Abstract
Three-dimensional nanoscale localization and tracking of dim single emitters can be obtained with a widefield fluorescence microscope exhibiting a double-helix point spread function (DH-PSF). We describe in detail how the localization precision quantitatively depends upon the number of photons detected and the z position of the nanoscale emitter, thereby showing a ~10 nm localization capability along x, y, and z in the limit of weak emitters. Experimental measurements are compared to Fisher information calculations of the ultimate localization precision inherent in the DH-PSF. The DH-PSF, for the first time, is used to track single quantum dots in aqueous solution and a quantum dot-labeled structure inside a living cell in three dimensions.
A critical problem in nanoscale science is the need to noninvasively and from a distance determine the location of a nanoscale object in three dimensions. Even though the diffraction limit of visible light is on the order of 200 nm, the two-dimensional (2D) image of a single molecule can be fit to find the x–y position of the molecule with nanometer precision1, 2. The localization precision scales roughly as σ/(N)1/2 where σ is the diffraction-limited standard deviation of the microscope point spread function (PSF) and N is the number of photons detected from a point-like emitter. This “super-localization” property, well-known in many areas of science 3, was used to great effect by many groups to track isolated point-like single-molecule emitters in a variety of contexts 4, 5. However, imaging a continuous structure requires a high density of fluorescent labels, and the nanoscale detail is lost because the PSFs overlap. By using photoswitchable or photoactivatable fluorophores to keep the concentration of active emitters low at any one time, super-resolution images can be extracted by successive single-molecule localizations 6–8. These methods were, until recently, mostly limited to 2D imaging. Unfortunately, three-dimensional (3D) position information is difficult to obtain using a diffraction-limited conventional fluorescence microscope for two reasons: (1) the PSF is symmetric about the focal plane meaning that a molecule × nm above the focal plane cannot be distinguished from a molecule × nm below the focal plane, and (2) the PSF contains little information about the axial position of the emitter for a few hundred nanometers about the focal plane 9, 10, meaning that it is quite difficult to super-localize molecules that are in the focal plane of the microscope. Recently, several solutions to these problems have been described including using astigmatism 11–13, imaging in two different focal planes 14, 10, 15, 16 and interferometry 17, 18. Fast 3D fluorescent particle tracking has also been performed using confocal imaging 19–21 and by using two rotating laser beams to track single quantum dots 21.
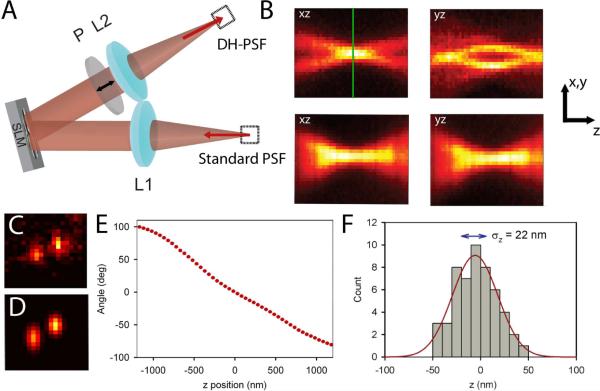
We recently demonstrated that a Double-Helix Point Spread Function (DH-PSF) can be used to super-localize single molecules in three dimensions 22. Combining the DH-PSF with photoactivatable single molecules enabled 3D super-resolution imaging, a method termed Double-Helix Photoactivated Localization Microscopy (DH-PALM). In a wide-field microscope exhibiting the DH-PSF, point sources appear on the imaging detector as two lobes instead of one, and the two lobes have a unique orientation depending on the z position of the emitter (see Figure 1BCD). The DH-PSF was developed and provided by R. Piestun and S.R.P. Pavani at the University of Colorado23.
Figure 1.
