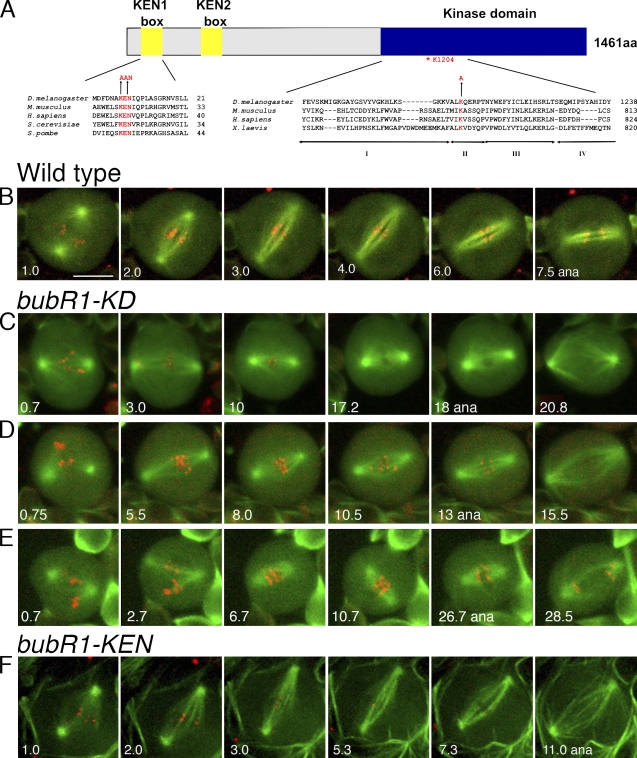
The BubR1 kinase domain controls spindle attachment to the kinetochores, whereas the KEN domain regulates activation of the spindle assembly checkpoint.
Abstract
BubR1 performs several roles during mitosis, affecting the spindle assembly checkpoint (SAC), mitotic timing, and spindle function, but the interdependence of these functions is unclear. We have analyzed in Drosophila melanogaster the mitotic phenotypes of kinase-dead (KD) BubR1 and BubR1 lacking the N-terminal KEN box. bubR1-KD individuals have a robust SAC but abnormal spindles with thin kinetochore fibers, suggesting that the kinase activity modulates microtubule capture and/or dynamics but is relatively dispensable for SAC function. In contrast, bubR1-KEN flies have normal spindles but no SAC. Nevertheless, mitotic timing is normal as long as Mad2 is present. Thus, the SAC, timer, and spindle functions of BubR1 are substantially separable. Timing is shorter in bubR1-KEN mad2 double mutants, yet in these flies, lacking both critical SAC components, chromosomes still segregate accurately, reconfirming that in Drosophila, reliable mitosis does not need the SAC.
Introduction
BubR1 is a key component of the spindle assembly checkpoint (SAC), the surveillance mechanism which detects improper kinetochore–spindle linkages and delays anaphase onset until proper attachments are established (Musacchio and Salmon, 2007). BubR1, like Mad2, binds directly to and inhibits Cdc20, the essential cofactor required by the anaphase-promoting complex/cyclosome (APC/C) to initiate mitotic exit. In addition to their roles in the SAC, BubR1 and Mad2 also determine basal mitotic timing, which is the minimum time that elapses between nuclear envelope breakdown (NEB) and anaphase onset even when there are no unattached kinetochores (i.e., no SAC delay; Meraldi et al., 2004). BubR1 also has a third function, promoting proper kinetochore–microtubule (MT [K–MT]) linkages (Lampson and Kapoor, 2005). This multifunctionality makes the phenotypes resulting from BubR1 inactivation difficult to interpret. For example, it is not known to what extent these different activities are interdependent. Is the SAC activity part of the activity-promoting K–MT linkages and vice versa? Is the timer function intrinsic to SAC function?
One way to address these questions is to identify specific mutations in BubR1 that affect one function but leave the others relatively intact. Such separation-of-function mutations would be invaluable for better understanding the contribution of BubR1 to mitosis and the SAC. The BubR1 protein has well-defined domains that might correlate with its presumed activities. The N-terminal half of metazoan BubR1 is similar to yeast MAD3, including a CDC20-binding region that encompasses two KEN boxes, both of which are critical for Mad3 binding to Cdc20 and consequently for SAC-mediated inhibition of the APC/C (Sczaniecka et al., 2008).
Unlike Mad3, the C-terminal half of BubR1 in most metazoans encodes a kinase whose role in the SAC, and in mitosis generally, is controversial. It has been reported to be both essential and dispensable for the SAC, depending on the study (Tang et al., 2001; Chen, 2002; Fang, 2002; Mao et al., 2003). It is also unclear whether the kinase activity is critical to promoting proper K–MT linkages (Harris et al., 2005; Zhang et al., 2007; Huang et al., 2008; Malureanu et al., 2009).
In Drosophila melanogaster, cells apparently do not need the SAC for accurate mitosis (Buffin et al., 2007), thus providing a clean baseline for assaying mitotic perturbations. Therefore, we examined in vivo mutations in fly BubR1 analogous to those described in the previous paragraphs in the kinase domain and the KEN box that would potentially separate its various activities.
Results and discussion
Inefficient spindle assembly in bubR1–kinase-dead (KD) cells
To determine what aspects of BubR1 function depend critically on the kinase, we generated a bubR1-KD allele (K1204A) and assayed its mitotic functions in vivo in larval neuroblasts (Fig. 1 A). bubR1-KD flies were viable and fertile, and only 1% of neuroblasts were aneuploid (Table I), which is far lower than the 25–50% seen in the genetic null bubR11 or other SAC mutations (Basu et al., 1999; Basto et al., 2000). Except for an increase in the prometaphase–metaphase/anaphase ratio in bubR1-KD cells (3.9 vs. 2.6 in wild type [WT]; unpublished data), mitosis appeared normal in fixed material.
Figure 1.
