Abstract
Synthetic biology aims to engineer novel cellular functions by assembling well-characterized molecular parts (i.e., nucleic acids and proteins) into biological “devices” that exhibit predictable behavior. Recently, efforts in eukaryotic synthetic biology have sprung from foundational work in bacteria. Designing synthetic circuits to operate reliably in the context of differentiating and morphologically complex cells presents unique challenges and opportunities for progress in the field. This review surveys recent advances in eukaryotic synthetic biology and describes how synthetic systems can be linked to natural cellular processes in order to manipulate cell behavior and to foster new discoveries in cell biology research.
Introduction
The analogy of cells as tiny machines has captured the attention and imagination of scientists from many disciplines, leading to the emergence of a research field known as synthetic biology. Synthetic biology encompasses a range of endeavors, such as chemical systems that mimic living systems (for review see Benner and Sismour 2005), novel functional assemblies of natural molecular components, simplified cells, and artificial ecosystems (for review see Purnick and Weiss, 2009). More conventional research disciplines define functions and interactions of cellular components based on the results of experiments where individual parts are perturbed or over-produced within a natural system or in vitro. Molecular characterizations are further supported by evolutionary studies that identify conserved motifs and by structural studies that reveal relationships between molecular shape and function. Synthetic biology applies this knowledge to rationally design and construct biological networks of interacting modules. Testing custom-built pathways in living cells provides a unique opportunity to observe the behavior of biological components in new contexts, revealing the extent of our understanding of natural systems.
Another integral aspect of synthetic biology is its relationship with mathematical modeling. Equations and computer simulations that model relationships between biological processes (e.g., gene activation, protein production, cell division, etc.) guide the design of synthetic systems by determining the vital parameters and parameter spaces in which the system will function as intended. Biological modules are then assembled in a manner consistent with the model (Elowitz and Leibler, 2000; Gardner et al., 2000; Ajo-Franklin et al., 2007; Brenner et al., 2007; Ellis et al., 2009). Once the assembly is tested and characterized in a living system, empirical data can provide new parameters to improve the mathematical model. In this way, iterative cycles of modeling, building, and testing lead to useful synthetic systems and new insights into living mechanisms.
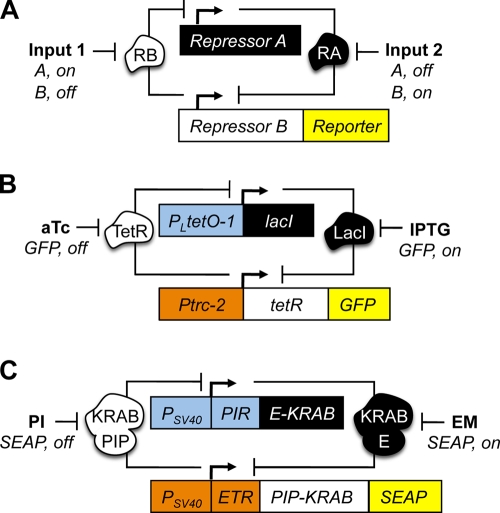
In this review, we focus on synthetic biology’s efforts to build novel biological pathways within single living cells. We use the term “synthetic circuit” or “synthetic device” to describe a system that is designed to execute a useful function (a bistable state, oscillation, pulse, etc.) in a predictable and reliable manner. Of particular interest are basic extensible functions with implications for biotechnology and gene therapy. For instance, a genetic toggle switch that responds to transient input signals, similar to how a lamp responds to a mechanical “on/off” switch, would allow long-term maintenance of protein expression without sustained drug administration. A simple bistable switch can be built using two mutually repressive genes (Fig. 1 A). When each gene encodes a transcriptional repressor of the other and each repressor is blocked by a chemical input, the system can be switched between two stable states (i.e., “gene 1 on, gene 2 off” vs. “gene 1 off, gene 2 on”). Gardner et al. (2000) used mathematical modeling to predict that bistability is possible with any set of promoters or repressors as long as they fulfill a minimum set of conditions (e.g., balanced promoter strengths). Using these predictions, they constructed and successfully operated a toggle switch in Escherichia coli (Fig. 1 B). Four years later, Kramer et al. (2004) reported that a similar device could be operated within a eukaryotic system (Chinese hamster ovary cells; Fig. 1 C).
Figure 1.
Synthetic genetic toggle switches. (A) A genetic toggle switch comprised of two genes (Repressor A and Repressor B) encoding repressor proteins (RA and RB) uses a mutual repression motif to achieve bistability. Transient exposure to Input 1 inhibits RB and switches the system’s stable state to Repressor A expression. Input 2 inhibits RA and activates Repressor B and a detectable output (Reporter). (B) Stable GFP output from a synthetic prokaryotic toggle switch is activated by isopropyl-β-d-thiogalactopyranoside (IPTG) and deactivated by anhydrotetracycline (aTc; Gardner et al., 2000). The LacI and TetR repressor proteins bind the Ptrc-2 and PLtetO-1 promoters, respectively. (C) In the mammalian toggle switch, secreted alkaline phosphatase (SEAP) output is activated erythromycin (EM) and deactivated by pristinamycin I (PI; Kramer et al., 2004). Repressor proteins, containing the KRAB (Kruppel-associated box) transcription repression domain fused with either the pristinamycin-induced transcription regulator protein (PIP) or the macrolide responsive MphR(A) protein (E) bind to promoters containing a simian virus 40 region (PSV40) and target sequences for PIP (PIR) or E (ETR).
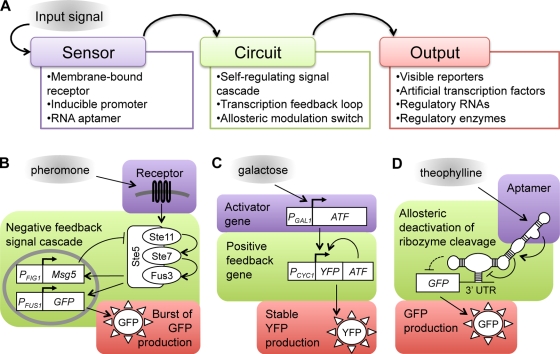
To generate devices that may be adapted to a variety of tasks, engineering principles such as modularity and standardization are applied to genetic construction. The mechanics of synthetic devices can be summarized as a three-level hierarchy of interchangeable functions (Fig. 2 A). A signal (e.g., a small molecule) is detected at the sensor level, the signal is then processed into a pattern of behavior at the circuit level, and expression of a reporter gene coincides with the behavior of the circuit at the output level. The overall design can be complex, containing multiple genes or proteins at each level of function (Fig. 2, B and C), or as simple as a single molecule that contains all three levels of function (Fig. 2 D). Step-wise progress in synthetic biology has yielded many advances that are focused on individual levels of function rather than a complete system. To illustrate how these engineering developments might be integrated into complete systems in the future, this review presents each within the framework of the sensor, circuit, and output levels of function.
