Abstract
During somatic hypermutation (SHM), B cells introduce mutations into their immunoglobulin genes to generate high affinity antibodies. Current models suggest a separation in the generation of G/C transversions by the Ung2-dependent pathway and the generation of A/T mutations by the Msh2/ubiquitinated proliferating cell nuclear antigen (PCNA-Ub)–dependent pathway. It is currently unknown whether these pathways compete to initiate mutagenesis and whether PCNA-Ub functions downstream of Ung2. Furthermore, these models do not explain why mice lacking Msh2 have a more than twofold reduction in the total mutation frequency. Our data indicate that PCNA-Ub is required for A/T mutagenesis downstream of both Msh2 and Ung2. Furthermore, we provide evidence that both pathways are noncompetitive to initiate mutagenesis and even collaborate to generate half of all G/C transversions. These findings significantly add to our understanding of SHM and necessitate an update of present SHM models.
To generate high affinity antibodies, germinal center (GC) B cells are enabled to introduce point mutations into the variable region of their rearranged Ig genes. This process of somatic hypermutation (SHM) occurs at an astonishing rate of one per thousand bases per generation, six orders of magnitude greater than spontaneous mutagenesis (Di Noia and Neuberger, 2007). SHM is initiated by the activation-induced cytidine deaminase (AID), an enzyme found to be differentially expressed in B cells of the GC (Muramatsu et al., 2000). AID deaminates C to U within single-stranded DNA, and targets both DNA strands in the variable and switch regions of Ig genes. To establish point mutations at and around the U, three alternative pathways can handle this initial lesion (Fig. 1). (a) Replication across a U instructs a template T to DNA polymerases and generates G/C to A/T transitions (Rada et al., 2004; Shen et al., 2006). (b) The U can be excised from the DNA backbone by the base excision repair (BER) protein Ung2, and an abasic or apyrimidinic (AP) site is generated, causing replicative DNA polymerases to stall. To continue replication, specialized translesion synthesis (TLS) polymerases can be recruited, enabling a direct replicative bypass of AP sites. As AP sites are noninstructive, these TLS polymerases generate G/C transversions and may contribute to G/C transitions (Ung2-dependent SHM). Accordingly, Ung mutant B cells lack most G/C transversions (Rada et al., 2002). (c) Alternatively, the U can be recognized as a U:G mismatch by the mismatch repair complex Msh2–Msh6, resulting in Exo-1 activation and the formation of a single-stranded gap around the initial mismatch. Interestingly, Msh2-, Msh6-, and Exo-1–deficient B cells lack 80–90% of all A/T mutations, suggesting that the gap-filling process is executed by TLS polymerases predominantly generating A/T mutations (Msh2-dependent SHM; Rada et al., 1998; Wiesendanger et al., 2000; Bardwell et al., 2004). As a significant fraction of A/T mutations (10–20%) are found in Msh2-deficient GC B cells but not in Ung2/Msh2 double-deficient GC B cells, Ung2-dependent mutagenesis generates the described fraction of A/T mutations (Rada et al., 2004). Whether Ung2-dependent A/T mutations are generated during long-patch BER (i.e., within the strand containing the AP site) or, alternatively, during the extension phase of TLS across the AP site is currently unknown. In summary, these data suggest a specific role of these pathways in recruiting and activating selective TLS polymerases to establish defined mutations. The combination of these pathways enables hypermutating B cells to generate the entire spectrum of nucleotide substitutions.
Figure 1.
Current model: pathways of SHM downstream of AID. The three pathways: (a) replication across U, (b) Ung2-dependent SHM, and (c) Msh2-dependent SHM. Known and unknown (?) polymerases involved in the generation of specific mutations are indicated. TS, transitions; TV, transversions.
In contrast to replicative DNA polymerases, TLS polymerases lack proofreading activity. The capacity of TLS polymerases to accommodate non–Watson–Crick base pairs within their catalytic center is beneficial regarding the accurate replication across modified bases, such as UV-C–induced cyclic pyrimidine dimers by polymerase η. However, TLS polymerases are highly mutagenic when replicating across undamaged DNA and defined lesions such as AP sites (Prakash et al., 2005; Jansen et al., 2007). Because each polymerase displays its own mutagenic signature, alterations in the mutation spectrum can often be attributed retrospectively to the absence of, or failure in activating, specific polymerases. The Y family of DNA polymerases comprises four members, of which at least polymerase η, Rev1, and to some degree polymerase κ are implicated in SHM. Rev1-deficient B cells display reduced frequencies of G/C to C/G transversions (Jansen et al., 2006; Ross and Sale, 2006), suggesting that Rev1 functions downstream of Ung2. In agreement, Rev1 is an effective cytidyl transferase when bypassing abasic sites in vitro (Nelson et al., 1996). In contrast, polymerase η is ineffective in handling abasic sites (Haracska et al., 2001) and preferentially inserts mismatched nucleotides opposite template T (Rogozin et al., 2001). Consistent with these in vitro data, the mutation spectra of polymerase η–deficient B cells from humans and mice lack a significant fraction of A/T mutations, suggesting polymerase η to be used mainly downstream of Msh2 (Zeng et al., 2001; Delbos et al., 2005; Martomo et al., 2005). In addition to its role downstream of Msh2, polymerase η was responsible for the remaining A/T mutations downstream of Ung2, as shown in Msh2-deficient mice (Delbos et al., 2007). Although the lack of polymerase κ had no effect on SHM (Schenten et al., 2002), polymerase κ was found to generate A/T mutations in case of polymerase η deficiency (Faili et al., 2009). Thus, polymerase κ can substitute polymerase η, whereas Rev1 cannot, as revealed by the normal generation of G to C transversions in polymerase η–deficient mice.