Structure of BubR1-KEN and BubR1-KD mutations and their effects on spindle function. (A) Alignment of the N-terminal KEN box (left) and the kinase domain (right), indicating in red the residues altered in bubR1-KEN and the conserved lysine altered in bubR1-KD. (B–E) Thin K-fibers and unstable spindle length in bubR1-KD cells. (B) WT neuroblast in mitosis. Within a few minutes of NEB, it has established a stable spindle, developed robust K-fibers, and aligned chromosomes on the metaphase plate. Anaphase (ana) occurs at ∼7 min after NEB. See Video 1. (C–E) Three different bubR1-KD neuroblasts displaying prolonged prometaphase. Chromosomes have difficulty congressing and remaining at the metaphase plate. The spindles appear diffuse, the K-fibers are poorly defined, and spindle length varies (compare the second and third frames of each series). Despite this aberrant behavior, each cell eventually enters anaphase, and the chromosomes segregate normally. See Fig. S1 and Videos 2 and 3. Videos in B–E are wide-field microscopy images of GFP-tubulin and mRFP1-Rod. (F) A bubR1-KEN mutant neuroblast with normal spindle morphology and dynamics. See Video 5. Spinning disk confocal microscope images of GFP-tubulin and mRFP1–BubR1-KEN marking kinetochores (which become difficult to detect as anaphase approaches) are shown. Bar, 5 µm.
Table I.
Analysis of the mitotic parameters in bubR1 mutant neuroblasts
| Straina | Time in colchicine | Mitotic densitybc | Relative mitotic density | Aneuploidyc | PSCSc | Notes |
| min | % | % | ||||
| WT (5) | 0 | 2.11 (0.26) | 1 | 0.2 (0.22)d | NA | viable |
| WT (5) | 30 | 3.36 (0.28) | 1.59 | NA | 0.17 (0.08) | NA |
| WT (5) | 60 | 5.73 (0.19) | 2.72 | NA | 0.20 (0.06) | NA |
| bubR1-KD (5) | 0 | 1.84 (0.12) | 1 | 0.97 (0.14) | NA | viable |
| bubR1-KD (5) | 30 | 2.67 (0.27) | 1.45 | NA | 0.58 (0.21) | NA |
| bubR1-KD (5) | 60 | 4.59 (0.30) | 2.50 | NA | 1.41 (0.72) | NA |
| bubR1-KEN (5) | 0 | 1.64 (0.10) | 1 | 0.67 (0.60) | NA | viable |
| bubR1-KEN (5) | 30 | 1.33 (0.23) | 0.81 | NA | 1.19 (0.47) | NA |
| bubR1-KEN (5) | 60 | 1.25 (0.17) | 0.77 | NA | 3.99 (1.48) | NA |
| bubR1-KEN asp (3) | 0 | 1.46 (0.10) | ND | 4.05 (0.73) | NA | lethal |
| asp (4) | 0 | 7.40 (0.21) | ND | 1.03 (0.26) | NA | lethal |
| bubR1-KD cnn (4) | 0 | 2.92 (0.31) | ND | 1.07 (0.31) | NA | viable |
| bubR1-KEN cnn (5) | 0 | 2.06 (0.20) | ND | 15.22 (6.26) | NA | lethal |
| cnn (3) | 0 | 2.65 (0.24) | ND | 0.83 (0.41) | NA | viable |
| bubR1-KD mad2P (4) | 0 | 1.93 (0.89) | 1 | 12.36 (4.22) | NA | lethal |
| bubR1-KD mad2P (5) | 30 | 1.33 (0.10) | 0.69 | NA | 2.68 (1.98) | NA |
| bubR1-KD mad2P (12) | 60 | 1.20 (0.13) | 0.62 | NA | 4.79 (2.73) | NA |
| bubR1-KEN mad2P (5) | 0 | 1.74 (0.18) | 1 | 0.53 (0.40) | NA | viable |
| bubR1-KEN mad2P (5) | 30 | 1.62 (0.12) | 0.93 | NA | 0.89 (0.17) | NA |
| bubR1-KEN mad2P (4) | 60 | 1.57 (0.12) | 0.91 | NA | 1.58 (0.92) | NA |
| mad2P (5) | 0 | 2.11 (0.26) | 1 | 0.77 (0.37) | NA | viable |
| mad2P (5) | 30 | 1.58 (0.12) | 0.75 | NA | 0.19 (0.18) | NA |
| mad2P (5) | 60 | 1.63 (0.19) | 0.77 | NA | 0.19 (0.20) | NA |
NA, not applicable.
The number of brains is shown in parentheses.
Mitotic density is defined as the mean number of cells in mitosis per optic field.
SD is shown in parentheses.
This value was taken from Buffin et al. (2007).
Live imaging of bubR1-KD neuroblasts confirmed a tendency to prolong prometaphase and revealed significant problems in spindle function. WT spindles (Fig. 1 B and Video 1) rapidly captured and aligned chromosomes, within 2–5 min, spindle length was stable, and kinetochore fibers (K-fibers) increased in thickness as anaphase approached, reflecting maturation of the K-fiber bundle (Maiato et al., 2004). In contrast, in bubR1-KD cells, the time elapsing from NEB to anaphase was longer, and the distribution was much broader (10–31 min; mean 16.5 min) compared with WT (6–11 min; mean 9.1 min; P < 0.0001; Fig. 1, C–E; and see Fig. 3 A). Chromosomes were slow to congress to the metaphase plate and sometimes had difficulty remaining aligned (Fig. 1, D and E; and Videos 2 and 3; and see Fig. 3 D). Spindle K-fibers were often far thinner than in WT and sometimes remained so right up to anaphase onset. Finally, the bubR1-KD spindle length was unstable during prometaphase and metaphase (Fig. 1, C–E; Fig. S1; and Videos 2 and 3), often shrinking soon after NEB. Some spindles reattained full length before anaphase onset. Of 19 spindles examined, 11 (55%) displayed this length instability versus 1/20 (5%) of WT. Despite these defects, chromosomes segregated accurately at anaphase (Fig. S2, A and B), a well-formed central spindle developed, and cytokinesis ensued without obvious difficulty (Videos 2 and 3).
Figure 3.
