Abstract
AAA+ proteins carry out diverse functions in cells. In most cases, their ATPase activity is very tightly regulated by protein partners and target ligands but the mechanism for this control has remained unclear. We have identified a conserved link between the ligand binding and ATPase sites in AAA+ proteins. This link, which we call the “Glutamate Switch”, regulates ATPase activity directly in response to binding of target ligands by controlling the orientation of the conserved glutamate residue in the DExx motif, switching it between active and inactive conformations. The reasons for this level of control of the ATPase activity are discussed in the context of the biological processes catalysed by AAA+ proteins.
ATPases Associated with a variety of cellular Activities (AAA+) proteins are distributed across all kingdoms of life 1-4 and have been classified into seven different clades based on structural and sequence information 3,4. They are involved in a multitude of diverse cellular processes including, but not limited to, protein degradation and chaperone activities, membrane fusion events, regulation of transcription, and DNA replication and repair. Many of the systems can be viewed as cargo delivery with a ligand being delivered to a site and then released before the protein returns to pick up another ligand (e.g. DNA polymerase processivity clamp loading). Other proteins are involved in events that “re-model” macromolecular ligands, converting them from one state to another (e.g. protein chaperones and transcriptional regulators). Possibly due to the complexity of the reactions they catalyse, AAA+ proteins rarely work in isolation and are frequently a part of multi-component assemblies. It is widely documented that the biochemical activities of the individual proteins are affected by interactions with other components but the molecular mechanisms of this regulation are poorly understood.
One initially puzzling feature of many AAA+ enzyme systems is the requirement for ATP in reactions that are energetically neutral or even favourable. For example, initiation of transcription of a number of genes in bacteria requires AAA+ transcriptional activators, such as PspF and NtrC, whereas transcription of most genes is not dependent upon these activators 5. Similarly, the requirement for ATP-dependent loading of DNA polymerase processivity clamps and ring helicases such as MCM is unclear since other ring helicases are able to load efficiently onto DNA without additional factors or ATP hydrolysis 6. Protease activity of AAA+ proteases is ATP-dependent whereas a plethora of other proteases do not require ATP because cleavage of the peptide bond is an energetically favourable process and total digestion of proteins is possible without a need for ATP.
To compound the puzzle, the rate of ATP turnover of AAA+ enzymes is frequently poor compared to other ATPases (typically, turnover numbers are 0.1 to a few per second for AAA+ enzymes compared to several hundred per second for ‘simple’ metabolic ATPases like hexokinase or pyruvate kinase and even for enzymes that catalyse more complex substrate remodelling reactions such as DNA helicases). To complicate the issue further, the ATPase activity is almost always regulated by binding of ligands, albeit in either a positive or negative manner, even for the same ligand. For example, binding of DNA to Replication Factor C (RFC) stimulates the ATPase activity of the protein by almost two orders of magnitude 7,8. By contrast, DNA binding to the Origin Recognition Complex (ORC) proteins represses ATPase activity 9. Despite the wide documentation of these effects on ATPase activity of AAA+ proteins, the molecular mechanism by which this regulation occurs has remained unclear but likely involves inhibition of the enzyme under certain conditions that is relieved to “activate” the ATPase. The ATPase active site of AAA+ proteins contains several motifs known to be important for many other ATPases including the so-called Walker A and B motifs 10 as well as several other important residues. Consequently, the reason for this low activity is unclear despite the availability of a considerable amount of structural information. Understanding this process is fundamental to understanding the mechanism and control of AAA+ proteins with implications across a wide range of cellular processes.
