Abstract
The transcription factor Ets1 contributes to the differentiation of CD8 lineage cells in the thymus, but how it does so is not understood. In this study, we demonstrate that Ets1 is required for the proper termination of CD4 expression during the differentiation of major histocompatability class 1 (MHC I)–restricted thymocytes, but not for other events associated with their positive selection, including the initiation of cytotoxic gene expression, corticomedullary migration, or thymus exit. We further show that Ets1 promotes expression of Runx3, a transcription factor important for CD8 T cell differentiation and the cessation of Cd4 gene expression. Enforced Runx3 expression in Ets1-deficient MHC I–restricted thymocytes largely rescued their impaired Cd4 silencing, indicating that Ets1 is not required for Runx3 function. Finally, we document that Ets1 binds at least two evolutionarily conserved regions within the Runx3 gene in vivo, supporting the possibility that Ets1 directly contributes to Runx3 transcription. These findings identify Ets1 as a key player during CD8 lineage differentiation and indicate that it acts, at least in part, by promoting Runx3 expression.
Thymocyte differentiation into the CD4 or CD8 lineages is a key event during the late steps of T cell development, in which precursors that have rearranged TCRβ and TCRα genes and express both CD4 and CD8 (double positive [DP]) are selected into mature CD4 T cells if MHC II–restricted, or CD8 T cells if MHC I–restricted (Starr et al., 2003; Bosselut, 2004; Singer and Bosselut, 2004). Lineage differentiation is defined by the onset of new programs of gene expression, most prominently the changes in Cd4 and Cd8 transcription from a DP to a single-positive (SP) CD4+CD8− or CD4−CD8+ pattern. Several transcription factors selectively promote the differentiation of either CD4 or CD8 T cells. The zinc finger proteins Gata3 and Thpok (also called cKrox or Zbtb7b) are necessary for the generation of CD4 cells (Hernández-Hoyos et al., 2003; Pai et al., 2003; He et al., 2005; Sun et al., 2005), whereas the transcription factor Runx3 is important for CD8 T cell development, notably by promoting the cessation of Cd4 expression (Taniuchi et al., 2002a; Ehlers et al., 2003; Woolf et al., 2003; Egawa et al., 2007). This function of Runx3 relies on the recruitment of Runx3 molecules to a cis-regulatory silencer element located in the first intron of the Cd4 gene (Taniuchi et al., 2002a, 2004). Runx3 has been shown to be up-regulated during the differentiation of DP thymocytes into CD8 cells in the thymus (Sato et al., 2005; Egawa et al., 2007; Egawa and Littman, 2008), but little is known about the transcriptional circuitry that controls its transcription.
Ets1 is the prototype of a family of transcription factors that bind specific DNA sequences typically centered over a GGAA tetranucleotide motif (Sharrocks, 2001; Verger and Duterque-Coquillaud, 2002). Multiple Ets factors are expressed in DP and SP thymocytes, including Ets1 and the related protein Ets2, both present throughout T cell development without marked preference for any T cell subset (Anderson et al., 1999). Despite this potential for functional redundancy, mice lacking Ets1 have impaired development of NK and T cells (Barton et al., 1998; Eyquem et al., 2004), and Ets1 is essential for Th1 effector differentiation (Grenningloh et al., 2005). Ets1 participates in two important aspects of early thymocyte development, allelic exclusion during TCRβ gene rearrangement and the survival of early (pre-DP) thymocytes (Eyquem et al., 2004). Although Ets1−/− mice have reduced thymocyte numbers as a result of these early effects, initial studies did not report major anomalies of late thymocyte development (Bories et al., 1995; Muthusamy et al., 1995; Barton et al., 1998). However, it was noticed that Ets1−/− CD8 SP cells maintained low-level CD4 expression (Barton et al., 1998), a finding confirmed by a more recent study that showed that this defect is cell autonomous (Clements et al., 2006). How Ets1 affects CD8 lineage differentiation has remained poorly understood. Because Ets1 was reported not to affect expression of Runx3, it was proposed that Ets1 disruption affected Runx3-mediated Cd4 silencing (Clements et al., 2006).
In this study, we have examined how Ets1 contributes to CD8 T cell differentiation. We show that Ets1 promotes the proper cessation of CD4 expression during the differentiation of MHC I–restricted thymocytes. However, Ets1 is not required for Runx3-mediated Cd4 silencing. Rather, Ets1 is important for Runx3 expression in these cells and binds at least two regions of the Runx3 gene. Our findings identify Ets1 as an important regulator of Runx3 expression and establish a novel connection in the network of transcription factors that control CD8 T cell differentiation in the thymus.
RESULTS
Ets1−/− mice contain an MHC I–restricted “maturelike” DP thymocyte population
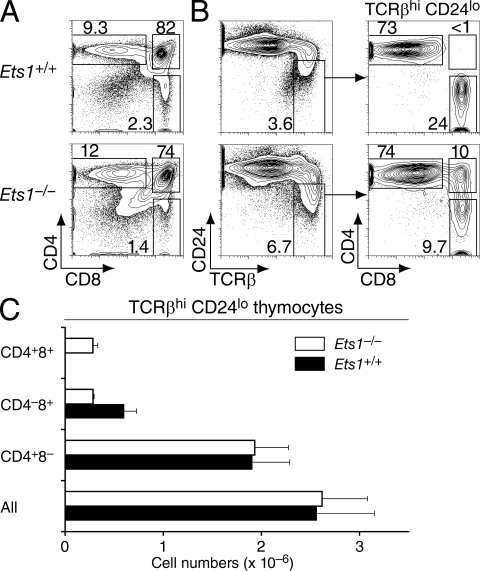
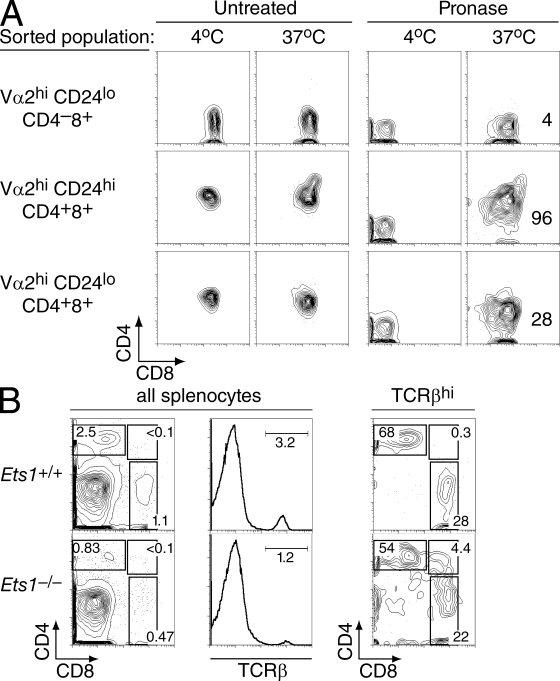
Consistent with previous studies (Barton et al., 1998; Eyquem et al., 2004; Clements et al., 2006), Ets1−/− thymi were hypocellular (40–50% of wild-type littermates; Table S1). Flow cytometric analyses of CD4 and CD8 expression showed a reduced frequency of CD8 SP thymocytes contrasting with a normal or increased representation of CD4 SP cells (Fig. 1 A). Given the low cellularity of Ets1−/− thymi, this resulted in substantially reduced numbers of CD8 SP thymocytes (25–30% of wild-type controls; Table S1). The most mature thymocyte subset, defined as TCRhi CD24lo, normally comprises CD4 or CD8 SP cells that have successfully completed positive selection and escaped negative selection (Fowlkes and Pardoll, 1989; Kishimoto and Sprent, 1999). In Ets1−/− mice, this subset included an unusual contingent of CD4+CD8+ thymocytes (Fig. 1, B and C). Such maturelike TCRhi CD24lo DP thymocytes were present in the thymus of Ets1-deficient newborn mice, indicating that this subset did not result from the accumulation over time of small numbers of long-lived thymocytes (Fig. S1). There was no maturelike DP subset in Ets1+/− thymi, which were phenotypically similar to their Ets1+/+ counterparts (unpublished data) and were used as controls in some experiments.
Figure 1.
Mature thymocytes fail to resolve into CD4 and CD8 SP populations in Ets1-deficient mice. (A and B) Thymocytes from Ets1+/+ (top) or Ets1−/− (bottom) mice were assessed by flow cytometry for surface expression of CD4, CD8, CD24, and TCRβ. Two-parameter contour plots are shown on all live cells for expression of CD4 and CD8 (A) or of CD24 and TCRβ (B, left); TCRhi CD24lo mature thymocytes are analyzed for CD4 and CD8 expression (B, right). Numbers next to boxes indicate the percentage of cells within that box. Note that the level of CD24 expression was not affected by Ets1 disruption by itself. Data are from more than three experiments. (C) Bar graphs (mean ± SEM; n = 5 for each genotype) represent the absolute numbers of thymocytes within each mature (TCRβhi CD24lo) subset.