The question remains how and when these error-prone polymerases become used to establish specific nucleotide substitutions. During replication, the DNA sliding clamp proliferating cell nuclear antigen (PCNA) functions as a critical processivity factor by tethering the DNA polymerases to the DNA template. Studies in yeast and subsequently in mammalian cells revealed an important role for site-specific monoubiquination of PCNA at lysine 164 (ubiquitinated PCNA [PCNA-Ub]) in recruiting and activating TLS polymerases upon replication fork stalling (Hoege et al., 2002; Stelter and Ulrich, 2003; Kannouche et al., 2004). In mice and chicken DT40 B cells homozygous for a PCNAK164R mutation, we and others recently demonstrated a critical role for PCNA-Ub in SHM (Arakawa et al., 2006; Langerak et al., 2007; Roa et al., 2008). As shown in memory B cells of PCNAK164R knockin mice, the failure to modify PCNA at lysine 164 caused a 10-fold decrease in mutations at template A/T, normally accounting for 50% of all mutations in WT mice. This decrease was accompanied by an overall 25% reduction in the mutation frequency. The lack of A/T mutations indicated a role for PCNA-Ub downstream of Msh2. In strong contrast, data from DT40 cells suggested a role for PCNA-Ub in generating the vast majority of G/C transversions, placing PCNA-Ub downstream of Ung2.
To dissect the role of PCNA-Ub in Ung2- and Msh2-dependent SHM, we intercrossed the PCNAK164R knockin strain of mice with mice deficient for Ung or Msh2. Comparing the efficacy of Ung2- and Msh2-dependent mutagenesis revealed that both pathways strongly depend on PCNA-Ub to generate mutations at template A/T but not at G/C. Furthermore, in addition to its role in the generation of A/T mutations, we identified a previously unknown role for Msh2 in generating 50% of all G/C transversions. Finally, we provide evidence for the employment of Ung2 and Msh2 in a noncompetitive manner in processing uracils downstream of AID, supporting the idea of a temporal separation of these pathways (Weill and Reynaud, 2008).
RESULTS AND DISCUSSION
We previously reported on the analysis of SHM in the JH4 intronic region of memory B cells from homozygous PCNAK164R mice (Langerak et al., 2007). As memory B cells compromised for SHM might resemble a selected pool of cells that have compensated their defect by entering additional rounds of GC reactions, the mutation frequency in these cells does not provide a good measure for the efficacy in generating point mutations during SHM. To study the impact of PCNA-Ub on the generation of somatic mutations more directly, we analyzed unselected Peyer's patch GC B cells of PCNAK164R mutant mice in this report. To dissect the contribution of PCNA-Ub in establishing mutations downstream of Ung2 and Msh2, we studied the intercrosses of PCNAK164R knockin mice with Ung or Msh2 mutant mice.
WT, single-, and double-mutant mice were analyzed for nonselected mutations in rearranged JH4 intronic regions. Compared with WT mice, both PCNAK164R and Msh2 single mutants showed a reduction of 36 and 61%, respectively, in the mutation frequency (0.94, 0.6, and 0.37%, respectively; Table I). The decreased mutation frequency is further revealed by the changes in mutation load between the different strains of mice (Fig. S1). Given the fact that both PCNA-Ub and Msh2 play a critical role in the generation of A/T mutations, the 1.7-fold difference in the mutation frequencies between PCNAK164R (0.6%) and Msh2 (0.37%) mutant B cells is quite striking. This difference might be attributed to a selective growth/survival disadvantage of Msh2-deficient B cells, to a significant contribution of Msh2 to G/C mutations, and/or to suppression of G/C mutations by PCNA-Ub (see Ung2-dependent G/C transversions… and PCNA-Ub selectively suppresses G/C transitions…, respectively). In addition, a marginal increase in the mutation frequency and mutation load in Ung-deficient GC B cells was detected (1.13 vs. 0.94%; Table I and Fig. S1). This latter finding is consistent with previous publications (Rada et al., 1998; Rada et al., 2002) and suggests that the 20% higher mutation frequency in Ung-deficient mice is caused by partial Ung-dependent BER in WT mice, implicating a role for Ung in error-free repair of AID-created uracils in Ig genes. Alternatively, accurate replication of a proportion of AP sites might occur, i.e., the insertion of a G opposite an AP site. Furthermore, the mutation frequency in GC B cells of PCNAK164R/Ung double-mutant mice resembled that of the PCNAK164R single mutant (0.63 vs. 0.6%), whereas the frequency detected in the PCNAK164R/Msh2 mutant mice resembled that of the Msh2 single mutants (0.4 vs. 0.37%).
