Abstract
Industrial processes to produce ethanol from lignocellulosic materials are available, but improved efficiency is necessary to make them economically viable. One of the limitations for lignocellulosic conversion to ethanol is the inaccessibility of the cellulose and hemicelluloses within the tight cell wall matrix. Ferulates (FA) can cross-link different arabinoxylan molecules in the cell wall of grasses via diferulate and oligoferulate bridges. This complex cross-linking is thought to be a key factor in limiting the biodegradability of grass cell walls and, therefore, the reduction in FA is an attractive target to improve enzyme accessibility to cellulose and hemicelluloses. Unfortunately, our knowledge of the genes responsible for the incorporation of FA to the cell wall is limited. A bioinformatics prediction based on the gene similarities and higher transcript abundance in grasses relative to dicot species suggested that genes from the pfam family PF02458 may act as arabinoxylan feruloyl transferases. We show here that the FA content in the cell walls and the transcript levels of rice genes Os05g08640, Os06g39470, Os01g09010 and Os06g39390, are both higher in the stems than in the leaves. In addition, an RNA interference (RNAi) construct that simultaneously down-regulates transcript levels of these four genes is associated with a significant reduction in FA of the cell walls from the leaves of the transgenic plants relative to the control (19% reduction, P < 0.0001). Therefore, our experimental results in rice support the bioinformatics prediction that members of family PF02458 are involved in the incorporation of FA into the cell wall in grasses.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-009-1077-1) contains supplementary material, which is available to authorized users.
Keywords: Biofuels, Digestibility, Glucuronoarabinoxylan, Hydroxycinnamates, Ferulate, Glycosyltransferase
Introduction
A major fraction of the calories consumed by humans are drawn from members of the Poaceae family, including rice (Oryza sativa), wheat (Triticum aestivum) and maize (Zea mays). These species are grown mainly for grain production and the residual crop biomass is a potential feedstock for biofuel production. It has been estimated that these crop residues could potentially produce 49.1 GL per year of ethanol (Kim and Dale 2004). Although methods for straw bioconversion to ethanol are known (Galbe and Zacchi 2007), the economics of the process depend on the international prices of ethanol relative to other fuel sources and on the efficiency of the bioconversion processes. Therefore, efforts to improve the efficiency of lignocellulosic production of ethanol are important to improve the profitability of the process and facilitate a broader adaptation of biofuels.
One of the limitations for lignocellulosic conversion to ethanol is the inaccessibility of cellulose and hemicellulosic materials within the tight cell wall matrix. Several methods have been designed to increase hydrolysis yield, including acidic and steam pre-treatments, and steam explosion (Nunes and Pourquie 1996). These treatments result in partial degradation of the cell wall polysaccharides and facilitate the access of enzymes to the cellulose fibers, but add additional costs to the ethanol production.
The main components of grass straw are cellulose (32–47%), hemicelluloses (19–27%) and lignin (5–24%) (Lequart et al. 1999). Glucuronoarabinoxylan (GAX), the major hemicellulose in grass cell walls, is composed of a β-1,4-linked xylosyl backbone with a single arabinosyl and glucuronic acid side chains primarily attached at the O-3, but also at the O-2 positions (see York and O’Neill 2008; Fincher 2009 for recent reviews). Grass primary and secondary cell walls also have high levels of hydroxycinnamates, particularly ferulate (FA up to 4%) and p-coumarate (pCA up to 3%) (Saulnier et al. 1999; Hatfield et al. 1999). Ferulates and low levels of pCA esters acylate arabinosyl residues of GAX. However, most of the pCA in grass cell walls is reported to be found acylating lignin side chains (Ishii 1997; Ralph et al. 2004; Hatfield et al. 2008). Ferulate side chains are susceptible to oxidative coupling and this oxidation enables the formation of cross-links between the polysaccharides, via diferulate (reviewed in Ishii 1997) and oligoferulate bridges (Burr and Fry 2009). These bridges also cross-link arabinoxylans to lignin through radical coupling reactions (Grabber et al. 2000; Lam et al. 1992; Ralph et al. 1995). Such cross-links are hypothesized to possess a number of important roles, such as the control of cell wall extensibility (Biggs and Fry 1987; Kondo et al. 1990), protection against pathogens (Jacquet et al. 1995; Ikegawa et al. 1996) and inhibition of the rate and extent of cell wall degradation by ruminant microbes (Eraso and Hartley 1990) and fungal enzymes (Grabber et al. 1998a, b). In addition, FA has been shown to have an inhibitory effect on yeast, requiring additional treatments to maintain ethanol production (Larsson et al. 2000; Persson et al. 2002).
Based on the previous observations, it has been proposed that the reduction in the degree of GAX feruloylation has the potential to alter GAX cross-linking and possibly improve the digestibility of grass cell walls (Grabber et al. 1998a, b; Dhugga 2007). Unfortunately, the determination of the genes involved in GAX biosynthesis and feruloylation has proven difficult to date. The genetic redundancy of many of the genes involved in the synthesis of cell wall components and the intrinsic plasticity of the cell wall assembly process has limited the success of classic mutant analysis (Vogel 2008). Frequently, non-mutagenized genes from the same family compensate for the mutated ones, masking the phenotyping effects. An alternative strategy to address this problem is the use of RNA interference (RNAi) targeting simultaneously multiple members of the gene family (Miki et al. 2005). This approach was followed in this study.
A candidate gene family for GAX feruloylation was proposed by Mitchell et al. (2007) based on a bioinformatics approach. They identified over represented ESTs in grass cDNA libraries when compared with dicot EST libraries based on the assumption that genes involved in GAX feruloylation would be more abundant in the grass EST libraries. Using this approach, they identified three families of glycosyl transferases as putative genes involved in GAX synthesis. In addition, they identified a family of 12 genes belonging to the CoA-acyl transferase superfamily (pfam PF02458) that were almost uniquely expressed in grasses. They hypothesized that members of the PF02458 family may encode for putative arabinoxylan feruloyl transferases (AFT) and be involved in acylating arabinoxylans with FA. Here, we provide experimental evidence supporting the association between the transcript levels of some members of this super family and differences in FA content in rice cell walls.
Materials and methods
Multiple alignment, tree construction and domain searching
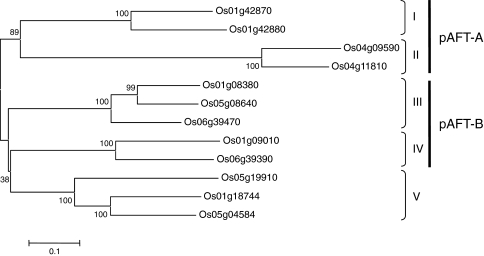
Twelve rice proteins from pfam family PF02458 (Mitchell et al. 2007) were first aligned using CLUSTALW (Thompson et al. 1994) (Supplementary Fig. S1). A neighbor-joining cluster analyses of the complete protein alignments was performed using the complete deletions option in MEGA4.1 (Tamura et al. 2007) and resulted in five clusters that were designated I–V (Fig. 1). The confidence of the different nodes was calculated using 1,000 bootstrap iterations (MEGA4.1).
Fig. 1.
Neighbor-joining tree generated from the alignment of the 12 proteins from O. sativa PF02458 family (putative arabinoxylan feruloyl transferase). The proteins were grouped into five clades (I–V) and the corresponding targets of the pAFT-A and pAFT-B constructs are indicated on the right. Numbers over the tree nodes are bootstrap confidence values based on 1,000 bootstrap iterations
Expressed sequence tags (ESTs) searches in GenBank were performed using BLASTN and limiting the searches to O. sativa. For domain and motif searches, the deduced protein sequences of the genes were searched against the pfam database http://pfam.sanger.ac.uk/search (Bateman et al. 2002) and against the InterPro database (Mulder et al. 2003) using InterProScan (Zdobnov and Apweiler 2001).
Constructs and transgenic plants
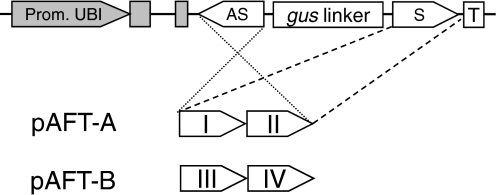
To simultaneously target multiple genes from PF02458, segments from two genes were cloned in tandem, in antisense and sense orientations in each RNAi construct (Fig. 2). Plasmid pAFT-A included segments from genes Os01g42880 (group I) and Os04g09590 (group II), whereas plasmid pAFT-B included segments from genes Os05g08640 (group III) and Os01g09010 (group IV) (Fig. 2). The location of these segments within the respective genes is provided in the legend of Fig. 2. The cloned segments have stretches of more than 21 nucleotides of perfect identity with the other genes from the same group and, therefore, each segment is expected to reduce the transcript levels of all members of its group.
