The PRELP heparin sulfate–binding protein translocates to the nucleus, where it impairs NF-κB transcriptional activity, which in turn regulates bone homeostasis.
Abstract
Proline/arginine-rich end leucine-rich repeat protein (PRELP) is a glycosaminoglycan (GAG)- and collagen-binding anchor protein highly expressed in cartilage, basement membranes, and developing bone. We observed that PRELP inhibited in vitro and in vivo mouse osteoclastogenesis through its GAG-binding domain (hbdPRELP), involving (a) cell internalization through a chondroitin sulfate– and annexin II–dependent mechanism, (b) nuclear translocation, (c) interaction with p65 nuclear factor κB (NF-κB) and inhibition of its DNA binding, and (d) impairment of NF-κB transcriptional activity and reduction of osteoclast-specific gene expression. hbdPRELP does not disrupt the mitogen-activated protein kinase signaling nor does it impair cell survival. hbdPRELP activity is cell type specific, given that it is internalized by the RAW264.7 osteoclast-like cell line but fails to affect calvarial osteoblasts, bone marrow macrophages, and epithelial cell lines. In vivo, hbdPRELP reduces osteoclast number and activity in ovariectomized mice, underlying its physiological and/or pathological importance in skeletal remodeling.
Introduction
The proline/arginine-rich end leucine-rich repeat (LRR) protein (PRELP) is a 58-kD heparin/heparan sulfate–binding protein first discovered in articular cartilage but present also in several connective tissue extracellular matrices. The protein comprises 382 aa residues, including a 20-residue signal peptide. It belongs to a subfamily of LRR proteins in the extracellular matrix. Members encompass several small LRR proteins (SLRRPs), including the chondroitin/dermatan sulfate proteoglycans decorin and biglycan and the keratan sulfate proteoglycans fibromodulin and lumican (Iozzo and Murdoch, 1996). 10–11 adjacent LRRs characterize this subfamily, flanked at either end by disulphide-bonded domains (Heinegård et al., 2002).
N-linked oligosaccharides are present in the central LRR domain of PRELP (Bengtsson et al., 1995), whose name reflects the abundance of proline and arginine in its N-terminal domain (Bengtsson et al., 1995). Compared with many of the other members of the SLRRP subfamily, PRELP has two atypical features. First, it does not contain glycosaminoglycan (GAG) chains; second, the N-terminal region, which is unique and conserved between rodents, bovine, and humans, binds heparin and heparan sulfate (Bengtsson et al., 2000). N-terminally truncated PRELP lacking this region cannot bind heparin, whereas a 6-mer heparin oligosaccharide is the smallest size showing some affinity to PRELP. Binding increases with length up to 18-mer and was found to depend on the degree of sulfation of heparin and heparan sulfate (Bengtsson et al., 2000). The protein binds collagens I and II with high affinity (Bengtsson et al., 2002) via its LRR domain, whereas the N-terminal part of PRELP can bind the heparan sulfate of perlecan or bind fibroblasts via surface heparan sulfate proteoglycans (Bengtsson, 1999), thus serving as a linker between these proteoglycans and the extracellular matrix.
The gene encoding PRELP maps at chromosome 1q32, and PRELP mRNA transcripts were found in articular chondrocytes, osteoblasts, and osteosarcoma cells of various species (Bengtsson et al., 2000). The protein was also found at basement membranes of skin, testis, and Bowman’s capsule of the kidney (Bengtsson et al., 2002). PRELP plays a role in eye and skin (Reardon et al., 2000; Grover et al., 2007). The protein is highly expressed in human sclera, and mutations have been found in advanced myopia (Majava et al., 2007). PRELP mutations are also involved in the pathogenesis of Hutchinson-Gilford progeria (Lewis, 2003), which is characterized, among other symptoms, by scleroderma, achondrogenesis, bone deformities, and osteoporosis (Hennekam, 2006).
Although PRELP was found in the skeleton expressed by chondrocytes and osteoblasts, there is no direct information regarding the role of the protein in skeletal remodeling. We sought to identify its role in bone homeostasis using an N-terminal peptide corresponding to the entire heparin-binding domain of PRELP (hbdPRELP). The peptide was tested in in vitro cultures of mouse osteoblasts and osteoclasts and in a mouse model of bone loss. Although hbdPRELP had no effect on osteoblasts and other cell types, it impaired osteoclastogenesis and bone resorption by a mechanism requiring its internalization, translocation to the nucleus, and inhibition of the transcription factor nuclear factor κB (NF-κB).
Results
Effect of hbdPRELP on osteoclastogenesis and bone resorption
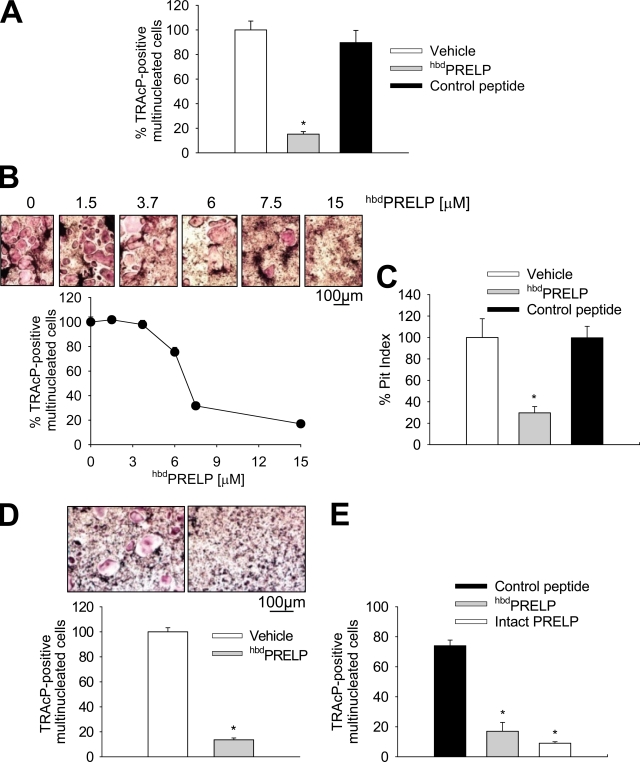
In vitro osteoclastogenesis assays showed that hbdPRELP but not our control heparin-binding peptide remarkably reduced osteoclast formation from unfractionated bone marrow cells treated with 1,25(OH)2VitaminD3 (Fig. 1 A). The hbdPRELP effect was concentration dependent, with a calculated IC50 of 7.3 µM and a narrow range of optimal concentrations (Fig. 1 B). Consistent with the impairment of osteoclast generation, hbdPRELP significantly reduced pit number (Fig. 1 C). Furthermore, hbdPRELP appeared to have a direct effect on the osteoclast lineage, as demonstrated by the inhibition of osteoclastogenesis in purified bone marrow macrophage cultures treated with macrophage colony-stimulating factor (M-CSF) and receptor activator of NF-κB (RANK) ligand (RANKL; Fig. 1 D). Notably, this effect was observed also using the intact PRELP protein, which showed a potency similar to that of the hbdPRELP peptide (Fig. 1 E). Finally, in osteoclasts previously differentiated by treatment with M-CSF and RANKL and then transferred onto bone slices and subsequently treated with hbdPRELP for 48 h, the peptide failed to affect pit formation, suggesting that the major effect is exerted on the mechanism of osteoclast formation rather than on that of bone resorption (Fig. S1 A).
Figure 1.
Effect of hbdPRELP on osteoclastogenesis and bone resorption. (A and B) Unfractionated mouse bone marrow cells were incubated in the presence of 1,25(OH)2VitaminD3 with vehicle, 15 µM hbdPRELP, or 15 µM of control peptide (A) or with the indicated concentrations of hbdPRELP (B). Osteoclasts were then stained for TRAcP, enumerated, and expressed as the percentage of vehicle treated. (C) Pit index of cells cultured as in A but onto bone slices. (D) Bone marrow macrophages were incubated for 6 d with 50 ng/ml M-CSF and 120 ng/ml RANKL plus vehicle or 15 µM hbdPRELP. Osteoclasts were then assessed by TRAcP staining (top) and enumerated (bottom). (E) Bone marrow macrophages were treated with 15 µM of control peptide, 15 µM hbdPRELP, or 15 µM of intact PRELP, and osteoclastogenesis was assessed as described in A. (A–E) Results are the mean ± SEM of three independent experiments (*, P < 0.01).