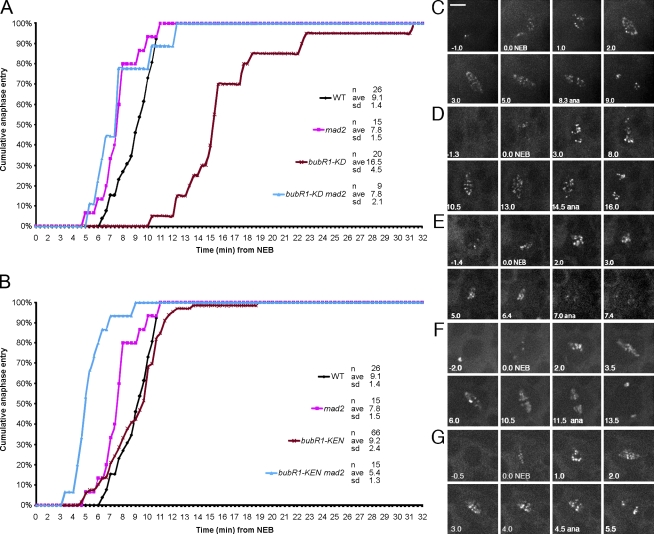
Mitotic timing in bubR1-KD, bubR1-KEN, and double mutants with mad2. (A) Comparative mitotic transit times for WT, bubR1-KD, mad2, and bubR1-KD mad2 double mutants. bubR1-KD cells are profoundly delayed, averaging 16.5 min versus 9.1 min in WT. In mad2 cells, anaphase is, on average, 2 min earlier than in WT (Buffin et al., 2007). bubR1-KD mad2 double mutant cells show the same timing as mad2 alone, revealing that the prometaphase delay in bubR1-KD is SAC dependent. (B) Comparative mitotic timing of bubR1-KEN and bubR1-KEN mad2 double mutant cells. bubR1-KEN cells show no change in timing relative to WT. In contrast, bubR1-KEN mad2 double mutant cells enter anaphase even earlier than mad2 mutant cells alone (5.4 min vs. 7.8 min; P < 0.005). (C–G) Frames from typical videos used to determine mitotic timing (NEB to anaphase [ana]). WT (C), bubR1-KD (D), bubR1-KD mad2 double mutants (E), bubR1-KEN (F), and bubR1-KEN mad2 double mutants (G) are shown. All cells but the one in E are marked with GFP-Rod. The cell in E is marked with GFP–BubR1-KD. See Videos 4, 6, and 7. Bar, 5 µm.
BubR1 is needed for correct K–MT attachments (Lampson and Kapoor, 2005), but the specific role of its kinase activity to this function is unclear. Zhang et al. (2007) and Huang et al. (2008) found that BubR1-KD was unable to promote efficient chromosome capture and congression. Elowe et al. (2007) and Malureanu et al. (2009) reported only minor defects of chromosome attachment. The different results may reflect different efficiencies of depleting the endogenous WT BubR1. Our genetic system (expressing mutant transgenes in the genetically null bubR1 background) avoids this potential pitfall.
The thin K-fibers, chromosome congression delays, and unstable metaphase alignments seen in our bubR1-KD cells support an important role for the kinase in promoting K–MT attachments and are similar to the phenotypes described in mammalian cells after siRNA treatment (Lampson and Kapoor, 2005). The unstable spindle length has not been previously described in bubR1-KD mutants in other model systems, but this too may be a consequence of defective K–MT attachments or, more specifically, alterations in the activities of kinetochore-associated factors influencing MT stability or dynamics. Depletion of proteins such as CLASP or EB1, which bind to and stabilize MT+ ends, leads to shortened spindles and congression problems (Maiato et al., 2002; Goshima et al., 2005). Such proteins or their regulators are thus potential candidates for BubR1 phosphorylation.
Robust SAC in bubR1-KD mutants
Because bubR1-KD cells with abnormal spindles delayed anaphase onset and had only low rates of aneuploidy, they appeared to have a functional SAC. To test whether this delay was indeed SAC dependent, we made double mutants of bubR1-KD and mad2P, a null mutation of mad2 which eliminates the SAC but is viable and generates little or no aneuploidy on its own (Buffin et al., 2007). These bubR1-KD mad2P cells were no longer delayed in prometaphase like bubR1-KD (mean 16.5 min) but instead showed the rapid mitotic transit time (mean 7.8 min) and early onset of cyclin B degradation (OCBD) typical of mad2P (Video 4; and see Fig. 3, A and E; and Fig. 4, B and C). Moreover, double mutant individuals were uniformly larval/pupal lethals, brains had a lower mitotic density, neuroblasts were much more often aneuploid (12%; Table I), and abnormal anaphases were more frequent (Fig. S2, C and D) than in either mutant alone. These results confirmed that the SAC was acting in bubR1-KD to protect the cells from premature anaphase. In a second test, bubR1-KD brains were treated with colchicine to depolymerize MTs. After 1 h, the mitotic density rose 2.5-fold, similar to WT (2.7-fold; Fig. 2, A and B; and Table I). Finally, we assayed the ability of bubR1-KD to provide the SAC activity needed for cnn (centrosomin) mutants to successfully complete mitosis. cnn cells have no functional centrosomes, and spindle assembly takes longer than WT. cnn cells are delayed in prometaphase by the SAC: removing Mad2 (in mad2 cnn double mutants) shortens prometaphase, raises aneuploidy rates from 1% in cnn alone to 18%, and kills the normally viable cnn flies (Buffin et al., 2007). In contrast, bubR1-KD cnn double mutants maintained the low aneuploid rates (around 1%) of bubR1-KD or cnn alone (Table I), and they survived to adulthood. By all of the aforementioned criteria, the bubR1-KD SAC is functional.
Figure 4.