Fig. 2.
Constructs for RNAi targeting multiple genes in the arabinoxylan feruloyl transferase family PF02458. The pANDA vectors carry a maize Ubiquitin1 promoter and first intron including splicing acceptor sites (Prom. UBI, gray), a nos terminator (T) and the targeted sequences in anti-sense (AS) and sense (S) orientations separated by a gus linker. pAFT-A region I includes a 333-bp segment from gene Os01g42880 (2,438–2,770); region II includes a 332-bp segment from Os04g09590 (6–337). pAFT-B region III includes a 306-bp segment from Os05g08640 (897–1,202); region IV includes a 305-bp segment from Os01g09010 (23–327). Positions are counted from the start codon based on the genomic DNA
Primers used to clone the different gene fragments were designed with Primer3 (http://frodo.wi.mit.edu/) and are listed in Supplementary Table S1. To develop PCR products including both selected targets, a 30-bp overlapping region was included in the reverse primer of the first gene and in the forward primer of the second gene. Equimolar amounts of the two overlapping PCR products were mixed and amplified with Pfu DNA polymerase (New England BioLabs, Ipswich, MA, USA). The inverted repeat regions were first cloned into the pENTR/D-TOPO vector (Invitrogen, Carlsbad, CA, USA) to generate the entry vectors and then transferred to pANDA vector (Miki and Shimamoto 2004; http://bsw3.naist.jp/simamoto/panda_vector.html) using an LR clonase reaction.
The pAFT-A and pAFT-B plasmids (Fig. 2) were transformed into rice (O. sativa L. ssp. japonica cv ‘Kitaake’) using Agrobacterium tumefaciens strain EHA 105 at the Ralph M. Parsons Foundation Plant Transformation Facility at UC Davis (http://ucdptf.ucdavis.edu/). Transgenic plants were regenerated from transformed rice calli by selecting for hygromycin resistance. Regenerated transgenic rice plants were grown in a greenhouse at 28°C, and screened for the presence of the transgene using primers AntOs01G42880F/AntOs04g09590R (for pAFT-A) and AntOs05g08640F/AntOs01g09010R (for pAFT-B, Supplementary Table S1).
When plants were 6 weeks old, samples were collected from the third, fourth and fifth (youngest) leaves and from the stems of both transgenic and control plants. Samples were collected from the primary transgenic (T0, preliminary experiment 1) and from their progeny (T1, experiment 2). In the first experiment, individual T0 plants from the different transgenic events were used as replications (5 independent transgenics for pAFT-A and 4 independent transgenics for pAFT-B). As negative controls, we used three untransformed Kitaake plants.
In the second experiment, two transgenic events were selected for pAFT-A (pAFT-A5 and AFT-A11) and two for pAFT-B (pAFT-B1 and pAFT-B11) based on the seed availability and results from experiment 1. Ten plants from each transgenic event were used as biological replicates. As negative control, we used ten non-transgenic T1 sister lines from the four transformation events (1 from AFT-A5, 2 from AFT-A11, 4 from AFT-B1, and 3 from AFT-B11). The objective of combining plants from the different transformation events in the negative control was to reduce the risk of introducing a systematic error due to somaclonal variation in a particular line. The stem samples used in the preliminary experiment 1 included the surrounding sheaths, whereas those from experiment 2 did not. In the second experiment, the sheaths were removed and samples for RNA extraction and for FA and pCA analyses were collected only from the stem internode immediately below the panicle (not fully mature). The proximal third of each leaf was used for expression studies and the central third for the determinations of the FA and pCA content analyses (the distal third was discarded).
cDNA synthesis and quantitative reverse transcription PCR (qRT-PCR)
Total RNA was isolated from the samples described above using Spectrum™ Plant Total RNA Kit (Sigma-Aldrich Crop, St. Louis, MO, USA). For the synthesis of first-strand cDNA, 2 μg of total RNA was reverse transcribed using an oligo (dT) primer and M-MLV reverse transcriptase (Promega, Madison, WI, USA) in 25 μL total volume according to the manufacturer’s instructions. cDNA samples were diluted with additional 50 μL of water.
Transcript levels of the different PF02458 genes were quantified by qRT-PCR on an ABI PRISM 7000 SDS (Applied Biosystems, Foster City, CA, USA) using SYBR® GREEN. The PCR conditions were 95°C for 10 min followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. For every experiment, 2 μL of diluted first-strand cDNA solution was used for PCR amplification in a 25 μl volume reaction with primers specific for each gene (Table S1). Primers for qRT-PCR were designed using Primer Express 2.0 (Applied Biosystems). Efficiencies of each pair of primers were calculated using four fourfold dilutions (1:1, 1:4, 1:16 and 1:64) in triplicate (Supplementary Table S1). Cycle thresholds (C T) were determined at the threshold fluorescence value of 0.2. Analyses of dissociation curves after the PCR amplification confirmed that all primer pairs amplified single PCR products.
A modified −ΔΔC T method (Livak and Schmittgen 2001) in which the primer efficiencies (Table S1) were used to adjust to linear values was used to normalize and calibrate transcript values relative to the endogenous control ELONGATION FACTOR 1-alpha (eEF-1a; GenBank accession number AK061464; Jain et al. 2006). The same calibrator was used for all genes within each experiment so that values could be compared across genes. The (1 + efficiency)−ΔΔCT values reflect the number of RNA copies in the reference sample per copy in the calibrator sample. In Fig. 3, the values in the Y axis were divided by 1,000 to simplify the figures. The significance of the differences in transcript levels between transgenics and controls, among tissues, and between transgenic events within each construct was determined using ANOVA (see the statistical analyses section below).
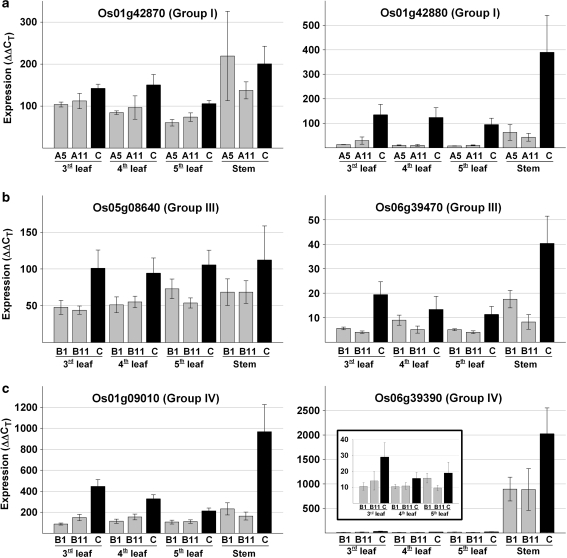
Fig. 3.
Transcript levels of genes targeted by a pAFT-A or b pAFT-B constructs in transgenic and control plants in experiment 2. a Transcript levels of genes from group I in the transgenic events A5 and A11 compared with the control. b, c The expression levels of groups III (b), IV (c) genes in the transgenic events B1 and B11 compared with the control. Values in the Y axes are normalized and calibrated values using the formula (1 + efficiency)−ΔΔCT method (Livak and Schmittgen 2001). All values were divided by 1,000 to simplify the graphs. The same calibrator was used for all genes so values are comparable across genes. Error bars are SE of the means
Analysis of cell wall hydroxycinnamate content
Isolation of cell wall material
Leaves and stems, collected as described above, were ground in a mortar with liquid nitrogen; 50–100 mg of ground samples was transferred to 2 ml tubes with 1.5 ml of 95% (v/v) ethanol. Tubes were mixed overnight on a rocking platform (LabNet International, Woodbridge, NJ, USA) at 20 rpm and room temperature (~25°C). The alcohol-insoluble material was recovered by centrifugation and mixed for 3 h with a 1:1 (v/v) mix of chloroform:methanol on a rocker. The tube was centrifuged to pellet the cell wall material, and the supernatant was discarded. The pellet was washed once with acetone and twice with distilled water. The resulting insoluble material was treated overnight at room temperature with 20 units of α-amylase (porcine pancreas from Sigma-Aldrich) and 10 units of pullulanase (Bacillus acidopullulyticus from Sigma-Aldrich) in 20 mM ammonium acetate (pH 6), washed three times with water, and finally dried by lyophilization to obtain the cell wall material.