Effect of hbdPRELP in vivo
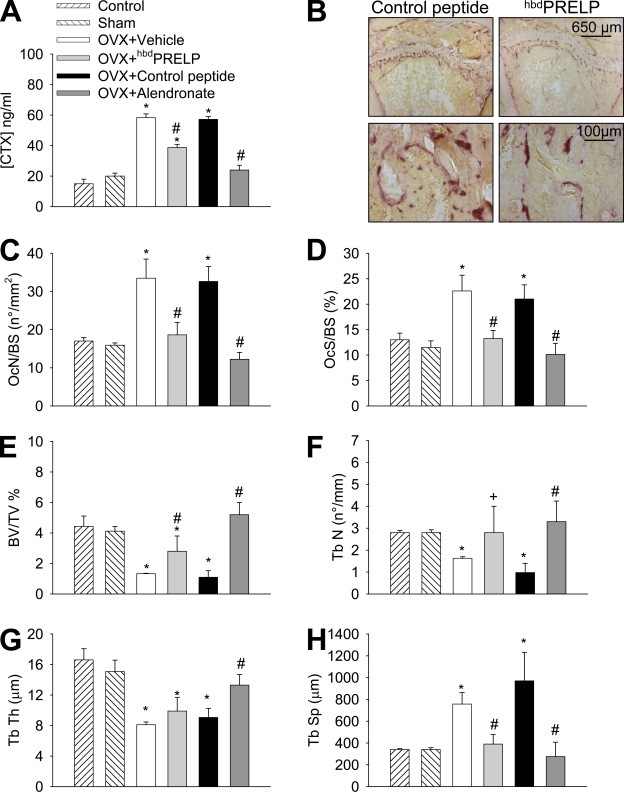
To assess whether hbdPRELP may also have a role in vivo, we increased osteoclast activity and induced bone loss in female mice by ovariectomy (Idris et al., 2008) and treated the animals with vehicle, hbdPRELP, or the control peptide and with the bisphosphonate alendronate as a reference drug. hbdPRELP significantly reduced the ovariectomy-induced increase of the urine bone resorption marker C-terminal collagen I cross-links (CTX; Fig. 2 A). In histological sections of proximal tibia, secondary spongiosa osteoclast number as well as osteoclast surface per bone surface area were decreased in ovariectomized mice treated with hbdPRELP versus vehicle- and control peptide–treated mice (Fig. 2, B–D). Consistently, the analysis of trabecular bone structural parameters (Fig. 2, E–H) showed the ability of hbdPRELP to significantly reduce the ovariectomy-induced bone loss.
Figure 2.
Effect of hbdPRELP in vivo. Ovariectomized (OVX) mice were treated with vehicle, 10 mg/kg body weight of hbdPRELP, or control peptide and 1 mg/kg body weight of alendronate as reference drug 5 d/wk for 5 wk. (A) Quantification of the bone resorption marker CTX in urine samples. (B) Tibial sections stained for TRAcP activity (purple stain) to evidence osteoclasts. (C–H) Measurement of osteoclast number/bone surface (OcN/BS; C), osteoclast surface/bone surface (OcS/BS; D), bone volume/tissue volume (BV/TV; E), trabecular number (Tb N; F), trabecular thickness (Tb Th; G), and trabecular separation (Tb Sp; H). (A and C–H) Data are the mean ± SD of five mice per group (*, P < 0.05 vs. sham; #, P < 0.05 vs. control peptide and vehicle; and +, P < 0.05 vs. control peptide).
Mechanism of action
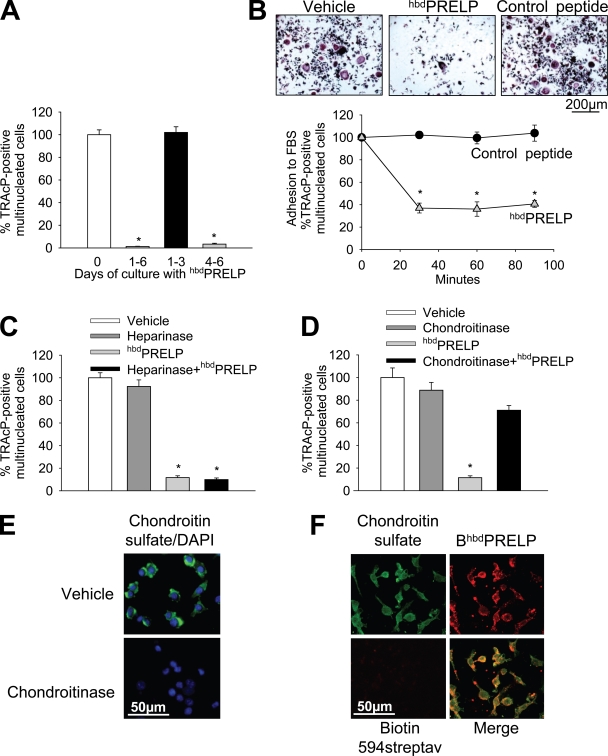
We next sought to establish when the peptide was active during in vitro osteoclast formation. Fig. 3 A shows that hbdPRELP reduced osteoclastogenesis both when given for the entire period of culture and during the last 3 d, whereas it was inactive when administered only throughout the first 3 d. The effect was irreversible in that treatment with 15 µM hbdPRELP for 5 d followed by hbdPRELP withdrawal, and continuation of the culture for a further 5 d did not result in the induction of osteoclastogenesis (unpublished data). Moreover, hbdPRELP significantly reduced adhesion when cells were pretreated in suspension with the peptide before seeding them in the culture dish (Fig. 3 B). This result suggests interference with cell surface molecules, perhaps proteoglycans.
Figure 3.
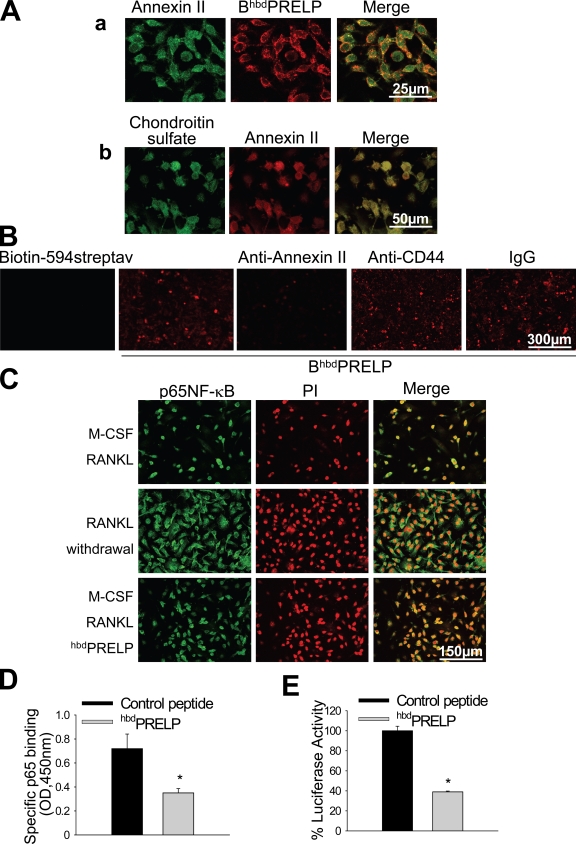
Role of cell surface proteoglycans in hbdPRELP-induced inhibition of osteoclastogenesis. (A) Purified mouse bone marrow macrophages, incubated with M-CSF and RANKL, were treated with vehicle (0) or 15 µM hbdPRELP for the entire time frame of the cultures (1–6 d), for the first 3 d (1–3), or for the last 3 d (4–6). (B) Osteoclast cultures, differentiated by M-CSF and RANKL, were trypsinized. Cells were then pretreated in suspension with vehicle, 15 µM hbdPRELP, or 15 µM of control peptide and allowed to adhere to substrate for the times indicated. Attached cells were stained for TRAcP (top) and enumerated (bottom). (C and D) Prefusion osteoclasts were pretreated with 2 U/ml heparinase III (C) or 0.45 U/ml chondroitinase ABC (D) before the addition of hbdPRELP, and then multinucleated osteoclasts were stained for TRAcP and enumerated. (E) Prefusion osteoclasts were pretreated with vehicle or with 0.45 U/ml chondroitinase ABC, fixed, and incubated with an anti–chondroitin sulfate antibody (green), with no permeabilization to assess the content of cell surface chondroitin sulfate. Cells were also stained with DAPI (blue) to detect nuclei. Pictures are the results of the merge between the two fluorescences. (F) Prefusion osteoclasts were treated with a biotin-tagged hbdPRELP (BhbdPRELP), fixed, permeabilized, and incubated with an anti–chondroitin sulfate antibody to detect colocalization of tagged hbdPRELP with cell surface and vesicular chondroitin sulfate (biotin 594-streptavidin [streptav] was used as a negative control). (A–D) Results are representative or the mean ± SEM of three independent experiments (*, P < 0.001).
Role of cell surface proteoglycans
To address whether cell surface proteoglycans carrying heparan sulfate chains were involved in the inhibitory effect by hbdPRELP on osteoclast formation, we performed osteoclastogenesis assays in the presence of heparinase III. Surprisingly, this treatment was unable to rescue osteoclast formation in the presence of hbdPRELP (Fig. 3 C). Therefore, we next investigated the involvement of cell surface proteoglycans substituted with chondroitin sulfate chains (Deepa et al., 2004). Indeed, we observed that pretreatment of prefusion osteoclast cultures with chondroitinase ABC prevented hbdPRELP from inhibiting osteoclast formation (Fig. 3 D). Confocal microscopy with a specific antibody confirmed that chondroitin sulfate was expressed by prefusion osteoclasts and was removed from the cell surface after treatment with chondroitinase (Fig. 3 E). Moreover, chondroitin sulfate largely colocalized with a hbdPRELP peptide tagged with biotin (BiotinhbdPRELP; Fig. 3 F).