Table I.
SHM frequencies in JH4 introns of GC B cells
| WT | K164R | Ung | K164R × Ung | Msh2 | K164R × Msh2 | |
| Number of mice | 5 | 5 | 2 | 2 | 3 | 3 |
| Number of mutated sequences | 234 | 183 | 76 | 90 | 114 | 90 |
| Total mutations | 1,083 | 563 | 442 | 290 | 216 | 181 |
| Total bp sequenced | 115,695 | 93,356 | 39,049 | 46,008 | 58,049 | 45,647 |
| Mutations/bp (%) | 0.9 (0.6–1.5) | 0.6 (0.4–0.7) | 1.1 (1–1.3) | 0.6 (0.6–0.7) | 0.4 (0.3–0.4) | 0.4 (0.3–0.5) |
A/T mutations downstream of Ung2 and Msh2 depend on PCNA-Ub
Comparing the mutation profiles (Fig. 2), both PCNAK164R and Msh2 mutant B cells exhibit a strong reduction in all A/T mutations, whereas Ung2-deficient B cells lacked most C/G transversion mutations, as previously described (Rada et al., 1998; Rada et al., 2002; Langerak et al., 2007). When comparing Msh2 to PCNAK164R/Msh2 mutant B cells, it becomes clear that Ung2-dependent A/T mutations, which have recently been shown to be introduced by polymerase η (Delbos et al., 2007), disappear almost completely in PCNAK164R/Msh2 mutant GC B cells, indicating that most Ung-dependent A/T mutations require PCNA-Ub to activate polymerase η (Fig. 2).
Figure 2.
Mutations in rearranged JH4 intronic sequences from WT and PCNAK164R, Msh2, Ung, PCNAK164R x Ung, and PCNAK164R x Msh2 mutant mice. (A) Pattern of nucleotide substitution. Values are expressed as the total number of mutations and the percentage of total mutations before and after correction for base composition. (B) Relative contribution of A/T mutations, G/C transitions, and G/C transversions. Values are expressed as the percentage of total mutations. The number of mice analyzed is indicated in Table I.
To analyze the role of PCNA-Ub in Msh2-dependent mutagenesis, we compared SHM in B cells from Ung mutant and PCNAK164R/Ung double-mutant mice. Although polymerase η has been proven to be responsible for the introduction of most A/T mutations downstream of Msh2, the lack of polymerase η revealed that the remaining A/T mutation activity depends on polymerase κ and at least a third as yet unidentified polymerase (Faili et al., 2009). The combined failure in removing uracils by Ung2 and ubiquinating PCNA resulted in a mutation spectrum in which almost all mutations (99.3%) were G/C transition mutations, a finding comparable to Ung/Msh2 double mutants previously described (Rada et al., 2004; Shen et al., 2006). These data indicate that during Msh2-dependent SHM, TLS polymerases η and κ both depend on PCNA-Ub to establish most A/T mutations. In summary, we suggest a model in which PCNA-Ub acts downstream of both Msh2 and Ung2 to ensure that TLS polymerase η (and in its absence also polymerase κ) is recruited to introduce mutations at template A/T, and high fidelity polymerases are prohibited from error-free replication of intact A/T templates. The remaining A/T mutations (3.4%) found in the PCNAK164R mutant might be generated in a PCNA-Ub–independent manner by polymerase η, polymerase κ, or an as yet unidentified polymerase. Future intercrosses between PCNAK164R mutant, polymerase η–, and polymerase κ–deficient mice should help to distinguish between these possibilities.
PCNA-Ub selectively suppresses G/C transitions but not transversions.