Saponification of hydroxycinnamate ester compounds
Ester-linked hydroxycinnamic acids were released from the isolated cell wall material by alkaline hydrolysis and extracted with ethyl acetate (Casler and Jung 1999). Briefly, the total ester-linked hydroxycinnamic acids were extracted from a 5 to 20 mg sample of isolated cell wall material with 2 ml of 2 N NaOH (previously degassed with helium); 20 μg of m-coumaric acid was added as a recovery standard. The tube was degassed briefly with helium, covered with foil to protect from light, and incubated for 18 h at 25°C. The tube was centrifuged and the supernatant was transferred to a new tube. The pellet was washed two additional times with 1 ml of water, and the supernatants were combined with the first one; 400 μl of concentrated HCl was added to lower the pH below 2. This was then extracted with ethyl acetate (3 times; 2, 1.5 and 1.5 ml), and centrifuged to break up the resulting emulsion. Extracts were combined and washed with 1 ml of saturated sodium chloride. The ethyl acetate extract was then evaporated with air, and re-dissolved in 200 μl of 50% (v/v) methanol for HPLC analysis. This method is expected to release only the ester-linked hydroxycinnamic acids, but not those linked by phenyl–phenyl or phenyl–ether linkages.
HPLC of hydroxycinnamic acid derivatives
Analyses of released pCA, m-coumaric, and FA monomers used an Agilent 1100 HPLC system, with DAD UV–Vis based on the method of Bunzel et al. (2002). A Varian Pursuit C18 5 mm 7.5 × 4.6 column was used at a flow rate of 1 ml min−1, and a gradient from 7% acetonitrile (ACN)—93% trifluoroacetic acid (TFA) to 9% ACN/6% methanol/85% TFA over 20 min, followed by 9% ACN/15% methanol/76% TFA over 15 min. All percentages are in v/v and the concentration of the TFA solution is 0.1%. Absorbance was monitored at 280 and 335 nm, and quantified using external standards. The results for pCA and FA were adjusted based on the recovery of m-coumaric acid. Phenolics were identified by comparison of their relative retention times and UV spectra with those of standard compounds. UV spectra were also used to judge peak purity. Ferulate dimers were not quantified in this study.
Statistical analysis
Analyses of variance were performed using the general linear model procedure of the SAS Version 9.1 program (SAS Institute 2006). To meet the assumptions of homogeneity of variances (tested by Levene’s test), and normality of residual (tested using Shapiro–Wilk test), the FA and pCA data were transformed using base 10 logarithms and the expression data using different power transformations. The presented P values correspond to the transformed data, but the adjusted means and their standard errors (as well as the percentage changes) are in the same scale as the original data (untransformed values). Adjusted least square means and type III sums of squares were used to account for missing data and unbalanced number of replications in the factorial analyses. The pCA and FA values from the individual transgenic events were compared with the control using contrasts. In experiment 1, the independent transgenic events of the same construct were used as replications (one T0 plant per construct), whereas in experiment 2, ten T1 plants from each of two selected transgenic events per construct were used as replications and ten non-transgenic sister lines were used as controls. Plants from the four different events were combined to generate a more stringent control (a larger variation within the control plants would require larger differences between treatments to detected significant results).
Results
Bioinformatics characterization of the genes
Searches against Pfam and InterPro indicated the presence of a transferase domain in all members of PF02458, confirming that this family is a member of the CoA-acyl transferase superfamily (clan CL0149). This family includes a number of CoA-dependent transferase enzymes that catalyze the transfer of acyl groups forming either esters or amides. This domain includes the HXXXD motif that has been proposed as the active site of the transferase domain (Supplementary Fig. S1).
A neighbor-joining tree clustered the 12 proteins from this family into five classes (Fig. 1), henceforth designated groups I–V. Protein sequences within groups are 64–80% similar, whereas genes from different groups were 40 and 61% similar. This high similarity among genes within families was also reflected at the DNA level, which facilitated the identification of regions including identical stretches of more than 21 bp. These regions with 21 or more identical base pairs within the RNAi constructs are expected to trigger simultaneous gene silencing of all members within a group (Miki et al. 2005). This strategy was used to maximize the probability of detecting a phenotypic effect even in the presence of genes with redundant function.
Genes from groups I to IV were selected as initial targets for the transgenic experiments based on the relationships inferred from the bioinformatics analyses and the results of the preliminary expression experiment (see below). Because the three genes from group V are expressed at low levels, are generally more abundant in the roots than in the vegetative tissues (see next section), and have available mutants, our transgenic resources were focused on genes from groups I to IV.
The longest stretch of identical nucleotides between members of group V and the antisense fragments used in the pAFT-A or pAFT-B constructs was 19-bp long, suggesting that these constructs do not have the ability to downregulate genes from group V.
The transgenic plants produced with chimeric constructs pAFT-A and pAFT-B were screened for the presence of the transgene by PCR. We obtained 14-positive rice transgenic plants for the pAFT-A construct and 12 for pAFT-B. Transgenic plants exhibited similar height, flowering time, straw strength, and overall morphology to untransformed plants and to T1 lines which were negative for the transgene.
EST counts and preliminary expression results for experiment 1
We searched the public rice dbEST database to determine the relative EST counts of the 12 genes from PF02458 across tissues. This search showed that the ESTs from the two members from group II and Os06g39470 (from group III) were the less abundant, whereas the ESTs from Os01g09010 (group IV) were the most abundant across all tissues (Table 1). ESTs for genes Os01g08380 (group III), Os05g19910 and Os01g18744 (group V) were more abundant in the roots than in the vegetative tissues, whereas the other genes showed the opposite trend (Table 1).
Table 1.
EST counts in different tissues compared with transcript levels estimated by qRT-PCR
| Group | Gene | EST counts (percentage of family total) | Transcripts qRT-PCR (percentage of family total) | Ratio | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Veg.a | %b | Repc | % | Root | % | Total | % | Leafd | % | Stem | % | Avg. alle | % | Stem/leaf | ||
| I | Os01g42870 | 13 | 7 | 4 | 3 | 5 | 7 | 22 | 5 | 9,249 | 17 | 29,770 | 7 | 14,380 | 10 | 3.2 |
| I | Os01g42880 | 32 | 16 | 31 | 22 | 4 | 5 | 67 | 17 | 10,197 | 18 | 51,886 | 12 | 20,619 | 14 | 5.1 |
| II | Os04g09590 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 38 | 0 | 222 | 0 | 84 | 0 | 5.8 |
| II | Os04g11810 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 37 | 0 | 38 | 0 | 38 | 0 | 1.0 |
| III | Os01g08380 | 5 | 3 | 17 | 12 | 14 | 19 | 36 | 9 | 0 | 0 | 4 | 0 | 1 | 0 | – |
| III | Os05g08640 | 12 | 7 | 0 | 0 | 0 | 0 | 12 | 3 | 15,760 | 29 | 41,341 | 10 | 22,155 | 15 | 2.6 |
| III | Os06g39470 | 3 | 2 | 2 | 1 | 1 | 1 | 6 | 1 | 552 | 1 | 7,759 | 2 | 2,354 | 2 | 14.1 |
| IV | Os01g09010 | 76 | 42 | 43 | 31 | 19 | 25 | 138 | 35 | 13,084 | 24 | 95,692 | 22 | 33,736 | 22 | 7.3 |
| IV | Os06g39390 | 7 | 4 | 12 | 9 | 0 | 0 | 19 | 5 | 3,552 | 6 | 176,553 | 41 | 46,803 | 31 | 49.7 |
| V | Os05g19910 | 5 | 3 | 4 | 3 | 7 | 9 | 16 | 4 | 279 | 1 | 563 | 0 | 350 | 0 | 2.0 |
| V | Os01g18744 | 13 | 7 | 5 | 3 | 26 | 34 | 44 | 11 | 179 | 0 | 12,338 | 3 | 3,219 | 2 | 68.9 |
| V | Os05g04584 | 17 | 9 | 22 | 16 | 0 | 0 | 39 | 10 | 2,427 | 4 | 14,142 | 3 | 5,356 | 4 | 5.8 |
| Total | 183 | 140 | 76 | 399 | 55,354 | 430,308 | 149,095 | |||||||||
The correlation between ESTs in vegetative tissues and relative transcript levels in leaves is R = 0.64 (P = 0.03)
aVegetative tissues (leaf and stem)
bPercentages are calculated over the total EST counts (or transcript levels) of the tissues indicated in each column for the complete gene family
cReproductive tissues (panicle and flowers)
dAverage of third, fourth and fifth leaves measurements
eAverage of third, fourth, fifth leaves and stem measurements
Within each tissue, the proportion of ESTs from the different genes relative to the total transcripts in the family was calculated (Table 1). The percentages of ESTs in vegetative tissues showed a significant correlation (R = 0.64, P = 0.03) with the experimental qRT-PCR determination of the relative transcript levels of the different genes in the leaves (Experiment 1; Table 1). The main difference between the EST counts and the qRT-PCR studies was the higher transcript levels observed in Os06g39390, which was mainly due to the differential expression of this gene in the stems, a result later confirmed in experiment 2 (see below). A simple explanation for this difference is the lack of ESTs from dedicated cDNA libraries for stems.
