Abstract
Infection by Toxoplasma gondii down-regulates the host innate immune responses, such as proinflammatory cytokine production, in a Stat3-dependent manner. A forward genetic approach recently demonstrated that the type II strain fails to suppress immune responses because of a potential defect in a highly polymorphic parasite-derived kinase, ROP16. We generated ROP16-deficient type I parasites by reverse genetics and found a severe defect in parasite-induced Stat3 activation, culminating in enhanced production of interleukin (IL) 6 and IL-12 p40 in the infected macrophages. Furthermore, overexpression of ROP16 but not ROP18 in mammalian cells resulted in Stat3 phosphorylation and strong activation of Stat3-dependent promoters. In addition, kinase-inactive ROP16 failed to activate Stat3. Comparison of type I and type II ROP16 revealed that a single amino acid substitution in the kinase domain determined the strain difference in terms of Stat3 activation. Moreover, ROP16 bound Stat3 and directly induced phosphorylation of this transcription factor. These results formally establish an essential and direct requirement of ROP16 in parasite-induced Stat3 activation and the significance of a single amino acid replacement in the function of type II ROP16.
Toxoplasma gondii is a eukaryotic pathogen that causes life threatening toxoplasmosis, a condition which includes encephalitis in immunocompromised individuals, such as those suffering from AIDS and congenital diseases or being treated by chemotherapy, upon primary infection during pregnancy in humans and animals (Montoya and Remington, 2008). T. gondii is an obligate intracellular eukaryotic parasite capable of proliferating exclusively inside a parasitophorous vacuole, which is formed during host cell invasion (Joynson and Wreghitt, 2001). Taxonomically, T. gondii belongs to the phylum Apicomplexa, which is defined by the presence of an apical complex including secretory organelles (Dubremetz, 2007). Among them, the large bulb-shaped organelles called rhoptries contain a variety of proteins, which are secreted into the host cytoplasm or in the forming parasitophorous vacuole during parasite entry to co-opt the host cell for growth and survival (Boothroyd and Dubremetz, 2008).
The host initiates a broad range of immune responses upon T. gondii infection. When infected by T. gondii, cells belonging to the innate immunity system, such as macrophages and dendritic cells, produce a set of proinflammatory cytokines, including IL-6, IL-12 p40, and nitric oxide, to induce the adaptive immune response or to directly eliminate the parasites (Denkers, 2003; Yap et al., 2006). In particular, toll-like receptor (TLR)–mediated MyD88-dependent innate and adaptive immune responses have been shown to be essential for the host defense against T. gondii (Gazzinelli et al., 2004). Recently, TLR11 has been shown to recognize the T. gondii profilin-like protein, resulting in the production of proinflammatory cytokine IL-12 by dendritic cells (Yarovinsky et al., 2005; Yarovinsky and Sher, 2006; Plattner et al., 2008). In addition to TLR11, TLR2 has also been implicated in parasite-induced immune responses (Scanga et al., 2002; Mun et al., 2003; Del Rio et al., 2004). In contrast, T. gondii–derived cyclophilin, C-18, has been shown to activate dendritic cells to produce IL-12 through a G protein–coupled chemokine receptor CCR5 in a TLR-independent fashion (Aliberti et al., 2003). This suggests that both TLR-dependent and -independent innate immune responses play important roles in the host defense against this parasite.
T. gondii is generally divided into three predominant clonal lineages (types I, II, and III) in addition to exotic strains (Ajzenberg et al., 2004). These clonal lineages correlate with differences in TLR-MyD88–dependent responses to T. gondii (Aliberti et al., 2003; Denkers et al., 2003; Robben et al., 2004; Kim et al., 2006). Compared with type I parasites, infection by type II parasites results in up-regulation of IL-12 p40 production in macrophages. The IL10-independent Stat3 activation, induced upon type I parasite infection, was previously shown to be defective in type II parasites (Butcher et al., 2005). A subsequent study involving a forward genetic approach, in which type II and III strains were intercrossed, identified a highly polymorphic rhoptry protein, ROP16, as a candidate gene product responsible for Stat3 activation. The introduction of type I ROP16 into type II parasites successfully induced prolonged Stat3 activation and suppression of IL-12 production (Saeij et al., 2006, 2007). However, the molecular mechanism by which ROP16 regulates Stat3 activation, leading to suppression of innate immune responses, and the basis for the strain differences between type I (or III) and II ROP16-dependent Stat3 activation are not understood.
In this paper, we have analyzed the physiological function of ROP16 by generating type I ROP16-deficient (rop16 KO) parasites. Stat3 activation is severely reduced in cells infected by type I rop16 KO parasites, leading to an up-regulation of IL-6 and IL-12 p40 production compared with cells infected by WT parasites. In addition, ectopic expression of ROP16 in mammalian cells dramatically increases Stat3 activation. Using an in vitro assay system, we demonstrate that the kinase activity of ROP16 is essential for Stat3 activation and that a single amino acid replacement resulting from the polymorphism between type I and II is responsible for the strain difference in terms of ROP16-regulated Stat3 activation. Furthermore the N-terminal portion of ROP16 interacts with Stat3 and ROP16 directly phosphorylates Stat3 tyrosine 705, indicating the direct and essential role of ROP16 in Stat3 activation.