Figure 2.
Three levels of modular functions within synthetic devices. (A) An input signal, such as a small molecule, ligand, or metabolite, is detected at the sensor level. The signal is then translated into a pattern of behavior at the circuit level. At the output level, reporter gene expression and/or effectors of downstream processes coincide with the behavior of the circuit. Each box lists examples of sensors, circuits, and outputs that are correspondingly shaded in B–D. (B) In a cell signaling–based device, pheromone input binds a receptor and triggers an engineered MAPK signal circuit that activates GFP expression and the negative regulator Msg5. Activation, negative feedback, then deactivation result in a burst of GFP output (Bashor et al., 2008). (C) In a transcription-based device, transient galactose input induces the expression of a PGAL1-driven ATF that initiates the stable transcription of a positive feedback gene and sustained YFP output (Ajo-Franklin et al., 2007). (D) Functions of a riboswitch are encoded within folded RNA domains. Theophylline input binds to the aptamer sensor region, causing a conformational change that is transduced down the molecule to disrupt ribozyme-mediated RNA cleavage and allow GFP expression (Win and Smolke 2007, 2008).
To date, advances in synthetic biology are largely represented by experiments in bacterial systems (for reviews see Sprinzak and Elowitz 2005; Drubin et al., 2007). The cytological simplicity and well-defined components of the bacterial cell make it an attractive system for operating synthetic circuits. Recently, however, synthetic biology researchers have developed devices that function successfully in structurally complex yeast, mammalian, and plant cells. The following survey of eukaryotic synthetic devices is accompanied by a discussion of approaches that link synthetic systems with natural cellular cues and phenotypes. We then discuss how eukaryotic cell structure can be harnessed to broaden the field of synthetic biology. We conclude with a speculation of synthetic biology’s contributions to cell biology research and medicine, as well as a discussion of current challenges of synthetic device engineering in eukaryotic systems.
Synthetic devices in eukaryotic cells
The first genetic circuits engineered in bacteria are elegant demonstrations of how molecular biological processes can be rationally constructed from predefined parts. Prokaryotic synthetic circuits have provided several exciting innovations, including light-responsive signal transduction systems (Levskaya et al., 2005) and oscillating transcription circuits (Elowitz and Leibler, 2000). Are similar advances in eukaryotic synthetic systems possible given the largely uncharacterized and complex infrastructure of eukaryotic cells, potential perturbations, and cross talk with endogenous factors? Indeed, successes in manipulating preexisting pathways and expressing synthetic gene networks in yeast, mammalian, and plant systems demonstrate the feasibility and usefulness of eukaryotic cells as platforms for synthetic biology.
Rewired cell-signaling pathways.
Cell-signaling pathways carry information from external stimuli across the membrane and into the cell via a cascade of interactions between protein modules (for review see Bhattacharyya et al., 2006). Synthetic biology researchers have used the well-characterized mating pheromone-induced MAPK signaling pathway from the budding yeast Saccharomyces cerevisiae to build and test synthetic auto-regulation. Ingolia and Murray (2007)placed a positive regulator of MAPK signaling under control of the MAPK signal cascade, creating a positive feedback loop that showed sustained output production after the stimulating pheromone was removed. A different approach used a modified membrane-associated Ste5 scaffold to activate gene expression of regulatory modules that either further induced or halted the MAPK signal cascade (Bashor et al., 2008). In the synthetic Ste5 systems, positive feedback changed signal output from a graded to a switch-like response, whereas negative feedback changed a sustained response to a pulsed or delayed response.
Engineered signal-processing cascades are circuit modules that can be linked to genetic and physiological outputs. The synthetic Ste5 MAPK signaling circuit (Bashor et al., 2008) could potentially regulate any gene of interest that is placed under control of the FUS1 promoter. Likewise, a plant system in which a Pho promoter is targeted by a histidine kinase-activated PhoB-VP64 transcription factor provides a means for linking cell signaling to gene transcription (Antunes et al., 2009). Physiological responses have been placed under the control of synthetic Fus3 and Hog1 MAPK proteins such that a pheromone input signal led to a hyper-osmolar response (and vice-versa) in yeast (Mody et al., 2009).
The sensor level of function in signaling circuits can be customized using engineered receptors that specifically bind distinct input signals. Early work in which the human G protein–coupled receptors were transferred to yeast pathways demonstrates the modularity of transmembrane sensors in eukaryotes (Strader et al., 1991; for review see Pausch, 1997). Since then, protein engineering via mutagenesis has produced a collection of “receptors activated solely by synthetic ligands” (RASSLs), which expands the number and variety of potential input stimuli for synthetic signaling circuits (for review see Conklin et al., 2008). Constitutive and deleterious stimulation observed in some RASSL systems could be fine-tuned by synthetic auto-regulatory circuits similar to the ones developed in yeast.
Synthetic regulation of gene transcription.
Gene regulation is a key end point for signal processing in living cells. Foundational work in molecular biology, such as the elucidation of the bacterial lac gene regulation system (Jacob and Monod, 1961), established a paradigm for genetic switches (for review see Wilson et al., 2007); an input signal modulates the binding of transcriptional regulator proteins at DNA docking sites near gene promoters, conferring a gene activity state (i.e., on or off). Following this design, artificial transcription factors (ATFs) consist of a protein domain, which binds to a specific DNA element near a minimal promoter, tethered to an effector domain (e.g., an activator or repressor) that regulates transcription. DNA binding domains, such as Gal4, lexA, and zinc finger motifs, are used to create locus-specific ATFs. Control of ATF function has also been accomplished with DNA binding domains that interact with their target sequences either in the presence or absence of specific small molecules (for review see Weber and Fussenegger, 2006).