The preliminary qRT-PCR results showed that the transcript levels of the members of this family were in generally higher in the stems–sheaths than in the leaves (average 15-fold higher, Table 1). This experiment also revealed that five highly expressed genes (2 from group I, 2 from group IV and Os05g08640 from group III) accounted for 92% of the transcripts from family PF02458 (Table 1). In general, these results suggest that EST counts are likely a good approximation to the relative transcript levels of the different genes within the family.
Expression results from experiment 2
Experiment 2 included ten biological replicates per event and per tissue and, therefore, provides a more precise estimation of the transcript levels of the different genes than the preliminary experiment 1. No significant differences in transcript levels of the highly expressed genes were detected between pAFT-A transgenic events A5 and A11 or between pAFT-B transgenic events B1 and B11, when the tissues were studied separately or when they were combined in the statistical analyses. Therefore, the following statistical analyses combine the two events examined for each construct, whereas the results for the individual events and tissues are presented in Fig. 3.
Transcript levels of the nine genes analyzed in experiment 2 were significantly correlated with the transcript levels of the same genes in experiment 1 (R = 0.95, P < 0.0001). Experiment 2 confirmed that the five highly expressed genes identified in experiment 1 account for most of the transcripts from family PF02458 in leaves and stems. The low expressing genes Os04g09590 and Os04g11810 (group II) and Os01g08380 (group III) represent together <1% of the transcripts detected in experiment 2. Therefore, the following description will be focused on the other six genes (values for all genes are available in Table 2).
Table 2.
Transcript levels of PF02458 genes in transgenic and control plants in experiment 2 as determined by qRT-PCR
| Group | Gene | Leaf | Stem | Total | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | Trans.a | %Trb | C versus Tr | Control | Trans.a | %Trb | C versus Tr | C versus Tr | Stem/leaves | Percentage of familyc | ||
| P | P | P | ||||||||||
| I | Os01g42870 | 132,345e | 88,423 | 67 | 0.0001 | 200,320 | 178,431 | 89 | 0.51 | 0.085 | 1.5 | 10.2 |
| Id | Os01g42880 | 116,988 | 13,134 | 11 | <0.0001 | 389,195 | 52,714 | 14 | 0.07 | <0.0001 | 3.3 | 12.6 |
| IId | Os04g09590 | 323 | 269 | 83 | 0.09 | 333 | 301 | 90 | 0.91 | 0.16 | 1.0 | 0.02 |
| II | Os04g11810 | 402 | 264 | 66 | 0.02 | 154 | 178 | 116 | 0.81 | 0.03 | 0.4 | 0.02 |
| III | Os01g08380 | ND | ND | – | – | ND | ND | – | – | – | – | ND |
| IIId | Os05g08640 | 100,185 | 54,188 | 54 | 0.0001 | 112,181 | 68,566 | 61 | 0.45 | 0.0003 | 1.1 | 7.0 |
| III | Os06g39470 | 14,659 | 5,492 | 37 | <0.0001 | 40,302 | 12,919 | 32 | 0.003 | <0.0001 | 2.7 | 1.4 |
| IVd | Os01g09010 | 327,193 | 120,411 | 37 | <0.0001 | 966,284 | 197,407 | 20 | 0.0004 | <0.0001 | 3.0 | 33.2 |
| IV | Os06g39390 | 21,116 | 11,865 | 56 | 0.01 | 2,021,531 | 890,640 | 44 | 0.04 | 0.0418 | 95.7 | 35.5 |
ND not detected
aGroups I and II transgenics are transformed with the pAFT-A construct and Groups III and IV with the pAFT-B construct
b%Tr = transcript levels in the transgenic plants as a percentage of the control
cPercentage of family = average of third, fourth, fifth leaves and stem measurements divided by the sum of the averages across all nine genes
dGene used as source of the anti-sense sequence in the RNAi construct
eThe 2−ΔΔCT values for all genes were calibrated using the same number and, therefore, can be compared across genes
Control plants showed consistently higher transcript levels in the stems than in the leaves (Tables 1 and 2) for the six genes with higher transcript levels. The statistical analyses from experiment 2 showed that these differences were significant for all genes from groups I to IV, but not for group III gene Os05g08640 (P = 0.64; stem/leaf ratio = 1.1). These results agree with those from experiment 1, except for gene Os05g08640 that had higher transcript levels in the stem relative to the leaves (stem–sheath/leaf ratio = 2.6, Table 1). The difference in the tissues collected for the stems in experiments 1 (stems and sheaths) and 2 (stems only) may explain these differences.
This group of highly expressed genes did not show consistent differences in transcript levels between young and old leaves, hence the average of the three leaves was used in the statistical analyses below (Table 2). The results for the individual leaves are presented in Fig. 3. It is important to note that merging the data from the three leaves resulted in a larger number of degrees of freedom and, therefore, a higher statistical power to detect significant differences in the leaves as compared to the stems.
Among the genes targeted by the pAFT-A construct, those in group I were expressed at much higher levels than those in group II in both leaves and stem (Table 2). Group I gene Os01g42880 showed the strongest down-regulation among all genes, with transcript levels in the transgenic plants being <15% of the values observed in the control plants (Table 2). Transcript levels of gene Os01g42870 in the leaves of transgenic plants were significantly lower than those in the control plants (67%, P < 0.001), but the differences in the stem were smaller and not significant (Table 2). A slight reduction in the transcript levels in the transgenic plants relative to the control was observed for group II genes in the leaves, despite the low transcript levels of these genes.
The plants transformed with the pAFT-B construct showed a significant reduction (P < 0.05) in transcript levels relative to the control in both leaves and stems for all the highly expressed genes from groups III and IV, with the exception of Os05gO8640 in the stem (Fig. 3; Table 2). Within group III, Os06g39470 showed a larger reduction in transcript levels than Os05g08640 in both leaves and stems (Table 2). Within the genes from group IV, a larger reduction in the transcript levels of the transgenic plants relative to the control was observed for Os01g09010 than for Os06g39390. For both genes, the reduction was larger in the stem than in the leaves (Fig. 3; Table 2), which may reflect the higher transcript levels of genes from group IV in the stems (see “Discussion”). The stem/leaf transcript ratio for Os06g39390 (95.7) was, by far, the largest among the four groups analyzed for this gene family (the next value was 3.3).
FA and pCA levels in transgenic plants
The effect of the down-regulation of the genes from groups I and II (construct pAFT-A) and from groups III and IV (construct pAFT-B) on the levels of FA and pCA in the cell walls was studied in two independent experiments. In each experiment, the variation of FA and pCA levels in different tissues from control plants are described first (Fig. 4), followed by the comparison between transgenic and control plants (Fig. 5). In the first experiment, a single T0 plants from independent transformation events were used as replicates (Fig. 5a), whereas in the second experiment 10 T1 plants from two independent transformation events per construct were used as replicates (Figs. 4, 5b, c). The selection of the transgenic events for the second experiment was based on the results of the first experiment and the availability of T1 seeds.
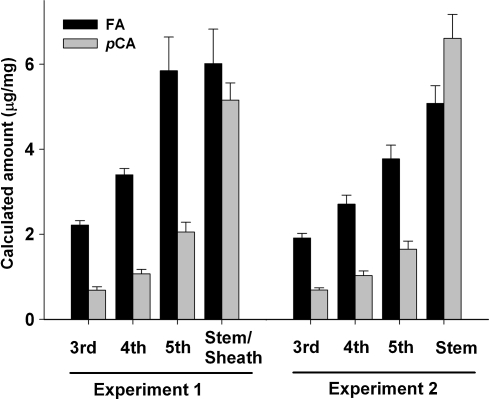
Fig. 4.