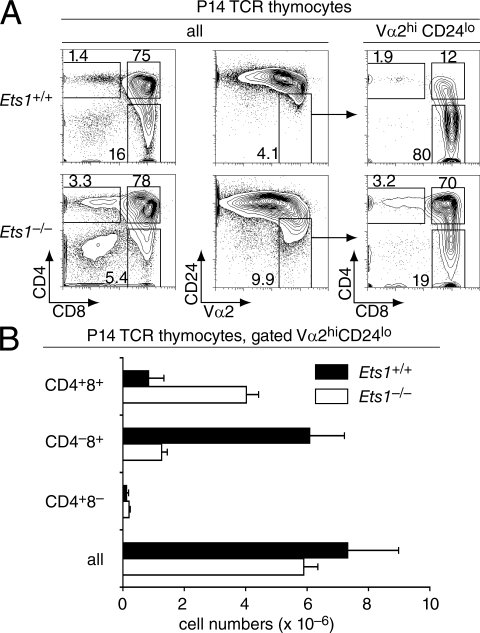
We next evaluated if this unusual maturelike DP population resulted from MHC I– or MHC II–induced positive selection by generating Ets1−/− mice carrying defined TCR specificities. In mice carrying the MHC I–restricted P14 TCR, which recognizes an LCMV-derived peptide presented by Kb, thymocytes being positively selected express high levels of the transgenic Vα2 and Vβ8 TCR chains (Pircher et al., 1989). The frequency of Vβ8+ thymocytes was lower in Ets1−/− than in Ets1+/+ P14 mice (Fig. S2 A), presumably reflecting the role of Ets1 in early thymocyte development and notably its requirement for efficient TCRβ allelic exclusion (Eyquem et al., 2004). Nonetheless, the correlation between Vβ8 and Vα2 expression was excellent in both genotypes, and we used either marker to characterize P14 thymocytes undergoing selection. Postselection Vα2hi CD24lo thymocytes were present in Ets1+/+ and Ets1−/− P14 mice in similar numbers (Fig. 2, A [middle] and B), and Ets1 disruption did not prevent the up-regulation of CD69, a surface molecule normally expressed in response to TCR signaling (Swat et al., 1993; Fig. S2 B); thus, Ets1 was not required for thymocytes to respond to positively selecting TCR engagements. However, postselection Vα2hi CD24lo Ets1−/− thymocytes were mostly CD4+CD8+, unlike their Ets1+/+ counterparts, which were predominantly CD4−CD8+ (Fig. 2, A [right] and B). Thus, MHC I–induced positive selection in the absence of Ets1 results in the generation of maturelike DP thymocytes. Correspondingly, the frequency and number of CD8 SP thymocytes were substantially reduced in Ets1−/− P14 TCR mice relative to their Ets1+/+ counterparts (Fig. 2, A [left] and B).
Figure 2.
Ets1−/− MHC I–restricted thymocytes fail to down-regulate CD4 during the late stages of positive selection. (A) Thymocytes from Ets1+/+ (top) or Ets1−/− (bottom) mice carrying the P14 TCR transgene were assessed for expression of CD4, CD8, CD24, and the transgenic Vα2 TCRα chain. Two-parameter contour plots of CD4 and CD8 expression (left) show a reduced frequency of CD8 SP cells in Ets1−/− thymi. The Vα2hi CD24lo subset is defined on two-parameter contour plots of CD24 and Vα2 expression (middle), and analyzed for CD4 and CD8 expression (right). Data are from more than three experiments. (B) Bar graphs (mean ± SEM) represent the absolute numbers of thymocytes within each mature (Vα2hi CD24lo) subset. Total thymocyte numbers (average ± SEM; × 10−6; n = 7) were 111 ± 23 and 43 ± 4.8 in Ets1+/+ and Ets1−/− P14 mice, respectively.
In contrast with these findings, positive selection by the MHC II-restricted OT-II TCR, which recognizes an ovalbumin-derived peptide presented by I-Ab (Hogquist et al., 1994), did not result in the presence of a maturelike DP population on the Ets1−/− background, and instead gave rise to a CD4-skewing of mature thymocytes similar to that on the Ets1+/+ background (Fig. S2 C).
Ets1−/− maturelike DP thymocytes are localized in the thymic medulla
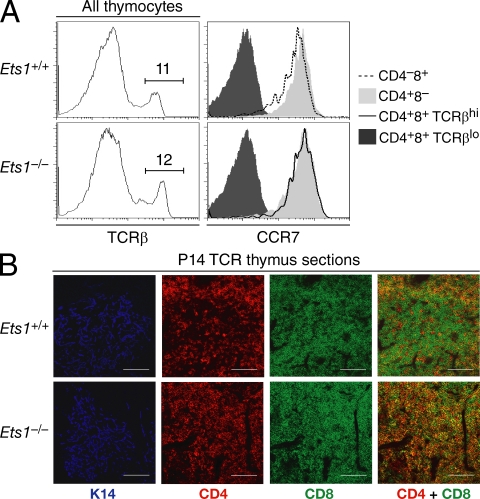
It was important to assess other indicators of differentiation in Ets1−/− DP TCRhi CD24lo thymocytes, notably to verify that the presence of this subset was not simply reflecting a requirement for Ets1 for CD24 expression. Normally, preselection TCRlo DP thymocytes reside in the thymic cortex, whereas positively selected thymocytes migrate to the medulla, as a result of their up-regulating the chemokine receptor CCR7 (Takahama, 2006). Flow cytometry analyses showed that Ets1−/− TCRβhi DP thymocytes were uniformly CCR7hi, expressing CCR7 levels similar to those seen in Ets1+/+ SP thymocytes and >10 times higher than those on TCRlo DP cells (either Ets1+/+ or Ets1−/−; Fig. 3 A). To evaluate if these CCR7hi DP thymocytes migrated to the medulla, we compared thymus sections of Ets1−/− and Ets1+/+ mice by immunohistological analysis of CD4 and CD8, and of the medullary cell marker cytokeratin 14 (K14; Klug et al., 1998). These experiments were performed on mice carrying the P14 TCR transgene in which thymocyte differentiation is normally skewed toward the CD8 lineage. K14+ medullary areas in P14 Ets1+/+ thymi contained mostly CD8 SP and only a few CD4 SP cells, whereas the K14-negative cortical areas were occupied by DP thymocytes (Fig. 3 B, top). K14-positive medullary areas were clearly defined in P14 Ets1−/− thymi; however, they were packed with DP thymocytes (Fig. 3 B, bottom) and could not be distinguished from the surrounding cortex on the basis of CD4 and CD8 expression alone, unlike the clear boundary seen in the P14 Ets1+/+ thymus. We conclude from these analyses that Ets1−/− maturelike DP thymocytes have the same medullary location as wild-type SP thymocytes.
Figure 3.
Ets1−/− MHC I–restricted maturelike DP thymocytes migrate to the medulla. (A) Thymocytes from Ets1+/+ and Ets1−/− mice were stained for surface expression of CD4, CD8, TCRβ and the chemokine receptor CCR7. Overlaid histograms (right plots) analyze expression of CCR7 on Ets1−/− DP and CD4 SP thymocytes (bottom graph), and on Ets1+/+ CD8 and CD4 SP thymocytes (top graph), all TCRβhi (as gated in left plots). Expression of CCR7 on TCRlo DP thymocytes is shown in both strains as a negative control (dark gray histograms). Data are representative of two experiments. (B) Frozen thymic sections were prepared from P14 transgenic Ets1+/+ or Ets1−/− mice, stained for cytokeratin 14 (K14, pseudo-colored as blue, defining medullary areas), CD4 (red), and CD8 (green). Overlaying CD4 and CD8 staining (right) shows exclusion of DP cells from medullary areas in Ets1+/+ but not in Ets1−/− mice. The red medullary staining in Ets1+/+ mice is contributed by the few CD4 SP cells that develop in these recombination-competent animals. Bars, 100 µm. Data are representative of three experiments.
Ets1−/− maturelike DP thymocytes are CD8 lineage cells
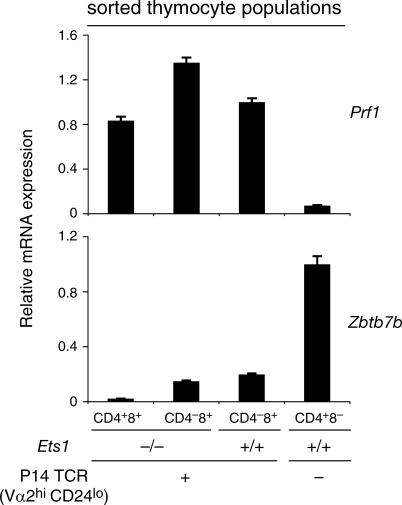
Because MHC I–induced selection normally gives rise to CD8-linage cells, we examined if Ets1−/− maturelike MHC I–restricted DP thymocytes were cells that undergo CD8 differentiation and fail to silence CD4, or cells that are redirected to the CD4 lineage and fail to silence CD8. We submitted maturelike Ets1−/− P14 DP thymocytes to real-time RT-PCR analysis of mRNAs encoding the transcription factor Thpok and the cytotoxic marker perforin, normally expressed in CD4 and CD8 lineage thymocytes, respectively (Fig. 4, rightmost two columns). As expected, Ets1−/− CD8 SP thymocytes expressed perforin but no Thpok (Fig. 4, second column). Importantly, the same was true of maturelike DP thymocytes (Vα2hi CD24lo DP cells from Ets1−/− P14 transgenic mice; Fig. 4, left column), indicating that these cells had a gene expression pattern typical of CD8 lineage cells.