Schematic and experimental characterization of the DH-PSF imaging system. (A) Schematic of the 4f setup that is attached to the detection path of a standard inverted microscope. See text for details. (B) Top panels are the xz and yz cross sections of the experimentally observed DH-PSF using a 200 nm fluorescent bead. For comparison, the same cross sections taken of the standard PSF of a conventional microscope are also shown in the lower panels. The green line indicates the focal plane, denoted as z = 0 in this paper. Note the gradual change of the standard PSF with z compared with the more dramatic change of the DH-PSF throughout the depth of field. The arrows correspond to scale bars that are 2 μm long in x–y and 700 nm long in z. The z axis has been lengthened for clarity. (C) Image of a single DCDHF-2V molecule in PMMA taken using the DH-PSF system. This molecule emitted on average 3100 photons per acquisition on top of a background noise of 16 photons/pixel per acquisition. (D) Output of the fitting algorithm for the data shown in (C). The fitting algorithm used in this paper fits the DH-PSF as the sum of two Gaussian functions. (E) A plot of the angle between the two lobes as a function of z position of the emitter that is used as a calibration curve. This curve was taken using a bright 200 nm fluorescent bead. (F) Histogram of 50 localizations in z of the molecule shown in (C) with a Gaussian fit shown in red. The localization precision, which is the standard deviation of the distribution, is explicitly shown on the plot.
Use of the DH-PSF has several advantages over other methods for 3D localization. The DH-PSF has a uniform and high Fisher information content about all three dimensions over a large range of z values, meaning that it should have very good localization precision that is relatively independent of the z position of the emitter 24. Another major advantage is that the shape of the DH-PSF is mostly invariant throughout the depth of field, meaning that x, y, z position can be extracted from the position and orientation of the two lobes rather than the precise shape of the image as is necessary in defocusing and astigmatism methods. This means that fitting the PSF can be more robust under low signal-to-noise conditions. Experiments showing the super-localization of extremely bright objects such as 1 μm fluorescent beads 25 and point scatterers 24 with the DH-PSF have been reported. However, many single-molecule PALM experiments are typically performed with fluorescent protein labels that emit an order of magnitude fewer photons than organic fluorophores, thus the performance of the DH-PSF for dim emitters is of particular interest. Here we examine in detail the localization precision of the DH-PSF in the limit of low numbers of photons detected. Comparisons with information theory calculations are presented, and then the performance of the system with low detected photon counts is demonstrated by widefield tracking of quantum dots with 7.5 ms resolution in both 80% glycerol/water and in live cells.
The DH-PSF is created by adding an all-optical 4f image processing section (Figure 1A) to the detection path of a conventional inverted microscope (Olympus IX71) with a 100X 1.4 NA oil-immersion objective (Olympus UPlanSApo 100x/1.40). The purpose of the 4f setup is to convolve the standard PSF with the DH-PSF. This convolution is performed by Fourier transforming the standard microscope image with a lens and multiplying the Fourier transform of that image with the Fourier transform of the DH-PSF, which is a phase-only function. The product of the two is then Fourier-transformed by a second lens onto the detector to restore a real-space image (Figure 1C). Two 15 cm focal length lenses (Edmund NT32-886) act as the Fourier transform elements. A phase-only reflective liquid crystal spatial light modulator (SLM, Boulder Nonlinear Systems XY Phase series) with a custom 500–700 nm reflective dielectric mirror is placed in the Fourier plane at a slight angle for optical convenience. Because the SLM only modulates the phase of one polarization of light, a linear polarizer is placed in the collection path to reject unmodulated light. This causes a loss in efficiency (50%) which might make this setup challenging to implement for dim emitters (see Supplementary Information (SI) for a discussion of the setup collection efficiency); nevertheless, this loss did not prevent 3D superresolution imaging with single organic fluorophores 22, and Figure 1C shows the DH-PSF image from a single molecule. It is important to note that the polarization loss is not a general requirement for imaging with a DH-PSF, as there are many ways to collect and modulate both polarizations. The goal of the present paper is to assess the DH-PSF purely on the photons collected, not the photons emitted, and addressing efficiency issues will be a subject of future work. The detector used in all experiments is an electron-multiplying Si CCD camera (Andor Ixon+) with the electron multiplication gain on the highest allowed setting (see SI for a detailed explanation of the gain calibration).