Cell wall ferulate (FA) and p-coumarate (pCA) content of individual leaves and stems from control plants in experiments 1 (stems include sheaths) and 2 (stems without sheaths). The third leaf is the oldest and the fifth leaf the youngest one. Values are microgram of FA and pCA per milligram of cell wall. Error bars are SE of the means
Fig. 5.
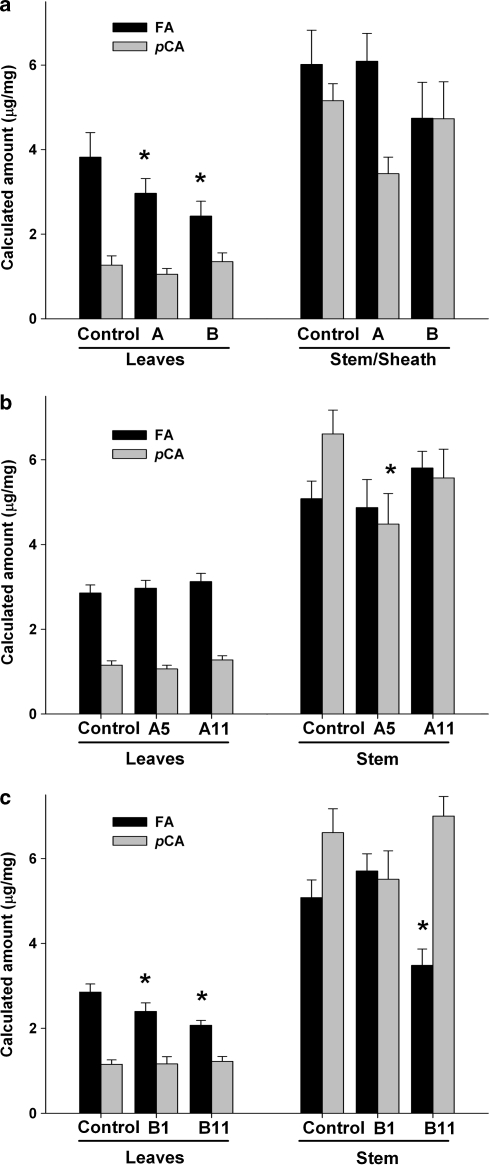
Cell wall ferulate (FA) and p-coumarate (pCA) content expressed as microgram of FA and pCA per milligram of cell wall. a Experiment 1: FA and pCA least square means of transgenic plants from independent transformation events with construct pAFT-A (label A average of 5 events) and pAFT-B (label B average of 4 events) when compared with the non-transgenic control (label C average of 3 plants) (b, c) Experiment 2: FA and pCA least square means of ten T1 plants from individual constructs. b pAFT-A5 and pAFT-A11 events for pAFT-A. c pAFT-B1 and pAFT-B11 events for construct pAFT-B. Asterisks indicate significant differences (P < 0.05) between transgenic lines and the corresponding control. Error bars are SE of the means
Because the levels of FA and pCA were higher in the stems than in the leaves, the two tissues were analyzed separately in both experiments. In addition, no significant interactions were detected in both experiments between leaves (3rd, 4th, and 5th) and genotype (control, pAFT-A, and pAFT-B) in two-way factorial ANOVAs. Therefore, the differences between transgenic and control plants were analyzed across the three leaves.
Experiment 1
Control plants
In the control plants, the levels of FA and pCA were higher in the stem–sheaths than in the leaves, and this difference was more evident for pCA (stem/leaf = 4.1) than for FA (stem/leaf = 1.6) (Fig. 4). Among the different leaves, both pCA and FA showed higher levels in the younger (5th) than in the older (3rd) leaves (P < 0.0001) and the ratios between youngest and oldest leaves were similar for FA (5th/3rd leaf = 2.6) and pCA (5th/3rd leaf = 3.0). This parallel variation in FA and pCA resulted in a significant correlation between the values of these two cell wall components across different tissues in the control plants (R = 0.75, P < 0.005). Control plants showed higher ratios of FA/pCA in the leaves (3.0) than in the stem–sheaths (1.2) (Fig. 4).
pAFT-A (groups I and II genes)
In the stems, the statistical contrasts between the transgenic plants and the non-transgenic controls showed no significant changes in FA (P = 0.99) or pCA (P = 0.07). In the leaves (main effect across the three leaves), a similar statistical analysis showed marginally significant differences between the pAFT-A transgenics and the control plants for FA (22% reduction, P = 0.04, Fig. 5a), but not for pCA (P = 0.18).
pAFT-B (groups III and IV genes)
In the stems, the contrasts between the pAFT-B transgenic and the non-transgenic control were not significant for either FA (P = 0.28) or pCA (P = 0.56). In the leaves, the same analysis revealed no significant differences in pCA (P = 0.17), but detected a 32% reduction in the levels of FA in the pAFT-B transgenic plants relative to the control (Fig. 5a) that was highly significantly (P = 0.003).
Experiment 2
The second experiment, using ten plants per event and two events per construct plus non-transgenic controls (including sister lines from each of the four transformation events) validated the results from experiment 1.
Control plants
In the control plants, the levels of FA and pCA were higher in the stem than in the leaves, with a higher difference for pCA (stem/leaf = 5.9) than for FA (stem/leaf = 1.8) (Fig. 4). As in experiment 1, higher levels of pCA and FA were found in the younger than in the older leaves (P < 0.0001), with slightly lower ratio for FA (5th/3rd leaf = 2.0) than for pCA (5th/3rd leaf = 2.4), and a significant correlation was detected between the FA and pCA values across tissues (R = 0.66, P < 0.0001, only control plants). As in experiment 1, higher ratios of FA/pCA were found in the leaves (2.5) than in the stems (0.8) of the control plants (Fig. 4).
pAFT-A (groups I and II genes)
In the stems, statistical contrasts combining the two different events (pAFT-A5 and pAFT-A11) showed no significant differences between transgenic and control plants for FA (P = 0.81), but marginally significant differences were detected for pCA (24% reduction, P = 0.04). Separate contrast between individual pAFT-A events and the control showed that the differences in pCA were driven by event pAFT-A5 (32% reduction, P = 0.01, Fig. 5b). However, event pAFT-A11 showed no significant pCA differences with the control (P = 0.35). None of the pAFT-A events showed significant differences in FA in the stem.
In the leaves, no significant differences for FA or pCA were detected in the statistical contrasts combining the two different pAFT-A events or in those testing each of the pAFT-A events separately (Fig. 5b). In summary, the pAFT-A transgenic events showed only a small effect on cell wall pCA in the stem, but the differences were only marginally significant.
pAFT-B (groups III and IV genes)
In the stem, no significant differences in cell wall FA (P = 0.28) or pCA (P = 0.68) were detected between the control and the combined transgenic events. The differences in pCA were also non-significant when each of the pAFT-B events was analyzed separately. However, the statistical tests for cell wall FA levels showed a significant decrease (31%, P < 0.01) in pAFT-B11 relative to the control (Fig. 5c). The same comparison was not significant for the pAFT-B1 event (P = 0.34).
In the leaves, no significant differences in cell wall pCA content were detected for the combined or individual pAFT-B events. However, as in experiment 1, a highly significant reduction in cell wall FA content was detected in the combined pAFT-B transgenic plants relative to the control (19% reduction, P < 0.0001). A separate analysis of the two individual pAFT-B transformation events revealed a consistent reduction in cell wall FA, although the differences were larger in pAFT-B11 (Fig. 5c). Line pAFT-B11 showed a 27% reduction in FA relative to the control and the differences were highly significant (P < 0.0001). Line pAFT-B1 showed a 12% significant reduction in FA across the leaves (P = 0.02).
Because the pAFT-A transgenic plants in this experiment showed no significant differences in the levels of FA with the control, it was possible to make a second statistical analysis comparing the individual pAFT-B events with the combined pAFT-A transgenic lines as an independent control. This analysis also showed a highly significant reduction in FA for both pAFT-B11 (P < 0.0001) and pAFT-B1 (P < 0.0001). In summary, the results across experiments, transgenic events, and statistical analyses showed a significant reduction in cell wall FA in the leaves of the rice pAFT-B transgenic plants relative to the controls.