Figure 4.
Ets1−/− MHC I–restricted maturelike DP thymocytes are CD8 lineage cells. Expression of the genes encoding Thpok (Zbtb7b) and perforin (Prf1) was analyzed by real-time RT-PCR on sorted Vα2hi CD24lo CD4+CD8+ and CD4−CD8+ thymocyte populations from Ets1+/+ and Ets1−/− mice carrying the P14 TCR transgene (left three columns) and on TCRhi CD4+CD8− thymocytes from wild-type mice (right column). mRNA levels, normalized on β-actin expression in the same sample, are shown relative to those in wild-type CD4+CD8− cells (Thpok) or Ets1+/+ P14 CD4−CD8+ cells (Prf1). Bars indicate the mean values derived from triplicate determination from a single sorted population; error bars show standard deviations. Data are representative of three or more independent sorted samples for each population.
We considered the possibility that maturelike DP thymocytes might be in the process of silencing Cd4, so that their expression of surface CD4 molecules would not be indicative of active Cd4 gene expression. To address this possibility, we measured CD4 and CD8 protein reexpression in sorted Ets1−/− P14 thymocytes that had been “stripped” of their surface coreceptor proteins by mild pronase digestion (Suzuki et al., 1995). Previous studies had documented that surface reexpression of coreceptor molecules in this assay is indicative of Cd4 and Cd8 gene expression (Brugnera et al., 2000; Yu et al., 2003). Most maturelike (Vα2hi CD24lo) DP thymocytes from P14 transgenic Ets1−/− mice reexpressed both CD4 and CD8 after pronase stripping, unlike their CD8 SP counterparts that only reexpressed CD8 (Fig. 5 A, bottom and top rows). However, CD4 reexpression levels were lower on maturelike DP than on their immature Vα2hi CD24hi counterparts (Fig. 5 A, bottom and middle rows). We draw two conclusions from these experiments. First, maturelike DP thymocytes actively express Cd4, indicating impaired silencing. Second, the lower CD4 reexpression by that population, compared with its CD24hi counterparts, suggest that some maturelike DP cells may eventually silence Cd4 and convert to a CD8 SP phenotype. However, the presence of DP T cells in the spleen of adult and neonate Ets1−/− mice (Fig. 5 B and not depicted) suggests that at least some maturelike DP thymocytes complete their intrathymic development before terminating Cd4 expression.
Figure 5.
Persistent CD4 expression in Ets1−/− maturelike DP thymocytes. (A) Thymocytes subsets from P14 TCR Ets1−/− mice were sorted as indicated in Fig. S3, stripped of their surface coreceptor molecules, and analyzed by flow cytometry for CD4 and CD8 surface expression after overnight single-cell suspension culture (right column). An aliquot of the pronase-treated cells was kept at 4°C and analyzed in parallel to verify the complete removal of CD4 and CD8 surface molecules after pronase digestion (third column). No change in surface coreceptor expression was seen in the absence of pronase treatment (two left columns). Data are representative of two separate experiments. Numbers in graphs indicate the mean fluorescence intensity of CD4 staining on CD8+ cells. (B) Splenocytes were prepared from 1-wk-old Ets1+/+ and Ets1−/− mice and analyzed as in Fig. 1 for expression of CD4, CD8, and TCRβ. CD4 versus CD8 two-parameter contour plots derived from TCRhi splenocytes show CD4+CD8+ splenocytes in Ets1−/− mice. Data are representative of six Ets1−/− and three Ets1+/+ neonates analyzed in two separate experiments.
Ets1 promotes Runx3 expression
We concluded from the previous findings that Ets1 disruption impaired the cessation of Cd4 expression during CD8 differentiation. The lineage specificity of Cd4 expression is determined by the Cd4 silencer, a 434-bp element located in the first intron of the Cd4 gene (Taniuchi et al., 2004). The silencer is activated in CD8-differentiating thymocytes, a process that normally requires the recruitment of the transcription factor Runx3, whose expression is up-regulated during CD8 differentiation (Taniuchi et al., 2002a; Woolf et al., 2003; Grueter et al., 2005). Thus, the impaired Cd4 silencing in Ets1−/− thymocytes indicated that Ets1 is important for the expression of Runx3 molecules, for their ability to repress Cd4 expression, or for both.
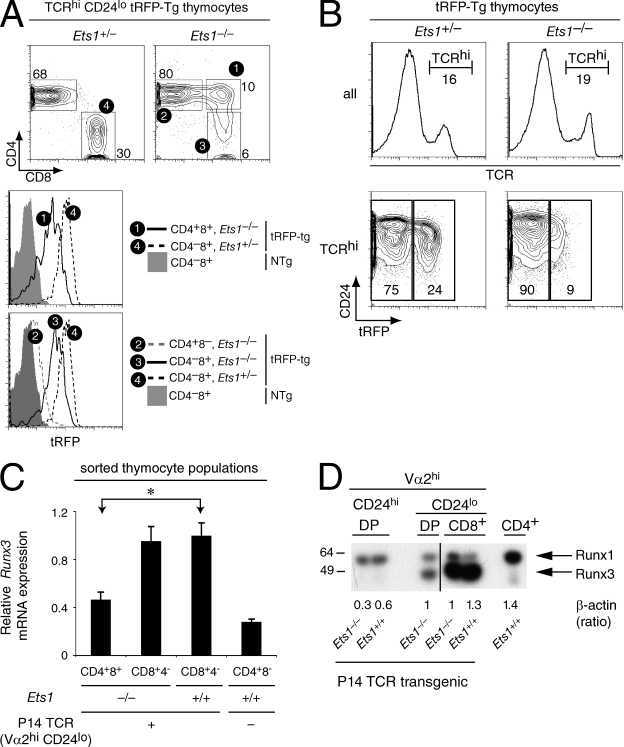
To distinguish between these possibilities, we first examined Runx3 expression in Ets1−/− thymocytes. In wild-type mice, there is little or no Runx3 gene expression in preselection DP thymocytes, and its preferential up-regulation during the DP to CD8 SP transition results in higher mRNA levels in CD8 than in CD4 SP thymocytes (Taniuchi et al., 2002a; Liu and Bosselut, 2004; Egawa et al., 2007). Analyses of Runx3 expression in maturelike DP thymocytes selected by endogenously rearranged TCRs are hampered by the small numbers of these cells; an additional level of complexity comes from alternative promoter usage in the Runx3 gene, resulting in mRNA species that appear to not equally contribute to Runx3 protein synthesis (Egawa et al., 2007; Egawa and Littman, 2008). To overcome these obstacles, we introduced into Ets1−/− mice a transgenic BAC reporter in which the sequence coding for the tandem-dimer-tomato red fluorescent protein (tRFP ; Shaner et al., 2005) had been inserted into the second exon of the Runx3 gene (Fig. S4 A and unpublished data). As the tRFP cDNA insertion respects all Runx3 noncoding sequences, and as tRFP translation is initiated from endogenous Runx3 ATG codons, expression of tRFP in the thymus matched expression of endogenous Runx3 protein (Woolf et al., 2003; Egawa et al., 2007). In wild-type thymi, we readily detected tRFP in a subset of DN cells and in CD8 lineage thymocytes, whereas little or no expression was seen in DP and CD4 lineage cells (Fig. S4 B and unpublished data). Similarly, there was little tRFP fluorescence in Ets1−/− CD4 lineage cells. However, fluorescence intensities in CD8 SP thymocytes were slightly lower in Ets1−/− than in their wild-type counterparts and tRFP expression in maturelike DP thymocytes was half of that in wild-type CD8 SP thymocytes (Fig. 6 A). In fact, the fraction of positively selected (TCRhi) thymocytes that expressed Runx3, as well as their level of expression, were lower in Ets1-deficient than Ets1-sufficient thymocytes, indicating that the low expression observed on maturelike DP cells did not result from a gating bias (Fig. 6 B). These experiments indicated that Ets1 is important for appropriate Runx3 expression.
Figure 6.