RESULTS
Defective Stat3 activation in rop16 KO type I parasite-infected cells
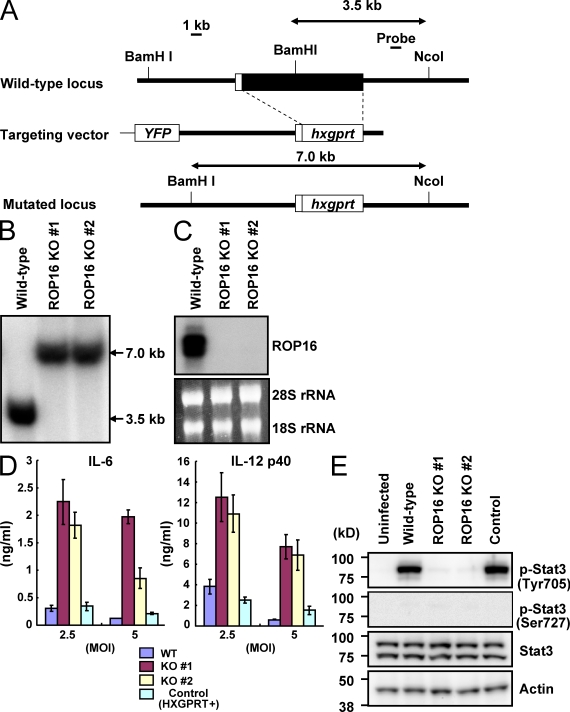
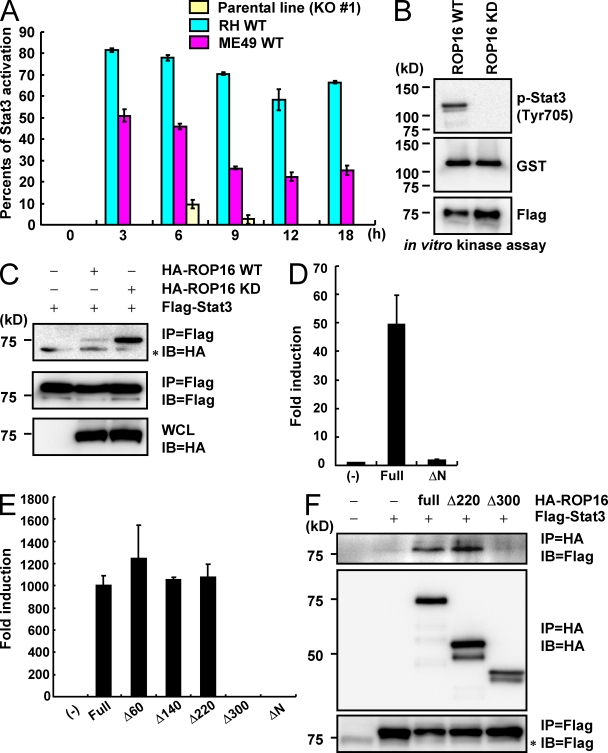
To examine the physiological role of ROP16 in the type I strain, rop16 KO parasites were generated by targeted gene disruption. The targeting vector contained flanking sequences of the ROP16 gene and the entire coding region of ROP16 was replaced with the hxgprt gene (Fig. 1 A). Upon selection for mycophenolic acid (MPA)/xanthine-resistant and yellow fluorescent protein (YFP)–negative parasites, we isolated two homologous recombinant mutants out of 47 clones (Fig. 1 B). Northern blot analysis confirmed that transcription of ROP16 messenger RNA in both mutant clones was completely abolished (Fig. 1 C). Intracellular replication of rop16 KO type I parasites in Vero cells and mouse embryonic fibroblasts (MEFs) was similar to that of WT parasites (unpublished data). Given the previous implication of ROP16 in the strain-dependent IL-12 p40 production and Stat3 activation (Saeij et al., 2007), we first assessed the effect of ROP16 deficiency in type I parasites on the parasite-induced proinflammatory cytokine production in mouse peritoneal macrophages (Fig. 1 D). Compared with WT (hxgprt-negative parental parasites) or hxgprt-positive control type I parasites, production of proinflammatory cytokines, such as IL-6 and IL-12 p40, was dramatically increased in macrophages infected by rop16 KO parasites. Stat3 activation has been shown to suppress proinflammatory cytokine production in cells infected with type I, but not type II, parasites (Kim et al., 2006). Therefore, we examined parasite-induced Stat3 activation in peritoneal macrophages or MEFs. Phosphorylation of Tyr705 on Stat3 was observed in cells infected with WT and control type I parasites in a time-dependent manner (Fig. 1 E; Fig. S1, A and B). In contrast, Stat3 phosphorylation of Tyr705 was almost absent in type I rop16 KO parasite-infected cells. Moreover, phosphorylation of Stat3 on Ser727, another Stat3 phosphorylation site which has been previously shown to play an important role in full activation of the transcription factor (Ihle, 2001), was not induced by parasite infection (Fig. 1 E). Collectively and as previously predicted (Saeij et al., 2006, 2007), these results demonstrate that ROP16 is indeed the determinant for the strain difference between type II and I (or III) with respect to Stat3 activation.
Figure 1.