Natural cellular processes such as differentiation have been emulated by synthetic transcription-based systems. Differentiation occurs within a homogenous population when a subset of cells experience a transient stimulus, trigger a switch in cellular physiology, and this change is maintained through mitosis in the cell lineage. Synthetic self-activating genes conferring long-term cellular memory have been constructed and tested in yeast (Becskei et al., 2001, Ajo-Franklin et al., 2007). Ajo-Franklin et al. (2007) used rational engineering to build a synthetic positive feedback system that converted a transient stimulus into sustained transcriptional activation. An inducible promoter (sensor) was designed to stimulate a two-gene circuit in which an ATF triggered a self-activating gene (Fig. 2 C). First, the Hill parameters (basal and maximal production rates, Hill cooperativity, and the concentration at half-maximum activity) were measured for two different ATFs in dividing cells. Then model circuits were tested in silico to determine which ATF would allow switch-like and sustained activation. As predicted by math modeling, the unsuitable ATF locked the gene circuit in the active state, whereas the suitable ATF allowed switching from an uninduced state to gene expression that remained active over several cell divisions.
Customizable control of protein production.
Translation is an attractive target for direct and tight regulation of protein concentrations. Synthetic biology achieves translational control by two strategies. First, RNA interference (RNAi) is used to fine-tune expression dynamics of transcription based systems (Deans et al., 2007; Greber et al., 2008; Tigges et al., 2009). Second, RNAi and functional RNAs are used as the basis for circuit behavior such as switch control and multiple input processing (Yen et al., 2004; Bayer and Smolke, 2005; Rinaudo et al., 2007; Win and Smolke, 2007, 2008; Beisel et al., 2008).
RNAi is mediated by sequence complementarity, allowing virtually any target messenger RNA (mRNA) to be silenced by custom-built sequences. A short interfering RNA (siRNA) or microRNA (miRNA), encoded in the transcript of a gene, targets the corresponding sequence of another transcript for degradation (via siRNA) or inhibition of translation (via miRNA). Synthetic biology researchers have used RNAi to reduce basal transcription, tightening control of synthetic gene networks (Deans et al., 2007; Greber et al., 2008). Deans et al. (2007) designed a modular IPTG-inducible switch that produces short hairpin RNAs (shRNAs) to prevent leaky expression of toxic transgenes in human and CHO cells. Greber et al. (2008) have recently proposed a general strategy in which intronic siRNAs diminish basal gene expression in the off state of inducible transcription systems and synthetic circuits.
In other synthetic devices, RNAi-based translational regulation serves as the pivotal function rather than a means to optimize performance. Synthetic ribozymes are functional RNA-based devices that mediate chemically induced mRNA self-cleavage in mammalian and yeast cells (Yen et al., 2004; Win and Smolke, 2007, 2008). “Antiswitch” (Bayer and Smolke, 2005) and “shRNA switch” (Beisel et al., 2008) systems control translation by regulated RNA duplex formation. Ribozymes, antiswitches, and shRNA switches contain an aptamer (sensor) module that specifically binds to a single small molecule, causing an allosteric modification that is propagated through a “transducer” module. Altered RNA folding of the device either permits or blocks translation of a target mRNA molecule that encodes a visible output signal (Fig. 2 D). Other RNAi-based synthetic devices can process combinations of inputs using Boolean logic. Ribozyme switches have been fitted with multiple aptamers to process the presence or absence of two small molecules in yeast (Win and Smolke, 2008). Rinaudo et al. (2007) designed a mammalian logic evaluator composed of genes with multiple RNAi target sequences in the 3′-untranslated region of each mRNA transcript. Fluorescent protein output in human kidney cell culture showed that the gene circuit correctly evaluated combinations of up to five transfected siRNA inputs. Modified systems in which ribozyme switches mediate the mammalian circuit will enable the detection of multiple chemical signals as well as siRNAs. Future efforts to engineer logic circuits that evaluate endogenous cellular inputs will lead the way to synthetic devices that can sense and correctly respond to complex physiological conditions.
Interfacing synthetic circuits with natural cellular processes
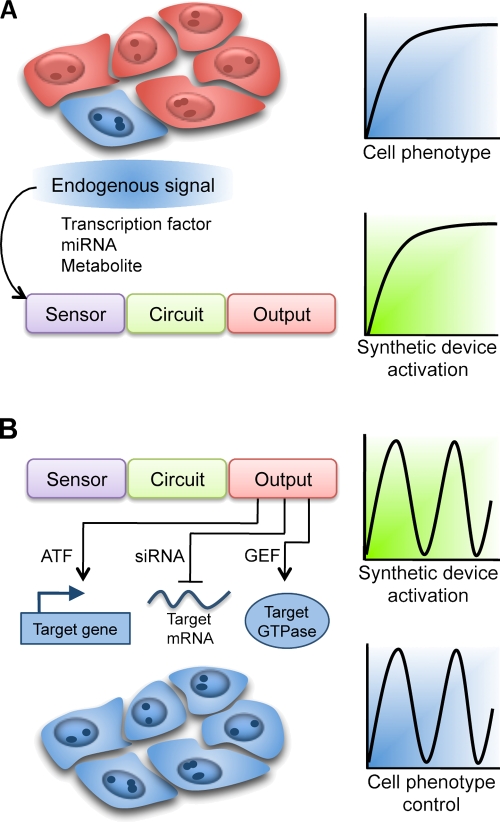
Engineering a synthetic device to sense changes in the intercellular environment (Fig. 3 A) synchronizes a visible reporter, and potentially a therapeutic product, with developmental and disease states. Conversely, devices can regulate cell behavior by interfacing synthetic device output with a cellular target, such as an endogenous gene, mRNA, or protein (Fig. 3 B). Successes in manipulating cytoskeletal, genetic, and physiological responses (Deans et al., 2007; Yeh et al., 2007; Mody et al., 2009) indicate progress toward interfacing synthetic systems with endogenous mechanisms. Guanine nucleotide exchange factors (GEFs) have been engineered to convert forskolin-induced protein kinase A (PKA) activation into cytoskeletal reorganization in mammalian cells (Yeh et al., 2007). In this study, a PKA-regulated protein domain was fused with a protein module that activated either Cdc42 or Rac1 GTPases, which induced broad membrane ruffles or thin spiked protrusions, respectively. The following discussion highlights examples of regulatory systems and strategies that could be used to couple cellular processes with synthetic devices at the sensor and output levels.
Figure 3.
Interfacing synthetic circuits with biological functions. (A) Synthetic devices can be placed under the control of sensors that sense inputs such as transcription factors, metabolites (Table I), and potentially miRNAs. In a population of cells that carry the circuit, the cell (shaded blue) that expresses certain developmental or disease cues triggers activity of the circuit. (B) At the output level, synthetic devices can be interfaced with cell phenotypes by placing endogenous targets under the control of circuit outputs: transcription factors target endogenous genes, small RNA silencers target transcripts, and guanine nucleotide exchange factors target GTPase enzymes that regulate cytoskeletal morphology. Consequently, all cells (shaded blue) that are exposed to the input stimulus undergo a programmed change in phenotype.