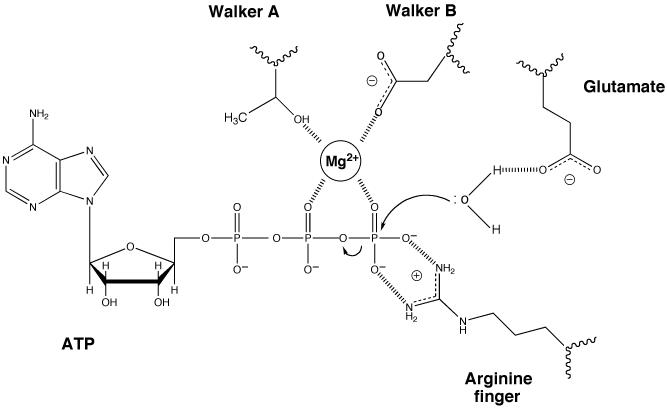
AAA+ enzymes have a set of conserved residues in the active site that promote ATP hydrolysis by a well established mechanism that is shared with ATPases from a variety of other protein families (Figure 1). Several residues interact with the bound ATP but do not play a direct role in catalysis so are not discussed here. However, key residues in the Walker A and Walker B motifs are involved in coordinating a magnesium ion that promotes catalysis. Another important residue that is often present is the “arginine finger” that reaches into the ATPase site, usually in trans from an additional subunit or domain, and plays a role in polarising the γ-phosphate to facilitate hydrolysis. There is also a conserved glutamate residue that is located within the Walker B motif in AAA+ and other DExx box proteins but is at the end of an adjacent β-strand in other ATPases. This residue serves to polarise a water molecule for in-line attack of the γ-phosphate during hydrolysis. Replacement of this residue with alanine creates proteins that are able to bind ATP but are severely impaired in hydrolysis activity often by several orders of magnitude (for example see 8,11,12). We reasoned that by examining structural information from a number of AAA+ systems we might be able to rationalise how these critical residues are affected to regulate catalysis. Consequently, we performed an analysis of the active site structures of all of the AAA+ proteins in the Protein Database. Although structures of all nucleotide and/or ligand bound states are not available for any single protein, comparison between different AAA+ systems revealed a mechanism for regulation of ATPase activity that appears to be shared across a wide range of AAA+ proteins.
Figure 1. Essential residues in the active site of ATPases.
ATP hydrolysis is promoted by several residues in this generalised active site of ATPases. Only residues with an established role in catalysis rather than binding are shown. When present, the “arginine finger” is usually provided in trans from an adjacent subunit or domain. The magnesium ion can be coordinated in different ways but that shown is one of the most common involving interactions with a threonine (or serine) from the Walker A motif, an aspartate from the Walker B motif and oxygens from the β and γ phosphates of the ATP. The role of the glutamate residue (in the DExx motif of AAA+ proteins) is to activate a water molecule by making the oxygen more electronegative and hence a better nucleophile for attack of the γ-phosphorus.
RESULTS
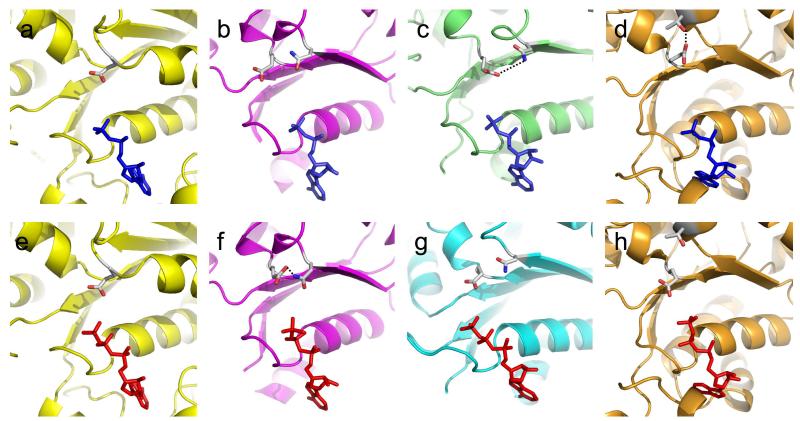
One of the few AAA+ proteins for which a large number of nucleotide complexes are available at high resolution is the bacterial transcription activator, PspF, a member of Clade 6 of AAA+ proteins 13. PspF activates σ54-dependent RNA polymerase (RNAP) by converting it from a closed (inactive) state to the open (active) conformation that initiates transcription. Structural information is available at high resolution for apo, ADP, ATP and ADPNP bound forms of the AAA+ domain of PspF 13. These studies provided the first insights into how ATPase activity is regulated. Comparison of the ADP and ATP complexes (Figure 2) reveals that the structures superimpose very well with the exception of a few regions that respond to the presence of the γ-phosphate. These changes are focussed in two main areas. The first of these is within the ATPase site itself. Although most of the active site residues alter little, there is a substantial movement of the glutamate residue of the DExx motif. In the ADP complex, the glutamate side chain is found in a position similar to that seen in most other ATPase active sites (e.g. F1 ATPase β-subunit 14) (Figure 2). By contrast, in the ATP complex, the glutamate is rotated approximately 100° from the conformation in the ADP complex and forms a hydrogen bond with an asparagine residue that is located on an adjacent β-strand (Figure 2). The effect of this rotation is to move the glutamate from a position in which it would be able to activate the in-coming attacking water molecule, to one in which it would not. Consequently, binding of ATP has the surprising effect of inactivating the ATPase activity by switching the active site from an active configuration to an inactive one. This “glutamate switch” converts the enzyme from a low energy ground state to a higher energy form that is now trapped in an inactive state. This conformation of the protein is stabilised by a set of water-mediated interactions that involve the γ-phosphate of the bound ATP as well as the glutamate and asparagine side chains and the magnesium ion. Consistent with these observations, the glutamate-asparagine pair is also present in the ATP-bound complex of the related σ54 activator ZraR 15 but is not formed in the ADP bound complex of another regulator, NtrC1 16.