Although the DH-PSF is a superposition of a number of Gauss-Laguerre modes derived from an optimization scheme, 23 the DH-PSF image for one z-position can be approximated reasonably well by the sum of two Gaussian functions (Figure 1D). The center x, y position of the emitter is found by finding the midpoint between the centers of the two lobes. The z position of the molecule is extracted by interpolation of the angle found between the lobes from a calibration curve of the type shown in Figure 1E. This calibration curve was extracted from images of a 200 nm fluorescent bead emitting 10,000 photons/frame at 645 nm, giving 5–10 nm localization precision for each z position. A piezoelectric objective z-positioner (Physik Instrumente PiFoc) was moved in known 50 nm steps, and 50 acquisitions at each z step were analyzed to find the z position. As with any 3D super-resolution technique based on molecular localization, the choice of fitting algorithm represents a tradeoff between increased localization precision and decreased computational efficiency 26, 27. In this work, we chose an algorithm that would provide reasonable localization precision, but not be so computationally expensive that it could not be performed on a standard desktop computer. The fitting algorithm used for all of the data draws a 19×19 pixel box around each emitter of interest and uses the MATLAB simplex optimization function, fminsearch, to minimize the least squares error between the sum of two Gaussians and the image. The double Gaussian model function has 11 independent parameters. A standard Intel Core Duo 2 desktop with 4 GB of RAM takes on the order of a second or less to fit each image. More sophisticated fitting schemes based on the observed PSF 26 could be implemented on a cluster or a GPU to increase fitting precision, but at the expense of computational speed. Results of 50 determinations of the z position of the same single molecule as in Figure 1C are shown in Figure 1F. This molecule emitted 3100 photons/frame on average on top of a background noise of 16 photons/pixel/frame. The resulting standard deviation of 22 nm is shown on the plot, illustrating how the localization precision is measured throughout this paper. Although it is useful to show that a single molecule can be localized with high precision in x, y, and z 22, it is far more useful to quantify the relationship between the localization precision and the number of photons detected as well as the dependence of the localization precision on the z position of an emitter, which will now be described.
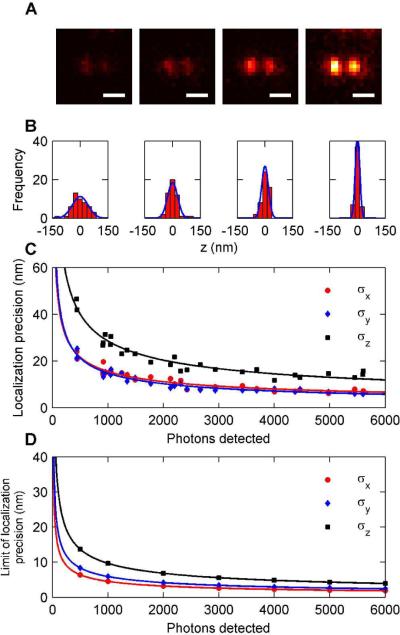
In order to effectively use the DH-PSF in a tracking or super-resolution experiment, it is necessary to know how the localization precision depends on the number of photons detected for a single acquisition (frame). We have determined the localization precision for different detected photon counts by measuring the positions of dim emitters many times. The sample consisted of 200 nm fluorescent beads emitting at 645 nm (Invitrogen, carboxylate modified) which were embedded in spincoated 1% poly(vinylalcohol) (72,000 g/mol, Carl Roth Chemicals) on Ar plasma-etched glass slides. The function of the thin PVA layer is to immobilize the beads on the coverslip. The beads were pumped at low intensities (0.1 to 1 W/cm2) by a 641 nm diode laser (Coherent CUBE 640-100C) in widefield mode to simulate the effect of a dim emitter. Emission from the beads was filtered through a 633 long pass dichroic filter (Omega XF2072), a long pass filter (Chroma HQ665LP), and a 670–690 nm bandpass filter (Chroma D670/690). Beads were chosen for these experiments because they have relatively stable emission with the majority of the fluctuations coming from shot noise. Single organic fluorophores and quantum dots can have photophysical intermittency effects that broaden the localization distribution and give deceptively large estimates for the localization precision. Immersion oil (n=1.518) was placed on top of the sample to prevent refractive index mismatches that could distort the DH-PSF or make accurate z estimates difficult. In Figure 2A, images of a fluorescent bead at the in-focus position emitting 500, 1000, 2000, and 4000 photons/frame are shown, with an average background noise of approximately 1–2 photons/pixel/frame. The exposure time for each image is 0.1 s. This level of background fluctuation is similar to that for single-molecule localization experiments with low excitation intensities. For each bead, and at each z position, 50 images were recorded to provide measurements of x, y, and z. An example of the distribution of localizations in the z direction is shown in Figure 2B along with fits to a Gaussian distribution. Note the dramatic narrowing of the distribution with more photons detected. Figure 2C is a plot of the localization precision for beads that emit across a range of 500 to 6000 photons/frame. The low end of the range represents detected photon counts from a typical photoswitchable fluorescent protein such as Dronpa or PAGFP under reasonable pumping conditions, while the high end represents the expected detected photon counts from a bright organic fluorophore such as a photoactivatable azido-DCDHF 28 or a photoswitchable Cy3–Cy5-thiol molecule 11, 29, 30. Naturally, it is possible to pump any emitter harder to obtain more photons/frame, within the limit provided by the total emitted photons available before the molecule photobleaches. For 1000 photons detected/frame, a typical number for a single EYFP molecule 31, it is possible to localize with 15 nm accuracy in x and y and 28 nm in z.