Inducible promoters serve as sensors in most transcription-based synthetic devices. Several artificial inducible transcription systems respond to endogenous physiological cues (for review see Weber and Fussenegger, 2006), including hormones (Webster et al., 1988; Braselmann et al., 1993), hypoxia (Tang et al., 2005), and low nutrient level–associated NADH depletion (Weber et al., 2006). In these systems, a signal-responsive transcription factor is fused to a DNA binding domain (e.g., Gal4) that binds to synthetic regulatory elements upstream of a minimal promoter and target gene (Table I). Sensors can also be built by assembling minimal promoters with natural regulatory DNA elements that are induced by endogenous transcription factors that respond to various stimuli (Strähle et al., 1987; Elledge and Davis, 1989; Ohno et al., 2008; Meijsing et al., 2009; for review see Guo et al., 2008). The cellular metabolite thiamine pyrophosphate (TPP) is sensed by synthetic riboswitches (Nomura and Yokobayashi, 2007; Yamauchi et al., 2008; Wieland et al., 2009). Other metabolites, including adenine, vitamin B12, flavin mononucleotide, and many others (for review see Blouin et al., 2009) remain to be tapped for riboswitch engineering. Development and disease-associated miRNAs are another potential input signal for synthetic systems (Brown et al., 2007). Plausibly, a complimentary target RNA sequence could be incorporated into the untranslated region of a repressor gene that holds a synthetic circuit in an off state. It would be interesting to see whether silencing of the repressor and activation of the device can reliably report miRNA expression in developing cells.
Table I.
Endogenous eukaryotic signals that stimulate synthetic sensors
| Input signal | Synthetic sensor | Sensor function |
| Estrogen | ATF: Gal4 (DBD), estrogen receptor (TAD) (Webster et al., 1988; Braselmann et al., 1993) | Transcription activation at the Gal4 element |
| Hypoxia | ATF: Gal4 (DBD), HIF-1α oxygen-dependent degradation domain, p65 (TAD) (Tang et al., 2005) | Transcription activation at the Gal4 element |
| l-arginine synthesis | ATF: ArgR (DBD), VP16 (TAD) (Hartenbach et al., 2007) | Transcription activation at the ARG box element |
| NADH depletion (low nutrient levels) | ATF: REX (DBD), VP16 (TAD) (Weber et al., 2006) | Transcription activation at the ROP element |
| Thiamine pyrophosphate | Riboswitch: thiamine pyrophosphate (TPP) aptamer (Nomura and Yokobayashi, 2007; Yamauchi et al., 2008; Wieland et al., 2009) | Translation control via mRNA cleavage |
Artificial transcription factor (ATF) fusion proteins and a synthetic riboswitch are described here. DBD = DNA binding domain; TAD = transcription activation domain.
Cell phenotypes can be regulated by synthetic circuit outputs that target endogenous genes and proteins (Fig. 3 B). In plants, RNAi knock-down and overexpression of a small set of endogenous genes leads to visible rapid depletion and restoration of chlorophyll (Antunes et al., 2006). These genes could be placed under the control of chemical-sensing circuits to create a living sentinel system for environmental hazards (Bowen et al., 2008). ATFs that contain Cys2-His2–type zinc-finger DNA binding domains have been targeted to specific mammalian genes (Sera, 2009), including the anti-angiogenic pigment epithelium derived factor (PEDF; Yokoi et al., 2007), utrophin (Desantis et al., 2009), and the breast tumor suppressor maspin (Beltran et al., 2007, 2008). Developing synthetic devices that produce gene-specific ATFs, siRNAs, and miRNAs may lead to innovative synthetic tools with significant impacts in environmental safety, experimental biology, and therapy.
Using the complexity of eukaryotic cells to create novel functions
Subcellular localization and compartmentalization.
Modular parts have been used to control the subcellular location of proteins and nucleic acids, enhancing proper assembly and function of synthetic devices. The signaling circuit developed by Bashor et al. (2008) exemplifies the effectiveness of cell membrane–bound protein scaffolds in controlling protein positioning, interactions, and circuit dynamics. Nuclear transport is commonly mediated by the SV40 nuclear localization sequence (NLS; Kalderon et al., 1984; Lanford and Butel, 1984) to enable ATFs to reach their gene targets. Other NLSs (McLane and Corbett, 2009) and nuclear export signals (NESs; Fischer et al., 1995; la Cour et al., 2003) with distinct translocation dynamics could be explored as ways to alter nuclear transport of synthetic device components. The mitochondria and ER have been used as localization sites for synthetic proteins tagged with Tom70 and Ubc6 peptides (Kornmann et al., 2009). Localization not only enhances synthetic device function, but can also define functional states. For instance, farnesylation-mediated positioning regulates a synthetic translation factor consisting of the eIF4G translation initiation factor, a viral RNA-binding domain, and a plasma membrane–targeting CAAX motif (Boutonnet et al., 2004). Inhibition of farnesylation releases the translation factor from the cell membrane, allowing it to specifically bind and translate mRNAs that contain a 21-nucleotide recognition site.
Toward controlling cell morphology.
Manipulating whole-cell morphology underlies the vision to build increasingly sophisticated synthetic systems that control important physiological processes. An initial step toward this aim is the use of synthetic signaling pathways to control distinct morphological changes in mammalian cells (Yeh et al., 2007; Levskaya et al., 2009). The realization of ambitions such as programmed cell differentiation and engineered cell motility may benefit from exploring cell polarity mechanisms that might be amenable to engineering. Cell polarity is established by asymmetrical distributions of mRNA and proteins during interphase and before cell division. Transcripts containing localization sequences, or “zipcodes,”are actively sorted in the cytoplasm for localized translation and protein function (for review see Paquin and Chartrand, 2008). RNA zipcodes suggest an approach for designing addressable synthetic mRNA molecules. Additionally, examples of rationally designed mRNA localization and transcription (Boutonnet et al., 2004) as well as scaffold-mediated protein activity (Bashor et al., 2008) may serve as a foundation for programmable cell polarity.
Contributions of synthetic biology to cell biology research and medicine
Synthetic biology as a research paradigm?
Construction and testing of synthetic devices in living cells can provide insight into cell biology by determining whether our mechanistic models can sufficiently describe natural systems or if there are still gaps in our understanding. In the context of research discovery, rational assembly of parts accomplishes three objectives: placing biological components into new contexts that may reveal previously unobserved characteristics, generating tools for analysis in vivo, and testing previously established models by reconstruction.