Figure 2. ATPase active sites showing the conformations of the glutamate.
(a) – F1 ATPase β-subunit active site with bound ADP (1BMF).
(b) – PspF complexed with ADP (2C98).
(c) – ORC1 complexed with ADP + DNA (2V1U).
(d) – HslU complexed with ADP (1G41).
(e) – F1 ATPase β-subunit active site with bound ADPNP (1BMF).
(f) – PspF complexed with ATP (2C96).
(g) – ORC2 complexed with ADPNP (1W5T).
(h) – HslU complexed with ATP (1DO0).
PDB ID codes are shown in parentheses. When formed, the glutamate switch is indicated by dotted lines (panels c, d, and f).
The second area of movement within the PspF complexes is remote from the active site and involves two loops (L1 and L2) that interact with σ54-RNAP 17. However, there is a direct peptide linkage between the E-N pair and these loops, providing communication between the ATPase and ligand-binding sites. Although there is no high resolution structure of the complex between PspF and σ54-RNAP, it is known that the complex is most stable when formed with the transition state analogue ADP-AlFx rather than ADP or ATP 18. The corollary of this observation is that the interaction between σ54-RNAP and PspF must stabilise the transition state and hence stimulate ATPase activity, most likely by releasing the glutamate in the ATPase active site from the inactive to the active configuration. However, confirmation of this proposal will require high resolution structural information of the PspF/σ54/RNAP complex.
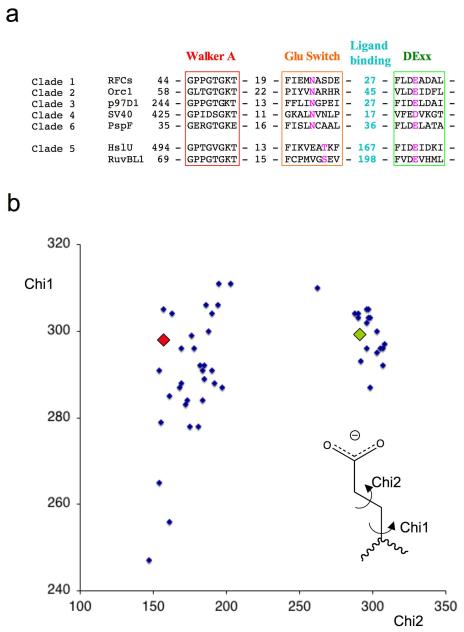
Analysis of the available crystal structures reveals that a glutamate switch operates in a similar manner in other AAA+ protein clades. In addition to PspF (Clade 6), other examples include DNA clamp loaders such as RFC (Clade 1) 19,20, DNA replication initiators such as ORC (Clade 2) 21,22, and the molecular chaperone p97 (Clade 3) 23. The HslUV/Clp family proteases and RuvB-like helicases (Clade 5) have an interesting variation in which the glutamate contacts a conserved threonine (or serine) residue that is placed two residues further along β-strand 2 24-26. Of course, similar pairings may be present in other systems for which there is insufficient structural information at present or for which the resolution of the structural data is too low for detailed analysis. In fact, sequence analysis reveals that examples of a conserved glutamate switch pair can be found in members of six of the seven AAA+ clades (Figure 3a). Due to its key role in promoting ATP hydrolysis, the glutamate residue is almost invariant across all AAA+ proteins but in rare cases can be an aspartate. Although also highly conserved, the asparagine can be replaced in some proteins by serine, threonine, or lysine, all residues that could make an interaction with a glutamate. We analysed 50 side chain conformations of the Glu residue from AAA+ protein structures in the database with a resolution better than 3.5Å. The side chain torsion angles (Chi1 & Chi2) show a clear bimodal distribution of Chi2 angles (Figure 3b), with approximately 100 degrees difference between the two clusters. This is in perfect agreement with what was observed in PspF between the ADP and ATP states described above. Interestingly, the “active” conformation, represented by the ADP state of PspF, shows a tighter clustering of Chi2 angles compared to that of the inactive conformation represented by the ATP state of PspF, consistent with the geometric constraints for positioning the Glu residue correctly for polarizing the attacking water molecule.