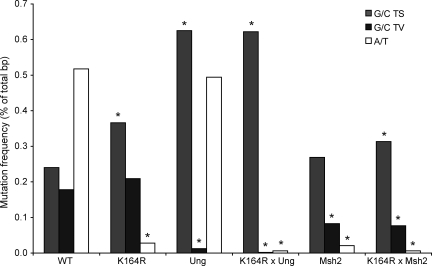
To determine the contribution of PCNA-Ub in generating G/C transitions and transversions, the mutation frequencies for each individual base substitution in GC B cells from WT and PCNAK164R mice were compared (Fig. 3 and Fig. S2). Interestingly, the generation of G/C transitions was increased in PCNAK164R mutant B cells, suggesting that PCNA-Ub normally counteracts/suppresses the accumulation of G/C transition mutations. These data suggest a model in which the U-containing strand is removed and PCNA-Ub is required to recruit a polymerase that inserts a C opposite the G on the other strand. In contrast to G/C transitions, the generation of G/C transversions occurred equally efficiently in PCNAK164R mutant GC B cells. Given the role for the TLS polymerase Rev1 in generating G to C transversions during SHM (Jansen et al., 2006; Ross and Sale, 2006), these findings exclude a role of PCNA-Ub in activating Rev1 and all other as yet unidentified G/C transverters during SHM in mammals. In agreement, damage tolerance mediated by Rev1 was found to be independent of PCNA-Ub in the chicken DT40 B cell line (Edmunds et al., 2008). Surprisingly, in PCNAK164R mutant DT40 cells, Rev1 depends on PCNA-Ub in generating G to C transversions during SHM (Arakawa et al., 2006). These results further indicate a specific role of PCNA-Ub in stimulating selectively A/T but not G/C mutations downstream of Ung2 and Msh2.
Figure 3.
Absolute frequency mutations grouped into G/C transitions, G/C transversions, and A/T mutations. Absolute mutation frequency of A/T mutations, G/C transitions (TS), and G/C transversions (TV). Values are expressed as the percentage of total sequenced base pairs from mutated clones. The contribution of single nucleotide substitutions is revealed in Fig. S2. Asterisks indicate significant changes compared with WT (χ2 test; all significant changes had a p-value <10−5, and nonsignificant changes had a p-value >0.1). The number of mice analyzed is indicated in Table I.
Distribution of mutations in GC B cells
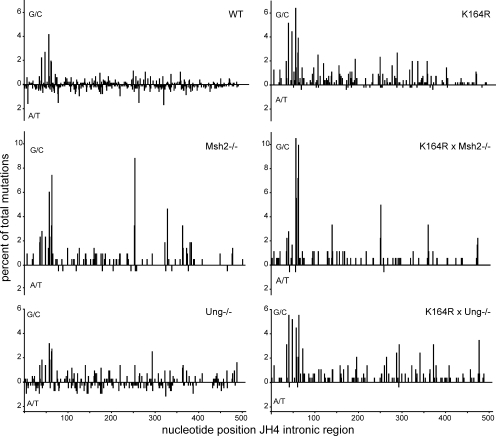
To compare the distribution of mutations of WT, single-, or double-mutant GC B cells, we plotted the frequency of G/C and A/T mutations as a percentage of total mutations against the nucleotide position within the JH4 intronic region (Fig. 4). As mentioned before, A/T mutations were strongly reduced in Msh2 and PCNAK164R single- and double-mutant B cells. Comparing the distribution of G/C mutations, except for some previously defined hotspots (Frey et al., 1998; Rada et al., 1998; Martomo et al., 2004; Delbos et al., 2007), no difference was observed regarding the overall distribution of the remaining G/C mutations in Msh2 and PCNAK164R/Msh2 mutant B cells. Furthermore, the failure to ubiquinate PCNA did not interfere with the distribution of G/C mutations, a finding consistent with the major role of PCNA-Ub in recruiting polymerases η and κ in establishing mutations at template A/T.
Figure 4.
Mutation distribution in the JH4 intronic region. The distribution of all G/C (above x axis) and A/T (below x axis) mutations along the JH4 intronic region starting from the splice donor are indicated as the percentage of total mutations. The genetic backgrounds are indicated. The number of mice analyzed is indicated in Table I.
Hotspot targeting in PCNAK164R mutant B cells
During SHM, mutations at template G/C are frequently found in RGYW/WRCY (W = A/T, R = A/G, and Y= C/T) hotspot motifs (Rogozin and Kolchanov, 1992). Consistent with this observation, AID was found to preferentially deaminate WRC motifs in vitro (Pham et al., 2003). To compare hotspot targeting between WT, single-, and double-mutant mice, we analyzed the percentage of mutations inside of RGYW/WRCY and GYW/WRC hotspots. Because of the lack of A/T mutations in PCNAK164R and Msh2 single and double mutants, there was a twofold increase of hotspot mutations in these mice when considering their relative contribution to all mutations. However, as hotspots overlap with AID deamination sites, we compared the targeting of G/C mutations (underlined) within RGYW/WRCY and GYW/WRC. No major differences were observed between WT and mutant GC B cells (Fig. S3), indicating that virtually all G/C mutations were generated directly downstream of AID or Ung2 during replication across U or AP sites, respectively.