Loss of ROP16 in type I parasites severely impairs Stat3 activation. (A) The structure of the ROP16 gene, the targeting vector and the predicted disrupted gene. Black boxes denote the exons. (B) Southern blot analysis of offspring from WT or two lines of ROP16-deficient parasites. 30 µg of total genomic DNA was extracted from parasites, digested with NcoI–BamHI, electrophoresed, and hybridized with the radiolabeled probe indicated in A. Southern blotting gave a single 3.5-kb band for WT and a 7.0-kb band for the disrupted locus. (C) Northern blot analysis on 10 µg of a total parasite RNA separated on gel, transferred to a nylon membrane, and hybridized with ROP16 probe. 28S and 18S ribosomal RNA was shown as the loading control (bottom). (D) Peritoneal macrophages from C57BL/6 mice were cultured with the indicated MOI of parasites in the presence of 30 ng/ml IFN-γ for 24 h. Concentrations of IL-6 and IL-12 p40 in the culture supernatants were measured by ELISA. Indicated values are means ± SD of triplicates. (E) Serum-starved peritoneal macrophages were infected with MOI = 10 of indicated parasites for 3 h. Activation of Stat3 was also determined by Western blot of cell extracts using anti–phospho-Stat3 Tyr705 or Ser727. Stat3 and actin levels are shown as loading controls. Data are representative of at least three (D) or two (B, C, and E) independent experiments.
Activation of Stat3 in mammalian cells ectopically expressing ROP16
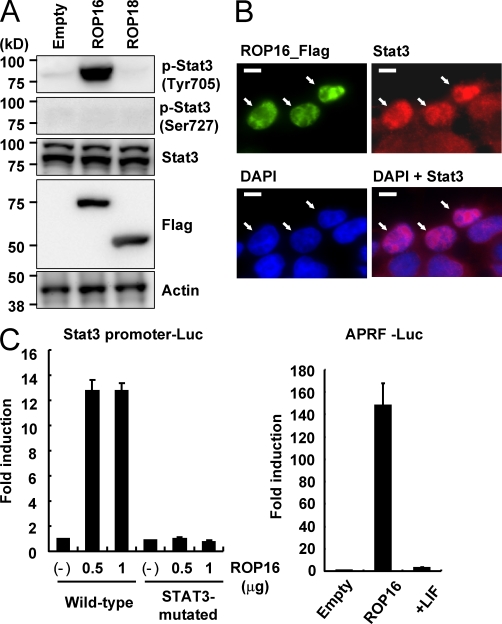
To explore the mechanism of ROP16-regulated Stat3 activation, we constructed vectors containing the ROP16 protein, lacking the N-terminal signal peptide, to enable expression in mammalian cells. The 293T cells were transfected with expression vectors for ROP16 or another rhoptry kinase family protein, ROP18, also lacking the N-terminal signal peptide. We then ectopically expressed these proteins and analyzed the effects on Stat3 phosphorylation. Strikingly, the overexpression of ROP16, but not ROP18, in 293T cells led to high levels of Stat3 phosphorylation of Tyr705 but not Ser727 (Fig. 2 A). Furthermore, indirect immunofluorescence analysis revealed that the ectopic expression of ROP16, but not ROP18, induced the nuclear translocation of Stat3 (Fig. 2 B and Fig. S2A), suggesting that ROP16 overexpression mediates Stat3 activation. To investigate this further, we performed a luciferase assay using reporters harboring Stat3-dependent promoters (Fig. 2 C). The presence of ROP16 caused the activation of luciferase when controlled by a WT Stat3-dependent promoter but not by a promoter mutated in the Stat3-binding sites. When APRF-luc, another Stat3-dependent luciferase reporter, was used, the ROP16-mediated activation was even more prominent. ROP16 has been shown to possess a nuclear localization signal (NLS; Saeij et al., 2007). When WT or NLS-mutated ROP16 expression vectors were introduced in 293T cells, Stat3 was activated in cells transduced with NLS-mutated as well as WT ROP16 (Fig. S2 B). This was in agreement with a previous finding that the NLS of ROP16 is not required for Stat3 and Stat6 activation (Saeij et al., 2007). Together, these results demonstrated that the cytosolic expression of ROP16 in mammalian cells is sufficient to functionally activate Stat3.
Figure 2.
Ectopic expression of ROP16 potentiates Stat3 activation in mammalian cells. (A) 293T cells were transfected with indicated expression vectors. 48 h after transfection, the cells were lysed and subjected to Western blotting. Activation of Stat3 was also determined by Western blot analysis of cell extracts using anti–phospho-Stat3 Tyr705 or Ser727. Stat3 and actin levels are shown as loading controls. Arrows indicate cells immunostained with anti-Flag in the top left and are placed at the same positions in the other photographs. (B) 293T cells were transfected with Flag-tagged ROP16 expression vectors. 48 h after transfection, the cells were fixed and stained with anti-STAT3 or anti-Flag, and then Alexa Fluor 488–conjugated anti–mouse IgG (green), Alexa Fluor 594–conjugated anti–rabbit IgG antibody (red), or DAPI (blue). Bars, 10 µm. (C) 293T cells were transfected with indicated Stat3-dependent luciferase reporters together with either indicated amounts (left) or 1 µg (right) Flag-tagged ROP16 or empty expression vectors. As a positive control, cells were treated with 100 ng/ml human leukemia inhibitory factor (LIF) for 12 h (right). Luciferase activities were expressed as fold increases over the background level shown by lysates prepared from mock-transfected cells. Indicated values are means ± the variation range of duplicates. Data are representative of at least three (A and C) or two (B) independent experiments.