Internalization of hbdPRELP
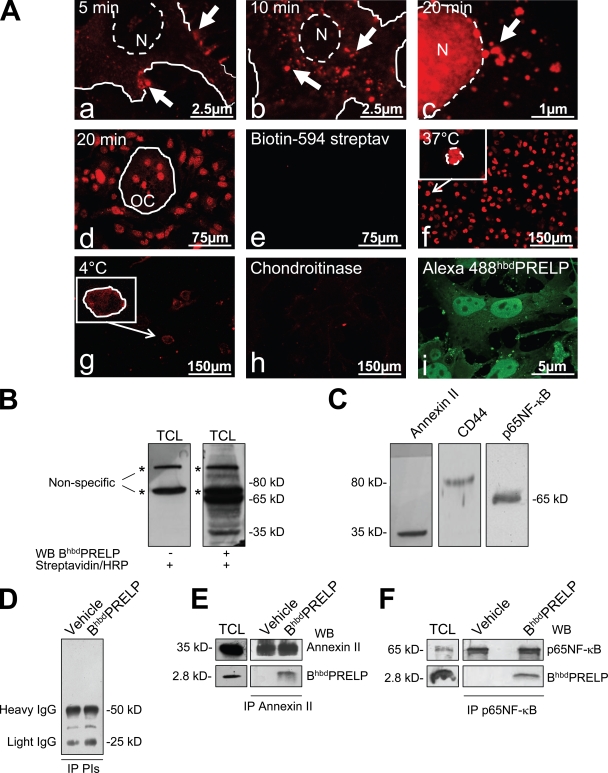
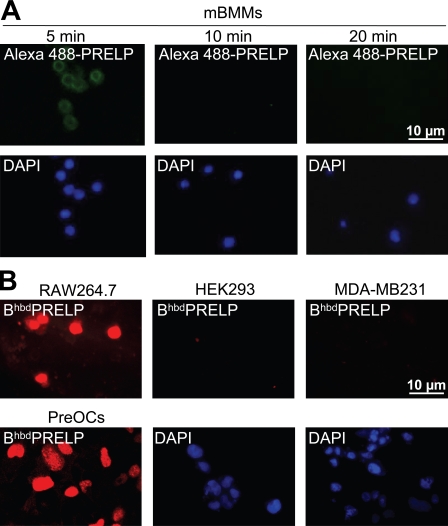
The hbdPRELP sequence contains eight arginine residues in motifs typical of viral proteins prone to be internalized by eukaryotic cells (Futaki et al., 2001). Therefore, we hypothesized that hbdPRELP may be internalized and affect osteoclastogenesis via an intracellular target. To address this aspect, we vitally incubated prefusion osteoclasts with BiotinhbdPRELP and monitored its uptake by confocal microscopy. After 5 min, BiotinhbdPRELP was mostly localized at the cell surface in structures reminiscent of membrane rafts (Fig. 4 A, a), whereas after 10 min, it was internalized in endosomal vesicles (Fig. 4 A, b). Interestingly, after 20 min, BiotinhbdPRELP was mainly found in the nucleus, and several endosomal vesicles appeared in the process of BiotinhbdPRELP transfer to the nuclear compartment (Fig. 4 A, c). Mature multinucleated osteoclasts were also able to internalize BiotinhbdPRELP and retained it in the nuclei after 20 min of vital incubation (Fig. 4 A, d). Control cultures incubated with biotin-594 streptavidin alone were negative (Fig. 4 A, e). Similar internalization was observed with another tagged peptide, Alexa Fluor 488–hbdPRELP (Fig. S1 B).
Figure 4.
Internalization of hbdPRELP and interacting proteins. (A) Vital incubation of prefusion (a–c) or mature mouse osteoclasts (OC; d) with BiotinhbdPRELP for the minutes indicated. Arrows indicate plasma membrane localization (a), localization in endosomal vesicles (b), and localization in vesicles in the vicinity of the nucleus (c). N, nuclei. (e) Negative control in the absence BiotinhbdPRELP. (f–h) Prefusion osteoclasts were incubated with BiotinhbdPRELP for 20 min at 37°C (f) or at 4°C (g) or were pretreated with 0.45 U/ml chondroitinase ABC (h) before the incubation with BiotinhbdPRELP. (f and g) Insets show higher magnification of the framed fields. (i) Fixed and permeabilized prefusion osteoclasts were incubated with Alexa Fluor 488–hbdPRELP. The solid lines indicate the cell surface, and the dashed lines indicate the nuclear envelope. (B) A prefusion osteoclast lysate was subjected to SDS-PAGE and processed with BiotinhbdPRELP (BhbdPRELP) as described in Materials and methods. Asterisks indicate nonspecific bands. (C) The filter shown in B was stripped and Western blotted for annexin II, CD44, and p65NF-κB. (D–F) Immunoprecipitation (IP) of BiotinhbdPRELP-treated prefusion osteoclast lysates with preimmune serum (D), an annexin II antibody (E), or a p65NF-κB antibody (F). Results are representative of three independent experiments. PIs, preimmune serum; TCL, total cell lysate; WB, Western blot.
BiotinhbdPRELP internalization required active cellular processes given that the events observed at 37°C (Fig. 4 A, f) were inhibited when the cells were incubated at 4°C (Fig. 4 A, g). Furthermore, pretreatment with chondroitinase ABC abolished BiotinhbdPRELP membrane binding and internalization (Fig. 4 A, h). Finally, we fixed and permeabilized prefusion osteoclasts and incubated these cells with Alexa Fluor 488–hbdPRELP. Indeed, we observed that fluorescent hbdPRELP not only decorated membrane and intracellular compartments but was also largely localized in the nuclei (Fig. 4 A, i).
Interaction with cellular proteins
To address which candidate protein could interact with BiotinhbdPRELP, we performed SDS-PAGE and blot transfer of untreated prefusion osteoclast total cell lysates, incubated the blots with BiotinhbdPRELP, and visualized the biotin by HRP-conjugated streptavidin and ECL. Indeed, several specific bands were revealed, among which, those most evident had apparent molecular masses of 35 and 65 kD (Fig. 4 B).
One important 35-kD protein known to have a central role in endocytosis is annexin II (Gerke et al., 2005). Indeed, by stripping the membrane and reprobing it with an annexin II antibody, we identified this protein at 35 kD (Fig. 4, compare B [right] with C [left]). CD44 represents another cell surface protein known to have roles in endocytosis in many cell types (Aguiar et al., 1999; Jiang et al., 2002) and in the fusion process of osteoclast precursors (Kania et al., 1997; Sterling et al., 1998; Suzuki et al., 2002). However, reprobing the membrane with an antibody recognizing CD44 revealed a band at 80 kD (Fig. 4 C, middle), which did not correspond to any of the specific BiotinhbdPRELP bands observed in Fig. 4 B.
NF-κB is a transcription factor crucial to osteoclast development and function. One of the components labeled by BiotinhbdPRELP in Fig. 4 B migrated at 65 kD, which corresponds to the molecular mass of the p65NF-κB subunit. Stripping and reprobing the membrane with a p65NF-κB subunit antibody indeed confirmed the presence of this subunit at 65 kD (Fig. 4, compare B [right] with C [right]). To assess specific protein–protein interactions of BiotinhbdPRELP with our candidate proteins, we performed immunoprecipitation assays in prefusion osteoclasts treated with vehicle or with BiotinhbdPRELP, demonstrating that indeed the tagged peptide coimmunoprecipitated in a complex with annexin II (Fig. 4 E) as well as in a complex with p65NF-κB (Fig. 4 F).
Role of annexin II
We next asked what the role could be of annexin II in hbdPRELP activity. Fig. 5 A (a) shows that BiotinhbdPRELP colocalized with annexin II at the cell surface and at endosomes. Interestingly, annexin II also fully colocalized with chondroitin sulfate (Fig. 5 A, b). Indeed, incubation with an anti–annexin II antibody reduced intracellular levels of BiotinhbdPRELP in prefusion osteoclasts vitally treated with the peptide (Fig. 5 B). Consistent with the lack of binding to CD44 (Fig. 4 C), an anti-CD44 antibody failed to affect the ability of prefusion osteoclasts to internalize BiotinhbdPRELP (Fig. 5 B).
Figure 5.
Role of annexin II and NF-κB. (A, a) Prefusion osteoclast incubation with BiotinhbdPRELP (BhbdPRELP; 10 min) and immunofluorescence detection of annexin II. (b) Immunofluorescence detection of chondroitin sulfate and annexin II. (B) Vital incubation of prefusion osteoclasts with BiotinhbdPRELP in the presence of anti-CD44, anti–annexin II antibody, and an irrelevant IgG. Biotin-594 streptavidin (streptav) served as a negative control in the absence of BiotinhbdPRELP. (C) Prefusion osteoclasts, treated as indicated, were incubated with an anti–p65NF-κB antibody followed by incubation with FITC-conjugated secondary antibody (NF-κB; left) and with propidium iodide (PI; middle) to detect nuclei. (D) Colorimetric p65NF-κB DNA binding assay of prefusion osteoclast lysates treated with vehicle or 15 µM hbdPRELP. (E) RAW264.7 cells were treated with control peptide or hbdPRELP and subjected to the luciferase assay as described in Materials and methods. (D and E) Results are the mean ± SEM of three independent experiments (*, P < 0.05).