Figure 2.
Characterizing the detected photon count dependence of 3D localization using the DH-PSF. (A) Images of a 200 nm fluorescent bead from which (from left to right) roughly 500, 1000, 2000, and 4000 photons per acquisition were detected on average (scale bar is 1 μm). (B) Histograms of z localizations for each of the beads in (A) that were used to extract localization precision. (C) Plot of the localization precision versus the number of photons detected for photon counts ranging from 400–5500 photons per acquisition. All three dimensions are plotted on the curve. Fits to the data give power law behavior of approximately (N)−1/2 where N is the number of photons detected. (D) Theoretical curves showing the best possible localization precision that could be obtained with our system for a given number of photons detected. Fits to the theoretical curves also give power law behavior of (N)−1/2. Note that the trend between the theoretical precision and the experimental precision is very similar although the values for experimental precision are higher.
All imaging systems have nonzero thermal and mechanical fluctuations; the variance of which adds to the square of the localization precision arising solely from the number of photons detected. At high photon counts and thus high precision, any systematic error arising from an imperfect setup can be seen more easily and must be removed. In our setup, and on the timescales of this experiment, this drift is typically less than 10 nm. The removal of drift at high photon counts is discussed in the SI. Least squares fits of the data to the general form σ = aNb gives values for b of −0.47, −0.52, and −0.49 for σx, σy, and σz, respectively. (N)−1/2 dependence is reasonable because we would expect the standard error of the mean to decrease in the same manner as for a normal distribution – more photons simply represent more sampling of the distribution.
Figure 2D shows the lower limit of the localization precision as derived from a Fisher information analysis for a DH-PSF that is carefully modeled after the experiments performed in Figure 2ABC. The curves in Figure 2D were calculating by computing the Fisher information of the theoretical PSF according to published methods 10. The Fisher information is a quantity that describes how much information a likelihood function contains about a parameter. In our case, the likelihood function is the DH-PSF (regarded as the probability distribution for detection of photons) and the parameters we are concerned about are the lateral and axial positions of the emitter. Although many information criteria exist, the Fisher analysis is especially pertinent in these experiments because the inverse of the Fisher information represents the best (or smallest) possible variance, regardless of the estimator, that can be reached when estimating the position of a point emitter with a given number of photons. The square root of this bound, called the Cramer-Rao lower bound, is the quantity labeled as the “limit of the localization precision” and plotted in Figure 2D. It is reasonable to assume that these numbers represent the best possible localization precision one could derive from the measurements in Figure 2B 2. (Please see the SI for a description of the Fisher information calculations as well as a detailed theoretical model of our optical system). Fits to the theoretical curves yield values for b of −0.5 for all three dimensions, which is consistent with the power law dependence of the experimental localization precision of our system. As is often the case, the numerical values in Figure 2D are much lower in all three dimensions than the experimental values in Figure 2C. One reason for this is the statistical inefficiency of our estimator. Another reason is that the gain stages of the camera add significant noise to the observed data 18, 32. It has been shown that in the limit of high gain, the multiplicative noise from the gain stages can effectively halve the number of photons detected. Another possibility is the lack of aberrations in the theoretical model which can diminish the information content of the PSF. Full accounting of all these additional possibilities should be a subject of future work intended to extract even more localization precision from the data.