Ung2 and Msh2 mutagenesis are noncompetitive
Whether the Ung2 and Msh2 pathways of SHM compete for the recognition of a U or act noncompetitively is a long-standing question (Di Noia and Neuberger, 2007; Weill and Reynaud, 2008). In the latter case, the common substrate U is expected to be processed in an error-free or error-prone manner to exclude that a U not processed by Ung2 enters Msh2-dependent mutagenesis and vice versa. To address this issue of SHM, it is essential to compare the absolute efficacies in generating defined nucleotide substitutions between the different mouse models. These absolute mutation frequencies of specific base substitution in GC B cells from WT as well as Ung, Msh2, and PCNAK164R single- and double-mutant mice are shown in Fig. 3 and Fig. S2.
As previously described, no major differences were found in the mutation frequencies of WT and Ung-deficient GC B cells (Rada et al., 2002), suggesting that the lack of Ung2-dependent mutagenesis can be compensated effectively by replication across U (G/C transitions) and/or Msh2-dependent mutagenesis. To determine whether Msh2 is capable of removing uracils that otherwise would be processed by Ung2, we compared the efficacy in the generation of A/T mutations between WT and Ung2-deficient B cells. If Msh2-dependent mutagenesis competes with Ung2 mutagenesis, one expects A/T mutations to increase in the absence of Ung2. When comparing the generation of A/T mutations, the lack of U removal by Ung2 did not increase the efficacy in generating A/T mutations (Fig. 3). These data reveal that Msh2-dependent SHM does not compete with Ung2-dependent mutagenesis, a finding consistent with the noncompetitive model of SHM. In line with this model, Msh2 appeared incapable of removing uracils usually processed by Ung2, as revealed by the selective and compensatory increase of G/C transitions as a consequence of direct replication over the U. This quantitative analysis provides direct evidence that uracils normally recognized and processed by Ung2 remain refractory to Msh2-dependent SHM in Ung2-deficient B cells.
To analyze the reverse situation, i.e., to determine whether Ung2 is capable of removing uracils normally processed by Msh2, we compared the efficacy in the generation of G/C transversions between WT and Msh2-deficient B cells. If Ung2 mutagenesis competes with Msh2, one expects G/C transversions to increase in the absence of Msh2. Strikingly, when comparing the generation of G/C transversions, the lack of mismatch recognition resulted in a twofold decrease in the frequency of these mutations. As nearly all G/C transversions depend on Ung2 as revealed in Ung2-deficient B cells, these data support not only the noncompetitive model but also indicate that 50% of all G/C transversions actually require a synergistic action of Msh2 and Ung2 (see following paragraph). Furthermore, the lack of mismatch recognition did not grossly change the efficacy in generating G/C transitions, suggesting that before replication uracils usually processed by Msh2 are now repaired by BER.
These selective changes in the mutation spectra between WT, Msh2, Ung2, and PCNAK164R mutant GC B cells further support the noncompetitive model between Ung2- and Msh2-dependent mutagenesis. This concept is in line with the model recently proposed by Weill and Reynaud (2008), in which both pathways are suggested to be active in S and G1, respectively.
Ung2-dependent G/C transversions downstream of Msh2
It has been reported that Msh2-deficient mice have a reduced mutation frequency that mainly has been attributed to a defect in establishing A/T mutations (Rada et al., 1998). Simultaneously, a relative increase of G/C transitions over G/C transversions was found in Msh2-deficient B cells. It has therefore been suggested that uracils usually processed by Msh2 are not repaired before replication, leading to the generation of transitions. However, as already mentioned, when comparing the efficacy in generating G/C transitions, no difference was found between WT and Msh2-deficient mice. Moreover, the lack of Msh2 resulted in a selective twofold reduction of G/C transversions as compared with WT and PCNAK164R mutant B cells. Therefore the 60% reduction in mutation frequency in Msh2-deficient GC B cells as compared with WT (0.37 vs. 0.94%) is likely explained by a defect in the generation of most A/T mutations and the 50% reduction in G/C transversions. In addition, the lower mutation frequency found in Msh2-deficient GC B cells as compared with PCNAK164R mutant GC B cells (0.37 vs. 0.6%) is likely explained by an increase in G/C transitions in PCNAK164R cells and a decrease in G/C transversions in Msh2 mutant B cells.
Concluding remarks
In conclusion, (a) PCNA-Ub acts downstream of Ung2 and Msh2 to generate A/T mutations by selectively activating the TLS polymerases η and κ, (b) PCNA-Ub can counteract G/C transitions but not G/C transversions, (c) about half of all G/C transversions depend on the synergistic action of Ung2 and Msh2, and (d) uracils are processed noncompetitively downstream of Msh2 and Ung2.