Role of the NF-κB transcription factor
The transcription factor NF-κB is a crucial determinant of osteoclastogenesis. In prefusion osteoclasts, obtained by incubation of bone marrow macrophages with M-CSF and RANKL for 4 d, p65NF-κB was translocated to the nucleus (Fig. 5 C, top), and, in contrast to RANKL withdrawal (Fig. 5 C, middle), treatment with hbdPRELP did not displace p65NF-κB from this localization (Fig. 5 C, bottom). This suggests that hbdPRELP does not affect p65NF-κB trafficking but may rather affect its transcriptional activity. To test this hypothesis, we evaluated p65NF-κB binding to DNA using a colorimetric assay (TransAM NF-κB p65 kit), which revealed that this binding was reduced by 50% in hbdPRELP-treated prefusion osteoclasts compared with control (Fig. 5 D). Accordingly, luciferase activity assay performed in the immortalized osteoclast precursors cell line RAW264.7, which also internalized BiotinhbdPRELP (see Fig. 8), evidenced a significant reduction of specific p65NF-κB transcriptional activity after treatment with hbdPRELP (Fig. 5 E).
Figure 8.
Specificity of the hbdPRELP effect. (A) Confocal microscopy of mouse bone marrow macrophages (mBMMs) vitally incubated for 5, 10, and 20 min with 15 µM Alexa Fluor 488–hbdPRELP. The bottom panels represent the nuclear DAPI staining of the cells present in the corresponding top panels. (B) Confocal microscopy of mouse osteoclast-like cells (RAW264.7), human epithelial kidney cells (HEK293), human breast cancer cells (MDA-MB231), and mouse prefusion osteoclasts (PreOCs) vitally incubated for 20 min with 15 µM BiotinhbdPRELP as described in A. The bottom middle and right panels represent the nuclear DAPI staining of the cells present in the corresponding top panels. Results are representative of three independent experiments.
Expression of osteoclast genes and activity of signal molecules
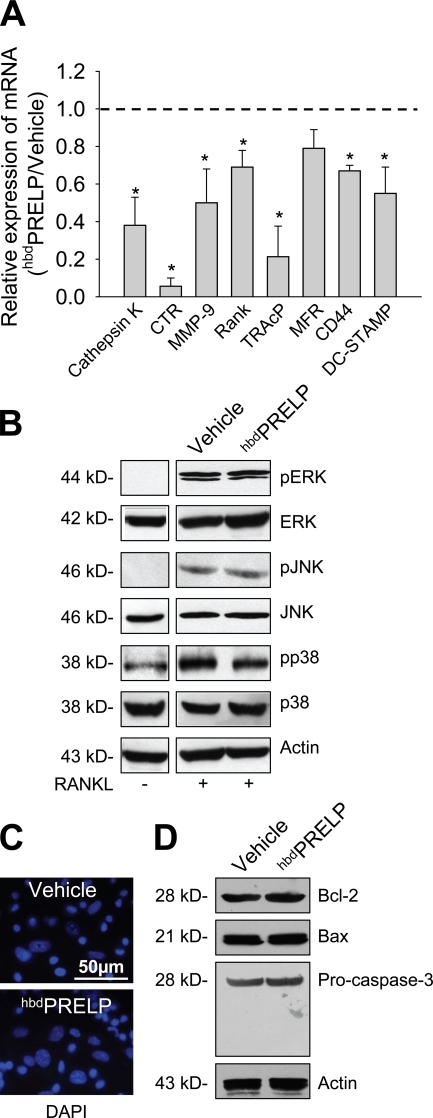
To confirm that NF-κB activity was impaired by the treatment with hbdPRELP, we evaluated the transcriptional expression of downstream osteoclast-specific genes. Real time RT-PCR showed down-regulation of cathepsin K, CTR (calcitonin receptor), MMP-9 (metalloproteinase-9), rank, and TRAcP (tartrate-resistant acid phosphatase) mRNAs (Fig. 6 A). Likewise, hbdPRELP reduced the expression of genes implicated in cell fusion such as DC-STAMP and CD44, with a modest effect also on MFR (macrophage fusion receptor; Fig. 6 A).
Figure 6.
Transcriptional regulation, signaling proteins, and apoptosis. (A) Real-time RT-PCR analyses of mRNA levels in hbdPRELP-treated purified prefusion osteoclasts relative to cultures treated with vehicle (set to 1; dashed line). Data, normalized versus the house-keeping gene Gapdh, are the mean ± SEM of three independent experiments (*, P < 0.05 vs. vehicle). (B) Purified bone marrow macrophages, treated for 4 d with M-CSF, were preincubated for 1 h with vehicle or 15 µM hbdPRELP and then with RANKL for 15 min. Cells were lysed, and Western blot analysis was performed for the indicated MAPKs. ERK, extracellular signal-regulated kinase; p, phospho. (C) Nuclei of prefusion osteoclasts treated with vehicle (top) or with hbdPRELP (bottom) were stained with DAPI to evaluate apoptosis. (D) Western blot analysis of prefusion osteoclasts for the detection of Bcl-2, Bax, and caspase 3 apoptosis-related proteins. Results are representative of three independent experiments.
We next evaluated the effect of hbdPRELP on MAPK signaling. Fig. 6 B shows that this treatment did not prevent RANKL-induced phosphorylation of the MAPKs ERK1/2, JNK, and p38, suggesting no effect of hbdPRELP on immediate cell signaling pathways triggered by RANK activation. hbdPRELP also failed to affect cell survival. In particular, the nuclei of hbdPRELP-treated prefusion osteoclasts, stained with DAPI, showed no nuclear fragmentation or altered morphology, indicating that apoptosis was not induced by treatment with hbdPRELP (Fig. 6 C). Consistently, Western blot analysis showed no changes in protein expression of the prosurvival factor Bcl-2 and of the proapoptotic factor Bax nor activation of procaspase 3 (Fig. 6 D).
Specificity
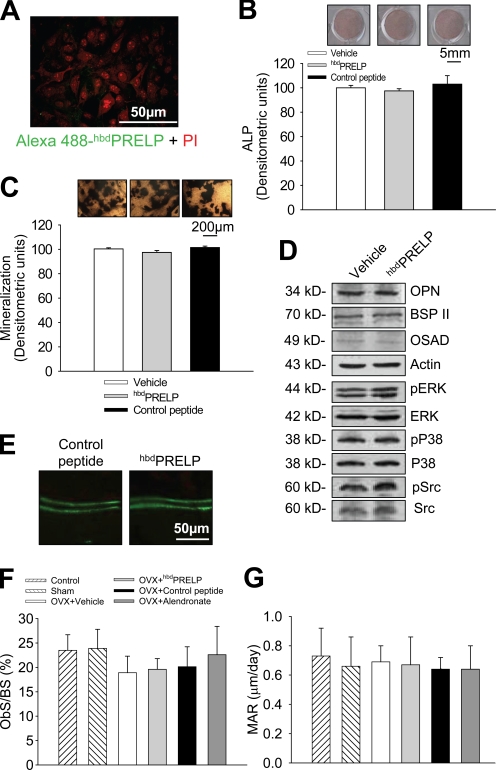
To assess species cross-reactivity, we evaluated the effect of hbdPRELP on mouse, rat, and human osteoclastogenesis. As shown in Fig. S2, prefusion osteoclasts of all three species showed similar nuclear internalization of Alexa Fluor 488–hbdPRELP (left) and inhibition of osteoclast formation (right). Fig. 7 (A–D) shows that hbdPRELP does not affect osteoblast activity. Alexa Fluor 488–hbdPRELP did not vitally bind to calvarial osteoblasts, nor did hbdPRELP affect alkaline phosphatase (ALP) activity, ability to mineralize, production of bone matrix proteins, and signaling protein phosphorylation. Moreover, the expression of genes representing different osteoblast functions was also unchanged (Table S1). Consistently, in vivo hbdPRELP failed to affect the osteoblast parameters in the tibial secondary spongiosa of ovariectomized mice (Fig. 7, E–G).
Figure 7.
Effect of hbdPRELP on osteoblasts. (A) Calvarial osteoblasts were fixed and incubated for 1 h with Alexa Fluor 488–hbdPRELP and propidium iodide (PI). (B) Calvarial osteoblasts were incubated for 4 d with vehicle, 15 µM hbdPRELP, or 15 µM of control peptide. Cells were fixed, and ALP was detected (top) and quantified (bottom). (C) Details of von Kossa staining of mineralized nodules in calvarial osteoblasts treated with vehicle, 15 µM hbdPRELP, or 15 µM of control peptide (top) and relative quantification of mineralization in the whole cultures (bottom). (D) Western blot analysis of calvarial osteoblast lysates for the proteins indicated. (A–D) Data are representative of the mean ± SEM of three independent experiments. ERK, extracellular signal-regulated kinase; OPN, osteopontin; OSAD, osteoadherin; p, phospho. (E–G) Histomorphometric analysis of ovariectomized (OVX) mice treated with vehicle, 10 mg/kg body weight of hbdPRELP or control peptide, and 1 mg/kg body weight of alendronate, as a reference drug, 5 d/wk for 5 wk. (E) Double in vivo calcein labeling. (E–G) Data are representative or the mean ± SD of five mice per group. ObS/BS, osteoblast surface/bone surface; MAR, mineral apposition rate.