Any 3D localization technique will depend not only on the photons detected but also on the z position of the emitter. This is because no imaging system demonstrated so far is truly invariant in z, and the form of the PSF, and hence the information content, can change dramatically with different axial positions 10, 18. Although our PSF is roughly invariant over a 2 μm axial range, it is still informative to study how the localization precision depends on the z position. To illustrate the shape of the PSF for emitters at z positions across the whole axial range, a bead emitting on average 2000 photons/frame is shown at five different axial positions in Figure 3A. Note that at extreme z values, more photons are diverted from the two main lobes, making it harder to fit the function with simple Gaussians. The localization precision for different z values and 500, 1000, and 2000 photons detected per frame are shown in Figure 3B. As can be seen from the plot, except for about 100 nm from the extreme edges of the depth of field, the localization precision depends fairly weakly on the axial position of the emitter, and typically has a shallow minimum near the focus for all three dimensions. The noise on the curve in the 500 photon plot is due simply to the low signal-to-noise of the measurement. As more photons are detected, the curves are smoother since higher signal-to-noise environments yield detected images that are more reproducible. With more photons detected the trend remains the same but the curves are simply translated vertically downward as (N)−1/2. Fisher information calculations of the lower limit of the localization precision for similar conditions for those of Figure 3B are shown in Figure 3C. As in Figure 2D, the theoretical limits are considerably lower than the observed localization precisions, with any discrepancy likely due to the same reasons discussed above. The theoretical curves follow similar trends to those observed in the experiment both in the shape of the curve and in the relative separation of the x, y curve and the z curve.
Figure 3.
Localization precision as a function of the z position of the emitter. (A) Images of a fluorescent bead with 2000 photons detected per acquisition at (from left to right) −960 nm, −510 nm, −20 nm, 510 nm, and 970 nm (scale bar is 1 μm). The z position when the two lobes are horizontal is considered to be z = 0. (B) Experimental determination of the localization precision in x, y, and z for (from left to right) 500, 1000, and 2000 photons/frame detected. Note that the localization precision stays relatively constant over a wide range of z values for the 2000 photon case. (C) Theoretical lower limit of the localization precision for (from left to right) 500, 1000 and 2000 photons detected per acquisition. The theoretical data has much lower precision values, but follows a very similar trend to that in the experimental curves.
To assess the ability of the DH-PSF system to extract 3D information in actual experiments with low numbers of detected photons, we have tracked and analyzed single quantum dots (QTracker 655, Invitrogen) freely diffusing in an aqueous environment. The goal of these experiments is twofold: the first being to push the system to the limit of low detected photons to see if meaningful information can still be obtained and also to assess whether the DH-PSF can work in an aqueous environment with the aberrations that result from imaging in water with an oil immersion objective. The quantum dots were diluted to a concentration of 0.1 nM in 80% glycerol/water and sonicated for 1 hr. Large aggregates of quantum dots were observed in the solution, so it was necessary to only analyze traces which showed digital blinking behavior characteristic of a single quantum dot. The solution was excited with an irradiance of 100 kW/cm2 with a 561 nm diode laser (Crystalaser, 25 mW) in order to increase the detected photon counts/frame and thus speed up the acquisition time. The emission light was further filtered by a long pass filter (Semrock BLP01-561R). The refractive index of an 80% glycerol solution is 1.44, while the immersion oil has an index of 1.518. We have rescaled the z values by the factor nsolution/noil to account for this index difference 33. Figure 4A shows a representative trajectory of a single quantum dot in solution with 7.5 ms exposure time. This trace is constructed of 326 frames, which corresponds to a total trace time of 2.44 seconds. Figure 4B shows images of the quantum dot with different z positions and photon counts taken at 22 ms, 0.25 s, 1.13 s, 1.68 s, and 1.96 s for images 1, 2, 3, 4, and 5, respectively. The track ended when the quantum dot either photobleached or diffused out of the field of view. Possibly because of the high excitation power, wildly fluctuating photon counts from the quantum dot were observed34. Throughout the trace an average of 640 +/− 330 photons/frame were detected. Figure 4C shows the detected photons as a function of time. Gaps in the plot indicate where the quantum dot blinked and could not be localized.
Figure 4.