Combining these and previous findings, we propose an updated model regarding the generation of base substitutions during SHM in which uracils are introduced on both strands at distinct phases of the cell cycle (Aoufouchi et al., 2008). U introduced during the S phase can directly act as template T during replication to generate G/C transitions or be converted to an AP site by Ung2. If not repaired, the AP site causes replicative polymerase to stall and activate a TLS polymerase to initiate the generation of both G/C transversions and transitions. In addition, we suggest that Ung2-dependent A/T mutations are generated by the combined action of PCNA-Ub and polymerase η during the extension phase of TLS across the AP site, or alternatively, during long-patch BER of the strand containing the AP site. Uracils generated outside the S phase would be recognized as a U:G mismatch by Msh2–Msh6, resulting in the formation of an Exo-1–generated single-stranded gap. The fill-in reaction requires the combined action of PCNA-Ub and polymerase η (or polymerase κ in the absence of polymerase η) to generate mutations at template A/T. We now suggest that during this gap-filling process polymerase η or high fidelity polymerases become stalled upon encountering an AP site. The subsequent activation of specialized TLS polymerases (such as Rev1) enables TLS across the AP site, explaining the proposed synergism between Msh2 and Ung2 in generating G/C transversions.
Three scenarios can explain the existence of Ung2-dependent AP sites within the Msh2/Exo-1–dependent single-stranded gap: (a) the AP site may preexist as a result of the combined action of AID and Ung2 before the gap formation or (b) a U exists in the single-stranded gap and as such is efficiently removed by Ung2, and/or (c) secondary deamination by AID takes place on the single-stranded gap, and the U is immediately processed into an AP site by Ung2. Further studies should reveal which of these sources of AP substrates contribute to the generation of Ung2/Msh2-dependent G/C transversions.
MATERIALS AND METHODS
Mice.
The generation and genotyping of PCNAK164R knockin, Ung-deficient (provided by D. Barnes and T. Lindahl, Cancer Research UK London Research Institute, London, England, UK), and Msh2-deficient (provided by H. te Riele, The Netherlands Cancer Institute, Amsterdam, Netherlands) mice and the Flpe deleter strain (provided by S. Dymecki, Harvard Medical School, Boston, MA) have been previously described (de Wind et al., 1995; Nilsen et al., 2000; Rodríguez et al., 2000; Langerak et al., 2005; Langerak et al., 2007). PCNAK164R mice were crossed to the Flpe deleter strain to delete the selection cassette from the targeted PCNA allele and crossed with Msh2 and Ung mutant mice to generate WT, single-, and double-mutant mice. Mice were maintained at the animal facility of the Netherlands Cancer Institute. All experiments were approved by an independent animal ethics committee of the Netherlands Cancer Institute.
Isolation of GC B cells and mutation analysis.
GC (CD19+, PNAhigh, CD95+) B cells were sorted from Peyer's patches of 2-mo-old animals. DNA was extracted using proteinase K treatment and ethanol precipitation. The JH4 3′ flanking intronic sequence of endogenous rearrangements of VHJ558 family members were amplified during 40 cycles of PCR using PfuUltra polymerase (Agilent Technologies; Jolly et al., 1997). PCR products were purified using a gel extraction kit (QIAquick; QIAGEN), cloned into a blunt vector (TOPO II; Invitrogen), and sequenced on a 3730 DNA Analyzer (Applied Biosystems). Sequence alignment was performed using Seqman software (DNASTAR). Data derived from WT and PCNAK164R littermates of Msh2 and Ung2 breedings with PCNAK164R were combined. Calculations exclude nonmutated sequences, insertions, and deletions. Clonally related sequences were counted only once.
Online supplemental material.
Fig. S1 depicts the accumulation of mutations in individual JH4 intronic sequences. Fig. S2 depicts the absolute mutation frequency of individual nucleotide substitutions. Fig. S3 depicts the hotspot targeting to RGYW/WRCY, and GYW/WRC motifs. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091707/DC1.
Acknowledgments
We thank F. van Diepen and A. Pfauth for cell sortings, the Netherlands Cancer Institute–Antoni van Leeuwenhoek Hospital (NKI-AVL) mouse facility for handling of the mice, and the NKI-AVL sequence facility for sequencing. Msh2-deficient mice were provided by H. te Riele, Ung-deficient mice were provided by D. Barnes and T. Lindahl through C.-A. Reynaud and J.-C. Weill, and Flpe deleter mice were provided by S. Dymecki. We also thank D. Withers, J. Borst, T. Sixma, and N. Wit for critically reading the manuscript. We apologize to everyone whose work could only be cited indirectly because of space limitations.
This project was supported by the Netherlands Organization for Scientific Research (Vidi grant NWO 917.56.328 to H. Jacobs).
The authors declare that they have no conflict of interest.