Ets1 promotes Runx3 expression. (A and B) Expression of Runx3 was evaluated in mice carrying a BAC transgene in which a tRFP cDNA has been inserted within the second exon of Runx3. (A) Two-parameter contour plots of CD4 and CD8 expression (top) are gated on TCRhi CD24lo thymocytes from Ets1+/− and Ets1−/− mice. Subsets defined by boxes are numbered and analyzed for tRFP expression. Overlaid histograms (bottom) depict tRFP fluorescence in indicated subsets of tRFP-transgenic Ets1+/− and Ets1−/− mice. Gray-shaded histogram show background fluorescence in CD8 SP thymocytes from control Ets1+/+ nontransgenic mice. The mean intensity of tRFP fluorescence in subset 1 (maturelike DP cells from Ets1−/− mice) was 49% of that in subset 4 (CD8 SP cells from tRFP-transgenic Ets1+/− controls; mean on all three experiments). (B) Two parameter plots of tRFP and CD24 expression (bottom) are shown on TCRhi gated cells (histograms, top). Data (A and B) is representative of three mice of each genotype analyzed in three separate experiments. (C) Expression of Runx3 was assessed as in Fig. 4 on the same mRNA preparations and is shown relative to that in Ets1+/+ P14 CD4−CD8+ cells. The difference between Ets1−/− Vα2hi CD24lo DP and Ets1+/+ CD8 SP thymocytes for Runx3 expression was statistically significant (*, P < 10−4, two tailed Student's t test). Data are from more than three experiments. (D) Expression of Runx proteins was assessed in sorted thymocyte subsets by immunoblotting with an antibody directed against the Runt domain and recognizing both Runx1 and Runx3. CD4 SP thymocytes were sorted from wild-type mice and used as positive and negative controls for Runx1 and Runx3 expression, respectively. MW marker sizes are indicated on the left. Numbers underneath indicate expression of β-actin in each samples, quantified on the same membrane and expressed relative to that of wild-type CD8 SP thymocyte. The β-actin signal was consistently lower in DP thymocytes than in other cell subsets, but was not reproducibly affected by Ets1 disruption. The figure is a composite of two parts of a single blot (separated as indicated by the vertical black bar). Data are from three determinations performed from two distinct sets of sorted cells.
We verified that reduced expression of the Runx3 reporter was indicative of reduced endogenous Runx3 expression using mice carrying the P14 TCR transgene. Runx3 mRNA expression was lower in Ets1−/− maturelike DP than in CD8 SP cells, whether wild-type or Ets1−/− (Fig. 6 C), and the same was true of Runx3 protein expression (Fig. 6 D), demonstrating that Ets1 disruption results in defective Runx3 up-regulation during the positive selection of MHC I–restricted thymocytes. This defect was specific to Runx3, as expression of Runx1 remained unchanged in all three subsets in Ets1−/− mice (Fig. 6 D). Consistent with analyses of Runx3 reporter mice, Ets1 was not strictly required for Runx3 up-regulation; in Ets1−/− mice Runx3 was detectable (although low) in maturelike DP thymocytes and present at subnormal levels in CD8 SP cells (Fig. 6, C and D). Thus, although Ets1 is not required for Runx3 expression, it is necessary for its proper up-regulation during CD8 lineage differentiation.
Enforced Runx3 expression restores CD4 silencing in Ets1−/− thymocytes
Having shown that Ets1 promotes Runx3 expression during CD8 cell differentiation, we next investigated if Ets1 was required for Runx3 to repress Cd4. To evaluate this possibility, we introduced into Ets1−/− mice a Runx3 transgene expressed throughout T cell development, starting at or before the preselection DP stage (Grueter et al., 2005). We reasoned that this transgene would fail to restore CD4 silencing in Ets1−/− mice if silencer activation by Runx3 molecules required Ets1 expression. Unlike in wild-type mice, and as previously shown (Grueter et al., 2005), Runx3 was expressed at all post-DN stages in the transgenic mice, including preselection DP thymocytes (CD69− cells) and CD4 lineage cells (Fig. S5 A). On a per-cell basis, expression of Runx3 molecules in transgenic CD69− cells was not greater than that of endogenous Runx3 in wild-type CD8 lineage cells, indicating that the transgene did not result in Runx3 overexpression (Grueter et al., 2005; Fig. S5 A). Furthermore, expression of the transgene was not affected by Ets1 disruption (Fig. S6).
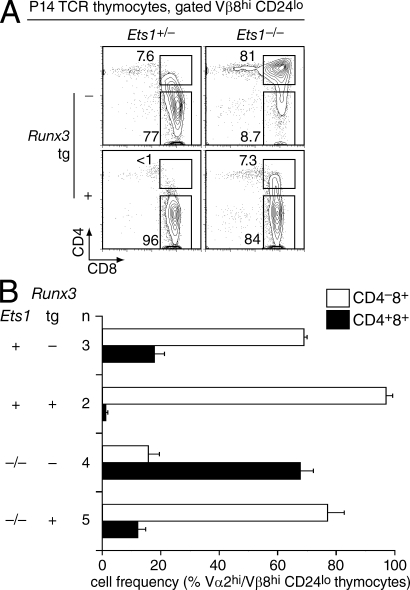
We introduced the Runx3 transgene into Ets1-sufficient and -deficient P14 TCR transgenic mice, and assessed the Vβ8hi (or Vα2hi) CD24lo subset for expression of CD4 and CD8. Expression of the Runx3 transgene on the P14 Ets1−/− background resulted in a substantial reduction in the size of the maturelike DP subset (Fig. 7 A, right), with ∼80% of the Vβ8hi CD24lo cells having down-regulated CD4 (Fig. 7 B). Transgenic Runx3 expression had previously been shown to cause CD4 down-regulation in preselection thymocytes (Telfer et al., 2004; Grueter et al., 2005; Kohu et al., 2005; Wildt et al., 2007), and analyses gated on all live cells showed that this was the case in Ets1-deficient thymocytes as well (Fig. S7). Because expression of transgenic Runx3 on a per-cell basis did not exceed that of endogenous Runx3 (Fig. S5 A), we interpret the early repression of Cd4 as reflecting the premature expression of transgenic compared with endogenous Runx3 rather than being caused by Runx3 overexpression. We conclude from these experiments that Runx3-mediated Cd4 silencing does not require Ets1, and that the impaired Cd4 silencing observed in Ets1−/− CD8 lineage cells is caused at least in part by their impaired Runx3 expression.
Figure 7.
Enforced Runx3 expression restores CD4 down-regulation in Ets1−/− MHC I–restricted thymocytes. (A) Thymocytes were prepared from Ets1+/− and Ets1−/− mice, carrying either the P14 TCR transgene only or both the P14 and Runx3 transgenes, and stained for CD4, CD8, CD24 and Vβ8. Two-parameter contour plots show CD4 and CD8 expression on gated Vβ8hi CD24lo thymocytes. (B) Bar graph indicates the percentages (average ± SEM; n: number of mice of each genotype) of Vα2hi or Vβ8hi CD24lo DP and CD8 SP thymocytes in each strain (all carrying the P14 TCR transgene). In these experiments, Ets1-competent control mice (Ets1+) were either Ets1+/− or Ets1+/+; both genotypes resulted in similar Ets1-sufficient phenotypes. Data in (A) and (B) is from more than three experiments.
In vivo recruitment of Ets1 to the Runx3 gene
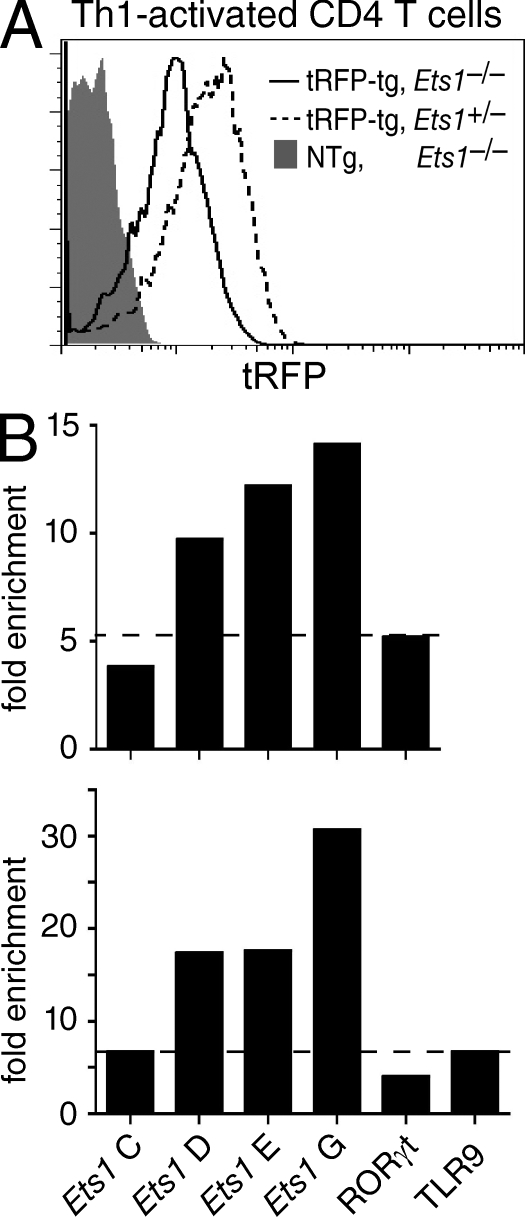
These findings prompted us to examine if Ets1 could directly promote Runx3 transcription by binding to specific sites within the Runx3 gene. To evaluate this possibility, we performed chromatin immunoprecipitation (ChIP) assays with an anti-Ets1 antibody that detects Ets1 binding to the IFN-γ promoter in Th1-differentiating effector T cells (Grenningloh et al., 2005). Anti-Ets1 ChIP did not detect any specific Ets1 binding when performed on P14 TCR transgenic thymocytes (unpublished data), possibly because of the low representation of CD8-differentiating thymocytes, even in P14 transgenic mice, or of an insufficient Ets1 protein contents in thymocytes. To overcome these limitations, we considered that, because both Ets1 and Runx3 are expressed in Th1-differentiating effectors (Grenningloh et al., 2005; Djuretic et al., 2007; Naoe et al., 2007), it was possible that Ets1 was also important for Runx3 expression in these cells. To evaluate this possibility, we assessed Th1 effectors carrying the Runx3-tRFP reporter. As expected, tRFP was expressed at greater levels in Th1 than in Th2 effectors (unpublished data); tRFP expression was lower in Ets1-deficient than in Ets1-sufficient Th1 effectors, indicating that Ets1 activation of Runx3 expression is not limited to CD8-differentiating thymocytes (Fig. 8 A).