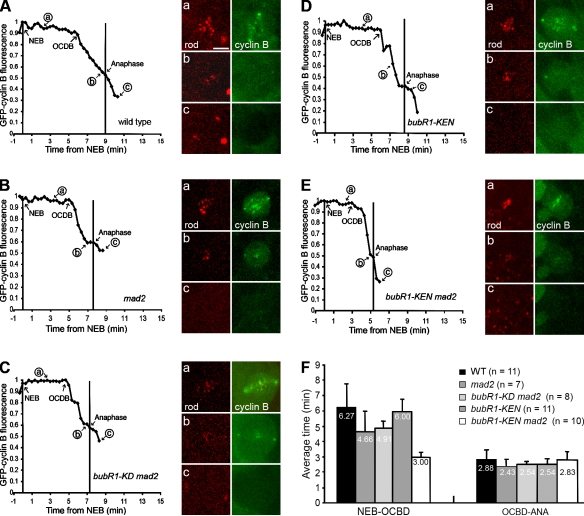
Cyclin B degradation profiles reflect the mitotic timing in bubR1-KEN and double mutant bubR1-KEN mad2 neuroblasts. Typical degradation profiles of GFP–cyclin B disappearance in single neuroblasts. (A–E) WT (A), mad2 (B), bubR1-KD mad2 (C), bubR1-KEN (D), and bubR1-KEN mad2 (E). The OCBD is unaffected by bubR1-KEN, beginning ∼6 min after NEB and 2–3 min before anaphase, as in WT. (F) Mean time of OCBD versus NEB (left) and anaphase (ANA) onset (right). OCBD begins 1.6 min earlier in mad2 or bubR1-KD mad2 mutant cells but 3 min earlier in bubR1-KEN mad2 mutant cells. Error bars indicate SD. See Videos 8–10 and Buffin et al. (2007). Bar, 5 µm.
Figure 2.
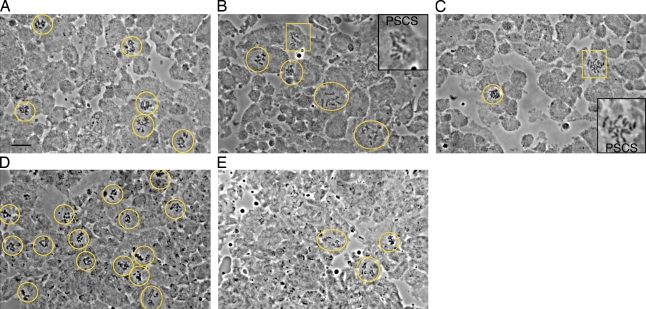
bubR1-KD mutants are checkpoint competent, but bubR1-KEN mutants are checkpoint defective. (A–C) Representative fields of cells from larval brains incubated in colchicine for 60 min. WT (A) or bubR1-KD mutant (B) cells accumulate in prometaphase, whereas bubR1-KEN mutants do not (C). However, PSCS is common in both mutants (boxed areas and insets in B and C). (D and E) The very high mitotic density caused by mutations in asp (D) is suppressed in bubR1-KEN asp double mutants (E). The number of anaphases (very rare in asp) also rises (oval in E). See Table I. Yellow circles and ovals mark mitotic cells in the fields. Orcein-stained preparations are shown. Bar, 10 µm.
However, the SAC in bubR1-KD may not be entirely normal. Premature sister chromatid separation (PSCS) rose sevenfold after 60 min in colchicine to 1.4% (vs. 0.2% in WT). PSCS is usually considered a sign of mitotic exit and therefore of SAC failure. These results might indicate some weakness in long-term maintenance of metaphase.
There is much conflicting data on the role of the BubR1 kinase in the SAC, in part because different assays for SAC activity are used. BubR1 protein bearing mutations (or deletions) of the kinase domain can inhibit APC/C (Chen, 2002; Yu, 2002), yet some studies in vivo or in mitotic extracts concluded that the kinase activity is critical (Mao et al., 2003; Kops et al., 2004), whereas others found it is important only for prolonged nocodazole-induced mitotic arrest (Huang et al., 2008; Malureanu et al., 2009).
Our data are consistent with this latter conclusion. However it is not clear that the inability to maintain prolonged mitotic arrest should be regarded as a weak SAC, which presumably would be less sensitive to unattached kinetochores than a strong SAC. It would then arrest cells with many unattached kinetochores (as after nocodazole treatment) but not cells with just a few. Studies measuring the duration of a SAC-mediated arrest in nocodazole may be measuring some other property unrelated to the SAC (for review see Rieder and Maiato, 2004).
The kinetochore protein CenpE reportedly binds to and regulates BubR1 kinase activity (Mao et al., 2003); thus, one might expect depletion of Cmet, the fly homologue of CenpE, to have a phenotype similar to bubR1-KD. Indeed, cmet mutant cells are delayed in prometaphase, but they exit mitosis in colchicine (Williams et al., 2003). However, unlike bubR1-KD, cmet mutants are highly aneuploid and lethal. Moreover, the spindle defects after cmet and bubR1 RNAi treatment differ (Maia et al., 2007). This suggests that Cmet may affect the SAC and the kinetochore independently of BubR1. Cmet itself is still recruited to bubR1-KD kinetochores (unpublished data).
To conclude, the kinase activity of BubR1 is substantially more important for spindle function than for the SAC. In bubR1-KD, spindles inefficiently make proper K–MT connections, whereas the SAC is relatively unaffected: it can arrest cells for up to1 h in colchicine, and when spindle function is compromised (as in bubR1-KD and cnn cells), it can delay anaphase to assure accurate chromosome segregation and low aneuploidy rates. Because in Drosophila the prometaphase–metaphase period typically lasts 9 min and an entire neuroblast cell cycle may last only 1 h (Truman and Bate, 1988), the value of a SAC capable of longer-term arrest is likely to be negligible.
No SAC but normal mitosis in bubR1-KEN and bubR1-KEN mad2 double mutant flies
To make a checkpoint-dead BubR1, we mutated the N-terminal KEN box (K7, E8, and N9) to AAN (Fig. 1 A). Flies with this mutation, called in this study bubR1-KEN, were viable and fertile, and aneuploidy in the larval neuroblasts was very low (0.7%; Table I), similar to that reported for the mad2P-null mutation (Buffin et al., 2007).