To assess hbdPRELP specificity on other cell types, we performed vital incubation of mouse bone marrow macrophages with Alexa Fluor 488–hbdPRELP, which was able to bind the cell surface at 5 min. However, this binding was not followed by peptide internalization and nuclear transfer at later times (Fig. 8 A). In contrast, we observed hbdPRELP internalization in RAW264.7 cells, representing mouse osteoclast precursors (Fig. 8 B). Vital incubation with tagged hbdPRELP of the immortalized cell lines HEK293 (human epithelial kidney) and MDA-MB231 (human breast cancer) also demonstrated no peptide internalization (Fig. 8 B). In addition, the MDA-MB231 cells treated with hbdPRELP showed no changes in proliferation, migration, and invasion ability compared with vehicle-treated cells (Fig. S3), and HEK293 cells subjected to luciferase assay showed no sensitivity of NF-κB transcriptional activity to hbdPRELP treatment (not depicted). These results suggest that hbdPRELP activity is specific only for certain cells such as osteoclasts of various species at particular stages of development.
Discussion
We believe that we have identified a novel mechanism controlling bone homeostasis, in which a domain of an extracellular matrix molecule modulates bone breakdown. The studied basic heparin- and, as identified in this study, chondroitin sulfate–binding N-terminal domain of PRELP was found to block osteoclast formation by a direct mechanism affecting prefusion osteoclasts through inhibition of p65NF-κB transcriptional activity. To the best of our knowledge, the activity of this matrix peptide appears novel, involving chondroitin sulfate chains at the cell surface, and may represent an important new determinant to control bone remodeling and homeostasis. Notably, the intact PRELP had similar effect on osteoclast formation as the hbdPRELP peptide, and the peptide showed antiresorptive activity in vivo. These observations have two important implications: (1) the protein is likely to play a physiological role in the control of bone remodeling, and (2) the peptide has the potential to represent a new antiresorptive biological agent valuable for therapy.
Intact PRELP was similarly active in impairing osteoclastogenesis as the hbdPRELP. We found that a cluster of enzymes, which cut proteoglycans of the cartilage and/or bone matrices, can potentially cleave the intact PRELP (Fig. S4; Breckon et al., 1999; Li et al., 2004; Nakamura et al., 2004; Young et al., 2005; Majumdar et al., 2008). Interestingly, the heparin-binding domain representing our hbdPRELP has no sites of cleavage by these enzymes, thus suggesting that the intact protein could be enzymatically processed and release active peptides encompassing the hbdPRELP sequence, which can then act as physiological regulators of bone resorption. Of course, we cannot rule out that the intact protein can be internalized as such, a circumstance addressable only in a systematic study as the simple use of a tagged intact PRELP cannot distinguish between the internalization of the entire sequence versus that of cleaved peptides.
The level of specificity for the effect of the hbdPRELP is interesting. A different peptide, representing the heparin-binding domain of chondroadherin, unexpectedly turned out to be ineffective on osteoclast development, albeit it bound to heparin and was eluted at higher salt concentrations (∼0.8 M NaCl) compared with the PRELP peptide (∼0.6 M NaCl; unpublished data). Thus, the chondroadherin peptide was chosen as an appropriate control for the specificity of the hbdPRELP peptide for a distinct class of cell surface GAG chains. This control peptide did not bind to the osteoclast precursor cells and did not affect osteoclastogenesis. Therefore, it is likely that the effect of the PRELP peptide is restricted to a limited number of cells with a specific cell surface ligand GAG. The exact specificity of the PRELP peptide for a distinct pattern of sulfation along the chondroitin sulfate chain is not known. In view of previous results showing tightest binding to optimally sulfated heparin (Bengtsson et al., 2000), it is likely that the interaction involves oversulfated disaccharides. Because the variability of the chondroitin sulfate chain is not known, the definition of binding parameters and the identification of the specific chondroitin sulfate chain involved are important tasks for future work.
PRELP is expressed by osteoblasts; however, consistent with its high specificity, at least the hbdPRELP peptide used in this study does not appear to have any autocrine effect on these cells. Similarly, bone marrow macrophages and epithelial cell lines were insensitive to hbdPRELP treatment. In contrast, the peptide directly affected osteoclast precursors at a late stage of differentiation. At this stage, a heparan sulfate proteoglycan, perlecan, not directly associated with the cell surface and with known ability to bind hbdPRELP was scanty or absent albeit expressed at earlier stages (unpublished data). In a previous study, heparan sulfate proteoglycans were found in mature osteoclasts (Nakano et al., 2004), but the lack of effect of heparinase III digestion in the osteoclastogenesis assays rules out a role of this GAG in the hbdPRELP peptide activities on osteoclast development.
However, we believe that our study has identified the mechanisms whereby hbdPRELP impairs osteoclast generation. We found that the peptide recognizes chondroitin sulfate chains of cell surface proteoglycans. In a separate series of experiments, we have used an Akubio RAPid-4 acoustic biosensor to show that chondroitin 6-sulfate (from human nucleus pulposus) indeed binds the BiotinhbdPRELP peptide (unpublished data). Internalization and nuclear localization of BiotinhbdPRELP has now been demonstrated in osteoclasts and depends on chondroitin sulfate because these processes are inhibited by the removal of these chains. Chondroitin sulfate proteoglycans have not been intensely investigated in osteoclasts (Li et al., 2002), and to the best of our knowledge, this is the first indication that they are involved in peptide internalization and endosome formation.
hbdPRELP is a highly cationic peptide with eight arginines distributed between aa 4 and 21, and this number of arginines was observed to be optimal for peptide internalization (Nakase et al., 2008). In fact, highly cationic peptides are able to enter into the cytoplasmic compartment and nucleus of cells from the extracellular environment (Futaki et al., 2002; Kosuge et al., 2008; El-Sayed et al., 2009), although they have no nuclear localization sequence. A recent study analyzing live cells demonstrated that these peptides enter through endocytosis and accumulate in endocytic vesicles without necessarily routing via the cytoplasm (Futaki et al., 2007). Arginine-rich peptides, including a basic peptide segment derived from the HIV-1 Tat protein, are categorized into one of the most frequently used peptide vectors (Futaki et al., 2007), and their uptake is dependent on heparan sulfate and chondroitin sulfate proteoglycans (Nakase et al., 2007). This is in line with our study and corroborates our observations that cell surface chondroitin sulfate chains are indispensable for the mechanism of action of hbdPRELP in osteoclasts.
In contrast, BiotinhbdPRELP internalization was totally independent of CD44 activity. This type I transmembrane glycoprotein is expressed by the osteoclast lineage, is involved in fusion of macrophages, and binds several matrix components, including hyaluronan, and its occupancy by matrix components prevents the formation of polykaria (Sterling et al., 1998). However, CD44 mRNA expression is impaired by hbdPRELP treatment, thus contributing to its inhibitory effect through a transcriptional route.
Importantly, our peptide appears to colocalize with annexin II, forming a complex indispensable for internalization in endosomes and transfer to the nucleus. Annexin II is a calcium-dependent phospholipid-binding protein that is involved in early endosomal organization (Harder et al., 1997). Menaa et al. (1999) suggested that annexin II stimulates osteoclastogenesis and bone resorption by activating T cells through a putative receptor to secreted granulocyte M-CSF. In contrast, in our study, we found a pivotal role for this protein in the inhibitory effect of hbdPRELP on osteoclast formation through its indispensible ability to cause peptide internalization. This discrepancy does not subtract from the results as in the two studies, the cell types and the phases of osteoclastogenesis in which annexin II was involved were different and the physiological control of bone resorption is known to require a balance between stimulatory and inhibitory stimuli. Collectively, our results point to chondroitin sulfate cell surface proteoglycans and annexin II as crucial determinants bringing about BiotinhbdPRELP endocytosis and trafficking.
A key element of our findings seems to be the ability of hbdPRELP to reach the nuclear compartment and form a complex with the p65 subunit of the transcription factor NF-κB. In bone marrow macrophages, p65NF-κB is translocated to the nucleus after RANK activation by RANKL. RANK appears in late osteoclast precursors (Teitelbaum and Ross, 2003), which is consistent with our observation that hbdPRELP is active only in late stages of osteoclastogenesis. Indeed, in our experimental conditions, BiotinhbdPRELP and p65NF-κB physically interact with a consequent reduction of NF-κB activity. This transcription factor is central to the osteoclastogenic process, and deletion of NF-κB subunits leads to blockage of osteoclast formation (Franzoso et al., 1997). Therefore, our observation favors the involvement of NF-κB in the mechanism whereby hbdPRELP blocks osteoclastogenesis. Consistently, no sign of apoptosis was observed in prefusion osteoclasts treated with hbdPRELP. This observation is coherent with the results by Franzoso et al. (1997), showing that p52 and p50NF-κB double gene inactivation in mice does not cause osteoclast death but rather halts the development of osteoclast precursors. It is interesting to note that treatment of prefusion osteoclasts with hbdPRELP does not affect RANKL-dependent MAPK phosphorylation, suggesting that the impairment of immediate cell signaling is not involved in its mechanism of action. It is also interesting that the suppression of NF-κB to around 50% was sufficient to strongly suppress osteoclastic differentiation, likely reflecting the fact that massive NF-κB activity is required for full induction of osteoclast formation. Furthermore, the PRELP peptide could exert additional roles at the cell surface by stimulating interactions with other cell surface molecules.
Notwithstanding the clear effect of hbdPRELP on osteoclast formation, we did not find any direct effect on in vitro bone resorption, as demonstrated in osteoclasts allowed to differentiate normally and then transferred onto bone slices and treated with hbdPRELP. In this regard, it has to be pointed out that, although the pivotal role of RANKL-induced NF-κB signaling is well established for osteoclast formation, the effect of this signaling on the mechanism of bone resorption remains to be fully elucidated (Teitelbaum and Ross, 2003).