Figure 8.
Ets1 binds the Runx3 locus. (A) Ets1 promotes Runx3 expression in Th1-differentiating CD4 effectors. Bead-purified CD4+CD8− LN T cells from tRFP transgenic Ets1−/− or Ets1+/− mice were activated under Th1 conditions and analyzed for tRFP expression 5 d later. Overlaid histogram show tRFP expression in gated CD4+CD8− Ets1−/− (plain line) and Ets1+/− (dashed line) effector cells. The gray-shaded histogram shows background fluorescence in nontransgenic Ets1−/− Th1 effectors activated in parallel. Data are from two mice of each genotype analyzed in two separate experiments. (B) Recruitment of Ets1 protein to the Runx3 gene in vivo was assessed by ChIP assays on Th1 effector T cells. Bar graphs represent fold-enrichment of segments C, D, E, and G from the Runx3 gene (Fig. S8 for location) in anti-Ets1 immunoprecipitates, expressed as indicated in Materials and methods. Horizontal bars depict background enrichment as detected in the RORγ and TLR9 genes. The top and bottom graphs represent two distinct experiments, each from a separate chromatin preparation.
In light of these results, we examined if we could detect Ets1 binding to the Runx3 promoter in Th1 effector cells. Indeed, ChIP assays demonstrated specific binding within a region surrounding the distal promoter, that is specifically active in CD8 lineage cells (Egawa et al., 2007; Egawa and Littman, 2008; amplified segments D and E; Fig. 8 B and Fig. S8 A). This evolutionary conserved region includes GGAA motifs compatible with Ets binding (Fig. S8 B). The enrichment of these segments in anti-Ets1 immunoprecipitates was two to three times greater than that of two irrelevant segments from the RORγ and TLR9 genes that we used as specificity controls (Fig. 8 B). Further supporting the conclusion that the binding around segments D and E was specific, little or no binding was detected to a region upstream of the promoter (amplified segment C; Fig. 8 B and Fig. S8, A and B). A search for additional conserved Ets motifs revealed potential binding sites in a region within the second Runx3 intron. Indeed, ChIP assays found strong Ets1 binding to a region (amplified segment G; Fig. 8 B and Fig. S8, A and C) containing a highly conserved AGGAAGY sequence that matches the consensus for Ets1 DNA binding (Sharrocks, 2001; Verger and Duterque-Coquillaud, 2002). We conclude from these experiments that Ets1 is recruited to multiple sites within the Runx3 locus in Th1 effectors; these findings support the possibility that Ets1 acts as a direct activator of Runx3 expression.
In summary, the present study demonstrates ChIP binding of Ets1 on Runx3, Ets1-dependence of Runx3 expression in two distinct cell types, and developmental rescue of Ets1 deficiency by Runx3 expression, and leads us to conclude that Ets1 promotes Cd4 silencing during CD8 lineage differentiation at least in part by promoting Runx3 expression.
DISCUSSION
The activation of Runx3 expression is a critical event during the differentiation of CD8 T cells from DP thymocytes, and signals the onset of Cd4 down-regulation (Egawa and Littman, 2008). However, the control of Runx3 expression in the thymus has remained largely mysterious. Although the CD4-differentiating factor Thpok represses Runx3 (Egawa and Littman, 2008; Muroi et al., 2008; Wang et al., 2008b), it is not yet known whether this effect is direct, and no factor has been shown to promote Runx3 expression in CD8-differentiating thymocytes. This study addressed this issue starting from the observation that the transcription factor Ets1 is required for the proper repression of CD4 during CD8 lineage differentiation. We show that Ets1 disruption impairs expression of Runx3 and we provide evidence that Ets1 directly contributes to Runx3 transcription. These findings identify Ets1 as a new node in the transcriptional circuitry that orchestrates CD4-CD8 differentiation (Wang and Bosselut, 2009). Although these findings contrast with the opposite conclusion reached by a previous work (Clements et al., 2006), that study evaluated Runx3 expression on unfractionated DP thymocytes expressing a diverse TCR repertoire. Both in wild-type and Ets1−/− mice, unfractionated DP thymocytes mostly comprise preselection cells that express little or no Runx3, presumably making that approach not sensitive enough to assess Runx3 gene expression in the relevant CD24lo TCRhi thymocyte population.
Deciphering the roles of Runx proteins in positive selection and lineage differentiation has been complicated by the functional redundancy between the two Runx genes expressed in developing T cells, Runx1 and Runx3 (Taniuchi et al., 2002a; Woolf et al., 2003; Egawa et al., 2007). Disruption of both genes in DP thymocytes prevents CD8 cell differentiation (Egawa et al., 2007). In post-DN thymocytes, expression of Runx1 is somewhat promiscuous, whereas expression of Runx3 is largely restricted to CD8-differentiating cells, suggesting that Runx3 is the main component of the Runx activity that promotes CD8 differentiation. Indeed, Runx1 inactivation does not affect CD4 expression during CD8 cell differentiation (Taniuchi et al., 2002a; Woolf et al., 2003; Egawa et al., 2007). However, the sole disruption of Runx3 impairs CD8 lineage differentiation only partially, and notably results in incomplete Cd4 derepression in CD8 lineage thymocytes (Taniuchi et al., 2002a; Woolf et al., 2003; Egawa et al., 2007). Disruption of Runx3 also results in increased Runx1 protein expression in CD8 lineage cells (Egawa and Littman, 2008), which is instrumental in attenuating the consequences of Runx3 disruption. Indeed, hemizygous inactivation of Runx1 in Runx3-deficient thymocytes completely abrogates Cd4 silencing by CD8 cells (Woolf et al., 2003), a result that is in line with other observations underscoring the sensitivity of Runx function to gene dosage (Barton and Nucifora, 2000).
Indirect comparisons with published studies (Taniuchi et al., 2002a; Woolf et al., 2003; Egawa et al., 2007) suggest that the loss of Cd4 silencing is greater in Ets1- than in Runx3-deficient CD8 lineage thymocytes, despite the residual Runx3 expression in the former. We see three potential explanations to this apparent paradox. First, unlike disruption of Runx3, disruption of Ets1 did not result in compensatory Runx1 up-regulation, consistent with a more pronounced effect on Cd4 silencing. Second, it is possible that Ets1 affects the expression of additional factors involved in Cd4 silencer function. Notably, two silencer DNA motifs, presumably recruiting thus far unknown factors, are required for Cd4 repression during CD8 lineage differentiation (Taniuchi et al., 2002b). It is conceivable that Ets1 is important for the expression of such additional silencer-binding proteins, thereby controling multiple key players of CD8 lineage differentiation. If that is the case, the rescue of Cd4 repression in Ets1−/− cells by Runx3 could indicate partial redundancy between such factors and Runx3. Alternatively, it is possible that the expression of these unknown silencer-binding factors is itself under the control of Runx3. In line with this possibility, Runx3 uses such a “feed-forward” loop to promote cytotoxic gene expression in effector CD8 T cells (Cruz-Guilloty et al., 2009).
Third, it has been proposed that Ets1 could bind the silencer and directly cooperate with Runx3 to repress Cd4 expression (Clements et al., 2006), in line with the in vitro synergy between Ets1 and Runx1 for TCR and BCR enhancer activation (Kim et al., 1999; Erman et al., 1998; Goetz et al., 2000; Gu et al., 2000). Specifically, a putative Ets1 motif exists between the two Runx binding sites of the silencer (Sawada et al., 1994; Taniuchi et al., 2002a) raising the possibility that Ets1 would bind the silencer and cooperate with Runx3 to promote Cd4 silencing. Although we did not observe any cooperative effect of Ets1 and Runx3 on silencer activity in cotransfection experiments (unpublished data), the present study does not directly evaluate this hypothesis. However, the fact that enforced Runx3 expression in Ets1−/− thymocytes results in efficient Cd4 silencing indicates that this function of Runx3 is not strictly Ets1-dependent.
Potential targets of Ets1, including CD5 or TCR genes, have been identified in vitro (Ho et al., 1990; Prosser et al., 1992; Tung et al., 2001; Arman et al., 2004) and in a large-scale ChIP study of Ets1 binding in the Jurkat human T cell line (Hollenhorst et al., 2007). However, only a few genes, including IFN-γ (Grenningloh et al., 2005) and Runx3 (this study) have been shown to both recruit and require Ets1 for their expression in vivo. We detected direct binding of Ets1 to the Runx3 locus in Th1 effectors, that, similar to CD8-differentiating thymocytes, express both Ets1 and Runx3 (Grenningloh et al., 2005; Djuretic et al., 2007; Naoe et al., 2007) and in which Ets1 similarly promotes Runx3 expression. These observations support the possibility that Ets1 directly promotes Runx3 expression by binding the Runx3 gene. While the two areas of Ets1 binding we identified on Runx3 are more than 30 kb apart on the sequence, it is possible that they are in close vicinity in the three-dimensional structure of the nucleus. That such an architecture is important for Ets1-mediated activation of Runx3 expression would be consistent with our observation that neither the distal promoter region nor the intronic Ets1 motif, when analyzed in isolation, respond to Ets1 in cotransfection experiments in T cell lines (unpublished data).