Colchicine failed to arrest bubR1-KEN cells in mitosis (Fig. 2 C and Table I), and PSCS was frequent, 20-fold higher than in WT. Likewise, the high mitotic density caused by the asp (abnormal spindle) mutation was reduced fivefold in bubR1-KEN asp double mutants (Fig. 2, D and E; and Table I), similar to the reduction seen when asp is combined with other SAC mutations (Basto et al., 2000; Buffin et al., 2007). Finally, and in contrast to bubR1-KD, double mutants of bubR1-KEN and cnn were pupal lethal with aneuploidy averaging 15% (Table I). bubR1-KEN neuroblasts showed no obvious defects in spindle structure or function (Fig. 1 F and Video 5). Stable spindles rapidly captured chromosomes and formed robust K-fibers, often within 3 min of NEB, as in WT. Anaphase figures presented normal symmetrical chromatids (Fig. S2, E and F). By all of these criteria, the mitotic phenotype of bubR1-KEN mutants is similar to mad2P (Buffin et al., 2007): viable, little aneuploidy, and normal spindle function but SAC defective.
Thus, the KEN box is critical to the SAC function of fly BubR1, as it is for yeast Mad3 (Sczaniecka et al., 2008). Malureanu et al. (2009) recently reported that expressing the N-terminal third of BubR1 (lacking the kinase domain and a second Cdc20-binding domain) restores substantial but not total SAC activity to bubR1-null mouse fibroblasts, and removing the N-terminal KEN box eliminates this activity and also perturbs K–MT attachments. Our BubR1-KEN mutant is a full-length protein, including the kinase domain, and its failure to provide any detectable SAC activity confirms and extends those observations (and suggests again that the kinase on its own can do little or nothing for the SAC). However, we see no effect of the KEN domain on K–MT attachments.
Much biochemical and genetic data argue that the SAC depends on both Mad2 and BubR1 in a single pathway (Musacchio and Salmon, 2007), but there are studies to the contrary. For example, Orr et al. (2007) found that Drosophila S2 cells could sustain a Mad2-independent (but BubR1 dependent) mitotic arrest under certain conditions. Skoufias et al. (2001) reported that Mad2 and BubR1 detect different kinds of K–MT problems. Moreover, the BubR1–Cdc20 complex on its own is a good in vitro inhibitor of the APC/C (Tang et al., 2001; Chen, 2002; Fang, 2002). Thus, it was just conceivable that the normal mitosis of mad2P flies described in Buffin et al. (2007) depended on a residual SAC activity provided by BubR1.
To test whether cells lacking both putative branches of the SAC could still divide accurately, we made bubR1-KEN mad2P double mutants. These flies were once again viable and fertile, aneuploidy was no higher than in bubR1-KEN or mad2P mutants alone (Table I), and anaphase figures appeared normal (Fig. S2, G and H). This confirms that mitosis in Drosophila is normally highly accurate even in the absence of a functional SAC (Buffin et al., 2007).
Separating the timer from the SAC
The mitotic timer regulates the interval elapsing between NEB and anaphase onset independently of any SAC-imposed delay caused by unattached kinetochores (Meraldi et al., 2004). Depletion of BubR1 or Mad2 (but not other SAC proteins) shortens this interval, and simultaneous depletion of both proteins accelerates the pace even further (Meraldi et al., 2004). This suggests that BubR1 and Mad2 have a timer function, possibly distinct from their checkpoint function, but because only these two SAC proteins bind Cdc20, the timer complex presumably also inhibits the APC/C and may be similar or identical to the SAC complex (Musacchio and Salmon, 2007; Nilsson et al., 2008; Kulukian et al., 2009).
Surprisingly, mitotic timing was normal in bubR1-KEN cells (Fig. 3 B). Whereas mad2P neuroblasts averaged 7.8 min from NEB to anaphase (vs. 9.1 min for WT; P < 0.001), bubR1-KEN timing was no different from WT (9.2 min; Fig. 3, B, C, and F; and Video 6). Thus, even though the SAC was entirely nonfunctional, the timer was intact. However, the timer made by BubR1-KEN required Mad2: in bubR1-KEN mad2P cells, timing averaged 5.4 min, 40% faster than bubR1-KEN or WT and 30% faster than mad2P cells (P < 0.005; Fig. 3, B, F, and G; and Videos 6 and 7).
This acceleration was accompanied by a correspondingly earlier OCBD, starting on average just 3 min after NEB (vs. 4.7 min for mad2; P < 0.02; Fig. 4, B, E, and F), whereas in bubR1-KEN cells, OCBD was similar to WT (6 min from NEB; Fig. 4, A, D, and F; and Videos 8–10). In contrast, the gap from OCBD to anaphase was relatively constant for all genotypes (Fig. 4). That OCBD is affected in the timer-defective mutants suggests that the timer is indeed regulating the activation of the APC/C.
Thus, the dichotomy between timer activity and checkpoint activity exists within BubR1 and can be separated. The KEN domain of BubR1 is specifically required to generate SAC inhibitor but is dispensable for the timer. This raises the possibility that the two complexes may be different biochemical entities or are generated by different pathways.
Alternatively, the difference between timer and checkpoint activity may be simply quantitative. The SAC may need a high inhibitor level, the timer only a low one. If BubR1-KEN still makes the inhibitor, but only inefficiently, it might (with the help of Mad2) still supply the low levels needed for normal timing, whereas the higher levels required for the SAC would need both Mad2 and intact BubR1. In contrast, intact BubR1 alone (in a mad2 mutant) would not make enough inhibitor even for the timer, and BubR1-KEN alone (in the double mutant bubR1-KEN mad2) would make even less, so timing would be faster still.
We have shown that the three mitotic functions of Drosophila BubR1 protein can be uncoupled, as assayed in vivo. The kinase domain mutation perturbs spindle function but has little, if any, effect on the SAC under normal conditions. The N-terminal KEN box mutation eliminates the SAC, but has no effect on the mitotic timer nor on spindle function. This reinforces the conclusion of Buffin et al. (2007) that the SAC is a checkpoint as defined by Hartwell and Weinert, (1989): it is not an intrinsic component of the mitotic machinery but just a surveillance mechanism. The checkpoint phenotype previously ascribed to BubR1 in RNAi knockdown or null mutations (Basu et al., 1999; Kops et al., 2004) is a composite phenotype, the consequences of perturbing spindle function, eliminating the SAC, and accelerating the clock. Being able to separate them should facilitate future study of BubR1’s roles at the kinetochore.