When device construction is guided by an engineering aim rather than a research goal, biological components are often juxtaposed in unique ways. Unexpected behavior of the synthetic device can reveal previously unknown characteristics of one or more components. Although it might be considered a setback to the synthetic device engineer, this type of outcome can spur important investigative research that enhances the body of scientific knowledge (Haynes et al., 2008).
In some cases, fusion proteins and synthetic circuits are designed as probes for studying molecular dynamics within living cells. Fusion proteins that artificially link cellular functions have been used in screens to probe phenotypes that are not accessible by more conventional genetic dissection (Sadler et al., 1989; Stirling et al., 1992; Kornmann et al., 2009). Kornmann et al. (2009) used a synthetic ER–mitochondria tethering protein to rescue a panel of mutations, allowing the identification of elusive natural components of ER–mitochondrial tethering in yeast. In a study of transcription dynamics, a synthetic circuit was used to directly test the hypothesis that long introns affect eukaryotic gene expression by increasing transcription elongation time (for review see Swinburne and Silver, 2008). Increased intron lengths within a self-inhibitory gene were accompanied by delayed pulses of protein expression in mouse cells (Swinburne et al., 2008). The genetic switch developed by Deans et al. (2007) allowed their group to observe the impact of graded bax gene induction on apoptosis. The results of this study indicate a possible high threshold level for Bax-induced death in mammalian cells.
Synthetic systems provide a simplified, well-defined framework for testing models of fundamental biological processes by reconstruction. The sufficiency of positive feedback to establish bistability, a property of cell cycle progression, has been demonstrated using synthetic circuits based on transcription and protein interactions (e.g., Becskei et al., 2001; Ingolia and Murray, 2007). Basic mechanisms underlying circadian oscillation have also been investigated using synthetic models (Ukai-Tadenuma et al., 2008; Tigges et al., 2009). Two natural phases of the circadian clock, daytime and nighttime expression, were reconstructed using artificial transcription activators and repressors under the control of endogenous clock–controlled DNA sequences. This outcome indicates that transcriptional regulation is a sufficient mechanism for circadian behavior (Ukai-Tadenuma et al., 2008). Synthetic circadian clock models provide a system for future studies to resolve the impact of other factors, such as post-translational protein modification, on circadian oscillation.
Implications for advances in medicine.
To date, a small number of direct contributions to medicine have emerged from advances in synthetic biology. Synthetic biology has been applied to the cost-effective production of an antimalarial drug precursor in yeast and bacteria (Ro et al., 2006; Anthony et al., 2009) and a new drug-screening technology in human cell culture (Weber et al., 2008). The latter effort focused on combating drug-resistant strains of Mycobacterium tuberculosis in which EthR represses ethA, a gene required for activation of the drug ethionamide. A synthetic system, in which EthR induced reporter gene activation in human cells, was used to identify noncytotoxic drugs that could cross the cell membrane and disrupt EthR function (Weber et al., 2008).
In developing therapeutic synthetic devices for human cells, it is crucial to anticipate potentially dangerous off-target effects and host cell gene disruption. One solution is to design synthetic devices that operate outside of human cells. To this end, E. coli have been engineered to carry a gene circuit that senses tumor-associated cues (i.e., hypoxia and bacterial aggregation near tumors) and expresses invasin, a protein that allows bacteria to enter mammalian cells (Anderson et al., 2006). Linking cytotoxin production with cell invasion will result in a probiotic strain that can seek out and destroy tumors. Still, creating human cells that secrete therapeutic products or regenerate tissues in a programmable way are attractive and powerful approaches to therapy. Tight artificial control of genes in response to combinatorial signals could allow tissue- and disease-specific expression and decrease toxicity. Furthermore, synthetic gene circuits that produce therapeutic products could translate a temporary dose of input signal into sustained output, circumventing the need for sustained and potentially toxic drug administration.
Current challenges and concluding remarks
Developing a large pool of quantitatively annotated, reliable, and modular biological parts could enhance progress in designing synthetic eukaryotic systems. Potentially, a multitude of novel parts can be culled from natural systems, built de novo based on evolutionarily conserved motifs, or generated by mutagenesis. Thorough testing of new modules is an important step in constructing functional systems. For instance, undesirable and unexpected performance of computer-designed zinc-finger modules (Ramirez et al., 2008) underscores the need for efficient ways to characterize in vivo libraries of putative new ATFs. Other reviews detail strategies to address the challenge of developing reliable standardized parts (for reviews see Andrianantoandro et al., 2006; Heinemann and Panke, 2006).
Synthetic biology researchers using eukaryotic systems should also consider the importance of stably integrating devices into the host cell genome. Expression of synthetic assemblies from transiently transfected plasmids is often used to quantify device behavior. For example, plasmid dosage of a synthetic circadian circuit (Tigges et al., 2009) appeared to influence the behavior of the device in some cells, but plasmid copy number variability from cell to cell cannot be directly determined. Stable transgene integration is a more reliable approach for altering and observing the impact of gene dosage in proliferating engineered cell populations (Ajo-Franklin et al., 2007).
As synthetic biology moves toward introducing more complex genetic systems into eukaryotic cells, tools should be developed to facilitate the manipulation of eukaryotic systems. Reproducible integration of synthetic devices and replacement of damaged devices could be accomplished using transgenic cells and organisms that carry standard integration sites (e.g., the Flp/FRT system from yeast). Additionally, insulator elements such as cHS4 (Yahata et al., 2007; Hanawa et al., 2009) could be used to shield synthetic genes from transcriptional perturbations and disruptive chromatin packaging. Mathematical modeling could be used to identify which eukaryotic cell features might have a significant impact on synthetic devices. Modeling the effect of large cytoplasmic and nuclear volumes, diffusion perturbed by membrane boundaries, and the likelihood of random nontarget ATF binding sites in large genomes may help to identify areas of focus for optimization of eukaryotic synthetic systems.
Synthetic biology research accomplishments thus far have demonstrated that custom-built systems can function within complex intercellular environments without being significantly hampered by unknown factors. Furthermore, eukaryotic pathways such as signaling cascades, nuclear transport, and subcellular localization can be harnessed to create circuits with diverse topologies. Unexplored eukaryotic cell components and pathways present a vast landscape with extensive potential for ongoing work in synthetic biology.
Acknowledgments
We thank the reviewers for their time and effort in helping to finalize our manuscript.