Figure 3. Conservation of the glutamate switch in AAA+ proteins.
(a) Sequence alignments of selected members of each clade for which crystal structures are known. The colour scheme follows that used in Figure 4. RFCs – Replication factor C small subunit A.fulgidus, Orc1 – Orc1 protein A.pernix, p97D1 - p97 D1 AAA+ domain M.musculus, SV40 – SV40 Large T antigen, PspF – PspF E.coli, HslU – HslU E.coli, RuvBL1 – RuvB-like 1 (TIP49a, Pontin) H.sapiens.
(b) Plot of side chain torsion angles for 50 active site glutamate (DExx box) residues from AAA+ protein structures in the protein database (www.rcsb.org) with a resolution better than 3.5Å. Only one copy from the asymmetric unit was used if angles were similar to reduce redundancy. The appropriate glutamate residue was selected from individual PDB files and the side chain torsion angles were calculated using the CCP4 program ANGLES 40 then normalized to 0-360 degrees for display. The values for the PspF-ADP complex (green diamond) and PspF-ATP complex (red diamond) are overlaid as examples of residues in the two conformations.
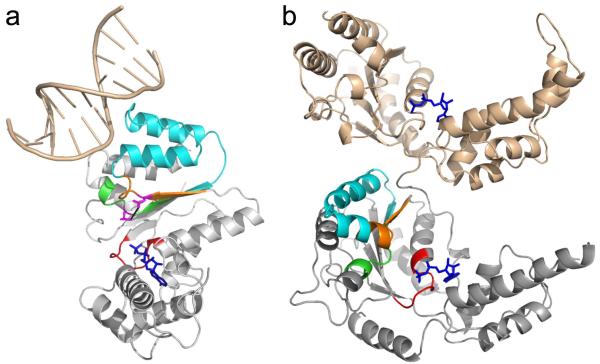
Furthermore, where it has been identified, the binding site for target ligands on the AAA+ domain involves the same region of the fold in all clades (Figure 4) suggesting a conservation of the linkage between ATPase and ligand binding sites. Interestingly, for type II AAA+ proteins such as p97, the second AAA+ domain (D2), which is not directly involved in substrate binding, interacts with the first AAA+ domain (D1) via similar regions in D2 (Figure 4) 27. Similar cooperativity between the two domains has also been proposed for other type II AAA+ proteins 28,29.
Figure 4. Link between the glutamate switch motif and ligand binding site.
(a) Glutamate switch pair location within a typical AAA+ domain and the linkage to the ligand binding site. The example shown is A.pernix ORC1 DNA complex (Clade 2, PDB ID code 2V1U). The Walker A motif is shown in red, the glutamate switch region in orange (with the E-N pair in magenta), the ligand binding site between the glutamate switch and the DExx box in cyan, the DExx box in green and the bound ADP in blue. A portion of the bound DNA is shown in wheat.
(b) D1 (wheat) and D2 (grey) domains of p97 (PDB ID code 3CF3) showing the interaction between the two ATPase domains in a type II AAA+ protein. The colouring of the D2 domain follows the same scheme as part (a).