Tracking of single quantum dot emitter in an 80% glycerol solution. (A) A representative 326 step trajectory taken with 7.5 ms integration time. (B) Images of quantum dots at times of 22 ms, 0.25 s, 1.13 s, 1.68 s, and 1.96 s for images 1, 2, 3, 4, and 5, respectively. (Scale bar is 1 μm) (C) Plot of the number of photons detected for each 7.5 ms time bin over the 2.43 s track. Strong blinking events are observed where the intensity drops to zero. (D) Plots of the mean-square displacement in x, y, and z for the average of 5 different single quantum dots. A fit to the MSD for displacement r gives a diffusion coefficient of 0.2 μm2/s.
Even though the photons detected/frame in the 3D diffusion track of the quantum dot are quite low, it is still possible to extract useful physical information from the data. The mean-squared displacement (MSD) of 5 quantum dots was found for short lag times in x, y, z and also the 3D Euclidean displacement vector r. For a lag time, tlag, the 3D MSD is defined by
Since we allowed the solution to come to equilibrium before taking measurements we can assume that the motion of the particles is purely diffusive. For purely diffusive motion with an uncertainty in the measurement σ at each time point (due mostly to limited photon counts and motion during the acquisition time), the MSD scaling versus tlag is given by
where D is the diffusion coefficient, n is the number of dimensions, and σi is the position uncertainty in each dimension 12, 19, 35, 36. Figure 4D shows a plot of the MSD in x, y, z, and r versus lag time averaged over the trajectories of 5 different quantum dots. The trends of the MSD with respect to x, y, and z are all linear and overlap with one another within the error bars. A fit to the MSD for r is shown and the trend is clearly linear. The D extracted from the fit was found to be 0.2 μm2/s. Using the viscosity of the 80% glycerol solution at 296 K of ~ 50 cP and the Einstein-Smoluchowski relation for a spherical particle, we obtained a hydrodynamic radius of 21 nm for the individual quantum dots. The large value for the radius is likely due to the proprietary coating on the quantum dots that is intended to ease entry into mammalian cells.
Imaging in live cells introduces many potential complications into a super-resolution tracking or imaging experiment. When using an oil immersion lens, significant aberrations are caused by focusing deep into water because of refractive index mismatch 37. Each compartment within a live mammalian cell has its own refractive index, which has the effect of making the light pass through many tiny optical elements 38. To assess whether useful DH-PSF images could be formed from the light coming from inside a live mammalian cell, we incubated COLO205 cells with 1 nM QTracker (Invitrogen) quantum dots and peptide carrier for 1 hour, followed by three washes with PBS pH 7.4 buffer. The COLO205 cells are approximately 8–10 μm in diameter, providing a much thicker sample than what has been utilized with the DH-PSF in prior work. Figure 5A shows a typical image of what is most likely a vesicle containing several endocytosed quantum dots. The imaging buffer is pH 7.4 PBS and the quantum dots are being excited with a 561 nm diode laser at an irradiance of ~10 kW/cm2. The motion of the intracellular structure typically followed a slow, nondiffusive path, thus data were taken with timelapse imaging with 0.1 s exposures every 1 s. On average 2400 photons were detected per frame from the structure on top of a background noise of 8 photons/pixel/frame. The track of the quantum dot was corrected for motion of the living cell by tracking a large fixed fluorescent aggregate also located in the cell. In Figure 5B, images at different time points displaying a well-formed DH-PSF are displayed. In Figure 5C the entire track, which lasted 300 s, is shown. Symbols representing the acquisitions in 5B are shown on the track in 5C. For this demonstration we assumed that the cell had the same refractive index as water to extract the z position of the emitter; since no quantification of the track dynamics is performed, this is a reasonable assumption. On the basis of this experiment it seems reasonable that the DH-PSF could be used effectively for live cell tracking experiments. Since we are using adaptive optics to form the DH-PSF, it should also be possible in the future to correct for any aberrations due to index mismatch within the sample using the SLM 38 to produce a more perfect DH-PSF.
Figure 5.
Tracking a quantum dot labeled structure in a live cell in 3D. (A) Fluorescence image of the emitter overlaid on a white light image of the COLO205 cell (scale bar is 2 μm). (B) Images taken of the structure at different points along the trajectory showing different z positions, the bright aggregate at the top edge should be ignored. An average of 2400 photons/frame with a background noise of 8 photons/pixel/frame was measured for this trajectory. (C) Three dimensional plot of the labeled structure's trajectory, showing a variety of diffusive and linear transport characteristics. Points on the curve that correspond to the images in (B) are shown by green shapes.