K.A. Haynes is supported by a National Institutes of Health (NIH) NRSA postdoctoral research fellowship. P.A. Silver is supported by grants from the NIH and the U.S. Department of Defense.
The authors declare no conflicts of interest.
Footnotes
Abbreviations used in this paper:
- ATF
- artificial transcription factor
- miRNA
- microRNA
- shRNA
- short hairpin RNA
- siRNA
- short interfering RNA
References
- Ajo-Franklin C.M., Drubin D.A., Eskin J.A., Gee E.P., Landgraf D., Phillips I., Silver P.A. 2007. Rational design of memory in eukaryotic cells. Genes Dev. 21:2271–2276 10.1101/gad.1586107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson J.C., Clarke E.J., Arkin A.P., Voigt C.A. 2006. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 355:619–627 10.1016/j.jmb.2005.10.076 [DOI] [PubMed] [Google Scholar]
- Andrianantoandro E., Basu S., Karig D.K., Weiss R. 2006. Synthetic biology: new engineering rules for an emerging discipline. Mol. Syst. Biol. 2:2006: 0028 10.1038/msb4100073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony J.R., Anthony L.C., Nowroozi F., Kwon G., Newman J.D., Keasling J.D. 2009. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab. Eng. 11:13–19 10.1016/j.ymben.2008.07.007 [DOI] [PubMed] [Google Scholar]
- Antunes M.S., Ha S.B., Tewari-Singh N., Morey K.J., Trofka A.M., Kugrens P., Deyholos M., Medford J.I. 2006. A synthetic de-greening gene circuit provides a reporting system that is remotely detectable and has a re-set capacity. Plant Biotechnol. J. 4:605–622 10.1111/j.1467-7652.2006.00205.x [DOI] [PubMed] [Google Scholar]
- Antunes M.S., Morey K.J., Tewari-Singh N., Bowen T.A., Smith J.J., Webb C.T., Hellinga H.W., Medford J.I. 2009. Engineering key components in a synthetic eukaryotic signal transduction pathway. Mol. Syst. Biol. 5:270 10.1038/msb.2009.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashor C.J., Helman N.C., Yan S., Lim W.A. 2008. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 319:1539–1543 10.1126/science.1151153 [DOI] [PubMed] [Google Scholar]
- Bayer T.S., Smolke C.D. 2005. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 23:337–343 [DOI] [PubMed] [Google Scholar]
- Becskei A., Séraphin B., Serrano L. 2001. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20:2528–2535 10.1093/emboj/20.10.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel C.L., Bayer T.S., Hoff K.G., Smolke C.D. 2008. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol. Syst. Biol. 4:224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran A., Parikh S., Liu Y., Cuevas B.D., Johnson G.L., Futscher B.W., Blancafort P. 2007. Re-activation of a dormant tumor suppressor gene maspin by designed transcription factors. Oncogene. 26:2791–2798 10.1038/sj.onc.1210072 [DOI] [PubMed] [Google Scholar]
- Beltran A.S., Sun X., Lizardi P.M., Blancafort P. 2008. Reprogramming epigenetic silencing: artificial transcription factors synergize with chromatin remodeling drugs to reactivate the tumor suppressor mammary serine protease inhibitor. Mol. Cancer Ther. 7:1080–1090 10.1158/1535-7163.MCT-07-0526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner S.A., Sismour A.M. 2005. Synthetic biology. Nat. Rev. Genet. 6:533–543 10.1038/nrg1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya R.P., Reményi A., Yeh B.J., Lim W.A. 2006. Domains, motifs, and scaffolds: the role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75:655–680 10.1146/annurev.biochem.75.103004.142710 [DOI] [PubMed] [Google Scholar]
- Blouin S., Mulhbacher J., Penedo J.C., Lafontaine D.A. 2009. Riboswitches: ancient and promising genetic regulators. ChemBioChem. 10:400–416 10.1002/cbic.200800593 [DOI] [PubMed] [Google Scholar]
- Boutonnet C., Boijoux O., Bernat S., Kharrat A., Favre G., Faye J.C., Vagner S. 2004. Pharmacological-based translational induction of transgene expression in mammalian cells. EMBO Rep. 5:721–727 10.1038/sj.embor.7400170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen T.A., Zdunek J.K., Medford J.I. 2008. Cultivating plant synthetic biology from systems biology. New Phytol. 179:583–587 10.1111/j.1469-8137.2008.02433.x [DOI] [PubMed] [Google Scholar]
- Braselmann S., Graninger P., Busslinger M. 1993. A selective transcriptional induction system for mammalian cells based on Gal4-estrogen receptor fusion proteins. Proc. Natl. Acad. Sci. USA. 90:1657–1661 10.1073/pnas.90.5.1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner K., Karig D.K., Weiss R., Arnold F.H. 2007. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc. Natl. Acad. Sci. USA. 104:17300–17304 10.1073/pnas.0704256104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown B.D., Gentner B., Cantore A., Colleoni S., Amendola M., Zingale A., Baccarini A., Lazzari G., Galli C., Naldini L. 2007. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat. Biotechnol. 25:1457–1467 [DOI] [PubMed] [Google Scholar]
- Conklin B.R., Hsiao E.C., Claeysen S., Dumuis A., Srinivasan S., Forsayeth J.R., Guettier J.M., Chang W.C., Pei Y., McCarthy K.D., et al. 2008. Engineering GPCR signaling pathways with RASSLs. Nat. Methods. 5:673–678 10.1038/nmeth.1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans T.L., Cantor C.R., Collins J.J. 2007. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 130:363–372 10.1016/j.cell.2007.05.045 [DOI] [PubMed] [Google Scholar]
- Desantis A., Onori A., Di Certo M.G., Mattei E., Fanciulli M., Passananti C., Corbi N. 2009. Novel activation domain derived from Che-1 cofactor coupled with the artificial protein Jazz drives utrophin upregulation. Neuromuscul. Disord. 19:158–162 10.1016/j.nmd.2008.11.005 [DOI] [PubMed] [Google Scholar]
- Drubin D.A., Way J.C., Silver P.A. 2007. Designing biological systems. Genes Dev. 21:242–254 10.1101/gad.1507207 [DOI] [PubMed] [Google Scholar]