The formation of the switch pair when ATP is bound offers a means to suppress the ATPase which can be alleviated upon ligand binding. However, in principle, there is no reason why this linkage might not be reversed. Indeed, this is the case for Clade 2 DNA initiator proteins, such as ORC and DnaA, the only clade for which there is also a crystal structure with the target ligand bound. In these proteins, the glutamate (aspartate in DnaA) remains in the active configuration in both the ADP and ATP analogue complexes in the absence of DNA 4,30,31 (Figure 2). However, this family of proteins is unusual in that ligand binding (in this case DNA) inhibits the ATPase activity rather than stimulating it 9. Recent structures of archaeal ORC proteins bound to their DNA targets 21,22 reveals that the E-N pair is formed in these complexes (Figure 2), precisely what would be expected for a system in which ligand binding inhibits, rather than stimulates, ATPase activity. Consequently, the glutamate switch can function in either direction, to stimulate or inhibit ATP hydrolysis at different stages within a complex assembly process. For example, ATPase activity of HslU is stimulated by binding of both HslV and substrate albeit by only 3-4 fold in each case 32. The structures show the glutamate is locked in the inactive configuration in the ADP complex but is free in the ATP bound form. In this case, the small stimulation compared to other systems may reflect a difference in equilibrium between “on” and “off“ states. Interestingly, in SV40 Large T antigen (LTag), which catalyses unwinding of long stretches of DNA during virus replication 33, the glutamate also remains in the active configuration in both ADP and ATP analogue complexes 34. However, full length LTag also contains a domain that binds to the SV40 replication origin. It is likely that the helicase activity needs to be restrained prior to origin firing and this may explain the presence of the E-N pair and its potential role as a switch upon replication origin binding. However, after origin firing, the enzyme needs to be a processive helicase and the switch is no longer required. Consistent with this suggestion, the ATPase activity of human papilloma virus E1 helicase (which is closely related to LTag and also has a conserved switch pair) is inhibited by the E2 origin binding protein35. Although the glutamate switch is widespread across AAA+ families, there are a few exceptions notably several proteins in Clade 7, a group with a wide variety of unrelated functions and for which there are few representative crystal structures 4.
Biochemical data are also consistent with the glutamate switch. The key role for the glutamate residue has been demonstrated for AAA+ proteins of every clade as well as for ATPases of many other families. Mutations at this position typically reduce ATPase activity by at least 1-2 orders of magnitude. However, more significantly, the link between ATPase activity and ligand binding should be broken in mutants lacking either the glutamate or the asparagine. Although there are very few cases where this has been properly explored, this is indeed the case for the archaeal clamp loader complex RFC for which it has been shown that the glutamate to alanine mutant not only has a substantially reduced ATPase activity but this basal activity of the complex is no longer stimulated by DNA 8. Although the effects of mutating the asparagine in RFC have not been reported, biochemical data for a number of substitutions at this residue in PspF have been reported recently 36. Mutating the asparagine in PspF abolishes the inhibitory effects of its negative regulator, PspA, consistent with the hypothesis that the E-N pair communicates ligand binding to ATPase activity. PspF hexamers are stabilised by ATP binding and mutants in which the switch pair can no longer form are defective in hexamerisation. Consequently, analysis of the data is complicated by these defects in hexamerisation (also required for maximal ATPase activity) and hence limits the information that can be drawn from these mutations. The only other protein in which the asparagine has been mutated is the Clade 2 protein, Cdc6, which has a role in the initiation of DNA replication in eukaryotes. Yeast cell lines expressing a mutant Cdc6 protein in which the asparagine, and the residues either side of it, were replaced with alanine residues showed a temperature-sensitive phenotype despite showing a wildtype level of expression of the mutant protein 37. The mutant cell line was defective in the rate of DNA replication and cells halted growth in S phase. However, detailed biochemistry of the isolated protein was not carried out.