Materials and methods
Flies
The bubR11 mutation (gift from M. Goldberg, Cornell University, Ithaca, New York) of bubR1 (CG7838) is genetic null (Basu et al., 1999; Logarinho et al., 2004). Flies expressing GFP-Rod, RFP-Rod, and the mutations mad2P, cnn allele cnnhk21, and asp allele aspE3 were described previously (Buffin et al., 2005, 2007). GFP-tubulin and GFP–cyclin B flies were a gift from J. Raff (Cancer Research UK Gurdon Institute, Cambridge, England, UK).
Construction of bubR1 K1204A (bubR1-KD) and bubR1-KEN
For BubR1-KD, mutagenizing primers were used to substitute an alanine for the nearly invariant lysine 1,204 (kinase subdomain II; Fig. 1 A). Mutations at the homologous position in vertebrate BubR1 (K795 in human) had already been used in several other studies (Tang et al., 2001; Chen, 2002; Mao et al., 2003; Kops et al., 2004; Harris et al., 2005; Zhao and Chen, 2006), some of which directly confirmed its loss of kinase activity. The mutation was put in both a WT BubR1–expressing P element transgene and a GFP-tagged version of BubR1 (controlled by the natural bubR1 promoter), which fully complements bubR11 and recapitulates endogenous BubR1 behavior during mitosis (Buffin et al., 2005). For BubR1-KEN, residues K7, E8, and N9 (corresponding to the Mad3 N-terminal KEN box shown to be critical to the SAC in yeast; King et al., 2007; Sczaniecka et al., 2008) were changed to A7, A8, and N9 (AAN) by using a mutagenizing primer in a monomeric RFP1 (mRFP1)–BubR1 construct (Buffin et al., 2005). The constructs were introduced into the germ line of w1118 flies by the services of BestGene, Inc. The transgenes (P[bubR1KD], P[GFP-bubR1KD], or P[RFP-bubR1KEN]) were then crossed into the bubR11 genetic background. These flies of genotype bubR11; P[bubR1KD] and bubR11; P[bubR1KEN] are called bubR1-KD and bubR1-KEN flies in this paper. Western blots of transgenic BubR1 variants (Fig. S3) were performed as previously described (Buffin et al., 2007) using the anti-BubR1 previously described (gift from C. Sunkel; Logarinho et al., 2004) and anti–α tubulin (Sigma-Aldrich). In brief, protein extracts from 50 brains of WT, BubR1-KD, BubR1-KEN (each expressed in the bubR11 mutant background), and bubR11 homozygous third instar larvae were loaded onto 8% SDS–acrylamide gels. Proteins were transferred to nitrocellulose membrane (Protran BA 85; Schleicher-Schuell) using an electrophoretic blotting device (Mini protean 3; Bio-Rad Laboratories). Membranes were blocked for 1 h in TBST (TBS and 0.1% Tween) with 5% dry milk and incubated for 1.5 h with anti-BubR1 diluted at 1:4,000 in TBST plus 1% milk. After washing in TBST, the blot was incubated for 1 h at room temperature with secondary antibody of goat anti–rabbit IgG conjugated with horseradish peroxidase (Bio-Rad Laboratories) diluted at 1:60,000. Immunodetection was performed with the SuperSignal kit (Thermo Fisher Scientific). Membranes were stripped and incubated with mouse anti–α-tubulin (Sigma-Aldrich) diluted at 1:1,000 to verify equal loading of proteins.
Cytology
For simple cytology of mitotic cells, third instar larval brains were fixed and stained in aceto-orcein as previously described (Buffin et al., 2007). Larvae were washed in water, and their brains were dissected in a drop of isotonic saline. The tissue was fixed by transferring it to a drop of 45% acetic acid for 30 s, followed by 60% acetic acid for an additional 15 s. Finally, the brains were stained with a drop of aceto-orcein (2% solution in 60% acetic acid) on a clean siliconized coverslip for 1 min. The coverslip was then picked up with a clean microscope slide, and the tissue was squashed with pressure applied from a thumb over the coverslip area. To determine the mitotic density (mean number of mitotic cells per microscopic field) in response to colchicine-induced depolymerization of MTs (Table I), brains were preincubated in 10−4 M colchicine in 0.7% NaCl for 0, 30, or 60 min and transferred to 0.5% Na citrate hypotonic solution for exactly 4 min before staining. The absolute frequency of PSCS was lower using these conditions for both WT and SAC-defective cells, but the relative increase was the same as reported previously using longer hypotonic incubations (Basu et al., 1999; Basto et al., 2000).
To determine aneuploidy, brains were preincubated in 10−4 M colchicine in 0.7% NaCl for 7 min, transferred to 0.5% Na citrate hypotonic solution for 4 min, and then fixed and stained. (This brief colchicine treatment aids in obtaining readable karyotypes; it is not used to test the SAC function.) Cells were observed with a microscope (Microphot; Nikon) and a 63× NA 1.4 phase contrast objective (Carl Zeiss, Inc.). A cell was considered aneuploid if it clearly had at least one extra chromosome.
In vivo imaging
Third instar larval brains were dissected in Shields and Sang M3 insect medium (Sigma-Aldrich) with 10% FBS and penicillin-streptomycin. Three to four larval brains were immediately transferred into 15 µl of M3 medium supplemented with 10% FBS, penicillin-streptomycin, 10 µg/ml insulin, and 5 µg/ml of fly extract and then placed on a standard membrane in a stainless steel slide, as described previously (Siller et al., 2005).