- Elledge S.J., Davis R.W. 1989. DNA damage induction of ribonucleotide reductase. Mol. Cell. Biol. 9:4932–4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis T., Wang X., Collins J.J. 2009. Diversity-based, model-guided construction of synthetic gene networks with predicted functions. Nat. Biotechnol. 27:465–471 10.1038/nbt.1536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz M.B., Leibler S. 2000. A synthetic oscillatory network of transcriptional regulators. Nature. 403:335–338 10.1038/35002125 [DOI] [PubMed] [Google Scholar]
- Fischer U., Huber J., Boelens W.C., Mattaj I.W., Lührmann R. 1995. The HIV-1 Rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 82:475–483 10.1016/0092-8674(95)90436-0 [DOI] [PubMed] [Google Scholar]
- Gardner T.S., Cantor C.R., Collins J.J. 2000. Construction of a genetic toggle switch in Escherichia coli. Nature. 403:339–342 10.1038/35002131 [DOI] [PubMed] [Google Scholar]
- Greber D., El-Baba M.D., Fussenegger M. 2008. Intronically encoded siRNAs improve dynamic range of mammalian gene regulation systems and toggle switch. Nucleic Acids Res. 36:e101 10.1093/nar/gkn443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z.S., Li Q., Bartlett D.L., Yang J.Y., Fang B. 2008. Gene transfer: the challenge of regulated gene expression. Trends Mol. Med. 14:410–418 10.1016/j.molmed.2008.07.003 [DOI] [PubMed] [Google Scholar]
- Hanawa H., Yamamoto M., Zhao H., Shimada T., Persons D.A. 2009. Optimized lentiviral vector design improves titer and transgene expression of vectors containing the chicken beta-globin locus HS4 insulator element. Mol. Ther. 17:667–674 10.1038/mt.2009.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartenbach S., Daoud-El Baba M., Weber W., Fussenegger M. 2007. An engineeredl-arginine sensor of Chlamydia pneumoniae enables arginine-adjustable transcription control in mammalian cells and mice. Nucleic Acids Res. 35:e136 10.1093/nar/gkm652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes K.A., Broderick M.L., Brown A.D., Butner T.L., Dickson T.O., Harden W.L., Heard L.H., Jessen E.L., Malloy K.J., Ogden B.J., et al. 2008. Engineering bacteria to solve the Burnt Pancake Problem. J. Biol. Eng. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinemann M., Panke S. 2006. Synthetic biology—putting engineering into biology. Bioinformatics. 22:2790–2799 10.1093/bioinformatics/btl469 [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Murray A.W. 2007. Positive-feedback loops as a flexible biological module. Curr. Biol. 17:668–677 10.1016/j.cub.2007.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob F., Monod J. 1961. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3:318–356 [DOI] [PubMed] [Google Scholar]
- Kalderon D., Richardson W.D., Markham A.F., Smith A.E. 1984. Sequence requirements for nuclear location of simian virus 40 large-T antigen. Nature. 311:33–38 10.1038/311033a0 [DOI] [PubMed] [Google Scholar]
- Kornmann B., Currie E., Collins S.R., Schuldiner M., Nunnari J., Weissman J.S., Walter P. 2009. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 325:477–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer B.P., Viretta A.U., Daoud-El-Baba M., Aubel D., Weber W., Fussenegger M. 2004. An engineered epigenetic transgene switch in mammalian cells. Nat. Biotechnol. 22:867–870 10.1038/nbt980 [DOI] [PubMed] [Google Scholar]
- la Cour T., Gupta R., Rapacki K., Skriver K., Poulsen F.M., Brunak S. 2003. NESbase version 1.0: a database of nuclear export signals. Nucleic Acids Res. 31:393–396 10.1093/nar/gkg101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanford R.E., Butel J.S. 1984. Construction and characterization of an SV40 mutant defective in nuclear transport of T antigen. Cell. 37:801–813 10.1016/0092-8674(84)90415-X [DOI] [PubMed] [Google Scholar]
- Levskaya A., Chevalier A.A., Tabor J.J., Simpson Z.B., Lavery L.A., Levy M., Davidson E.A., Scouras A., Ellington A.D., Marcotte E.M., Voigt C.A. 2005. Synthetic biology: engineering Escherichia coli to see light. Nature. 438:441–442 10.1038/nature04405 [DOI] [PubMed] [Google Scholar]
- Levskaya A., Weiner O.D., Lim W.A., Voigt C.A. 2009. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 461:997–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLane L.M., Corbett A.H. 2009. Nuclear localization signals and human disease. IUBMB Life. 61:697–706 10.1002/iub.194 [DOI] [PubMed] [Google Scholar]
- Meijsing S.H., Pufall M.A., So A.Y., Bates D.L., Chen L., Yamamoto K.R. 2009. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 324:407–410 10.1126/science.1164265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mody A., Weiner J., Ramanathan S. 2009. Modularity of MAP kinases allows deformation of their signalling pathways. Nat. Cell Biol. 11:484–491 10.1038/ncb1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomura Y., Yokobayashi Y. 2007. Reengineering a natural riboswitch by dual genetic selection. J. Am. Chem. Soc. 129:13814–13815 10.1021/ja076298b [DOI] [PubMed] [Google Scholar]
- Ohno K., Ishihata K., Tanaka-Azuma Y., Yamada T. 2008. A genotoxicity test system based on p53R2 gene expression in human cells: assessment of its reactivity to various classes of genotoxic chemicals. Mutat. Res. 656:27–35 [DOI] [PubMed] [Google Scholar]
- Paquin N., Chartrand P. 2008. Local regulation of mRNA translation: new insights from the bud. Trends Cell Biol. 18:105–111 10.1016/j.tcb.2007.12.004 [DOI] [PubMed] [Google Scholar]
- Pausch M.H. 1997. G-protein-coupled receptors in Saccharomyces cerevisiae: high-throughput screening assays for drug discovery. Trends Biotechnol. 15:487–494 10.1016/S0167-7799(97)01119-0 [DOI] [PubMed] [Google Scholar]
- Purnick P.E., Weiss R. 2009. The second wave of synthetic biology: from modules to systems. Nat. Rev. Mol. Cell Biol. 10:410–422 10.1038/nrm2698 [DOI] [PubMed] [Google Scholar]