DISCUSSION
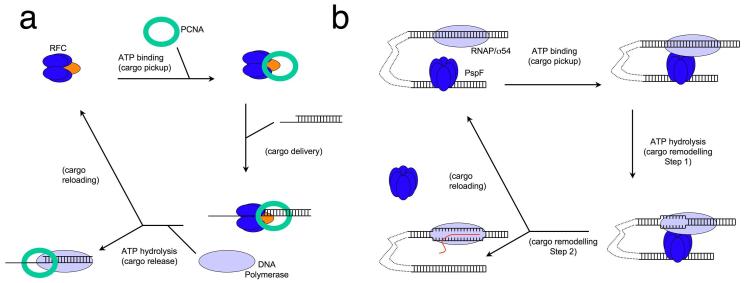
Why is ATPase activity so tightly regulated in most AAA+ enzymes? The clue comes from looking at the functions of the proteins. Many ATPases utilise the free energy of hydrolysis of ATP to drive reactions, either by transferring chemical energy from one form to another or converting it into mechanical energy. By contrast, the sorts of reactions catalysed by AAA+ enzymes are frequently different in a subtle way. Inhibition of ATPase activity is found in systems with complex assembly pathways and multiple intermediate steps. Typically, ATP hydrolysis is suppressed (e.g. through ATP binding in RFC or ligand binding as in ORC) until the system is fully assembled, but ATP turnover then allows completion of the reaction and/or recycling of components. Many reactions, such as cargo delivery or loading, involve single events on a particular macromolecular substrate requiring a series of steps that need to occur in a precise pathway. In these systems, ATP is used to control the directionality of the reaction rather than simply in a catalytic role, usually by regulating the assembly of a multi-component system. The role of ATP is to ensure the correct assembly of the components rather than as a means to lower the activation energy of the reaction. As the system begins to assemble, ATP binds but hydrolysis is suppressed until the system is fully competent. There are several well characterised examples of these principles across many clades of AAA+ proteins. One of these is synaptic vesicle fusion in which the complex between NSF, SNAPs and SNAREs is stabilised on ATP binding but disassembles when ATP is hydrolysed38. The loading of PCNA rings onto DNA primer junctions by RFC is another example (Figure 5). In this case, ATP binding is sufficient to stabilise the RFC/PCNA complex and even to support loading of the clamp around DNA but DNA-stimulated hydrolysis of ATP releases the PCNA onto the DNA and recycles the RFC components to allow them to pick up another PCNA ring8. Yet another example is the activation of RNA polymerase/σ54 by transcriptional activators such as PspF or NtrC39 (Figure 5). RNAP/σ54 binds to the promoter site and forms a stable closed complex that is unable to initiate transcription without being remodelled by activators. These activators bind to a specific activation sequence, located 80-150 bp upstream from the transcriptional starting site. ATP binding is required for stable hexamer formation of the activators and their interactions with RNAP/σ54 through DNA looping, a process facilitated by DNA bending proteins such as Integrative Host Factor (IHF). ATP hydrolysis releases the activators from the activated RNAP/σ54 allowing transcription to proceed.
Figure 5. Overall mechanism of two well characterised AAA+ protein catalysed reactions.
(a) PCNA clamp loading by Replication Factor C. Upon ATP binding, RFC forms a stable complex with PCNA (cargo pickup) that is competent to load onto DNA at primer-template junctions (cargo delivery). ATP hydrolyis by the small subunits (blue) releases PCNA (cargo release) which is then bound by DNA polymerase. Finally, ATP hydrolysis at the large subunit (orange) recycles the RFC and allows pick up of the next PCNA (cargo reloading).
(b) Bacterial transcription activation by PspF. Upon ATP binding, PspF, which binds to the DNA sequence upstream of transcription start site, interacts with RNAP/σ54 through DNA looping (cargo pickup). At the point of ATP hydrolysis, PspF forms a stable complex with RNAP/σ54 and initial remodelling of RNAP/σ54/DNA occurs (cargo remodelling). Upon the completion of ATP hydrolysis, transcription proceeds and PspF dissociates from the complex (cargo remodelling and reloading).
In all of these examples, ATP hydrolysis serves as a switch to control the process and recycle the components after completion of the reaction. The coupling to ATP hydrolysis allows control of the process and explains the requirement for ATP in processes that, formally, are energetically favourable or neutral. For example, some ring helicases can load themsleves onto DNA but the involvement of ATP-dependent helicase loaders (such as ORC and DnaC) provides controlled loading at specific sites and times during the cell cycle rather than at random sites over the genome. Similarly, although cleavage of the peptide bond is favourable, controlled proteolysis by AAA+ proteases, such as ClpXP and HslUV, is a much more discriminating process that only digests proteins that are selectively delivered to the protease by unfolding them in situ. The small energetic cost of utilising ATP as a switch is outweighed by the advantages of assembling the systems correctly and thereby controlling reactions that might otherwise have drastic consequences for the cell.
Acknowledgements
This work was funded by Wellcome Trust and BBSRC (XZ) Cancer Research UK (DBW).