That Ets1 promotes Runx3 expression is consistent with the expression pattern of these two genes. Ets1 and Runx3 are coexpressed at multiple stages of T cell differentiation, including in early DN thymocytes, in CD8 lineage cells, in Th1 effectors, and in NK T cells (Anderson et al., 1999; Taniuchi et al., 2002a; Woolf et al., 2003). However, although expression of Runx3 in the T cell lineage is stage specific, expression of Ets1 is more promiscuous and is not restricted to CD8 lineage cells during positive selection. This brings two possibilities as to the function of Ets1 in Runx3 expression. First, it is possible that Ets1 serves as a “permissive” or priming factor, that makes thymocytes competent to express Runx3 but would not serve to convert extra-cellular clues into Runx3 expression. Second, intrathymic signals could trigger post-translational modifications that constrain Ets1 activity, thereby making Ets1 a “signal-sensor” that contributes to adjust Runx3 levels in response to environmental cues. Ets1 DNA binding in vitro is inhibited by the calcium-induced phosphorylation of serines encoded within its exon 7 (Pognonec et al., 1990; Pufall et al., 2005). Although these modifications do not appear essential for Ets1 functions during Th1 effector differentiation (Grenningloh et al., 2008), there is genetic evidence that they affect Ets1 activity in thymocytes (Higuchi et al., 2007).
The possibility that the intracellular calcium concentration, and thereby TCR signals, affects Ets1 activation of Runx3 expression raises a provocative correlate with the biology of CD4-CD8 lineage choice. Current models propose that TCR signals are of longer duration in MHC II– than in MHC I–signaled thymocytes, and thereby promote CD4 over CD8 lineage choice (Singer and Bosselut, 2004). In this perspective, it is conceivable, although at present speculative, that persistent TCR signals in MHC II–restricted thymocytes would result in sustained Ets1 phosphorylation, which in turn would minimize Ets1 recruitment to the Runx3 gene and contribute to limit its expression. It is likely that multiple mechanisms contribute to match lineage choice to MHC specificity in the thymus and affect Runx3 expression (Singer et al., 2008), and further work will be needed to evaluate the potential role of Ets1 phosphorylation in this process. However, mice genetically engineered to express only Ets1 molecules lacking exon 7–encoded sequences (and therefore not subject to phosphorylation-induced inhibition of DNA binding) have a slightly increased frequency of CD8 SP thymocytes (Higuchi et al., 2007), consistent with the possibility that increased Ets1 DNA binding would favor Runx3 expression and CD8 cell differentiation.
Because Runx activity also contributes to repress the CD4-committing factor Thpok (Setoguchi et al., 2008), it could be envisioned that impaired Runx3 expression as a result of Ets1 disruption would cause MHC I–restricted thymocytes to up-regulate Thpok and therefore to fail CD8 differentiation or to be redirected into the CD4 lineage. However, we did not detect Thpok expression in Ets1−/− MHC I–restricted thymocytes, possibly because Runx1 expression in these cells was sufficient to prevent their up-regulation of Thpok.
The heterogeneity of CD4 and Runx3 expression in Ets1−/− MHC I–restricted thymocytes is reminiscent of the variegated CD4 expression by Runx3-deficient CD8 lineage cells, which include maturelike DP thymocytes similar to those found in Ets1−/− mice (Taniuchi et al., 2002a; Woolf et al., 2003). Expression of Cd4 and Cd8 genes is also subject to variegation as a result of mutations of the Cd4 silencer and of Cd8 enhancers, respectively (Kioussis and Ellmeier, 2002; Taniuchi et al., 2004). “Pronase stripping” analyses suggest a second source for the heterogeneity of Cd4 expression by Ets1−/− CD8 lineage cells, namely that Ets1 disruption delays, rather than prevents, Cd4 silencing (and presumably Runx3 up-regulation). It is also important to note that the pleiotropic effects of Ets1 disruption, including on TCRβ allelic exclusion (Eyquem et al., 2004), may indirectly affect the sequence of developmental events that normally characterize positive selection. It is possible that such changes feedback on lineage-specific gene expression programs, and, combined with environmental constraints unique to each cell, contribute to the heterogeneous Runx3 and Cd4 expression of Ets1−/− thymocytes.
In summary, the present study demonstrates the transcription factor Ets1 is required for the proper cessation of Cd4 expression during the intrathymic development of MHC I–restricted CD8 lineage cells, and that it acts at least in part by promoting the expression of the Cd4 repressor Runx3.
MATERIALS AND METHODS
Mice.
Wild-type C57BL/6 (B6) mice were obtained from the National Cancer Institute (NCI) animal production facility. Mice carrying a disrupted Ets1 locus (Barton et al., 1998) were maintained heterozygous and intercrossed to obtain Ets1−/− mice. Mice carrying the P14 TCR transgene (Pircher et al., 1989; originally obtained from Taconic) or the OT-II TCR transgene (Barnden et al., 1998; originally obtained from Jax), and mice carrying a Runx3 transgene (Grueter et al., 2005) were intercrossed with Ets1+/− animals to generate Ets1−/− mice with the desired transgene combination. Mice were housed in a specific pathogen–free facility and were analyzed between 6 and 12 wk of age, unless otherwise indicated. The BAC reporter transgene for Runx3 expression was prepared as previously described (Wang et al., 2008a) by inserting a tRFP cDNA (tdTomato, a gift from R. Tsien, University of California, San Diego, La Jolla, CA; Shaner et al., 2005), using recombineering technology (http://recombineering.ncifcrf.gov/), and microinjected into C57BL/6 fertilized oocytes. Resulting animals (I5-founder–derived line) were backcrossed to Ets1−/− mice. Animal procedures were approved by the NCI Animal Care and Use Committee.
Cell preparation, staining, and analyses of gene expression.
Single-cell suspensions of thymocytes and splenocytes were prepared and stained as described previously (Liu et al., 2003). Flow cytometry data were acquired either on a two-laser FACSCalibur or on an LSR-II cytometer (both from BD) using the software and configuration provided by the manufacturer. Data were analyzed with FlowJo Software (Tree Star, Inc.). Dead cells were excluded from analyses on the basis of Forward Light Scatter and either propidium iodide, DAPI, or 7-AAD gating. Cell sorting was performed on a FACSVantage SE (BD). RNA was extracted from sorted cells with TRIzol (Invitrogen), reverse-transcribed from oligo-dT primers, and analyzed by quantitative real time PCR as previously described (Jenkinson et al., 2007), using an ABI PRISM 7900HT sequence detection system (Applied Biosystems) and previously published primer and probe sets (Jenkinson et al., 2007). Gene expression values are normalized to β-actin in the same sample. Expression of Runx proteins was analyzed by immunoblotting of whole-cell lysates with an antibody that recognizes Runx1 and Runx3, a generous gift of M. Satake, Tohoku University, Sendai, Japan (Sato et al., 2005); expression of β-actin on the same membrane was assessed as a loading control, and quantified using the Odyssey system (Li-Cor) where indicated. Analyses of CD4 and CD8 reexpression after pronase stripping were conducted as previously described (Brugnera et al., 2000).
Immunohistology.
OCT-embedded frozen tissue sections were air-dried 15 min before acetone fixation. For costaining, sections were incubated simultaneously with optimal dilutions of polyclonal anti-mouse keratin 14 (Covance Research), FITC anti–mouse CD8 (clone 53–6.7), and anti–mouse CD4 (clone H129-19; BD). Immunoreactivity to CD4 was enhanced by tyramide amplification (PerkinElmer). Controls included slides incubated with normal rabbit IgG or isotype-matched rat IgG. Microscopic analysis was performed with a LSM 510 confocal microscope (Carl Zeiss, Inc.).
ChIP assays.
ChIP was performed from Ets1+/+ Th1 effector CD4+ cells as previously described (Grenningloh et al., 2005). A detailed protocol is available upon request. The following antibodies were used: rabbit anti-Ets1 (C-20) and control rabbit IgG (both from Santa Cruz Biotechnology, Inc.). Isolated DNA fragments were amplified by quantitative PCR (Mx300P, Stratagene) using the following primers: Segment C (F, 5′-GTTGACTGGTGGGAATAAAG-3′; R, 5′-AGGGTTTGGCACATACTG-3′), segment D (F, 5′-AACACCCTAAGAGCATCAAA-3′; R, 5′-TTTATGGGAGTTGGGATTTA-3′), segments E (F, 5′-ATCCACAAACAGAAAGCCTA-3′; R, 5′-TGTCAACCCAATCTCACAT-3′), and segment G (F, 5′-TAACCGGTAACTGGGATG-3′; R, 5′-CGCTGAGGTTGAGAGTGT-3′). For each target segment, fold enrichment was defined as the ratio of the target in the anti-Ets1 immunoprecipitates relative to the control IgG immunoprecipitate, calculated as 2([anti-Ets1 cycle number] − [control cycle number]).