Footnotes
Abbreviations used:
- AID
- activation-induced cytidine deaminase
- AP
- apyrimidinic
- BER
- base excision repair
- GC
- germinal center
- PCNA-Ub
- ubiquitinated proliferating cell nuclear antigen
- SHM
- somatic hypermutation
- TLS
- translesion synthesis
References
- Aoufouchi S., Faili A., Zober C., D'Orlando O., Weller S., Weill J.C., Reynaud C.A. 2008. Proteasomal degradation restricts the nuclear lifespan of AID. J. Exp. Med. 205:1357–1368 10.1084/jem.20070950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakawa H., Moldovan G.L., Saribasak H., Saribasak N.N., Jentsch S., Buerstedde J.M. 2006. A role for PCNA ubiquitination in immunoglobulin hypermutation. PLoS Biol. 4:e366 10.1371/journal.pbio.0040366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardwell P.D., Woo C.J., Wei K., Li Z., Martin A., Sack S.Z., Parris T., Edelmann W., Scharff M.D. 2004. Altered somatic hypermutation and reduced class-switch recombination in exonuclease 1-mutant mice. Nat. Immunol. 5:224–229 10.1038/ni1031 [DOI] [PubMed] [Google Scholar]
- Delbos F., De Smet A., Faili A., Aoufouchi S., Weill J.C., Reynaud C.A. 2005. Contribution of DNA polymerase η to immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 201:1191–1196 10.1084/jem.20050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delbos F., Aoufouchi S., Faili A., Weill J.C., Reynaud C.A. 2007. DNA polymerase η is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 204:17–23 10.1084/jem.20062131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wind N., Dekker M., Berns A., Radman M., te Riele H. 1995. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 82:321–330 [DOI] [PubMed] [Google Scholar]
- Di Noia J.M., Neuberger M.S. 2007. Molecular mechanisms of antibody somatic hypermutation. Annu. Rev. Biochem. 76:1–22 10.1146/annurev.biochem.76.061705.090740 [DOI] [PubMed] [Google Scholar]
- Edmunds C.E., Simpson L.J., Sale J.E. 2008. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 30:519–529 10.1016/j.molcel.2008.03.024 [DOI] [PubMed] [Google Scholar]
- Faili A., Stary A., Delbos F., Weller S., Aoufouchi S., Sarasin A., Weill J.C., Reynaud C.A. 2009. A backup role of DNA polymerase kappa in Ig gene hypermutation only takes place in the complete absence of DNA polymerase eta. J. Immunol. 182:6353–6359 10.4049/jimmunol.0900177 [DOI] [PubMed] [Google Scholar]
- Frey S., Bertocci B., Delbos F., Quint L., Weill J.C., Reynaud C.A. 1998. Mismatch repair deficiency interferes with the accumulation of mutations in chronically stimulated B cells and not with the hypermutation process. Immunity. 9:127–134 10.1016/S1074-7613(00)80594-4 [DOI] [PubMed] [Google Scholar]
- Haracska L., Washington M.T., Prakash S., Prakash L. 2001. Inefficient bypass of an abasic site by DNA polymerase eta. J. Biol. Chem. 276:6861–6866 10.1074/jbc.M008021200 [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- Jansen J.G., Langerak P., Tsaalbi-Shtylik A., van den Berk P., Jacobs H., de Wind N. 2006. Strand-biased defect in C/G transversions in hypermutating immunoglobulin genes in Rev1-deficient mice. J. Exp. Med. 203:319–323 10.1084/jem.20052227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J.G., Fousteri M.I., de Wind N. 2007. Send in the clamps: control of DNA translesion synthesis in eukaryotes. Mol. Cell. 28:522–529 10.1016/j.molcel.2007.11.005 [DOI] [PubMed] [Google Scholar]
- Jolly C.J., Klix N., Neuberger M.S. 1997. Rapid methods for the analysis of immunoglobulin gene hypermutation: application to transgenic and gene targeted mice. Nucleic Acids Res. 25:1913–1919 10.1093/nar/25.10.1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannouche P.L., Wing J., Lehmann A.R. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 14:491–500 10.1016/S1097-2765(04)00259-X [DOI] [PubMed] [Google Scholar]
- Langerak P., Nygren A.O., Schouten J.P., Jacobs H. 2005. Rapid and quantitative detection of homologous and non-homologous recombination events using three oligonucleotide MLPA. Nucleic Acids Res. 33:e188 10.1093/nar/gni187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langerak P., Nygren A.O., Krijger P.H., van den Berk P.C., Jacobs H. 2007. A/T mutagenesis in hypermutated immunoglobulin genes strongly depends on PCNAK164 modification. J. Exp. Med. 204:1989–1998 10.1084/jem.20070902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Gearhart P.J. 2004. A role for Msh6 but not Msh3 in somatic hypermutation and class switch recombination. J. Exp. Med. 200:61–68 10.1084/jem.20040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martomo S.A., Yang W.W., Wersto R.P., Ohkumo T., Kondo Y., Yokoi M., Masutani C., Hanaoka F., Gearhart P.J. 2005. Different mutation signatures in DNA polymerase eta- and MSH6-deficient mice suggest separate roles in antibody diversification. Proc. Natl. Acad. Sci. USA. 102:8656–8661 10.1073/pnas.0501852102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M., Kinoshita K., Fagarasan S., Yamada S., Shinkai Y., Honjo T. 2000. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 102:553–563 10.1016/S0092-8674(00)00078-7 [DOI] [PubMed] [Google Scholar]