Discussion
Candidate genes for the incorporation of pCA and FA to grass cell walls
In grasses, hydroxycinnamates pCA and FA acylate cell wall polymers (Ishii 1997; Ralph et al. 2004; Grabber et al. 2004). FA acylates the C5 hydroxyl of α-l-arabinosyl side chains of arabinoxylans, whereas pCA mainly acylates lignin and only small quantities acylate arabinoxylans in immature tissues (Grabber et al. 2004).
The role of pCA is less understood than the role of FA. The accumulation of pCA occurs parallel to lignin deposition and some studies suggest that syringyl units are enzymatically pre-acylated with pCA before the incorporation to lignin, so sinapyl p-coumarate is the logical precursor incorporated into lignin (Ralph et al. 1994a; Lu and Ralph 2008; Grabber et al. 2004). In this study, we observed significantly higher pCA content in the stem than in the leaves, which may reflect the higher lignification of the stem relative to the leaves. Because the stem samples were from immature stems, the high pCA content may reflect its role as precursor in the incorporation of lignin. It would be worth monitoring the changes in pCA during the maturation of the stems in future studies.
The ester links between FA and arabinoxylans facilitate the cross-linking of xylans through the formation of diferulate (Ralph et al. 1994b; Ishii 1997; Ralph et al. 2004; Grabber et al. 1995) and oligoferulate bridges (Burr and Fry 2009). In addition, these bridges copolymerize and cross-link the arabinoxylans to lignin (Grabber et al. 2000; Lam et al. 1992; Ralph et al. 1995). The enzyme responsible for the formation of the ester links between the FA and the arabinoxylans has been designated as arabinoxylan feruloyl transferase (AFT). Several studies have demonstrated that the cross-linking of arabinoxylan to lignin by ferulate monomers and dimers reduces cell wall degradability by hydrolytic enzymes (Jung and Allen 1995; Grabber et al. 1995, 1998a, b). Therefore, the reduction in FA is an interesting target to increase the degradability of grass cell walls, both for biofuel production or to improve the quality of forage grasses for ruminants.
A better understanding of the genes responsible for the incorporation of FA to the cell wall is required for an intelligent manipulation of the grass cell walls. Although no AFT grass gene is currently known, Mitchell et al. (2007) hypothesized that the CoA-acyl transferase family PF02458 was a strong candidate for AFT because no other acyl transferase group was differentially expressed to such a large extent between cereals and dicots. This large difference is expected for genes with AFT activity because the presence of feruloyl groups on arabinoxylans is common in the cell wall of grasses, but these groups are absent or present at low levels in most dicots (Bacic et al. 1988). The PF02458 family includes genes encoding hydroxycinnamoyl transferases (Yang et al. 1997) supporting Mitchell et al. (2007) hypothesis, but it also includes genes encoding acetyl transferases that may be associated with the acetylation of arabinoxylans (Carpita 1996). These diverse results complicate the prediction of the function of the different members of the PF02458 family solely based on the homology to other genes.
This study provides experimental evidence supporting the role of genes in the PF02458 family in the incorporation of hydroxycinnamic acids to the rice cell walls.
Correlation between the expression of PF02458 genes and the incorporation of FA and pCA into cell walls
The FA and pCA contents in leaves and stems showed a positive correlation with the transcript levels of most of the highly expressed genes from family PF02458 (Table 3). Except for Os05g08640, all other members from groups I, III, and IV (members of group II were excluded because of their low transcript levels) showed significant correlations with the cell wall contents of at least one of the two hydroxycinnamates (Table 3).
Table 3.
Correlation between gene transcript levels and pCA and FA content in control lines
| Group | Gene | pCA | FA | ||
|---|---|---|---|---|---|
| R | P | R | P | ||
| I | Os01g42870 | 0.42 | 0.04 | 0.17 | 0.42 |
| I | Os01g42880 | 0.60 | 0.002 | 0.55 | 0.005 |
| III | Os05g08640 | 0.04 | 0.80 | 0.01 | 0.93 |
| III | Os06g39470 | 0.44 | 0.005 | 0.19 | 0.23 |
| IV | Os01g09010 | 0.56 | <0.0001 | 0.42 | 0.009 |
| IV | Os06g39390 | 0.64 | <0.0001 | 0.48 | 0.002 |
However, when the stems were eliminated from the analyses, none of the highly expressed PF02458 genes showed a significant positive correlation with the FA or pCA cell wall content. This suggests that the correlations presented in Table 3 are driven mainly by the large differences between leaves and stems both for FA and pCA content and for transcript levels. The fact that the stem/leaf ratio for pCA was higher than that for FA might be interpreted as reflecting a higher proportion of lignin in the stems relative to the leaves due to the preferential association of pCA with lignin (Grabber et al. 2004).
This correlation between pCA/FA levels in the rice cell walls and transcript levels of several of the genes from family PF02458 provides indirect support to the hypothesis that members of this family may have an AFT function (Mitchell et al. 2007). More direct support for this hypothesis was obtained in this study from the comparison between gene expression and hydroxycinnamate levels in RNAi transgenic and non-transgenic control plants.
RNAi down-regulation efficiency
To understand the effect of RNAi constructs pAFT-A and pAFT-B on FA and pCA content, it is necessary to discuss first the level of down-regulation achieved for the different targeted genes in the different transformation events. Among the genes targeted by the pAFT-A construct, the gene representing the source of the antisense sequence, Os01g42880 (group I) showed the highest reduction in transcript levels in the transgenic plants relative to the control, whereas the effect on the other gene from group I (Os01g42870) was smaller and significant only in the leaves. The genes from group IV showed a similar pattern, with a higher reduction in the transcript levels of the gene used as the antisense target in pAFT-B (Os01g09010) than of the secondary target (Os06g39390). Both genes from group IV showed a larger reduction in the stem relative to the leaves (Table 2), which may be related with the fact that both genes (particularly Os06g39390) have higher transcript levels in the stems than in the leaves. The concentration of the target mRNA has been suggested as a factor that sometimes affects the silencing efficiency, with genes transcribed at higher levels silenced better than those with lower transcript levels (Miki et al. 2005; Hu et al. 2004).
The relative transcript levels of the genes from group III targeted by pAFT-B did not follow the general rules described above. Os05g08640, which was the source of the antisense segment in pAFT-B and that was expressed at higher levels, showed a smaller proportional reduction in transcript levels in the transgenic plants relative to the control than the secondary target Os06g39470. These differences were consistent for the two transgenic events B1 and B11 (Fig. 3), and both tissues and experiments (Tables 1 and 2). We currently do not know the cause of the more efficient down-regulation of Os06g39470 relative to Os05g08640 in the pAFT-B transgenic plants.
Using the different genes as replications, the level of down-regulation in the leaves was significantly correlated with the corresponding values in the stems (R = 0.86, P = 0.007) suggesting that, in general, the two transformation events resulted in consistent levels of down-regulation among tissues. This observation is consistent with the presence of a UBIQUITIN promoter in the transgenes, which is expected to be expressed at high levels in both tissues.
In summary, both constructs were effective in down-regulating the most highly expressed targeted genes and, therefore, they provide a valid tool to determine if these genes are associated with the incorporation of FA and pCA into the rice cell walls. In general, pAFT-B seemed more effective in targeting multiple highly expressed genes, whereas pAFT-A was effective in down-regulating one of the genes from group I and only partially the other gene.
Effect of the RNAi reduction in genes from family PF02458 on FA and pCA
The broader silencing effect of the pAFT-B construct relative to pAFT-A provides a tentative explanation for the larger effects on ester-linked cell wall FA content observed in plants transformed with pAFT-B. Down-regulation of different members of the PF02459 family had different effects on FA and pCA content. The reduction in transcript levels of groups III and IV genes in pAFT-B transgenic lines had a strong effect on FA, but not on pCA cell wall content, whereas down-regulation of groups I and II genes in pAFT-A transgenic lines had a slight effect on pCA in the stem. Thus, it is tempting to speculate that the processes affecting the incorporation of these two hydroxycinnamate acids into the rice cell walls may not be necessarily coupled, and that different members of the PF02458 gene family may control these processes separately.
pAFT-A (groups I and II genes)
The reduction in transcript levels of groups I and II genes in pAFT-A transgenic lines was associated with a reduction in cell wall pCA in the stems that was similar across the two experiments (Fig. 5a, b). However, the probability associated with both tests was too close to the threshold significance level to draw any strong conclusion. The 33% reduction in pCA observed in experiment 1 was borderline significant (P = 0.07), whereas the 24% reduction detected in experiment 2 was only marginally significant (P = 0.04) when the two constructs pAFT-A5 and pAFT-A11 were analyzed together.