Kinase activity of ROP16 is essential for Stat3 activation
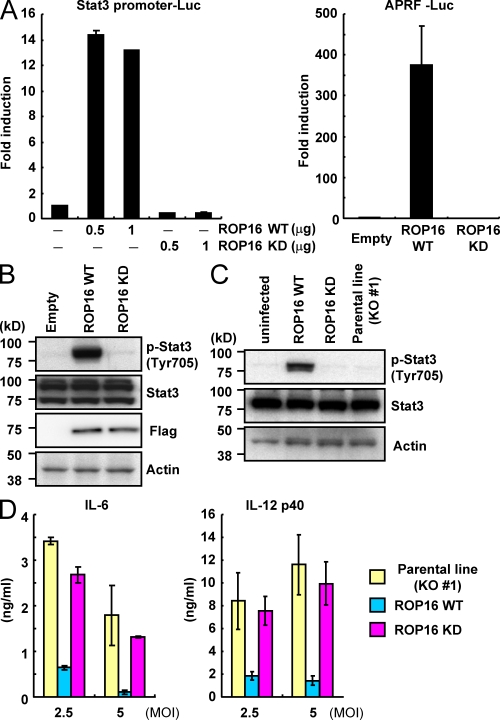
Although the kinase activity of ROP18 has been shown to be essential for the virulence of type III parasites (Saeij et al., 2006; Taylor et al., 2006), the biological significance of the kinase activity of ROP16 remains unclear. To address this point, we first constructed a kinase-inactive mutant of ROP16 (ROP16D539A) and expressed it in 293T cells to determine whether the kinase activity of ROP16 plays an important role in Stat3 activation (Fig. 3 A). In contrast to the strong activation of the Stat3-dependent promoters in WT ROP16-transduced cells, activation of Stat3-dependent promoters was not detected in ROP16D539A-transfected cells. In addition, and unlike WT ROP16, ROP16D539A failed to induce Stat3 Tyr705 phosphorylation (Fig. 3 B). To assess the importance of the ROP16 kinase activity under physiological conditions, we performed a functional complementation of rop16 KO parasites with either WT ROP16 (ROP16WT) or ROP16D539A (ROP16KD) containing the N-terminal signal peptide (Fig. S3 A). Whereas parasites complemented with ROP16WT parasites provoked a significant Stat3 phosphorylation in the MEFs, this effect was completely abolished in cells infected with ROP16KD parasites (Fig. 3 C). Furthermore, T. gondii–induced IL-6 and IL-12 p40 production was severely reduced when macrophages were infected with ROP16WT but not with ROP16KD or control parasites (Fig. 3 D). Collectively, these results suggested that the kinase activity of ROP16 plays a pivotal role in the function of ROP16 in terms of Stat3 activation and suppression of proinflammatory cytokine production.
Figure 3.
The kinase activity of ROP16 is essential for Stat3 activation. (A) 293T cells were transfected with the indicated Stat3-dependent luciferase reporters together with indicated amounts of Flag-tagged WT or kinase-inactive ROP16 (ROP16WT or ROP16KD, respectively) or empty expression plasmids. Luciferase activities were expressed as fold increases over the background levels shown by lysates prepared from mock-transfected cells. (B) 293T cells were transfected with the indicated expression vectors. 48 h after transfection, the cells were lysed and subjected to Western blotting. Activation of Stat3 was also determined by Western blot analysis of cell extracts using anti–phospho-Stat3. (C) Serum-starved MEFs were infected with an MOI = 10 of the indicated parasites for 18 h. Activation of Stat3 was also determined by Western blot analysis of cell extracts using anti–phospho-Stat3 Tyr 705. Stat3 and actin levels are shown as loading controls. (D) Peritoneal macrophages from C57BL/6 mice were cultured with the indicated MOI of parasites in presence of 30 ng/ml IFN-γ for 24 h. Concentrations of IL-6 and IL-12 p40 in the culture supernatants were measured by ELISA. Indicated values are means ± SD of triplicates. Data are representative of three (A and C), four (B), or two (D) independent experiments.
A single amino acid polymorphism in ROP16 determines its Stat3-activating potency
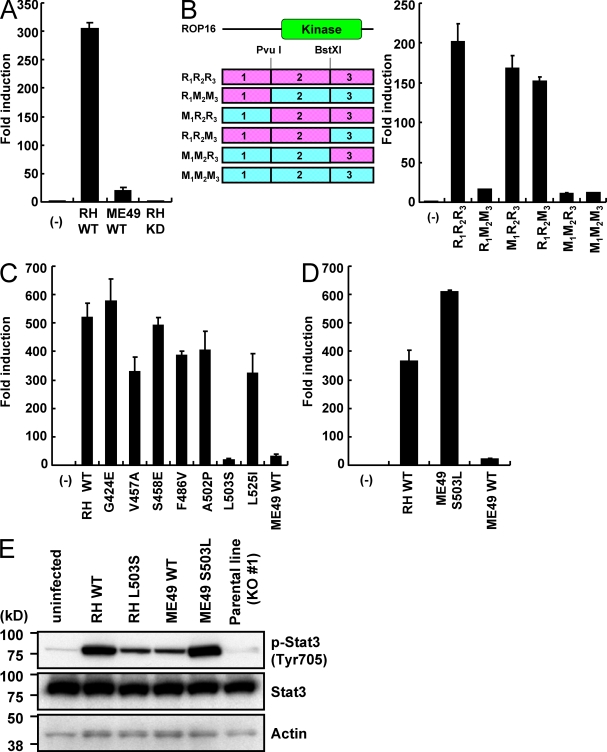
ROP16 is highly polymorphic between type I (or III) and II parasites (Saeij et al., 2006, 2007). Given that the relative expression levels among the three strains from the quantitative RT-PCR data deposited in ToxoDB are similar (unpublished data), the polymorphism might be responsible for the defective Stat3 activation in cells infected with type II parasites. However, the precise molecular mechanism involved remains to be elucidated. To address this issue, we cloned type II (ME49) ROP16 complementary DNA from a type II strain, ME49, into a mammalian expression vector. In 293T cells expressing ME49 ROP16, activation of the Stat3-dependent promoters was only slightly induced, as compared with cells expressing kinase-inactive ROP16D539A. However, it was dramatically lower compared with cells expressing type I (RH) ROP16, indicating that potency of ME49 ROP16 in Stat3 activation may be profoundly but not totally impaired (Fig. 4 A). Next, to identify which regions of the ME49 ROP16 protein are responsible for the defective Stat3 activation, a series of chimeric ROP16 constructs were generated. Domains of ROP16 were swapped between RH and ME49 ROP16, taking advantage of the presence of conserved restriction enzyme sites, and the resulting chimeras were assessed for Stat3 activation in 293T cells (Fig. 4 B). The results revealed a severe defect in Stat3 activation conferred by replacement of the central domain R2 in RH ROP16 with M2 of ME49 ROP16. This region exhibits nine polymorphic amino acid substitutions, and each of them were replaced in RH ROP16 with the corresponding sequence of ME49 and tested for Stat3-dependent reporter activation (Saeij et al., 2007; Fig. S4). Among them, the substitution of a leucine residue at position 503 to a serine (L503S) located in the kinase domain resulted