In conclusion, the heparin-binding domain of PRELP is a novel direct negative regulator of osteoclast generation, which is also effective in vivo. It inhibits NF-κB signaling in late-stage prefusion committed osteoclast precursors, with no effect on osteoblasts and other cell types tested. These observations could open new avenues for the understanding of the biology of osteoclasts and the involvement of matrix components in the regulation of bone development and remodeling and could also have implications for the treatment of bone diseases.
Materials and methods
Peptides
Intact PRELP was extracted and purified from bovine nasal cartilage (Bengtsson et al., 2000). A synthetic human peptide (NH2-QPTRRPRPGTGPGRRPRPRPRPTPC-COOH) corresponding to the 24 N-terminal aa of PRELP and representing the active part of the heparin-binding domain (hbdPRELP) was synthesized by Schafer-N with an additional cysteine residue in its C-terminal end. One preparation of the peptide was synthesized with a biotin at the C-terminal cysteine (BiotinhbdPRELP), whereas another preparation was reacted with Alexa Fluor 488 C5 maleimide (Alexa Fluor 488–hbdPRELP; Invitrogen) according to the manufacturer’s instructions. The degree of derivatization was estimated to be in excess of 90% and was verified by using MALDI-TOF (matrix-assisted laser desorption/ionization time of flight) mass spectrometry. In pilot experiments, we tested a different heparin-binding peptide from chondroadherin of 14 aa (NH2-CKFPTKRSKKAGRH-COOH), corresponding to the C-terminal part of chondroadherin. This peptide turned out to be ineffective on osteoclast development and was used as a control.
Animals
Procedures involving animals and their care were conducted in conformity with national and international laws and policies (European Economic Community Council Directive 86/609, OJ L 358, 1, December 12, 1987; Italian Legislative Decree 116/92, Gazzetta Ufficiale della Repubblica Italiana no. 40, February 18, 1992; National Institutes of Health guide for the Care and Use of Laboratory Animals, National Institutes of Health Publication no. 85-23, 1985) and were approved by the Institutional Review Board of the University of L’Aquila.
In vivo study
8-wk-old female C57Bl6/J mice were ovariectomized and, after 3 d, were treated with vehicle (PBS), control peptide (10 mg/kg body weight), hbdPRELP (10 mg/kg body weight), or alendronate (1 mg/kg body weight) by i.p. injection 5 d/wk for 5 wk (number of animals/group = 5). At the end of the experiment, urine samples were collected for detection of CTX assay by the Ratlaps ELISA kit (IDS Nordic Bioscience) according to the manufacturer’s instructions. Animals were then sacrificed, and the hindlimb long bones were removed and fixed in 4% paraformaldehyde.
Histomorphometric analysis
Tibiae, embedded in glycol-methacrylate (Technovit 9100 New; Heraeus Kulzer GmbH), were sectioned longitudinally through the frontal plate, and sections (2 µm thick) were subjected to TRAcP staining. Osteoclast number/bone surface (number/square millimeters), osteoclast surface/bone surface (percentage), osteoblast surface/bone surface (percentage), and structural parameters, including bone volume/total volume (percentage), trabecular number (number/millimeter), trabecular thickness (micrometers), and trabecular separation (micrometers), were measured in a metaphyseal region extending at least 100 µm away from the distal end of the growth plate and excluding the endocortical surfaces (secondary spongiosa; Parfitt et al., 1987; Marzia et al., 2000). Dynamic assessment of the mineral apposition rate was calculated after double injection of calcein 10 and 3 d before animal sacrifice. Histomorphometric measurements were performed with an interactive image analysis system (IAS 2000; Delta Sistemi) consisting of a color video–equipped computer linked to the microscope by a video camera.
Osteoclast cultures
Bone marrow was flushed from the long bones of 7-d-old CD1 mice. Cells were recovered and cultured in DME plus 10% FBS up to 6 d in the presence of 10−8 M 1,25(OH)2VitaminD3. To obtain purified bone marrow macrophages, bone marrow cells from 7-d-old mice or rats were diluted 1:1 in Hanks’ balanced salt solution, layered over Histopaque 1077, and centrifuged at 400 g for 30 min. Cells recovered were resuspended in DME containing 10% FBS and plated. After 3 h, cultures were extensively washed to remove nonadherent cells, and then DME supplemented with 10% FBS, 50 ng/ml M-CSF, and 120 ng/ml RANKL was added to adherent cells to induce osteoclastogenesis. Human osteoclasts were differentiated from the peripheral blood mononuclear cells. In brief, diluted blood (1:1 in Hanks’ solution) was layered over Ficoll/Histopaque 1077 solution and centrifuged at 400 g for 30 min. Buffy-coat cells thus isolated were washed twice with Hanks’ solution, resuspended in DME containing 10% FBS, and plated. After 3 h, cells were rinsed to remove nonadherent cells and cultured in the same medium in the presence of 50 ng/ml M-CSF and 30 ng/ml RANKL. Unless otherwise stated, treatment of osteoclast cultures with hbdPRELP or control peptide started the third day of culture, affecting only adherent cells referred to as prefusion osteoclasts, which are defined as TRAcP-positive mononuclear cells. The peptide was replaced at each change of medium.
Enzymatic treatments
To assess the involvement of cell surface proteoglycans carrying heparin sulfate or chondroitin sulfate chains in the inhibitory effect of PRELP, osteoclastogenesis was performed in the presence of 2 U/ml heparinase III or 0.45 U/ml chondroitinase ABC, respectively. The enzymes were replaced at each change of medium. At the end of the experiment, enzymatic activity was terminated by extensive washing of the cultures, which were then fixed in 4% paraformaldehyde.
TRAcP activity
Cells were fixed in 4% paraformaldehyde, and then TRAcP activity was evaluated histochemically using the Sigma-Aldrich kit #386 according to the manufacturer’s instruction.
Bone resorption
Osteoclasts were differentiated as described in the Osteoclast cultures section onto bone slices or were differentiated in plastic dishes, detached by trypsin procedure, and cultured onto bone slices for 48 h. Slices were then fixed in 4% paraformaldehyde, ultrasonicated in 1% sodium hypochlorite to remove the cells, and stained with 1% toluidine blue. Pit index was computed according to Caselli et al. (1997). In brief, the resorption pits were divided in three visual categories according to their diameter: small, <10 µm; medium, 10–30 µm; and large, >30 µm. The numbers of pits per each category were scored by multiplying by a different factor according to their dimensions: for small pits, 0.3; for medium pits, 1; and for large pits, 3. The sum of the three scores gave the pit index.
Osteoblast cultures
Calvariae were removed from 7-d-old CD1 mice and digested three times with 1 mg/ml Clostridium histolyticum type IV collagenase and 0.25% trypsin for 20 min at 37°C. Cells from the second and third digestions were grown in DME plus 10% FBS and checked for the expression of the osteoblast markers ALP, Runx2 (Runt-related transcription factor-2), PTHrP (parathyroid hormone/parathyroid hormone–related peptide receptor), type I collagen, and osteocalcin (Marzia et al., 2000).
ALP activity
Osteoblasts were fixed in 4% paraformaldehyde, and then ALP activity was evaluated histochemically using the Sigma-Aldrich kit #85 according to the manufacturer’s instruction.
Mineralization assay
Osteoblast standard medium was supplemented with 10 mM β-glycerophosphate and 50 µg/ml ascorbate. Osteoblasts were cultured for 3 wk before characterization of mineralization by von Kossa staining.
Adhesion assay
Osteoclast-enriched bone marrow cultures were trypsinized and treated in suspension for 30 min with vehicle alone or with 15 µM peptide in FBS-free DME. Cells were then plated in wells coated with 20% FBS. At the end of the incubation, attached cells were fixed and subjected to TRAcP histochemical staining. Multinucleated TRAcP-positive osteoclasts were then enumerated.
Real-time RT-PCR
Total RNA was extracted using the TRIZOL, and then 1 µg was reverse transcribed, and the equivalent of 0.1 µg was used for the PCR reactions using the Brilliant SYBR green QPCR master mix (Agilent Technologies). Primer sequences and real-time conditions are listed in Table S2.
Immunoprecipitation and Western blotting
Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer, and protein content was measured by the Bradford method. For immunoprecipitation, 5 µg of specific antibodies or preimmune serum was incubated for 2 h at 4°C with protein G–conjugated agarose beads. After five washes in RIPA buffer, 1 mg of protein of each sample was added and incubated overnight at 4°C. Samples were then washed with RIPA buffer, resuspended in 2× reducing Laemmli sample buffer, and boiled before SDS-PAGE.
For Western blots, proteins resolved on a 10% SDS-PAGE were trans-blotted to nitrocellulose membranes and probed with the primary antibody for 1 h at room temperature, washed, and incubated with the appropriate HRP-conjugated secondary antibody for 1 h at room temperature. In Fig. 4 B, blots were first incubated with BiotinhbdPRELP and then washed and incubated with streptavidin/HRP. Protein bands were revealed by ECL.
p65NF-κB binding activity
p65NF-κB binding to DNA was evaluated using the ELISA TransAM NF-κB p65 kit #40096 (Active Motif) according to the manufacturer’s instructions. In brief, the kit consists of a 96-well plate with an immobilized oligonucleotide containing a p65NF-κB consensus binding site. The activated p65NF-κB contained in cell extracts specifically bound to this oligonucleotide, and then the complex was detected by an antibody directed against the p65NF-κB subunit. The addition of an HRP-conjugated secondary antibody provided sensitive colorimetric readout.