Brains were imaged at room temperature with a spinning disk confocal head (Ultraview; PerkinElmer) mounted on an inverted microscope (DMI6000; Leica) with a 100× NA 1.4 lens and a camera (QuantEM 512SC; Photometrics), all piloted by MetaMorph 7 (MDS Analytical Technologies). At 20-s intervals, a z series of images consisting of seven 1-µm steps was acquired with 1× binning. Confocal video frames are maximum intensity projections. Time-lapse image series were converted into videos with ImageJ software (National Institutes of Health), and still images were processed using Photoshop (Adobe). For mitotic timing, NEB was defined as when the RFP- or GFP-Rod signal began to be visible on kinetochores. Anaphase onset was defined as the moment sister kinetochores (marked with Rod) began to separate. In some films, we followed RFP-BubR1, in which case NEB was defined as the moment the RFP signal on kinetochores began to rapidly intensify (because some signal is present even in prophase; Buffin et al., 2005). To be consistent, only neuroblasts were scored (recognizable by their size and their asymmetric division) and not other cells. For GFP–cyclin B dynamics, neuroblasts expressing one copy of GFP–cyclin B and RFP-Rod were filmed as above. GFP fluorescence in each z section was quantified for the whole cell and for a central region containing the kinetochores and most of the spindle. This gave a more robust measurement of the OCBD because spindle-associated cyclin B is the first to be degraded during metaphase (Buffin et al., 2007). The signal was adjusted for background and for bleaching relative to the signal of a neighboring nonmitotic cell (assumed to be constant). In the graphs for Fig. 4, the signal levels for the whole cell are displayed as normalized signal relative to the maximal intensity measured for the cell.
For the observations of GFP-tubulin in living neuroblasts, brains were gently squashed in a drop of 0.7% saline between a coverslip and a slide. Excess liquid was removed with a paper tissue while observing the flattening of the brain under a dissecting microscope. Once the desired flatness was obtained (the brain typically flattens to a disk ∼2 mm in diameter), the preparation was sealed by drawing a drop of halocarbon oil around the periphery of the coverslip (Buffin et al., 2005). Fluorescent time-lapse videos were acquired with an inverted microscope (IX-70; Olympus), xenon lamp, and camera (OrcaER; Hamamatsu Photonics), piloted by the Cell-R hardware and software system (Olympus). Acquisition times per frame were 100 ms for GFP and 300 ms for mRFP1. Images were collected at 10-, 15-, or 20-s intervals with a 60× NA 1.4 objective and 1× binning.
Online supplemental material
Fig. S1 shows the evolution of neuroblast spindle length as a function of time from NEB. Fig. S2 shows representative anaphase figures from WT and mutant neuroblasts. Fig. S3 shows Western blots comparing expression levels of endogenous BubR1 (WT) with homozygous transgenic lines expressing BubR1-KD and BubR1-KEN in the bubR11 mutant background. Videos 1–3 and 5 show spindle assembly (GFP-tubulin) and mitosis in WT (Video 1), in two examples of bubR1-KD mutant (Videos 2 and 3), and in an example of bubR1-KEN mutant (Video 5) neuroblasts, corresponding to Fig. 1 (B and D–F). Videos 4, 6, and 7 show examples of mitotic timing in bubR1-KD mad2P, bubR1-KEN, and bubR1-KEN mad2P double mutant neuroblasts, respectively, corresponding to Fig. 3 (E–G). Videos 8–10 show GFP–cyclin B degradation during mitosis in WT, bubR1-KEN, and bubR1-KEN mad2P neuroblasts, respectively, corresponding to Fig. 4 (A, D, and E). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200905026/DC1.
Acknowledgments
We thank the Institut Jacques Monod Imagery department for help in confocal acquisition.
Support came in part by funds to R.E. Karess and Z. Rahmani from the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche (grant ANR-08-BLAN-0006-01), the Association pour la Recherche sur le Cancer (ARC), and La Ligue Nationale Contre le Cancer. M.E. Gagou was supported by the Research Training Network program of the European Union, C. Lefebvre was supported by the ARC, and D. Emre was supported by the Ministère de l’Education Nationale et de la Recherche et Technologie.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- KD
- kinase dead
- K-fiber
- kinetochore fiber
- K–MT
- kinetochore–MT
- mRFP1
- monomeric RFP1
- MT
- microtubule
- NEB
- nuclear envelope breakdown
- OCBD
- onset of cyclin B degradation
- PSCS
- premature sister chromatid separation
- SAC
- spindle assembly checkpoint
- WT
- wild type
References
- Basto R., Gomes R., Karess R.E. 2000. Rough deal and Zw10 are required for the metaphase checkpoint in Drosophila. Nat. Cell Biol. 2:939–943 10.1038/35046592 [DOI] [PubMed] [Google Scholar]
- Basu J., Bousbaa H., Logarinho E., Li Z., Williams B.C., Lopes C., Sunkel C.E., Goldberg M.L. 1999. Mutations in the essential spindle checkpoint gene bub1 cause chromosome missegregation and fail to block apoptosis in Drosophila. J. Cell Biol. 146:13–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex. Curr. Biol. 15:856–861 10.1016/j.cub.2005.03.052 [DOI] [PubMed] [Google Scholar]
- Buffin E., Emre D., Karess R.E. 2007. Flies without a spindle checkpoint. Nat. Cell Biol. 9:565–572 10.1038/ncb1570 [DOI] [PubMed] [Google Scholar]
- Chen R.H. 2002. BubR1 is essential for kinetochore localization of other spindle checkpoint proteins and its phosphorylation requires Mad1. J. Cell Biol. 158:487–496 10.1083/jcb.200204048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowe S., Hümmer S., Uldschmid A., Li X., Nigg E.A. 2007. Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21:2205–2219 10.1101/gad.436007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G. 2002. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol. Biol. Cell. 13:755–766 10.1091/mbc.01-09-0437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goshima G., Nédélec F., Vale R.D. 2005. Mechanisms for focusing mitotic spindle poles by minus end–directed motor proteins. J. Cell Biol. 171:229–240 10.1083/jcb.200505107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris L., Davenport J., Neale G., Goorha R. 2005. The mitotic checkpoint gene BubR1 has two distinct functions in mitosis. Exp. Cell Res. 308:85–100 10.1016/j.yexcr.2005.03.036 [DOI] [PubMed] [Google Scholar]