Experiments including additional transgenic events or a much larger number of replications will be required to confirm the preliminary association described here between the down-regulation of the genes from groups I and II and the reduced levels of pCA in the stems.
pAFT-B (groups III and IV genes)
The most consistent and relevant result from this study was the FA reduction observed in the transgenic lines transformed with the RNAi construct pAFT-B. This construct generated reductions of 40–60% in the transcripts levels of three highly expressed genes, which together account for approximately 70% of the total transcripts from family PF02458. This large reduction in transcript levels was associated with a consistent reduction in cell wall FA in the leaves of the pAFT-B transgenic plants relative to the control both in experiment 1 (32% reduction, P = 0.004) and in experiment 2 (19% reduction, P < 0.0001).
The reduction in FA in the leaves relative to the control was also consistent between transgenic events, but the differences observed for pAFT-B11 (27%, P < 0.0001) were larger than those in pAFT-B1 (12%, P = 0.02). The cause of the stronger phenotypic effect of the pAFT-B11 event relative to the pAFT-B1 event is currently unknown, particularly since the reductions in the transcript levels of the four pAFT-B target genes were not significantly different between events. Only Os06g39470 showed a slightly stronger reduction in transcript level in pAFT-B11 relative to pAFT-B1 (Fig. 3), but these differences seem too small to explain the larger differences in FA. The stronger effect of the pAFT-B11 event relative to the pAFT-B1 in the leaves may also explain why the differences in cell wall FA in the stem were significant for the first event, but not for the second one (Fig. 5c).
The significant reduction in the ester-linked cell wall FA monomers in two experiments and in two independent RNAi transgenic events that significantly down-regulated rice genes Os05g08640, Os06g39470, Os01g09010, and Os06g39390 provide strong support to the hypothesis that one or more of these genes are involved in the incorporation of FA to the cell walls in the leaves of rice. This conclusion was also indirectly supported by the correspondence in the differences between transcript levels of these genes in leaves and stems and the differences in the levels of cell wall FA in the same tissues. These results also validated the strategy of simultaneously silencing multiple genes within a multigene family to address the problem of redundancy in functional studies.
Conclusions and future directions
Because FA dimers were not quantified in this study, our conclusions are limited to the ester-linked FA monomers. However, FA dimers represent only a small portion of the ferulates present in grass cell walls and, therefore, the results presented here are likely representative of the overall effect of the RNAi down-regulation of members of the PF02458 gene family on cell wall FA. Obel et al. (2002) found that only 10–20% of the FA present in the cell walls of leaves from young wheat seedlings was present as dimers. A similar value (22%) was reported by Grabber et al. (1998b) for cell walls from maize suspension cultures.
The significant reduction in cell wall ester-linked FA monomers observed in plants with reduced transcript levels of genes from PF02459 family groups III and IV support the hypothesis that some of these genes may be associated with the incorporation of FA to the cell wall. Because the level of GAX feruloylation has been correlated with cell wall digestibility (Grabber et al. 1998b; Dhugga 2007), the reduction in FA has the potential to increase cell wall digestibility and increase the efficiency of ethanol production. A reduction in cell wall FA may be also beneficial to improve the quality of forage grasses, but may have negative effects on the ability of grass plants to resist some pathogen attacks. Although further experiments are required to test these hypotheses, the identification of a group of genes affecting the FA content of the rice cell wall provides a valuable entry point to study the effect of FA on cell wall structure and digestibility.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
We thank Daisuke Miki and Ko Shimamoto for providing the pANDA vectors, Dr. D. M. Tricoli at the Ralph M. Parsons Foundation Plant Transformation Facility at UC Davis for the production of the rice transgenic plants, and Chevron for funding this research.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- ACN
Acetonitrile
- AFT
Arabinoxylan feruloyl transferases
- cDNA
Complementary DNA
- CT
Cycle thresholds
- EST
Expressed sequence tags
- FA
Ferulates
- GAX
Glucuronoarabinoxylan
- gus
Beta-glucuronidase
- HPLC
High-performance liquid chromatography
- nos
Nopaline synthase
- pCA
p-Coumarate
- PCR
Polymerase chain reaction
- qRT-PCR
Quantitative reverse transcription PCR
- RNAi
RNA interference
- SE
Standard error
- TFA
Trifluoroacetic acid
References
- Bacic A, Harris PJ, Stone BA. Structure and function of plant cell walls. In: Preiss J, editor. The biochemistry of plants. San Diego: Academic Press; 1988. pp. 297–372. [Google Scholar]
- Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam protein families database. Nucleic Acids Res. 2002;30:276. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs KJ, Fry SC. Phenolic cross-linking in the plant cell wall. In: Cosgrove DJ, Knievel DP, editors. Physiology of cell expansion during plant growth. Edinburgh, UK: The American Society of Plant Physiologists; 1987. pp. 46–57. [Google Scholar]
- Bunzel M, Allerdings E, Sinwell V, Ralph J, Steinhart H. Cell wall hydroxycinnamates in wild rice (Zizania aquatica L.) insoluble dietary fibre. Eur Food Res Technol. 2002;214:482–488. doi: 10.1007/s00217-002-0512-3. [DOI] [Google Scholar]
- Burr SJ, Fry SC. Extracellular cross-linking of maize arabinoxylans by oxidation of feruloyl esters to form oligoferuloyl esters and ether-like bonds. Plant J. 2009;58:554–567. doi: 10.1111/j.1365-313X.2009.03800.x. [DOI] [PubMed] [Google Scholar]
- Carpita NC. Structure and biogenesis of the cell walls of grasses. Ann Rev Plant Physiol Plant Mol Biol. 1996;47:445–476. doi: 10.1146/annurev.arplant.47.1.445. [DOI] [PubMed] [Google Scholar]
- Casler MD, Jung HG. Selection and evaluation of smooth bromegrass clones with divergent lignin or etherified ferulic acid concentration. Crop Sci. 1999;39:1866–1873. doi: 10.2135/cropsci1999.3961866x. [DOI] [Google Scholar]
- Dhugga KS. Maize biomass yield and composition for biofuels. Crop Sci. 2007;47:2211–2227. doi: 10.2135/cropsci2007.05.0299. [DOI] [Google Scholar]
- Eraso F, Hartley RD. Monomeric and dimeric phenolic constituents of plant cell walls-possible factors influencing wall biodegradability. J Sci Food Agric. 1990;51:163–170. doi: 10.1002/jsfa.2740510203. [DOI] [Google Scholar]
- Fincher GB. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiol. 2009;149:27–37. doi: 10.1104/pp.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbe M, Zacchi G. Pretreatment of lignocellulosic materials for efficient bioethanol production. In: Olsson L, Ahring BK, editors. Biofuels. Berlin: Springer; 2007. pp. 41–65. [DOI] [PubMed] [Google Scholar]
- Grabber JH, Hatfield RD, Ralph J, Zon J, Amrhein N. Ferulate cross-linking in cell walls isolated from maize cell suspensions. Phytochemistry. 1995;40:1077–1082. doi: 10.1016/0031-9422(95)00413-2. [DOI] [Google Scholar]
- Grabber JH, Hatfield RD, Ralph J. Diferulate cross-links impede the enzymatic degradation of non-lignified maize walls. J Sci Food Agric. 1998;77:193–200. doi: 10.1002/(SICI)1097-0010(199806)77:2<193::AID-JSFA25>3.0.CO;2-A. [DOI] [Google Scholar]