Luciferase assay
RAW264.7 cells were transfected with 1.35 µg of pNF-κB–Luc vector (Takara Bio Inc.), 1.35 µg of pMT2T-p65 vector, and 0.3 µg of pRL-TK vector using Lipofectamine 2000 (Invitrogen). After 36 h, cells were harvested, and lysates were incubated with the Luciferase Assay Substrate by using the Dual-Luciferase Reporter Assay system (Promega) according to the manufacturer’s instruction. Firefly luciferase activity was normalized to Renilla luciferase activity.
Proliferation, migration, and invasion assays
Proliferation of MDA-MB231 cells was evaluated using the CellTiter 96 Aqueous One Solution Cell Proliferation Assay (MTS) from Promega. Migration assay was performed by the modified Boyden chamber method (Albini et al., 1987). Polycarbonate filters were coated with 45 µg/cm2 gelatin in the upper compartment of the trans-well chambers, and MDA-MB231 cells were added and allowed to migrate for 12 h in the presence of NIH3T3 cell–conditioned media. Filters were then stained with hematoxylin/eosin. Invasion assay was performed in a similar manner except that the filters were coated with 35 µg/cm2 of reconstituted matrigel and processed after 24 h.
Fluorescence and confocal microscopy
Cells were fixed with 4% paraformaldehyde and incubated for 1 h at room temperature with Alexa Fluor 488–conjugated hbdPRELP (Alexa Fluor 488–hbdPRELP) or primary antibodies followed by FITC- or TRITC-conjugated secondary antibody. Cells were also incubated vitally with BiotinhbdPRELP, which was revealed by fluorescent biotin 594-streptavidin. To detect nuclei, cells were stained with DAPI (blue fluorescence) or with propidium iodide (red fluorescence). Cells were then observed at room temperature by conventional epifluorescence using a microscope (Axioplan; Carl Zeiss, Inc.) or by confocal microscopy using a confocal microscope (FluoView IX81 FVBF; Olympus). For fluorescence microscopy, we used 2.5× NA 0.075, 10× NA 0.30, 20× NA 0.5, and 40× NA 0.75 Plan-Neofluar objective lenses. Images were captured with a camera (AxioCam MRC5; Carl Zeiss, Inc.) using the AxioVs 40 version 4.7.1.0 software (Carl Zeiss, Inc.). For confocal microscopy, we used 10× NA 0.30 and 40× NA 0.85 UPlan-Apochromat or 60× NA 1.4 oil Plan-Apochromat objective lenses. Images were captured using FluoView 500 software (Olympus).
Digital images
Light, fluorescence, and confocal microscopy pictures were captured as JPEG files as specified in the previous section. Western blots images were captured by the Molecular Analyst software for the model 670 scanning densitometer (Bio-Rad Laboratories) as JPEG files. The areas of interest were selected and reproduced for documentation by Photoshop version 6 or 7 software (Adobe).
Statistics
All experiments were performed in triplicates and repeated at least three times. Data are expressed as the mean ± SEM. Statistical analysis was performed by one-way analysis of variance, followed by unpaired Student’s t test. A p-value of <0.05 was conventionally considered statistically significant.
Online supplemental material
Fig. S1 shows the effect of hbdPRELP on bone resorption and evaluation of its internalization. Fig. S2 shows the effect of hbdPRELP on mice, rat, and human osteoclast formation. Fig. S3 shows the effect of hbdPRELP on MDA-MB231 cell proliferation, migration, and invasion. Fig. S4 contains a schematic representation of the intact PRELP protein. Table S1 shows the effect of hbdPRELP on the transcriptional expression of osteoblast genes. Table S2 lists the primer sequences and real-time conditions used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200906014/DC1.
Acknowledgments
We are indebted with Dr. Rita Di Massimo for her excellent assistance in writing this manuscript.
This work was supported by the European Commission grant OSTEOGENE (contract no. LSHM-CT-2003-502941) to A. Teti and D. Heinegård, by the Swedish Research Council and King Gustaf V's 80-year fund to D. Heinegård, and by grants from the Italian Association for Cancer Research and from the Swiss Bridge Foundation to A. Teti. N. Rucci is the recipient of the Robert Schenk Research Prize 2009. M. Capulli is the recipient of a Federazione Italiana della Ricerca sul Cancro fellowship. A. Del Fattore is the recipient of a European Calcified Tissue Society/Amgen fellowship.
Footnotes
Abbreviations used in this paper:
- ALP
- alkaline phosphatase
- CTX
- C-terminal collagen I cross-links
- GAG
- glycosaminoglycan
- LRR
- leucine-rich repeat
- M-CSF
- macrophage colony-stimulating factor
- NF-κB
- nuclear factor κB
- PRELP
- proline/arginine-rich end LRR protein
- RANK
- receptor activator of NF-κB
- RANKL
- RANK ligand
- RIPA
- radioimmunoprecipitation assay
- SLRRP
- small LRR protein
- TRAcP
- tartrate-resistant acid phosphatase
References
- Aguiar D.J., Knudson W., Knudson C.B. 1999. Internalization of the hyaluronan receptor CD44 by chondrocytes. Exp. Cell Res. 252:292–302 10.1006/excr.1999.4641 [DOI] [PubMed] [Google Scholar]
- Albini A., Iwamoto Y., Kleinman H.K., Martin G.R., Aaronson S.A., Kozlowski J.M., McEwan R.N. 1987. A rapid in vitro assay for quantitating the invasive potential of tumor cells. Cancer Res. 47:3239–3245 [PubMed] [Google Scholar]
- Bengtsson E. 1999. Structure and interactions of the extracellular matrix protein PRELP. PhD thesis. Lund University, S-221 00 Lund, Sweden: 163 pp [Google Scholar]
- Bengtsson E., Neame P.J., Heinegård D., Sommarin Y. 1995. The primary structure of a basic leucine-rich repeat protein, PRELP, found in connective tissues. J. Biol. Chem. 270:25639–25644 10.1074/jbc.270.43.25639 [DOI] [PubMed] [Google Scholar]
- Bengtsson E., Aspberg A., Heinegård D., Sommarin Y., Spillmann D. 2000. The amino-terminal part of PRELP binds to heparin and heparan sulfate. J. Biol. Chem. 275:40695–40702 10.1074/jbc.M007917200 [DOI] [PubMed] [Google Scholar]
- Bengtsson E., Mörgelin M., Sasaki T., Timpl R., Heinegård D., Aspberg A. 2002. The leucine-rich repeat protein PRELP binds perlecan and collagens and may function as a basement membrane anchor. J. Biol. Chem. 277:15061–15068 10.1074/jbc.M108285200 [DOI] [PubMed] [Google Scholar]
- Breckon J.J., Papaioannou S., Kon L.W., Tumber A., Hembry R.M., Murphy G., Reynolds J.J., Meikle M.C. 1999. Stromelysin (MMP-3) synthesis is up-regulated in estrogen-deficient mouse osteoblasts in vivo and in vitro. J. Bone Miner. Res. 14:1880–1890 10.1359/jbmr.1999.14.11.1880 [DOI] [PubMed] [Google Scholar]
- Caselli G., Mantovanini M., Gandolfi C.A., Allegretti M., Fiorentino S., Pellegrini L., Melillo G., Bertini R., Sabbatini W., Anacardio R., et al. 1997. Tartronates: a new generation of drugs affecting bone metabolism. J. Bone Miner. Res. 12:972–981 10.1359/jbmr.1997.12.6.972 [DOI] [PubMed] [Google Scholar]
- Deepa S.S., Yamada S., Zako M., Goldberger O., Sugahara K. 2004. Chondroitin sulfate chains on syndecan-1 and syndecan-4 from normal murine mammary gland epithelial cells are structurally and functionally distinct and cooperate with heparan sulfate chains to bind growth factors. A novel function to control binding of midkine, pleiotrophin, and basic fibroblast growth factor. J. Biol. Chem. 279:37368–37376 10.1074/jbc.M403031200 [DOI] [PubMed] [Google Scholar]
- El-Sayed A., Futaki S., Harashima H. 2009. Delivery of macromolecules using arginine-rich cell-penetrating peptides: ways to overcome endosomal entrapment. AAPS J. 11:13–22 10.1208/s12248-008-9071-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franzoso G., Carlson L., Xing L., Poljak L., Shores E.W., Brown K.D., Leonardi A., Tran T., Boyce B.F., Siebenlist U. 1997. Requirement for NF-kappaB in osteoclast and B-cell development. Genes Dev. 11:3482–3496 10.1101/gad.11.24.3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futaki S., Suzuki T., Ohashi W., Yagami T., Tanaka S., Ueda K., Sugiura Y. 2001. Arginine-rich peptides. An abundant source of membrane-permeable peptides having potential as carriers for intracellular protein delivery. J. Biol. Chem. 276:5836–5840 10.1074/jbc.M007540200 [DOI] [PubMed] [Google Scholar]