- Ramirez C.L., Foley J.E., Wright D.A., Müller-Lerch F., Rahman S.H., Cornu T.I., Winfrey R.J., Sander J.D., Fu F., Townsend J.A., et al. 2008. Unexpected failure rates for modular assembly of engineered zinc fingers. Nat. Methods. 5:374–375 10.1038/nmeth0508-374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinaudo K., Bleris L., Maddamsetti R., Subramanian S., Weiss R., Benenson Y. 2007. A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 25:795–801 10.1038/nbt1307 [DOI] [PubMed] [Google Scholar]
- Ro D.K., Paradise E.M., Ouellet M., Fisher K.J., Newman K.L., Ndungu J.M., Ho K.A., Eachus R.A., Ham T.S., Kirby J., et al. 2006. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 440:940–943 10.1038/nature04640 [DOI] [PubMed] [Google Scholar]
- Sadler I., Chiang A., Kurihara T., Rothblatt J., Way J., Silver P. 1989. A yeast gene important for protein assembly into the endoplasmic reticulum and the nucleus has homology to DnaJ, an Escherichia coli heat shock protein. J. Cell Biol. 109:2665–2675 10.1083/jcb.109.6.2665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sera T. 2009. Zinc-finger-based artificial transcription factors and their applications. Adv. Drug Deliv. Rev. 61:513–526 10.1016/j.addr.2009.03.012 [DOI] [PubMed] [Google Scholar]
- Sprinzak D., Elowitz M.B. 2005. Reconstruction of genetic circuits. Nature. 438:443–448 10.1038/nature04335 [DOI] [PubMed] [Google Scholar]
- Stirling C.J., Rothblatt J., Hosobuchi M., Deshaies R., Schekman R. 1992. Protein translocation mutants defective in the insertion of integral membrane proteins into the endoplasmic reticulum. Mol. Biol. Cell. 3:129–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader C.D., Gaffney T., Sugg E.E., Candelore M.R., Keys R., Patchett A.A., Dixon R.A. 1991. Allele-specific activation of genetically engineered receptors. J. Biol. Chem. 266:5–8 [PubMed] [Google Scholar]
- Strähle U., Klock G., Schütz G. 1987. A DNA sequence of 15 base pairs is sufficient to mediate both glucocorticoid and progesterone induction of gene expression. Proc. Natl. Acad. Sci. USA. 84:7871–7875 10.1073/pnas.84.22.7871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne I.A., Silver P.A. 2008. Intron delays and transcriptional timing during development. Dev. Cell. 14:324–330 10.1016/j.devcel.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne I.A., Miguez D.G., Landgraf D., Silver P.A. 2008. Intron length increases oscillatory periods of gene expression in animal cells. Genes Dev. 22:2342–2346 10.1101/gad.1696108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.L., Tang Y., Zhang Y.C., Agarwal A., Kasahara H., Qian K., Shen L., Phillips M.I. 2005. A hypoxia-inducible vigilant vector system for activating therapeutic genes in ischemia. Gene Ther. 12:1163–1170 10.1038/sj.gt.3302513 [DOI] [PubMed] [Google Scholar]
- Tigges M., Marquez-Lago T.T., Stelling J., Fussenegger M. 2009. A tunable synthetic mammalian oscillator. Nature. 457:309–312 [DOI] [PubMed] [Google Scholar]
- Ukai-Tadenuma M., Kasukawa T., Ueda H.R. 2008. Proof-by-synthesis of the transcriptional logic of mammalian circadian clocks. Nat. Cell Biol. 10:1154–1163 10.1038/ncb1775 [DOI] [PubMed] [Google Scholar]
- Weber W., Fussenegger M. 2006. Pharmacologic transgene control systems for gene therapy. J. Gene Med. 8:535–556 10.1002/jgm.903 [DOI] [PubMed] [Google Scholar]
- Weber W., Link N., Fussenegger M. 2006. A genetic redox sensor for mammalian cells. Metab. Eng. 8:273–280 10.1016/j.ymben.2005.12.004 [DOI] [PubMed] [Google Scholar]
- Weber W., Schoenmakers R., Keller B., Gitzinger M., Grau T., Daoud-El Baba M., Sander P., Fussenegger M. 2008. A synthetic mammalian gene circuit reveals antituberculosis compounds. Proc. Natl. Acad. Sci. USA. 105:9994–9998 10.1073/pnas.0800663105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster N.J., Green S., Jin J.R., Chambon P. 1988. The hormone-binding domains of the estrogen and glucocorticoid receptors contain an inducible transcription activation function. Cell. 54:199–207 10.1016/0092-8674(88)90552-1 [DOI] [PubMed] [Google Scholar]
- Wieland M., Benz A., Klauser B., Hartig J.S. 2009. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed. Engl. 48:2715–2718 10.1002/anie.200805311 [DOI] [PubMed] [Google Scholar]
- Wilson C.J., Zhan H., Swint-Kruse L., Matthews K.S. 2007. The lactose repressor system: paradigms for regulation, allosteric behavior and protein folding. Cell. Mol. Life Sci. 64:3–16 10.1007/s00018-006-6296-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win M.N., Smolke C.D. 2007. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. USA. 104:14283–14288 10.1073/pnas.0703961104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win M.N., Smolke C.D. 2008. Higher-order cellular information processing with synthetic RNA devices. Science. 322:456–460 10.1126/science.1160311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yahata K., Maeshima K., Sone T., Ando T., Okabe M., Imamoto N., Imamoto F. 2007. cHS4 insulator-mediated alleviation of promoter interference during cell-based expression of tandemly associated transgenes. J. Mol. Biol. 374:580–590 10.1016/j.jmb.2007.09.054 [DOI] [PubMed] [Google Scholar]
- Yamauchi T., Miyoshi D., Kubodera T., Ban M., Nishimura A., Sugimoto N. 2008. Riboswitches for enhancing target gene expression in eukaryotes. ChemBioChem. 9:1040–1043 10.1002/cbic.200700782 [DOI] [PubMed] [Google Scholar]
- Yeh B.J., Rutigliano R.J., Deb A., Bar-Sagi D., Lim W.A. 2007. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature. 447:596–600 10.1038/nature05851 [DOI] [PubMed] [Google Scholar]
- Yen L., Svendsen J., Lee J.S., Gray J.T., Magnier M., Baba T., D’Amato R.J., Mulligan R.C. 2004. Exogenous control of mammalian gene expression through modulation of RNA self-cleavage. Nature. 431:471–476 10.1038/nature02844 [DOI] [PubMed] [Google Scholar]
- Yokoi K., Zhang H.S., Kachi S., Balaggan K.S., Yu Q., Guschin D., Kunis M., Surosky R., Africa L.M., Bainbridge J.W., et al. 2007. Gene transfer of an engineered zinc finger protein enhances the anti-angiogenic defense system. Mol. Ther. 15:1917–1923 10.1038/sj.mt.6300280 [DOI] [PubMed] [Google Scholar]