in a dramatic decrease in Stat3 activation to a level comparable to that mediated by ME49 ROP16 (Fig. 4 C). Next, we assessed whether the single reverse mutation of the serine residue at position 503 to a leucine (S503L) in ME49 ROP16 restored Stat3 activation. We found that ME49 ROP16 S503L resulted in full restoration of Stat3 activation (Fig. 4 D). These findings indicated that a single amino acid of ROP16 may determine the strain difference between RH and ME49 strains. Finally, we generated transgenic parasite strains expressing WT or L503S-type RH ROP16 (RHWT or RHL503S, respectively) or WT or S503L-type ME49 ROP16 (ME49WT or ME49S503L, respectively) in rop16 KO parasites to assess their Stat3 activation in vivo (Fig. S3 B). In perfect agreement with a previous observation (Saeij et al., 2007), Stat3 activation, as measured by phosphorylation, was significantly increased by RHWT and ME49S503L and induced, but markedly reduced, by RHL503S or ME49WT (Fig. 4 E). These results indicate that WT ME49 (type II) ROP16 and L503S-type RH (type I) ROP16 mutants are impaired but not completely defective in Stat3 activation in vivo. Collectively, these results clearly demonstrated that the single amino acid polymorphism at position 503 in ROP16 determines the strain difference between type I and II strains in terms of Stat3 activation.
Figure 4.
The L503S polymorphic mutation in type II ROP16 determines its defective Stat3 activation. (A–D) 293T cells were transfected with the APRE-luc plasmid together with the indicated ROP16 expression vectors. Luciferase activities were expressed as fold increases over the background levels shown by lysates prepared from mock-transfected cells. Indicated values are means ± the variation range of duplicates. A list of the chimeric ROP16 expression vectors is shown (B, left). R, RH (pink); M, ME49 (blue). (E) Serum-starved MEFs were infected with an MOI = 10 of the indicated parasites for 18 h. Activation of Stat3 was also determined by Western blot analysis of cell extracts using anti–phospho-Stat3 Tyr 705. Stat3 and actin levels are shown as loading controls. Data are representative of three (A and E) or two (B–D) independent experiments.
Direct Stat3 activation by ROP16
Regarding the molecular basis of ROP16-mediated Stat3 activation, a previous finding suggested that ROP16 may indirectly activate Stat3, based on Stat3 phosphorylation in the very early time points of infection (Saeij et al., 2007). Indeed, as observed for 293T cells, when we infected MEFs with transgenic parasites expressing type I or type II ROP16, and a parental line, we observed moderate Stat3 phosphorylation in cells infected with type II ROP16-expressing parasites, albeit significantly reduced compared with that induced by the infection of type I ROP16-expressing parasites. In sharp contrast, Stat3 activation in cells infected with rop16 KO parasites was nearly abolished (Fig. 5 A and Fig. S5), indicating that ROP16 might directly activate Stat3. To challenge this possibility, we next performed an in vitro kinase assay using recombinant GST-tagged Stat3 as the substrate. Flag-tagged ROP16WT and ROP16KD were expressed in 293T cells and cell lysates were immunoprecipitated with anti-Flag. We detected phosphorylation of Stat3 Tyr705 in immunoprecipitates of ROP16WT, but not ROP16KD (Fig. 5 B), suggesting that Stat3 is directly phosphorylated by ROP16. Next, the interaction of Stat3 with ROP16 was investigated by a series of coimmunoprecipitation experiments. 293T cells were transiently transfected with plasmids encoding HA-tagged ROP16WT or ROP16KD along with Flag-tagged Stat3. HA-tagged WT or kinase-inactive ROP16 were coimmunoprecipitated with anti-Flag, although relatively less ROP16WT was copurified compared with ROP16KD (Fig. 5 C), indicating the existence of an interaction between ROP16 and Stat3. We then examined which regions of ROP16 are required for the interaction with Stat3. When we transiently expressed the C-terminal portion of ROP16 that contained only the kinase domain (residues 345–727) in 293T cells, we failed to observe activation of the Stat3-dependent promoter (Fig. 5 D and Fig. S6). We constructed a series of deletion mutants of ROP16 (amino acids deleted: Δ60, 1–63; Δ140, 1–143; Δ220, 1–223; and Δ300, 1–303) and tested them for Stat3 activation (Fig. S6). Overexpression of full-length ROP16, and ROP16 deletion mutants Δ60, Δ140, and Δ220, but not deletion mutant Δ300, led to activation of the Stat3-dependent promoter (Fig. 5 E). We examined the interaction of Stat3 with full-length ROP16 and ROP16 deletion mutants Δ220 and Δ300. Flag-tagged Stat3 proteins were coprecipitated with anti-HA in cells coexpressing HA-tagged full-length ROP16 or ROP16 deletion mutant Δ220, but not Δ300 (Fig. 5 F). Collectively, these results demonstrate that the N-terminal region between aa 223 and 303 of ROP16 may be required for the interaction with Stat3 (Fig. S6).
