;88:122–9. doi: 10.1128/jb.88.1.122-129.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen LS, Stellrecht CM, Gandhi V. RNA-directed agent, cordycepin, induces cell death in multiple myeloma cells. Br J Haematol. 2008;140:682–391. doi: 10.1111/j.1365-2141.2007.06955.x. [DOI] [PubMed] [Google Scholar]
- 29.Monkkonen H, Auriola S, Lehenkari P, et al. A new endogenous ATP analog (ApppI) inhibits the mitochondrial adenine nucleotide translocase (ANT) and is responsible for the apoptosis induced by nitrogen-containing bisphosphonates. Br J Pharmacol. 2006;147:437–45. doi: 10.1038/sj.bjp.0706628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monkkonen H, Kuokkanen J, Holen I, et al. Bisphosphonate-induced ATP analog formation and its effect on inhibition of cancer cell growth. Anticancer Drugs. 2008;19:391–9. doi: 10.1097/CAD.0b013e3282f632bf. [DOI] [PubMed] [Google Scholar]
- 31.Balakrishnan K, Stellrecht CM, Genini D, et al. Cell death of bioenergetically compromised and transcriptionally challenged CLL lymphocytes by chlorinated ATP. Blood. 2005;105:4455–62. doi: 10.1182/blood-2004-05-1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dennison JB, Balakrishnan K, Gandhi V. Preclinical activity of 8-chloroadenosine with mantle cell lymphoma: Roles of energy depletion and inhibition of DNA and RNA synthesis. British Journal Haematology. 2009 doi: 10.1111/j.1365-2141.2009.07850.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen LS, Nowak BJ, Ayres ML, et al. Inhibition of ATP synthase by chlorinated adenosine analogue. Biochem Pharmacol. 2009;78:583–91. doi: 10.1016/j.bcp.2009.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corden JL. Tails of RNA polymerase II. Trends Biochem Sci. 1990;15:383–7. doi: 10.1016/0968-0004(90)90236-5. [DOI] [PubMed] [Google Scholar]
- 35.Zhang J, Corden JL. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J Biol Chem. 1991;266:2290–6. [PubMed] [Google Scholar]
- 36.O’Brien T, Hardin S, Greenleaf A, Lis JT. Phosphorylation of RNA polymerase II C-terminal domain and transcriptional elongation. Nature. 1994;370:75–7. doi: 10.1038/370075a0. [DOI] [PubMed] [Google Scholar]
- 37.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–60. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sims RJ, 3rd, Belotserkovskaya R, Reinberg D. Elongation by RNA polymerase II: the short and long of it. Genes Dev. 2004;18:2437–68. doi: 10.1101/gad.1235904. [DOI] [PubMed] [Google Scholar]
- 39.Dreyer C, Hausen P. Inhibition of mammalian RNA polymerase by 5,6-dichlororibofuranosylbenzimidazole (DRB) and DRB triphosphate. Nucleic Acids Res. 1978;5:3325–35. doi: 10.1093/nar/5.9.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamaguchi Y, Wada T, Handa H. Interplay between positive and negative elongation factors: drawing a new view of DRB. Genes Cells. 1998;3:9–15. doi: 10.1046/j.1365-2443.1998.00162.x. [DOI] [PubMed] [Google Scholar]
- 41.Sehgal PB, Derman E, Molloy GR, Tamm I, Darnell JE. 5,6-Dichloro-1-Beta-D-ribofuranosylbenzimidazole inhibits initiation of nuclear heterogeneous RNA chains in HeLa cells. Science. 1976;194:431–3. doi: 10.1126/science.982026. [DOI] [PubMed] [Google Scholar]
- 42.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 43.Baumli S, Lolli G, Lowe ED, et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. Embo J. 2008;27:1907–18. doi: 10.1038/emboj.2008.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Conroy A, Stockett DE, Walker D, et al. SNS-032 is a potent and selective CDK 2, 7 and 9 inhibitor that drives target modulation in patient samples. Cancer Chemother Pharmacol. 2009 doi: 10.1007/s00280-008-0921-5. [DOI] [PubMed] [Google Scholar]
- 45.Ljungman M, Paulsen MT. The cyclin-dependent kinase inhibitor roscovitine inhibits RNA synthesis and triggers nuclear accumulation of p53 that is unmodified at Ser15 and Lys382. Mol Pharmacol. 2001;60:785–9. [PubMed] [Google Scholar]
- 46.Lacrima K, Valentini A, Lambertini C, et al. In vitro activity of cyclin-dependent kinase inhibitor CYC202 (Seliciclib, R-roscovitine) in mantle cell lymphomas. Ann Oncol. 2005;16:1169–76. doi: 10.1093/annonc/mdi217. [DOI] [PubMed] [Google Scholar]
- 47.MacCallum DE, Melville J, Frame S, et al. Seliciclib (CYC202, R-Roscovitine) induces cell death in multiple myeloma cells by inhibition of RNA polymerase II-dependent transcription and down-regulation of Mcl-1. Cancer Res. 2005;65:5399–407. doi: 10.1158/0008-5472.CAN-05-0233. [DOI] [PubMed] [Google Scholar]
- 48.Poole AM, Logan DT. Modern mRNA proofreading and repair: clues that the last universal common ancestor possessed an RNA genome? Mol Biol Evol. 2005;22:1444–55. doi: 10.1093/molbev/msi132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fish RN, Kane CM. Promoting elongation with transcript cleavage stimulatory factors. Biochim Biophys Acta. 2002;1577:287–307. doi: 10.1016/s0167-4781(02)00459-1. [DOI] [PubMed] [Google Scholar]
- 50.Chen L, Vethantham V, Manley J, Gandhi V. Inhibition of mammalian polyadenylation by ATP analogs. AACR Meeting Abstracts; 2007. p. 3203. [Google Scholar]