References
- 1.Kunau WH, et al. Two complementary approaches to study peroxisome biogenesis in Saccharomyces cerevisiae: forward and reversed genetics. Biochimie. 1993;75:209–24. doi: 10.1016/0300-9084(93)90079-8. [DOI] [PubMed] [Google Scholar]
- 2.Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 3.Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 4.Erzberger JP, Berger JM. Evolutionary relationships and structural mechanisms of AAA+ proteins. Annu Rev Biophys Biomol Struct. 2006;35:93–114. doi: 10.1146/annurev.biophys.35.040405.101933. [DOI] [PubMed] [Google Scholar]
- 5.Browning DF, Busby SJ. The regulation of bacterial transcription initiation. Nat Rev Microbiol. 2004;2:57–65. doi: 10.1038/nrmicro787. [DOI] [PubMed] [Google Scholar]
- 6.Ahnert P, Picha KM, Patel SS. A ring-opening mechanism for DNA binding in the central channel of the T7 helicase-primase protein. Embo J. 2000;19:3418–27. doi: 10.1093/emboj/19.13.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoder BL, Burgers PM. Saccharomyces cerevisiae replication factor C. I. Purification and characterization of its ATPase activity. J Biol Chem. 1991;266:22689–97. [PubMed] [Google Scholar]
- 8.Seybert A, Wigley DB. Distinct roles for ATP binding and hydrolysis at individual subunits of an archaeal clamp loader. Embo J. 2004;23:1360–71. doi: 10.1038/sj.emboj.7600130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klemm RD, Austin RJ, Bell SP. Coordinate binding of ATP and origin DNA regulates the ATPase activity of the origin recognition complex. Cell. 1997;88:493–502. doi: 10.1016/s0092-8674(00)81889-9. [DOI] [PubMed] [Google Scholar]
- 10.Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. Embo J. 1982;1:945–51. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mogk A, et al. Roles of individual domains and conserved motifs of the AAA+ chaperone ClpB in oligomerization, ATP hydrolysis, and chaperone activity. J Biol Chem. 2003;278:17615–24. doi: 10.1074/jbc.M209686200. [DOI] [PubMed] [Google Scholar]
- 12.Joly N, Rappas M, Wigneshweraraj SR, Zhang X, Buck M. Coupling nucleotide hydrolysis to transcription activation performance in a bacterial enhancer binding protein. Mol Microbiol. 2007;66:583–95. doi: 10.1111/j.1365-2958.2007.05901.x. [DOI] [PubMed] [Google Scholar]
- 13.Rappas M, Schumacher J, Niwa H, Buck M, Zhang X. Structural basis of the nucleotide driven conformational changes in the AAA+ domain of transcription activator PspF. J Mol Biol. 2006;357:481–92. doi: 10.1016/j.jmb.2005.12.052. [DOI] [PubMed] [Google Scholar]
- 14.Abrahams JP, Leslie AG, Lutter R, Walker JE. Structure at 2.8 Å resolution of F1-ATPase from bovine heart mitochondria. Nature. 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 15.Sallai L, Tucker PA. Crystal structure of the central and C-terminal domain of the sigma(54)-activator ZraR. J Struct Biol. 2005;151:160–70. doi: 10.1016/j.jsb.2005.05.006. [DOI] [PubMed] [Google Scholar]
- 16.Lee SY, et al. Regulation of the transcriptional activator NtrC1: structural studies of the regulatory and AAA+ ATPase domains. Genes Dev. 2003;17:2552–63. doi: 10.1101/gad.1125603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rappas M, et al. Structural insights into the activity of enhancer-binding proteins. Science. 2005;307:1972–5. doi: 10.1126/science.1105932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaney M, et al. Binding of transcriptional activators to sigma 54 in the presence of the transition state analog ADP-aluminum fluoride: insights into activator mechanochemical action. Genes Dev. 2001;15:2282–94. doi: 10.1101/gad.205501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bowman GD, O’Donnell M, Kuriyan J. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature. 2004;429:724–30. doi: 10.1038/nature02585. [DOI] [PubMed] [Google Scholar]
- 20.Seybert A, Singleton MR, Cook N, Hall DR, Wigley DB. Communication between subunits within an archaeal clamp-loader complex. Embo J. 2006;25:2209–18. doi: 10.1038/sj.emboj.7601093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaudier M, Schuwirth BS, Westcott SL, Wigley DB. Structural basis of DNA replication origin recognition by an ORC protein. Science. 2007;317:1213–6. doi: 10.1126/science.1143664. [DOI] [PubMed] [Google Scholar]