In conclusion, we have analyzed in detail how the 3D localization ability for single nanoscale emitters of the DH-PSF imaging system depends both upon the number of photons detected and the axial position of the emitter. It was found that the localization precision scales as (N)−1/2 as expected, and that the localization precision is a relatively flat function of the z position of the emitter, meaning that the DH-PSF can resolve molecules well over the whole 2 μm depth of field. Information theory calculations show similar trends to those observed in the experiments and suggest that further improvements in localization precision are likely with improved estimators. To prove that the DH-PSF can be useful in low photon count experiments, single quantum dots were tracked in aqueous solution and their diffusion statistics analyzed. The expected Brownian behavior was observed and physically relevant quantities were extracted with good precision. Finally, as a proof-of-principle that the DH-PSF can localize single emitters in live cells, we imaged and tracked quantum dots inside a living mammalian cell. Based on the evidence above, we believe that the DH-PSF has a significant role to play in 3D super-resolution imaging and tracking experiments of single molecules.
Supplementary Material
Acknowledgement
We thank R. Piestun and S. R. P. Pavani for providing the DH-PSF phase mask. We thank Julie Biteen, Randall Goldsmith, and Steven Lee for helpful discussions. Hsiao-lu Lee cultured the COLO205 cells used in this work. This work was supported in part by Grant Number R01GM085437 from the National Institute of General Medical Sciences. MAT and MDL acknowledge support from National Science Foundation Graduate Research Fellowships. MAT is also supported by a Bert and DeeDee McMurtry Stanford Graduate Fellowship, and MDL is also supported by a 3Com Corporation Stanford Graduate Fellowship.
References
- 1.Thompson RE, Larson DR, Webb WW. Biophys. J. 2002:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ober RJ, Ram S, Ward ES. Biophys. J. 2004:1185–1200. doi: 10.1016/S0006-3495(04)74193-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bobroff N. Rev. Sci. Instrum. 1986:1152–1157. [Google Scholar]
- 4.Yildiz A, Forkey JN, McKinner SA, Ha T, Goldman YE, Selvin PR. Science. 2003:2061–2065. doi: 10.1126/science.1084398. [DOI] [PubMed] [Google Scholar]
- 5.Kim SY, Gitai Z, Kinkhabwala A, Shapiro L, Moerner WE. Proc. Nat. Acad. Sci. U. S. A. 2006;29:10929–10934. doi: 10.1073/pnas.0604503103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rust MJ, Bates M, Zhuang X. Nat. Methods. 2006:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betzig E, Patterson GH, Sougrat R, Lindwasser OW, Olenych S, Bonifacino JS, Davidson MW, Lippincott-Schwartz J, Hess HF. Science. 2006;5793:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 8.Hess ST, Girirajan TPK, Mason MD. Biophys. J. 2006:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greengard A, Schechner YY, Piestun R. Opt. Lett. 2006;2:181–183. doi: 10.1364/ol.31.000181. [DOI] [PubMed] [Google Scholar]
- 10.Ram S, Prabhat P, Chao J, Ward ES, Ober RJ. Biophys. J. 2008:6025–6043. doi: 10.1529/biophysj.108.140392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang B, Wang W, Bates M, Zhuang X. Science. 2008:810–813. doi: 10.1126/science.1153529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holtzer L, Meckel T, Schmidt T. Appl. Phys. Lett. 2007:053902. [Google Scholar]