- Hartwell L.H., Weinert T.A. 1989. Checkpoints: controls that ensure the order of cell cycle events. Science. 246:629–634 10.1126/science.2683079 [DOI] [PubMed] [Google Scholar]
- Huang H., Hittle J., Zappacosta F., Annan R.S., Hershko A., Yen T.J. 2008. Phosphorylation sites in BubR1 that regulate kinetochore attachment, tension, and mitotic exit. J. Cell Biol. 183:667–680 10.1083/jcb.200805163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King E.M., van der Sar S.J., Hardwick K.G. 2007. Mad3 KEN boxes mediate both Cdc20 and Mad3 turnover, and are critical for the spindle checkpoint. PLoS One. 2:e342 10.1371/journal.pone.0000342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kops G.J., Foltz D.R., Cleveland D.W. 2004. Lethality to human cancer cells through massive chromosome loss by inhibition of the mitotic checkpoint. Proc. Natl. Acad. Sci. USA. 101:8699–8704 10.1073/pnas.0401142101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulukian A., Han J.S., Cleveland D.W. 2009. Unattached kinetochores catalyze production of an anaphase inhibitor that requires a Mad2 template to prime Cdc20 for BubR1 binding. Dev. Cell. 16:105–117 10.1016/j.devcel.2008.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M.A., Kapoor T.M. 2005. The human mitotic checkpoint protein BubR1 regulates chromosome-spindle attachments. Nat. Cell Biol. 7:93–98 10.1038/ncb1208 [DOI] [PubMed] [Google Scholar]
- Logarinho E., Bousbaa H., Dias J.M., Lopes C., Amorim I., Antunes-Martins A., Sunkel C.E. 2004. Different spindle checkpoint proteins monitor microtubule attachment and tension at kinetochores in Drosophila cells. J. Cell Sci. 117:1757–1771 10.1242/jcs.01033 [DOI] [PubMed] [Google Scholar]
- Maia A.F., Lopes C.S., Sunkel C.E. 2007. BubR1 and CENP-E have antagonistic effects upon the stability of microtubule-kinetochore attachments in Drosophila S2 cell mitosis. Cell Cycle. 6:1367–1378 [DOI] [PubMed] [Google Scholar]
- Maiato H., Sampaio P., Lemos C.L., Findlay J., Carmena M., Earnshaw W.C., Sunkel C.E. 2002. MAST/Orbit has a role in microtubule–kinetochore attachment and is essential for chromosome alignment and maintenance of spindle bipolarity. J. Cell Biol. 157:749–760 10.1083/jcb.200201101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Rieder C.L., Khodjakov A. 2004. Kinetochore-driven formation of kinetochore fibers contributes to spindle assembly during animal mitosis. J. Cell Biol. 167:831–840 10.1083/jcb.200407090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malureanu L.A., Jeganathan K.B., Hamada M., Wasilewski L., Davenport J., van Deursen J.M. 2009. BubR1 N terminus acts as a soluble inhibitor of cyclin B degradation by APC/C(Cdc20) in interphase. Dev. Cell. 16:118–131 10.1016/j.devcel.2008.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y., Abrieu A., Cleveland D.W. 2003. Activating and silencing the mitotic checkpoint through CENP-E-dependent activation/inactivation of BubR1. Cell. 114:87–98 10.1016/S0092-8674(03)00475-6 [DOI] [PubMed] [Google Scholar]
- Meraldi P., Draviam V.M., Sorger P.K. 2004. Timing and checkpoints in the regulation of mitotic progression. Dev. Cell. 7:45–60 10.1016/j.devcel.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nilsson J., Yekezare M., Minshull J., Pines J. 2008. The APC/C maintains the spindle assembly checkpoint by targeting Cdc20 for destruction. Nat. Cell Biol. 10:1411–1420 10.1038/ncb1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr B., Bousbaa H., Sunkel C.E. 2007. Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells. Mol. Biol. Cell. 18:850–863 10.1091/mbc.E06-07-0587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Maiato H. 2004. Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell. 7:637–651 10.1016/j.devcel.2004.09.002 [DOI] [PubMed] [Google Scholar]
- Sczaniecka M., Feoktistova A., May K.M., Chen J.S., Blyth J., Gould K.L., Hardwick K.G. 2008. The spindle checkpoint functions of Mad3 and Mad2 depend on a Mad3 KEN box-mediated interaction with Cdc20-anaphase-promoting complex (APC/C). J. Biol. Chem. 283:23039–23047 10.1074/jbc.M803594200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siller K.H., Serr M., Steward R., Hays T.S., Doe C.Q. 2005. Live imaging of Drosophila brain neuroblasts reveals a role for Lis1/dynactin in spindle assembly and mitotic checkpoint control. Mol. Biol. Cell. 16:5127–5140 10.1091/mbc.E05-04-0338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoufias D.A., Andreassen P.R., Lacroix F.B., Wilson L., Margolis R.L. 2001. Mammalian mad2 and bub1/bubR1 recognize distinct spindle-attachment and kinetochore-tension checkpoints. Proc. Natl. Acad. Sci. USA. 98:4492–4497 10.1073/pnas.081076898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Bharadwaj R., Li B., Yu H. 2001. Mad2-Independent inhibition of APCCdc20 by the mitotic checkpoint protein BubR1. Dev. Cell. 1:227–237 10.1016/S1534-5807(01)00019-3 [DOI] [PubMed] [Google Scholar]
- Truman J.W., Bate M. 1988. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev. Biol. 125:145–157 10.1016/0012-1606(88)90067-X [DOI] [PubMed] [Google Scholar]
- Williams B.C., Li Z., Liu S., Williams E.V., Leung G., Yen T.J., Goldberg M.L. 2003. Zwilch, a new component of the ZW10/ROD complex required for kinetochore functions. Mol. Biol. Cell. 14:1379–1391 10.1091/mbc.E02-09-0624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H. 2002. Regulation of APC-Cdc20 by the spindle checkpoint. Curr. Opin. Cell Biol. 14:706–714 10.1016/S0955-0674(02)00382-4 [DOI] [PubMed] [Google Scholar]
- Zhang J., Ahmad S., Mao Y. 2007. BubR1 and APC/EB1 cooperate to maintain metaphase chromosome alignment. J. Cell Biol. 178:773–784 10.1083/jcb.200702138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y., Chen R.H. 2006. Mps1 phosphorylation by MAP kinase is required for kinetochore localization of spindle-checkpoint proteins. Curr. Biol. 16:1764–1769 10.1016/j.cub.2006.07.058 [DOI] [PubMed] [Google Scholar]