Online supplemental material.
Fig. S1 shows thymocyte subsets in Ets1−/− newborn mice. Fig. S2 analyzes T cell selection in TCR transgenic Ets1−/− thymi. Fig. S3 displays the sorting strategy used in the coreceptor reexpression assay (Fig. 5 A). Fig. S4 shows the schematic structure and expression pattern of the Runx3 tRFP BAC reporter. Fig. S5 shows expression of the Runx3 transgene in thymocyte subsets. Fig. S6 documents that Ets1 disruption does not affect expression of the Runx3 transgene. Fig. S7 shows that Ets1 is not required for Runx3-mediated CD4 repression. Fig. S8 displays the location and sequence of Ets1 binding regions detected by ChIP analyses on the Runx3 gene. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092024/DC1.
Acknowledgments
We thank M. Satake for the anti-Runx antiserum, Barbara Taylor, and Subhadra Banerjee for cell sorting; Susan Garfield for assistance with confocal microscopy; Ehydel Castro, Peter Mercado, and Genevieve Sanchez for expert mouse care; Al Singer for insightful discussions; and Jon Ashwell, Anne Gégonne, Ranjan Sen, and Al Singer for critical reading of the manuscript.
This work was supported in part by the Intramural Research Program of the National Cancer Institute, Center for Cancer Research, National Institutes of Health (NIH), and by NIH grant AI081052. M. Ehlers is a fellow of the Claussen-Simon-Foundation and supported by the Max-Planck-Institute for Infection Biology, Germany.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- DP
- double positive
- SP
- single positive
- tRFP
- tandem-dimer-tomato red fluorescent protein
References
- Anderson M.K., Hernandez-Hoyos G., Diamond R.A., Rothenberg E.V. 1999. Precise developmental regulation of Ets family transcription factors during specification and commitment to the T cell lineage. Development. 126:3131–3148 [DOI] [PubMed] [Google Scholar]
- Arman M., Calvo J., Trojanowska M.E., Cockerill P.N., Santana M., López-Cabrera M., Vives J., Lozano F. 2004. Transcriptional regulation of human CD5: important role of Ets transcription factors in CD5 expression in T cells. J. Immunol. 172:7519–7529 [DOI] [PubMed] [Google Scholar]
- Barnden M.J., Allison J., Heath W.R., Carbone F.R. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34–40 10.1046/j.1440-1711.1998.00709.x [DOI] [PubMed] [Google Scholar]
- Barton K., Nucifora G. 2000. AML1 haploinsufficiency, gene dosage, and the predisposition to acute leukemia. Bioessays. 22:214–218 [DOI] [PubMed] [Google Scholar]
- Barton K., Muthusamy N., Fischer C., Ting C.N., Walunas T.L., Lanier L.L., Leiden J.M. 1998. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 9:555–563 10.1016/S1074-7613(00)80638-X [DOI] [PubMed] [Google Scholar]
- Bories J.C., Willerford D.M., Grévin D., Davidson L., Camus A., Martin P., Stéhelin D., Alt F.W. 1995. Increased T-cell apoptosis and terminal B-cell differentiation induced by inactivation of the Ets-1 proto-oncogene. Nature. 377:635–638 10.1038/377635a0 [DOI] [PubMed] [Google Scholar]
- Bosselut R. 2004. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat. Rev. Immunol. 4:529–540 10.1038/nri1392 [DOI] [PubMed] [Google Scholar]
- Brugnera E., Bhandoola A., Cibotti R., Yu Q., Guinter T.I., Yamashita Y., Sharrow S.O., Singer A. 2000. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 13:59–71 10.1016/S1074-7613(00)00008-X [DOI] [PubMed] [Google Scholar]
- Clements J.L., John S.A., Garrett-Sinha L.A. 2006. Impaired generation of CD8+ thymocytes in Ets-1-deficient mice. J. Immunol. 177:905–912 [DOI] [PubMed] [Google Scholar]
- Cruz-Guilloty F., Pipkin M.E., Djuretic I.M., Levanon D., Lotem J., Lichtenheld M.G., Groner Y., Rao A. 2009. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 206:51–59 10.1084/jem.20081242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djuretic I.M., Levanon D., Negreanu V., Groner Y., Rao A., Ansel K.M. 2007. Transcription factors T-bet and Runx3 cooperate to activate Ifng and silence Il4 in T helper type 1 cells. Nat. Immunol. 8:145–153 10.1038/ni1424 [DOI] [PubMed] [Google Scholar]
- Egawa T., Littman D.R. 2008. ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat. Immunol. 9:1131–1139 10.1038/ni.1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa T., Tillman R.E., Naoe Y., Taniuchi I., Littman D.R. 2007. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med. 204:1945–1957 10.1084/jem.20070133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers M., Laule-Kilian K., Petter M., Aldrian C.J., Grueter B., Würch A., Yoshida N., Watanabe T., Satake M., Steimle V. 2003. Morpholino antisense oligonucleotide-mediated gene knockdown during thymocyte development reveals role for Runx3 transcription factor in CD4 silencing during development of CD4-/CD8+ thymocytes. J. Immunol. 171:3594–3604 [DOI] [PubMed] [Google Scholar]
- Erman B., Cortes M., Nikolajczyk B.S., Speck N.A., Sen R. 1998. ETS-core binding factor: a common composite motif in antigen receptor gene enhancers. Mol. Cell. Biol. 18:1322–1330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyquem S., Chemin K., Fasseu M., Bories J.C. 2004. The Ets-1 transcription factor is required for complete pre-T cell receptor function and allelic exclusion at the T cell receptor beta locus. Proc. Natl. Acad. Sci. USA. 101:15712–15717 10.1073/pnas.0405546101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowlkes B.J., Pardoll D.M. 1989. Molecular and cellular events of T cell development. Adv. Immunol. 44:207–264 10.1016/S0065-2776(08)60643-4 [DOI] [PubMed] [Google Scholar]
- Goetz T.L., Gu T.L., Speck N.A., Graves B.J. 2000. Auto-inhibition of Ets-1 is counteracted by DNA binding cooperativity with core-binding factor alpha2. Mol. Cell. Biol. 20:81–90 10.1128/MCB.20.1.81-90.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh R., Kang B.Y., Ho I.C. 2005. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 201:615–626 10.1084/jem.20041330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenningloh R., Miaw S.C., Moisan J., Graves B.J., Ho I.C. 2008. Role of Ets-1 phosphorylation in the effector function of Th cells. Eur. J. Immunol. 38:1700–1705 10.1002/eji.200738112 [DOI] [PubMed] [Google Scholar]
- Grueter B., Petter M., Egawa T., Laule-Kilian K., Aldrian C.J., Wuerch A., Ludwig Y., Fukuyama H., Wardemann H., Waldschuetz R., et al. 2005. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J. Immunol. 175:1694–1705 [DOI] [PubMed] [Google Scholar]
- Gu T.L., Goetz T.L., Graves B.J., Speck N.A. 2000. Auto-inhibition and partner proteins, core-binding factor beta (CBFbeta) and Ets-1, modulate DNA binding by CBFalpha2 (AML1). Mol. Cell. Biol. 20:91–103 10.1128/MCB.20.1.91-103.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., He X., Dave V.P., Zhang Y., Hua X., Nicolas E., Xu W., Roe B.A., Kappes D.J. 2005. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 433:826–833 10.1038/nature03338 [DOI] [PubMed] [Google Scholar]
- Hernández-Hoyos G., Anderson M.K., Wang C., Rothenberg E.V., Alberola-Ila J. 2003. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 19:83–94 10.1016/S1074-7613(03)00176-6 [DOI] [PubMed] [Google Scholar]
- Higuchi T., Bartel F.O., Masuya M., Deguchi T., Henderson K.W., Li R., Muise-Helmericks R.C., Kern M.J., Watson D.K., Spyropoulos D.D. 2007. Thymomegaly, microsplenia, and defective homeostatic proliferation of peripheral lymphocytes in p51-Ets1 isoform-specific null mice. Mol. Cell. Biol. 27:3353–3366 10.1128/MCB.01871-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho I.C., Bhat N.K., Gottschalk L.R., Lindsten T., Thompson C.B., Papas T.S., Leiden J.M. 1990. Sequence-specific binding of human Ets-1 to the T cell receptor alpha gene enhancer. Science. 250:814–818 10.1126/science.2237431 [DOI] [PubMed] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., Carbone F.R. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Hollenhorst P.C., Shah A.A., Hopkins C., Graves B.J. 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21:1882–1894 10.1101/gad.1561707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson S.R., Intlekofer A.M., Sun G., Feigenbaum L., Reiner S.L., Bosselut R. 2007. Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. J. Exp. Med. 204:267–272 10.1084/jem.20061982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W.Y., Sieweke M., Ogawa E., Wee H.J., Englmeier U., Graf T., Ito Y. 1999. Mutual activation of Ets-1 and AML1 DNA binding by direct interaction of their autoinhibitory domains. EMBO J. 18:1609–1620 10.1093/emboj/18.6.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kioussis D., Ellmeier W. 2002. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat. Rev. Immunol. 2:909–919 10.1038/nri952 [DOI] [PubMed] [Google Scholar]