Figure 5.
Direct Stat3 activation by ROP16. (A) Relative Stat3 phosphorylation levels. Serum-starved MEFs were infected with an MOI = 10 of the indicated parasites for indicated times. The intensity of Stat3 phosphorylation observed by immunoblotting with anti–phospho-Stat3 antibody was quantitated using a phosphoimager and normalized to images of total Stat3, as shown in Fig. S5. Each count from which that of the uninfected point was subtracted was shown. (B) 293T cells were transiently transfected with Flag-tagged ROP16WT or FLAG kinase–inactive ROP16KD. Cell lysates were immunoprecipitated with anti-FLAG and subjected to an in vitro kinase reaction in the presence of GST–Stat3. Proteins were separated on SDS-PAGE, followed by Western blotting to analyze Stat3 phosphorylation, GST-Stat3, and Flag-tagged ROP16WT or ROP16KD by anti–phospho-Stat3 (Tyr705), anti-GST, and anti-Flag, respectively. (C) Lysates of 293T cells transiently cotransfected with 2 µg Flag-tagged Stat3 and/or 2 µg HA-tagged ROP16WT or ROP16KD expression vectors were immunoprecipitated with the indicated antibodies. *, nonspecific bands. (D and E) 293T cells were transfected with the indicated Stat3-dependent luciferase reporters together with indicated expression vectors. Luciferase activities were expressed as fold increases over the background levels shown by lysates prepared from mock-transfected cells. (F) Lysates of 293T cells transiently cotransfected with 2 µg Flag-tagged Stat3 and/or 2 µg HA-tagged ROP16 full-length, Δ200, or Δ280 expression vectors were immunoprecipitated with the indicated antibodies. *, nonspecific bands. Indicated values are means ± the variation range of duplicates. Data are representative of three (A, C, and E) or two (B, D, and F) independent experiments.
Profile of the kinase domain and activity of ROP16
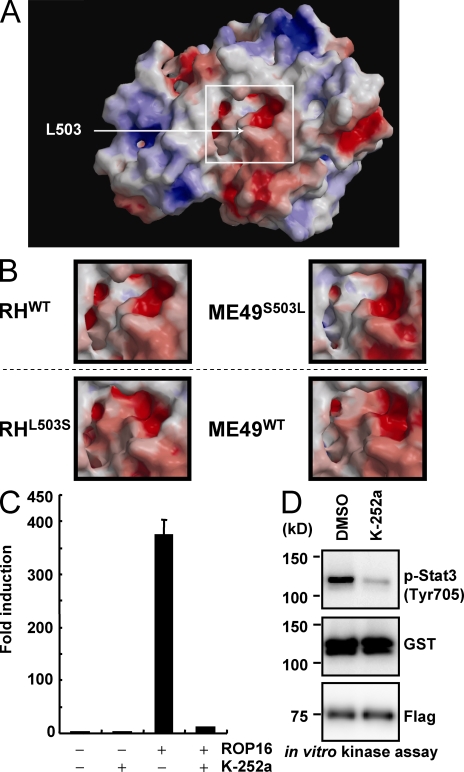
Next we built in silico structural models for the kinase domains of ROP16 variants to assess whether ROP16 presents the signature of a kinase capable of phosphorylating tyrosine residues (Fig. 6 A). The in silico model of the kinase domain of ROP16 was structurally aligned to known kinases and then ranked using a combined structure and sequence-profile-profile similarity score. This analysis indicated that the highest similarity was to dual-specificity mitogen-activated protein kinase kinases (Table S1). In addition, we compared the kinase domains of RHWT, RHL503S, ME49WT, and ME49S503L to examine the significance of residue position 503 (Fig. 6 B and Fig. S7 A). These models indicated that residue 503 is completely solvent inaccessible and may act to stabilize the C-terminal surface of the cavity formed by the N and C-terminal subdomains (Fig. 6 A). In addition, we found that both the modeled RHL503S and ME49WT cavities were larger and contained lower electrostatic potentials than did RHWT or ME49S503L (Fig. 6 B and Fig. S7 A). The corresponding cavity has been shown to form the active site in the most similar known kinases (Ohren et al., 2004; Zhao et al., 2007; Table S1), suggesting that the replacement of leucine with serine in type I ROP16 may result in an active site shape that is similar to that of type II ROP16. Finally we performed luciferase reporter and in vitro kinase assays to characterize the kinase activity of ROP16 by kinase inhibitors. 293T cells were transfected with empty or ROP16 expression vectors in the presence or absence of various kinase inhibitors together with the Stat3-dependent luciferase reporter (Fig. 6 C and Fig. S7 B). Among them, we found that treatment with K-252a severely inhibited the ROP16-mediated activation of the Stat3-dependent promoter (Fig. 6 C and Fig. S7 B). Furthermore, K-252a treatment markedly reduced in vitro ROP16-mediated phosphorylation (Fig. 6 D). Collectively, these results show that the kinase activity of ROP16 is K-252a sensitive and that the amino acid substitution of a leucine at position 503 may affect the active cavity of the kinase domain.