- Grabber JH, Ralph J, Hatfield RD. Ferulate cross-links limit the enzymatic degradation of synthetically lignified primary walls of maize. J Agric Food Chem. 1998;46:2609–2614. doi: 10.1021/jf9800099. [DOI] [Google Scholar]
- Grabber JH, Ralph J, Hatfield RD. Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. J Agric Food Chem. 2000;48:6106–6113. doi: 10.1021/jf0006978. [DOI] [PubMed] [Google Scholar]
- Grabber JH, Ralph J, Lapierre C, Barrière Y. Genetic and molecular basis of grass cell-wall degradability. I. Lignin-cell wall matrix interactions. Comptes Rendus Biol. 2004;327:455–465. doi: 10.1016/j.crvi.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Hatfield RD, Wilson JR, Mertens DR. Composition of cell walls isolated from cell types of grain sorghum stems. J Sci Food Agric. 1999;79:891–899. doi: 10.1002/(SICI)1097-0010(19990501)79:6<891::AID-JSFA304>3.0.CO;2-#. [DOI] [Google Scholar]
- Hatfield RD, Ralph J, Grabber JH. A potential role for sinapyl p-coumarate as a radical transfer mechanism in grass lignin formation. Planta. 2008;228:919–928. doi: 10.1007/s00425-008-0791-4. [DOI] [PubMed] [Google Scholar]
- Hu X, Hipolito S, Lynn R, Abraham V, Ramos S, Wong-Staal F. Relative gene-silencing efficiencies of small interfering RNAs targeting sense and antisense transcripts from the same genetic locus. Nucleic Acids Res. 2004;32:4609–4617. doi: 10.1093/nar/gkh790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikegawa T, Mayama S, Nakayashiki H, Kato H. Accumulation of diferulic acid during the hypersensitive response of oat leaves to Puccinia coronata f.sp. avenae and its role in the resistance of oat tissues to cell wall degrading enzymes. Physiol Mol Plant Pathol. 1996;48:245–256. doi: 10.1006/pmpp.1996.0021. [DOI] [Google Scholar]
- Ishii T. Structure and functions of feruloylated polysaccharides. Plant Sci. 1997;127:111–127. doi: 10.1016/S0168-9452(97)00130-1. [DOI] [Google Scholar]
- Jacquet G, Pollet B, Lapierre C, Mhamdi F, Rolando C. New ether-linked ferulic acid-coniferyl alcohol dimers identified in grass straws. J Agric Food Chem. 1995;43:2746–2751. doi: 10.1021/jf00058a037. [DOI] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Jung HG, Allen MS. Characteristics of plant cell walls affecting intake and digestibility of forages by ruminants. J Anim Sci. 1995;73:2774–2790. doi: 10.2527/1995.7392774x. [DOI] [PubMed] [Google Scholar]
- Kim S, Dale BE. Global potential bioethanol production from wasted crops and crop residues. Biomass Bioenergy. 2004;26:361–375. doi: 10.1016/j.biombioe.2003.08.002. [DOI] [Google Scholar]
- Kondo T, Mizuno K, Kato T. Cell wall-bound p-coumaric and ferulic acids in Italian ryegrass. Can J Plant Sci. 1990;71:495–499. doi: 10.4141/cjps90-058. [DOI] [Google Scholar]
- Lam TBT, Iiyama K, Stone BA. Cinnamic acid bridges between cell wall polymers in wheat and phalaris internodes. Phytochemistry. 1992;31:1179–1183. doi: 10.1016/0031-9422(92)80256-E. [DOI] [Google Scholar]
- Larsson S, Quintana-Sáinz A, Reimann A, Nilvebrant NO, Jönsson L. Influence of lignocellulose-derived aromatic compounds on oxygen-limited growth and ethanolic fermentation by Saccharomyces cerevisiae. Appl Biochem Biotechnol. 2000;84–86:617–632. doi: 10.1385/ABAB:84-86:1-9:617. [DOI] [PubMed] [Google Scholar]
- Lequart C, Nuzillard JM, Kurek B, Debeire P. Hydrolysis of wheat bran and straw by an endoxylanase: production and structural characterization of cinnamoyl-oligosaccharides. Carbohydr Res. 1999;319:102–111. doi: 10.1016/S0008-6215(99)00110-X. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lu FC, Ralph J. Novel tetrahydrofuran structures derived from beta–beta-coupling reactions involving sinapyl acetate in Kenaf lignins. Org Biomol Chem. 2008;6:3681–3694. doi: 10.1039/b809464k. [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K. Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol. 2004;45:490–495. doi: 10.1093/pcp/pch048. [DOI] [PubMed] [Google Scholar]
- Miki D, Itoh R, Shimamoto K. RNA silencing of single and multiple members in a gene family of rice. Plant Physiol. 2005;138:1903–1913. doi: 10.1104/pp.105.063933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell RAC, Dupree P, Shewry PR. A novel bioinformatics approach identifies candidate genes for the synthesis and feruloylation of arabinoxylan. Plant Physiol. 2007;144:43–53. doi: 10.1104/pp.106.094995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R, Attwood TK, Bairoch A, Barrell D, Bateman A, Binns D, Biswas M, Bradley P, Bork P. The InterPro Database 2003 brings increased coverage and new features. Nucleic Acids Res. 2003;31:315–318. doi: 10.1093/nar/gkg046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes AP, Pourquie J. Steam explosion pretreatment and enzymatic hydrolysis of eucalyptus wood. Bioresour Technol. 1996;57:107–110. doi: 10.1016/0960-8524(96)00019-3. [DOI] [Google Scholar]
- Obel N, Porchia AC, Scheller HV. Dynamic changes in cell wall polysaccharides during wheat seedling development. Phytochemistry. 2002;60:603–610. doi: 10.1016/S0031-9422(02)00148-6. [DOI] [PubMed] [Google Scholar]
- Persson P, Andersson J, Gorton L, Larsson S, Nilvebrant NO, Jonsson LJ. Effect of different forms of alkali treatment on specific fermentation inhibitors and on the fermentability of lignocellulose hydrolysates for production of fuel ethanol. J Agric Food Chem. 2002;50:5318–5325. doi: 10.1021/jf025565o. [DOI] [PubMed] [Google Scholar]
- Ralph J, Hatfield RD, Quideau S, Helm RF, Grabber JH, Jung H-JG. Pathway of p-coumaric acid incorporation into maize lignin as revealed by NMR. J Am Chem Soc. 1994;116:9448–9456. doi: 10.1021/ja00100a006. [DOI] [Google Scholar]
- Ralph J, Quideau S, Grabber JH, Hatfield RD. Identification and synthesis of new ferulic acid dehydrodimers present in grass cell walls. J Chem Soc Perkin Trans. 1994;1:3485–3498. doi: 10.1039/p19940003485. [DOI] [Google Scholar]
- Ralph J, Grabber JH, Hatfield RD. Lignin-ferulate crosslinks in grasses: active incorporation of ferulate polysaccharide esters into ryegrass lignins. Carbohydr Res. 1995;275:167–178. doi: 10.1016/0008-6215(95)00237-N. [DOI] [Google Scholar]
- Ralph J, Bunzel M, Marita JM, Hatfield RD, Lu F, Kim H, Schatz PF, Grabber JH, Steinhart H. Peroxidase-dependent cross-linking reactions of p-hydroxycinnamates in plant cell walls. Phytochem Rev. 2004;3:79–96. doi: 10.1023/B:PHYT.0000047811.13837.fb. [DOI] [Google Scholar]
- SAS Institute (2006) SAS user’s guide, version 9.1. In: SAS Institute, Inc., Cary, New York
- Saulnier L, Crépeau MJ, Lahaye M, Thibault JF, Garcia-Conesa MT, Kroon PA, Williamson G. Isolation and structural determination of two 5, 5′-diferuloyl oligosaccharides indicate that maize heteroxylans are covalently cross-linked by oxidatively coupled ferulates. Carbohydr Res. 1999;320:82–92. doi: 10.1016/S0008-6215(99)00152-4. [DOI] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis software (MEGA) Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J. Unique aspects of the grass cell wall. Curr Opin Plant Biol. 2008;11:301–307. doi: 10.1016/j.pbi.2008.03.002. [DOI] [PubMed] [Google Scholar]
- Yang Q, Reinhard K, Schiltz E, Matern U. Characterization and heterologous expression of hydroxycinnamoyl/benzoyl-CoA:anthranilate N-hydroxycinnamoyl/benzoyltransferase from elicited cell cultures of carnation, Dianthus caryophyllus L. Plant Mol Biol. 1997;35:777–789. doi: 10.1023/A:1005878622437. [DOI] [PubMed] [Google Scholar]
- York WS, O’Neill MA. Biochemical control of xylan biosynthesis: which end is up? Curr Opin Plant Biol. 2008;11:258–265. doi: 10.1016/j.pbi.2008.02.007. [DOI] [PubMed] [Google Scholar]
- Zdobnov EM, Apweiler R. InterProScan-an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.