- Kishimoto H., Sprent J. 1999. Several different cell surface molecules control negative selection of medullary thymocytes. J. Exp. Med. 190:65–73 10.1084/jem.190.1.65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug D.B., Carter C., Crouch E., Roop D., Conti C.J., Richie E.R. 1998. Interdependence of cortical thymic epithelial cell differentiation and T-lineage commitment. Proc. Natl. Acad. Sci. USA. 95:11822–11827 10.1073/pnas.95.20.11822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohu K., Sato T., Ohno S., Hayashi K., Uchino R., Abe N., Nakazato M., Yoshida N., Kikuchi T., Iwakura Y., et al. 2005. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. J. Immunol. 174:2627–2636 [DOI] [PubMed] [Google Scholar]
- Liu X., Bosselut R. 2004. Duration of TCR signaling controls CD4-CD8 lineage differentiation in vivo. Nat. Immunol. 5:280–288 10.1038/ni1040 [DOI] [PubMed] [Google Scholar]
- Liu X., Adams A., Wildt K.F., Aronow B., Feigenbaum L., Bosselut R. 2003. Restricting Zap70 expression to CD4+CD8+ thymocytes reveals a T cell receptor-dependent proofreading mechanism controlling the completion of positive selection. J. Exp. Med. 197:363–373 10.1084/jem.20021698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muroi S., Naoe Y., Miyamoto C., Akiyama K., Ikawa T., Masuda K., Kawamoto H., Taniuchi I. 2008. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat. Immunol. 9:1113–1121 10.1038/ni.1650 [DOI] [PubMed] [Google Scholar]
- Muthusamy N., Barton K., Leiden J.M. 1995. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 377:639–642 10.1038/377639a0 [DOI] [PubMed] [Google Scholar]
- Naoe Y., Setoguchi R., Akiyama K., Muroi S., Kuroda M., Hatam F., Littman D.R., Taniuchi I. 2007. Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J. Exp. Med. 204:1749–1755 10.1084/jem.20062456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pai S.Y., Truitt M.L., Ting C.N., Leiden J.M., Glimcher L.H., Ho I.C. 2003. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 19:863–875 10.1016/S1074-7613(03)00328-5 [DOI] [PubMed] [Google Scholar]
- Pircher H., Bürki K., Lang R., Hengartner H., Zinkernagel R.M. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 342:559–561 10.1038/342559a0 [DOI] [PubMed] [Google Scholar]
- Pognonec P., Boulukos K.E., Bosselut R., Boyer C., Schmitt-Verhulst A.M., Ghysdael J. 1990. Identification of a Ets1 variant protein unaffected in its chromatin and in vitro DNA binding capacities by T cell antigen receptor triggering and intracellular calcium rises. Oncogene. 5:603–610 [PubMed] [Google Scholar]
- Prosser H.M., Wotton D., Gegonne A., Ghysdael J., Wang S., Speck N.A., Owen M.J. 1992. A phorbol ester response element within the human T-cell receptor beta-chain enhancer. Proc. Natl. Acad. Sci. USA. 89:9934–9938 10.1073/pnas.89.20.9934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pufall M.A., Lee G.M., Nelson M.L., Kang H.S., Velyvis A., Kay L.E., McIntosh L.P., Graves B.J. 2005. Variable control of Ets-1 DNA binding by multiple phosphates in an unstructured region. Science. 309:142–145 10.1126/science.1111915 [DOI] [PubMed] [Google Scholar]
- Sato T., Ohno S., Hayashi T., Sato C., Kohu K., Satake M., Habu S. 2005. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 22:317–328 10.1016/j.immuni.2005.01.012 [DOI] [PubMed] [Google Scholar]
- Sawada S., Scarborough J.D., Killeen N., Littman D.R. 1994. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 77:917–929 10.1016/0092-8674(94)90140-6 [DOI] [PubMed] [Google Scholar]
- Setoguchi R., Tachibana M., Naoe Y., Muroi S., Akiyama K., Tezuka C., Okuda T., Taniuchi I. 2008. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 319:822–825 10.1126/science.1151844 [DOI] [PubMed] [Google Scholar]
- Shaner N.C., Steinbach P.A., Tsien R.Y. 2005. A guide to choosing fluorescent proteins. Nat. Methods. 2:905–909 10.1038/nmeth819 [DOI] [PubMed] [Google Scholar]
- Sharrocks A.D. 2001. The ETS-domain transcription factor family. Nat. Rev. Mol. Cell Biol. 2:827–837 10.1038/35099076 [DOI] [PubMed] [Google Scholar]
- Singer A., Bosselut R. 2004. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv. Immunol. 83:91–131 10.1016/S0065-2776(04)83003-7 [DOI] [PubMed] [Google Scholar]
- Singer A., Adoro S., Park J.H. 2008. Lineage fate and intense debate: myths, models and mechanisms of CD4- versus CD8-lineage choice. Nat. Rev. Immunol. 8:788–801 10.1038/nri2416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr T.K., Jameson S.C., Hogquist K.A. 2003. Positive and negative selection of T cells. Annu. Rev. Immunol. 21:139–176 10.1146/annurev.immunol.21.120601.141107 [DOI] [PubMed] [Google Scholar]
- Sun G., Liu X., Mercado P., Jenkinson S.R., Kypriotou M., Feigenbaum L., Galéra P., Bosselut R. 2005. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat. Immunol. 6:373–381 10.1038/ni1183 [DOI] [PubMed] [Google Scholar]
- Suzuki H., Punt J.A., Granger L.G., Singer A. 1995. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity. 2:413–425 10.1016/1074-7613(95)90149-3 [DOI] [PubMed] [Google Scholar]
- Swat W., Dessing M., von Boehmer H., Kisielow P. 1993. CD69 expression during selection and maturation of CD4+8+ thymocytes. Eur. J. Immunol. 23:739–746 10.1002/eji.1830230326 [DOI] [PubMed] [Google Scholar]
- Takahama Y. 2006. Journey through the thymus: stromal guides for T-cell development and selection. Nat. Rev. Immunol. 6:127–135 10.1038/nri1781 [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Osato M., Egawa T., Sunshine M.J., Bae S.C., Komori T., Ito Y., Littman D.R. 2002a. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 111:621–633 10.1016/S0092-8674(02)01111-X [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Sunshine M.J., Festenstein R., Littman D.R. 2002b. Evidence for distinct CD4 silencer functions at different stages of thymocyte differentiation. Mol. Cell. 10:1083–1096 [DOI] [PubMed] [Google Scholar]
- Taniuchi I., Ellmeier W., Littman D.R. 2004. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv. Immunol. 83:55–89 10.1016/S0065-2776(04)83002-5 [DOI] [PubMed] [Google Scholar]
- Telfer J.C., Hedblom E.E., Anderson M.K., Laurent M.N., Rothenberg E.V. 2004. Localization of the domains in Runx transcription factors required for the repression of CD4 in thymocytes. J. Immunol. 172:4359–4370 [DOI] [PubMed] [Google Scholar]
- Tung J.W., Kunnavatana S.S., Herzenberg L.A., Herzenberg L.A. 2001. The regulation of CD5 expression in murine T cells. BMC Mol. Biol. 2:5 10.1186/1471-2199-2-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A., Duterque-Coquillaud M. 2002. When Ets transcription factors meet their partners. Bioessays. 24:362–370 10.1002/bies.10068 [DOI] [PubMed] [Google Scholar]
- Wang L., Bosselut R. 2009. CD4-CD8 lineage differentiation: Thpok-ing into the nucleus. J. Immunol. 183:2903–2910 10.4049/jimmunol.0901041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wildt K.F., Zhu J., Zhang X., Feigenbaum L., Tessarollo L., Paul W.E., Fowlkes B.J., Bosselut R. 2008a. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat. Immunol. 9:1122–1130 10.1038/ni.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wildt K.F., Castro E., Xiong Y., Feigenbaum L., Tessarollo L., Bosselut R. 2008b. The zinc finger transcription factor Zbtb7b represses CD8-lineage gene expression in peripheral CD4+ T cells. Immunity. 29:876–887 10.1016/j.immuni.2008.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildt K.F., Sun G., Grueter B., Fischer M., Zamisch M., Ehlers M., Bosselut R. 2007. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J. Immunol. 179:4405–4414 [DOI] [PubMed] [Google Scholar]
- Woolf E., Xiao C., Fainaru O., Lotem J., Rosen D., Negreanu V., Bernstein Y., Goldenberg D., Brenner O., Berke G., et al. 2003. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc. Natl. Acad. Sci. USA. 100:7731–7736 10.1073/pnas.1232420100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q., Erman B., Bhandoola A., Sharrow S.O., Singer A. 2003. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J. Exp. Med. 197:475–487 10.1084/jem.20021765 [DOI] [PMC free article] [PubMed] [Google Scholar]