Figure 6.
In silico, in vitro, and in vivo profiling of the ROP16 kinase domain. (A) The molecular surface of the kinase domain of RHWT, colored by electrostatic potential, with red (or blue) representing the most negative (or positive) values. The active site region is framed by a white box, and the approximate location of the solvent inaccessible residue 503 is indicated by an arrow. (B) Close-up views of the active site regions, corresponding to the box in A, for RHWT, ME49S503L, RHL503S, and ME49WT. Top (RHWT and ME49S503L) and bottom (RHL503S and ME49WT) types, distinguished by a dashed line, represent high and low potencies for Stat3 activation, respectively. (C) 293T cells were transfected with the APRF-luc reporters together with 1 µg of empty or Flag-tagged ROP16 (ROP16WT) expression vectors in the presence of 5 µM K-252a or DMSO (control). Luciferase activities were expressed as fold increases over the background levels shown by lysates prepared from mock-transfected cells. Indicated values are means ± the variation range of duplicates. (D) 293T cells were transiently transfected with Flag-tagged ROP16WT. Cell lysates were immunoprecipitated with anti-FLAG and subjected to an in vitro kinase reaction by using GST–Stat3 as the substrate in the presence of 5 µM K-252a or DMSO (control). Proteins were separated on SDS-PAGE, followed by Western blotting to analyze Stat3 phosphorylation, GST-Stat3, and Flag-tagged ROP16WT by anti–phospho-Stat3 (Tyr705), anti-GST, and anti-Flag, respectively. Data are representative of two independent calculations (A and B) or three independent experiments (C and D).
DISCUSSION
In this study, we first confirmed by reverse genetics that a product of T. gondii ROP16 gene is responsible for Stat3 activation and suppression of T. gondii–induced proinflammatory cytokines induced by type I strains. Indeed, disruption of the ROP16 gene in the type I strain converted the parasite into to a type II phenotype regarding Stat3 phosphorylation and activation. Consistent with a direct role played by ROP16, the complementation of rop16 KO parasites with type I ROP16, but not with type II ROP16, fully rescued the phenotype.
In addition, we elucidated the molecular mechanisms underlying the strain difference between type I (or III) and II ROP16 through an in vitro assay system using mammalian cells that allowed us to precisely characterize chimeric and site-specific mutants of ROP16 affected in Stat3 activation. This led us to the identification of a crucial polymorphic leucine/serine residue at position 503 in ROP16 that determines the strain difference with respect to Stat3 activation. Furthermore, we demonstrated that ROP16 directly phosphorylates Stat3 and that the N-terminal portion of ROP16 is essential for ROP16–Stat3 interaction. A previous study showed that infection of type II, as well as type I and III, parasites induced early Stat3 activation, suggesting that ROP16 may play a role in sustaining, but not initiating, Stat3 (and Stat6) activation (Saeij et al., 2007). In contrast, our results using rop16 KO type I parasites exhibited almost complete loss of Stat3 activation throughout the time course of infection (Fig. S1 B), indicating that ROP16 may be required for onset, as well as for sustaining Stat3 activation.
Furthermore, Leu503 in ROP16 might be required for sustaining Stat3 phosphorylation at later time points because Stat3 phosphorylation decreased more rapidly in cells infected with transgenic parasites expressing type II ROP16 than those expressing type I ROP16 (Fig. 5 A and Fig. S5). Considering the in silico modeling of RHWT, RHL503S, ME49WT, and ME49S503L, the amino acid substitution observed in the kinase domain of type II ROP16 may distort the active site and possibly result in impaired substrate binding, culminating in the low capacity for Stat3 activation. However, the formal proof of this prediction should be performed by crystallizing the structures of the ROP16 variants. Moreover, we showed that the NLS on ROP16 was not essential for Stat3 activation, which is consistent with a previous study (Saeij et al., 2007). Given that Stat3 phosphorylation presumably occurs in the cytoplasm (Bromberg and Darnell, 2000; Kishimoto, 2005), cytoplasmic, but not nuclear localized, ROP16 may be more suitable for Stat3 activation. In addition, evidence for an interaction between Stat3 and ROP16 was obtained by coimmunoprecipitation experiments in 293T cells. Interestingly, ROP16KD appears to form a more stable interaction with Stat3 than with ROP16WT (Fig. 5 C), possibly because it fails to phosphorylate the substrate and, thus, liberate it from the catalytic site.
Regarding the time scale of Stat3 phosphorylation, the T. gondii–induced activation occurs later than that in response to cytokines such as IL-6 or IL-10 (Montag and Lotze, 2006; and Fig. S1 B). The disparities between two modes of Stat3 activation might be a result of the time lag required for the parasite to inject sufficient amounts of ROP16 for full activation of Stat3 into host cells, or to the functional insufficiency of ROP16 as a tyrosine kinase in comparison to typical Stat3 kinases such as JAK1 or Src (Ihle, 2001).