- 22.Dueber EL, Corn JE, Bell SD, Berger JM. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317:1210–3. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 23.Zhang X, et al. Structure of the AAA ATPase p97. Mol Cell. 2000;6:1473–84. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- 24.Matias PM, Gorynia S, Donner P, Carrondo MA. Crystal structure of the human AAA+ protein RuvBL1. J Biol Chem. 2006;281:38918–29. doi: 10.1074/jbc.M605625200. [DOI] [PubMed] [Google Scholar]
- 25.Bochtler M, et al. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–5. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 26.Trame CB, McKay DB. Structure of Haemophilus influenzae HslU protein in crystals with one-dimensional disorder twinning. Acta Crystallogr D Biol Crystallogr. 2001;57:1079–90. doi: 10.1107/s0907444901007673. [DOI] [PubMed] [Google Scholar]
- 27.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–26. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 28.Schlee S, Groemping Y, Herde P, Seidel R, Reinstein J. The chaperone function of ClpB from Thermus thermophilus depends on allosteric interactions of its two ATP-binding sites. J Mol Biol. 2001;306:889–99. doi: 10.1006/jmbi.2001.4455. [DOI] [PubMed] [Google Scholar]
- 29.Hattendorf DA, Lindquist SL. Cooperative kinetics of both Hsp104 ATPase domains and interdomain communication revealed by AAA sensor-1 mutants. Embo J. 2002;21:12–21. doi: 10.1093/emboj/21.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singleton MR, et al. Conformational changes induced by nucleotide binding in Cdc6/ORC from Aeropyrum pernix. J Mol Biol. 2004;343:547–57. doi: 10.1016/j.jmb.2004.08.044. [DOI] [PubMed] [Google Scholar]
- 31.Erzberger JP, Pirruccello MM, Berger JM. The structure of bacterial DnaA: implications for general mechanisms underlying DNA replication initiation. Embo J. 2002;21:4763–73. doi: 10.1093/emboj/cdf496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoo SJ, et al. Purification and characterization of the heat shock proteins HslV and HslU that form a new ATP-dependent protease in Escherichia coli. J Biol Chem. 1996;271:14035–40. doi: 10.1074/jbc.271.24.14035. [DOI] [PubMed] [Google Scholar]
- 33.Dean FB, et al. Simian virus 40 (SV40) DNA replication: SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc Natl Acad Sci U S A. 1987;84:16–20. doi: 10.1073/pnas.84.1.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gai D, Zhao R, Li D, Finkielstein CV, Chen XS. Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell. 2004;119:47–60. doi: 10.1016/j.cell.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 35.White PW, et al. Characterization of recombinant HPV6 and 11 E1 helicases: effect of ATP on the interaction of E1 with E2 and mapping of a minimal helicase domain. J Biol Chem. 2001;276:22426–38. doi: 10.1074/jbc.M101932200. [DOI] [PubMed] [Google Scholar]
- 36.Joly N, Burrows PC, Buck M. An intramolecular route for coupling ATPase activity in AAA+ proteins for transcription activation. J Biol Chem. 2008;283:13725–35. doi: 10.1074/jbc.M800801200. [DOI] [PubMed] [Google Scholar]
- 37.Schepers A, Diffley JF. Mutational analysis of conserved sequence motifs in the budding yeast Cdc6 protein. J Mol Biol. 2001;308:597–608. doi: 10.1006/jmbi.2001.4637. [DOI] [PubMed] [Google Scholar]
- 38.Sollner T, Bennett MK, Whiteheart SW, Scheller RH, Rothman JE. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion. Cell. 1993;75:409–18. doi: 10.1016/0092-8674(93)90376-2. [DOI] [PubMed] [Google Scholar]
- 39.Rappas M, Bose D, Zhang X. Bacterial enhancer-binding proteins: unlocking sigma54-dependent gene transcription. Curr Opin Struct Biol. 2007;17:110–6. doi: 10.1016/j.sbi.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 40.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–3. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]