- 13.Kao HP, Verkman AS. Biophysical Journal. 1994:1291–1300. doi: 10.1016/S0006-3495(94)80601-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ram S, Chao J, Prabhat P, Ward ES, Ober RJ. Proc. SPIE. 2007:64430D-1–64430D-7. [Google Scholar]
- 15.Juette MF, Gould TJ, Lessard MD, Mlodzianoski MJ, Nagpure BS, Bennett BT, Hess ST, Bewersdorf J. Nat. Meth. 2008:527–529. doi: 10.1038/nmeth.1211. [DOI] [PubMed] [Google Scholar]
- 16.Sun Y, McKenna JD, Murray JM, Ostap EM, Goldman YE. Nano Letters. 2009;7:2676–2682. doi: 10.1021/nl901129j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shtengel G, Galbraith JA, Galbraith CG, Lippincott-Schwartz J, Gillette JM, Manley S, Sougrat R, Waterman CM, Kanchanawong P, Davidson MW, Fetter RD, Hess HF. Proceedings of the National Academy of Sciences. 2009;9:3125–3130. doi: 10.1073/pnas.0813131106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.v. Middendorff C, Egner A, Geisler C, Hell SW, Schönle A. Opt. Express. 2008;25:20774–20788. doi: 10.1364/oe.16.020774. [DOI] [PubMed] [Google Scholar]
- 19.Cang H, Wong CM, Xu CS, Rizvi AH, Yang H. Appl. Phys. Lett. 2006:223901. [Google Scholar]
- 20.Lessard GA, Goodwin PM, Werner JH. Appl. Phys. Lett. 2007;22:224106–3. [Google Scholar]
- 21.McHale K, Berglund AJ, Mabuchi H. Nano Letters. 2007;11:3535–3539. doi: 10.1021/nl0723376. [DOI] [PubMed] [Google Scholar]
- 22.Pavani SRP, Thompson MA, Biteen JS, Lord SJ, Liu N, Twieg RJ, Piestun R, Moerner WE. Proc. Nat. Acad. Sci. U. S. A. 2009;9:2995–2999. doi: 10.1073/pnas.0900245106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pavani SRP, Piestun R. Opt. Express. 2008:3484–3489. doi: 10.1364/oe.16.003484. [DOI] [PubMed] [Google Scholar]
- 24.Pavani SRP, Greengard A, Piestun R. Appl. Phys. Lett. 2009:021103–1–021103–3. [Google Scholar]
- 25.Pavani SRP, Piestun R. Opt. Express. 2008:22048–22057. doi: 10.1364/oe.16.022048. [DOI] [PubMed] [Google Scholar]
- 26.Mlodzianoski MJ, Juette MF, Beane GL, Bewersdorf J. Opt. Express. 2009;10:8264–8277. doi: 10.1364/oe.17.008264. [DOI] [PubMed] [Google Scholar]
- 27.Huang B, Jones SA, Brandenburg B, Zhuang X. Nat Meth. 2008;12:1047–1052. doi: 10.1038/nmeth.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lord SJ, Conley NR, Lee H-D, Samuel R, Liu N, Twieg RJ, Moerner WE. J. Am. Chem. Soc. 2008;29:9204–9205. doi: 10.1021/ja802883k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Conley NR, Biteen JS, Moerner WE. J. Phys. Chem. B. 2008:11878–11880. doi: 10.1021/jp806698p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bates M, Huang B, Dempsey GT, Zhuang X. Science. 2007:1749–1753. doi: 10.1126/science.1146598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Biteen JS, Thompson MA, Tselentis NK, Bowman GR, Shapiro L, Moerner WE. Nat. Methods. 2008;11:947–949. doi: 10.1038/NMETH.1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Basden AG, Haniff CA, Mackay CD. Monthly Notices of the Royal Astronomical Society. 2003;3:985–991. [Google Scholar]
- 33.Wiersma SH, Torok P, Visser TD, Varga P. J. Opt. Soc. Am. A. 1997;7:1482–1490. [Google Scholar]
- 34.Lee SF, Osborne MA. ChemPhysChem. 2009;13:2174–2191. doi: 10.1002/cphc.200900200. [DOI] [PubMed] [Google Scholar]
- 35.Werner J, Guberman J, Gitai Z. ASCB 47th Annual meeting abstract; Washington, DC. 2007. [Google Scholar]
- 36.Xu CS, Cang H, Montiel D, Yang H. J. Phys. Chem. C. 2007;1:32–35. [Google Scholar]
- 37.Hell SW, Reiner G, Cremer C, Stelzer EHK. Journal of Microscopy. 1993;169:391–14. [Google Scholar]
- 38.Kam Z, Hanser B, Gustafsson MGL, Agard DA, Sedat JW. Proc. Natl. Acad. Sci. U. S. A. 2001;7:3790–3795. doi: 10.1073/pnas.071275698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.