- Nelson J.R., Lawrence C.W., Hinkle D.C. 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature. 382:729–731 10.1038/382729a0 [DOI] [PubMed] [Google Scholar]
- Nilsen H., Rosewell I., Robins P., Skjelbred C.F., Andersen S., Slupphaug G., Daly G., Krokan H.E., Lindahl T., Barnes D.E. 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol. Cell. 5:1059–1065 10.1016/S1097-2765(00)80271-3 [DOI] [PubMed] [Google Scholar]
- Pham P., Bransteitter R., Petruska J., Goodman M.F. 2003. Processive AID-catalysed cytosine deamination on single-stranded DNA simulates somatic hypermutation. Nature. 424:103–107 10.1038/nature01760 [DOI] [PubMed] [Google Scholar]
- Prakash S., Johnson R.E., Prakash L. 2005. Eukaryotic translesion synthesis DNA polymerases: specificity of structure and function. Annu. Rev. Biochem. 74:317–353 10.1146/annurev.biochem.74.082803.133250 [DOI] [PubMed] [Google Scholar]
- Rada C., Ehrenstein M.R., Neuberger M.S., Milstein C. 1998. Hot spot focusing of somatic hypermutation in MSH2-deficient mice suggests two stages of mutational targeting. Immunity. 9:135–141 10.1016/S1074-7613(00)80595-6 [DOI] [PubMed] [Google Scholar]
- Rada C., Williams G.T., Nilsen H., Barnes D.E., Lindahl T., Neuberger M.S. 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 12:1748–1755 10.1016/S0960-9822(02)01215-0 [DOI] [PubMed] [Google Scholar]
- Rada C., Di Noia J.M., Neuberger M.S. 2004. Mismatch recognition and uracil excision provide complementary paths to both Ig switching and the A/T-focused phase of somatic mutation. Mol. Cell. 16:163–171 10.1016/j.molcel.2004.10.011 [DOI] [PubMed] [Google Scholar]
- Roa S., Avdievich E., Peled J.U., Maccarthy T., Werling U., Kuang F.L., Kan R., Zhao C., Bergman A., Cohen P.E., et al. 2008. Ubiquitylated PCNA plays a role in somatic hypermutation and class-switch recombination and is required for meiotic progression. Proc. Natl. Acad. Sci. USA. 105:16248–16253 10.1073/pnas.0808182105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez C.I., Buchholz F., Galloway J., Sequerra R., Kasper J., Ayala R., Stewart A.F., Dymecki S.M. 2000. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat. Genet. 25:139–140 10.1038/75973 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Kolchanov N.A. 1992. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1171:11–18 [DOI] [PubMed] [Google Scholar]
- Rogozin I.B., Pavlov Y.I., Bebenek K., Matsuda T., Kunkel T.A. 2001. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2:530–536 10.1038/88732 [DOI] [PubMed] [Google Scholar]
- Ross A.L., Sale J.E. 2006. The catalytic activity of REV1 is employed during immunoglobulin gene diversification in DT40. Mol. Immunol. 43:1587–1594 10.1016/j.molimm.2005.09.017 [DOI] [PubMed] [Google Scholar]
- Schenten D., Gerlach V.L., Guo C., Velasco-Miguel S., Hladik C.L., White C.L., Friedberg E.C., Rajewsky K., Esposito G. 2002. DNA polymerase kappa deficiency does not affect somatic hypermutation in mice. Eur. J. Immunol. 32:3152–3160 [DOI] [PubMed] [Google Scholar]
- Shen H.M., Tanaka A., Bozek G., Nicolae D., Storb U. 2006. Somatic hypermutation and class switch recombination in Msh6(−/−)Ung(−/−) double-knockout mice. J. Immunol. 177:5386–5392 [DOI] [PubMed] [Google Scholar]
- Stelter P., Ulrich H.D. 2003. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 425:188–191 10.1038/nature01965 [DOI] [PubMed] [Google Scholar]
- Weill J.C., Reynaud C.A. 2008. DNA polymerases in adaptive immunity. Nat. Rev. Immunol. 8:302–312 10.1038/nri2281 [DOI] [PubMed] [Google Scholar]
- Wiesendanger M., Kneitz B., Edelmann W., Scharff M.D. 2000. Somatic hypermutation in MutS homologue (MSH)3-, MSH6-, and MSH3/MSH6-deficient mice reveals a role for the MSH2–MSH6 heterodimer in modulating the base substitution pattern. J. Exp. Med. 191:579–584 10.1084/jem.191.3.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X., Winter D.B., Kasmer C., Kraemer K.H., Lehmann A.R., Gearhart P.J. 2001. DNA polymerase eta is an A-T mutator in somatic hypermutation of immunoglobulin variable genes. Nat. Immunol. 2:537–541 10.1038/88740 [DOI] [PubMed] [Google Scholar]