- Futaki S., Nakase I., Suzuki T., Youjun Z., Sugiura Y. 2002. Translocation of branched-chain arginine peptides through cell membranes: flexibility in the spatial disposition of positive charges in membrane-permeable peptides. Biochemistry. 41:7925–7930 10.1021/bi0256173 [DOI] [PubMed] [Google Scholar]
- Futaki S., Nakase I., Tadokoro A., Takeuchi T., Jones A.T. 2007. Arginine-rich peptides and their internalization mechanisms. Biochem. Soc. Trans. 35:784–787 10.1042/BST0350784 [DOI] [PubMed] [Google Scholar]
- Gerke V., Creutz C.E., Moss S.E. 2005. Annexins: linking Ca2+ signalling to membrane dynamics. Nat. Rev. Mol. Cell Biol. 6:449–461 10.1038/nrm1661 [DOI] [PubMed] [Google Scholar]
- Grover J., Lee E.R., Mounkes L.C., Stewart C.L., Roughley P.J. 2007. The consequence of PRELP overexpression on skin. Matrix Biol. 26:140–143 10.1016/j.matbio.2006.10.005 [DOI] [PubMed] [Google Scholar]
- Harder T., Kellner R., Parton R.G., Gruenberg J. 1997. Specific release of membrane-bound annexin II and cortical cytoskeletal elements by sequestration of membrane cholesterol. Mol. Biol. Cell. 8:533–545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinegård D., Aspberg A., Franzén A., Lorenzo P. 2002. Glycosylated matrix proteins. Connective Tissue and Its Heritable Disorders: Molecular, Genetic, and Medical Aspects. Second edition Royce P.M., Steinmann B., John Wiley & Sons Inc, New York: 271–291 [Google Scholar]
- Hennekam R.C. 2006. Hutchinson-Gilford progeria syndrome: review of the phenotype. Am. J. Med. Genet. A. 140:2603–2624 [DOI] [PubMed] [Google Scholar]
- Idris A.I., Sophocleous A., Landao-Bassonga E., van't Hof R.J., Ralston S.H. 2008. Regulation of bone mass, osteoclast function, and ovariectomy-induced bone loss by the type 2 cannabinoid receptor. Endocrinology. 149:5619–5626 10.1210/en.2008-0150 [DOI] [PubMed] [Google Scholar]
- Iozzo R.V., Murdoch A.D. 1996. Proteoglycans of the extracellular environment: clues from the gene and protein side offer novel perspectives in molecular diversity and function. FASEB J. 10:598–614 [PubMed] [Google Scholar]
- Jiang H., Peterson R.S., Wang W., Bartnik E., Knudson C.B., Knudson W. 2002. A requirement for the CD44 cytoplasmic domain for hyaluronan binding, pericellular matrix assembly, and receptor-mediated endocytosis in COS-7 cells. J. Biol. Chem. 277:10531–10538 10.1074/jbc.M108654200 [DOI] [PubMed] [Google Scholar]
- Kania J.R., Kehat-Stadler T., Kupfer S.R. 1997. CD44 antibodies inhibit osteoclast formation. J. Bone Miner. Res. 12:1155–1164 10.1359/jbmr.1997.12.8.1155 [DOI] [PubMed] [Google Scholar]
- Kosuge M., Takeuchi T., Nakase I., Jones A.T., Futaki S. 2008. Cellular internalization and distribution of arginine-rich peptides as a function of extracellular peptide concentration, serum, and plasma membrane associated proteoglycans. Bioconjug. Chem. 19:656–664 10.1021/bc700289w [DOI] [PubMed] [Google Scholar]
- Lewis M. 2003. PRELP, collagen, and a theory of Hutchinson-Gilford progeria. Ageing Res. Rev. 2:95–105 10.1016/S1568-1637(02)00044-2 [DOI] [PubMed] [Google Scholar]
- Li Z., Hou W.S., Escalante-Torres C.R., Gelb B.D., Bromme D. 2002. Collagenase activity of cathepsin K depends on complex formation with chondroitin sulfate. J. Biol. Chem. 277:28669–28676 10.1074/jbc.M204004200 [DOI] [PubMed] [Google Scholar]
- Li J., Liao E.Y., Dai R.C., Wei Q.Y., Luo X.H. 2004. Effects of 17 beta-estradiol on the expression of interstitial collagenases-8 and -13 (MMP-8 and MMP-13) and tissue inhibitor of metalloproteinase-1 (TIMP-1) in ovariectomized rat osteoblastic cells. J. Mol. Histol. 35:723–731 10.1007/s10735-004-6206-3 [DOI] [PubMed] [Google Scholar]
- Majava M., Bishop P.N., Hägg P., Scott P.G., Rice A., Inglehearn C., Hammond C.J., Spector T.D., Ala-Kokko L., Männikkö M. 2007. Novel mutations in the small leucine-rich repeat protein/proteoglycan (SLRP) genes in high myopia. Hum. Mutat. 28:336–344 10.1002/humu.20444 [DOI] [PubMed] [Google Scholar]
- Majumdar M.K., Chockalingam P.S., Bhat R.A., Sheldon R., Keohan C., Blanchet T., Glasson S., Morris E.A. 2008. Immortalized mouse articular cartilage cell lines retain chondrocyte phenotype and respond to both anabolic factor BMP-2 and pro-inflammatory factor IL-1. J. Cell. Physiol. 215:68–76 10.1002/jcp.21282 [DOI] [PubMed] [Google Scholar]
- Marzia M., Sims N.A., Voit S., Migliaccio S., Taranta A., Bernardini S., Faraggiana T., Yoneda T., Mundy G.R., Boyce B.F., et al. 2000. Decreased c-Src expression enhances osteoblast differentiation and bone formation. J. Cell Biol. 151:311–320 10.1083/jcb.151.2.311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menaa C., Devlin R.D., Reddy S.V., Gazitt Y., Choi S.J., Roodman G.D. 1999. Annexin II increases osteoclast formation by stimulating the proliferation of osteoclast precursors in human marrow cultures. J. Clin. Invest. 103:1605–1613 10.1172/JCI6374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura H., Sato G., Hirata A., Yamamoto T. 2004. Immunolocalization of matrix metalloproteinase-13 on bone surface under osteoclasts in rat tibia. Bone. 34:48–56 10.1016/j.bone.2003.09.001 [DOI] [PubMed] [Google Scholar]
- Nakano K., Okada Y., Saito K., Tanaka Y. 2004. Induction of RANKL expression and osteoclast maturation by the binding of fibroblast growth factor 2 to heparan sulfate proteoglycan on rheumatoid synovial fibroblasts. Arthritis Rheum. 50:2450–2458 10.1002/art.20367 [DOI] [PubMed] [Google Scholar]
- Nakase I., Tadokoro A., Kawabata N., Takeuchi T., Katoh H., Hiramoto K., Negishi M., Nomizu M., Sugiura Y., Futaki S. 2007. Interaction of arginine-rich peptides with membrane-associated proteoglycans is crucial for induction of actin organization and macropinocytosis. Biochemistry. 46:492–501 10.1021/bi0612824 [DOI] [PubMed] [Google Scholar]
- Nakase I., Takeuchi T., Tanaka G., Futaki S. 2008. Methodological and cellular aspects that govern the internalization mechanisms of arginine-rich cell-penetrating peptides. Adv. Drug Deliv. Rev. 60:598–607 10.1016/j.addr.2007.10.006 [DOI] [PubMed] [Google Scholar]
- Parfitt A.M., Drezner M.K., Glorieux F.H., Kanis J.A., Malluche H., Meunier P.J., Ott S.M., Recker R.R. 1987. Bone histomorphometry: standardization of nomenclature, symbols, and units. Report of the ASBMR Histomorphometry Nomenclature Committee. J. Bone Miner. Res. 2:595–610 [DOI] [PubMed] [Google Scholar]
- Reardon A.J., Le Goff M., Briggs M.D., McLeod D., Sheehan J.K., Thornton D.J., Bishop P.N. 2000. Identification in vitreous and molecular cloning of opticin, a novel member of the family of leucine-rich repeat proteins of the extracellular matrix. J. Biol. Chem. 275:2123–2129 10.1074/jbc.275.3.2123 [DOI] [PubMed] [Google Scholar]
- Sterling H., Saginario C., Vignery A. 1998. CD44 occupancy prevents macrophage multinucleation. J. Cell Biol. 143:837–847 10.1083/jcb.143.3.837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K., Zhu B., Rittling S.R., Denhardt D.T., Goldberg H.A., McCulloch C.A., Sodek J. 2002. Colocalization of intracellular osteopontin with CD44 is associated with migration, cell fusion, and resorption in osteoclasts. J. Bone Miner. Res. 17:1486–1497 10.1359/jbmr.2002.17.8.1486 [DOI] [PubMed] [Google Scholar]
- Teitelbaum S.L., Ross F.P. 2003. Genetic regulation of osteoclast development and function. Nat. Rev. Genet. 4:638–649 10.1038/nrg1122 [DOI] [PubMed] [Google Scholar]
- Young D.A., Lakey R.L., Pennington C.J., Jones D., Kevorkian L., Edwards D.R., Cawston T.E., Clark I.M. 2005. Histone deacetylase inhibitors modulate metalloproteinase gene expression in chondrocytes and block cartilage resorption. Arthritis Res. Ther. 7:R503–R512 10.1186/ar1702 [DOI] [PMC free article] [PubMed] [Google Scholar]