Another Stat family member, Stat6, is also activated during the early phase of T. gondii infection (Saeij et al., 2007). Activation of Stat3 and Stat6 in the parasite-infected macrophages might lead to suppression of IL-12, which is essential for the development of Th1-prone immune responses, and to differentiation of the infected cells into immune suppressive macrophages, respectively (Mordue and Sibley, 2003; Trinchieri, 2003; Robben et al., 2005; Sinha et al., 2005). Whether ROP16 directly phosphorylates Stat6 as well as Stat3 can be addressed in the future using the rop16 KO and applying similar in vitro assay systems.
ROP16 was originally reported to be a serine/threonine kinase (PS/TK; Saeij et al., 2006, 2007; El Hajj et al., 2006; Dubremetz, 2007). Rather unexpectedly, we found that Tyr705 of recombinant Stat3 is phosphorylated by ROP16 in an in vitro kinase assay. Therefore, although there is no experimental evidence showing that ROP16 phosphorylates the serine or threonine residue of known substrates, ROP16 might be capable of phosphorylating not only serine/threonine but also tyrosine residues. Sequence analysis of the kinase domain of ROP16, using the Conserved Domain Search Program at the National Center for Biotechnology Information database, revealed a rather weak level of conservation compared with typical PS/TKs (e.g., e-values for ROP16 and a typical PS/TK IKK-β are 4e-09 and 4-e52, respectively). Moreover, in silico structural analysis revealed similarity to dual-specificity mitogen-activated kinase kinases, which are shown to be tyrosine/threonine kinases (Crews et al., 1992), for the kinase domain of ROP16. This, along with the observation that a tyrosine kinase inhibitor K-252a treatment severely impaired the kinase activity of ROP16 for Stat3 Tyr705 phosphorylation in in vitro and in vivo assays, suggests that ROP16 might potentially present the signature of a kinase capable of phosphorylating tyrosine residues (Fig. 6, C and D; Table S1). Given that protein tyrosine kinases are considered to have originated from within the superfamily of PS/TKs during the evolution of sponges (Kruse et al., 1997), PS/TKs in protozoa belonging to the phylum Apicomplexa, which includes T. gondii, might possess the characteristics of dual-specific kinases. A structural approach, in which three-dimensional structures of ROP2 and ROP8 were recently resolved (Qiu et al., 2009), might possibly apply to the case of ROP16 and lead to the key catalytic site for Stat3 phosphorylation in future studies.
Collectively, this study establishes that ROP16 directly mediates Stat3 phosphorylation and leads to the identification of single amino acid polymorphism in ROP16 that is responsible for its dramatically reduced activity in type II strain. The rop16 KO parasites will further assist in uncovering the physiological functions of ROP16 including the biological significance of the nuclear translocation.
MATERIALS AND METHODS
Cells, mice, and parasites.
C57BL/6 mice (6–8 wk of age) were obtained from SLC. Peritoneal macrophages were collected from peritoneal cavities 96 h after 4% thioglycollate injection. All animal experiments were conducted with the approval of the Animal Research Committee of Graduate School of Medicine in Osaka University. RHΔhxgprt and ME49 tachyzoites of T. gondii were maintained on Vero or MEFs by biweekly passage in RPMI (Nacalai Tesque) supplemented with 2% heat-inactivated FCS (JRH Biosciences), 100 U/ml penicillin, and 0.1 mg/ml streptomycin (Invitrogen).
Generation of ROP16-deficient type I T. gondii.
A genomic DNA containing the ROP16 gene was isolated from PCR amplification using primers 5′-GCGGCCGCTGTCTGTCCTTCCATCGGCGTGTATT-3′ and 5′-AGATCTTGCGACAAACAAGATCACAG-3′ to generate a 5.0-kb-long fragment and primers 5′-GTCGACGGTGTAAGGTTCCCACCTTAACACC-3′ and 5′-CTCGAGATATCAATTAACGCACACTTGAAGGTCGC-3′ to generate a 1.0-kb-short fragment. The gene encoding T. gondii ROP16 consists of a single exon. The targeting vector was constructed by replacing entire coding sequence of ROP16 gene with the HXGPRT gene expression cassette (p2855). Outside the targeting vector, YFP expression vector containing the 1.0-kb SAG1 promoter (amplified using primers 5′-GAATTCACTCGTCAAAAAACCAGAAGAAA-3′ and 5′-CTCGAGATGCTTGCGTAGGTTGACCTCTGA-3′) and the poly A additional signal from DHFR gene (sag1-YFP) was ligated using a NotI site for the negative selection of random integration (Mazumdar et al., 2006). 100 µg of the targeting vector linearized by EcoRV were transfected into tachyzoites of the RHΔhxgprt parental strain as previously described (Donald et al., 1996). After 25 µg/ml MPA (Sigma-Aldrich) and 25 µg/ml xanthine (Wako Chemicals USA, Inc.) selection for 14 d, MPA/xanthine-resistant colonies were sorted using FACSAria (BD) to isolate YFP-negative parasites. Then, the MPA/xanthine-resistant and YFP-negative parasites were subjected to limiting dilution to isolate the clones. A total of 47 clones were selected and screened by PCR for detecting homologous recombinants using primers 5′-CGGGTTTGAATGCAAGGTTTCGTGCTG-3′ (from the DHFR promoter of the HXGPRT expression vector) and 5′-CAATGGCGCGTGTGTGTTCAAAC-3′ (genomic sequence outside the short fragment of the ROP16 locus) to detect homologous recombinants. This resulted in isolation of two homologous recombinants. Subsequently, genomic DNA of WT and ROP16-deficeint parasites was extracted and subjected to Southern blot analysis using DNA probe, which was generated by PCR (using primers 5′-GGAACGTCACCTTAATACGT-3′ and 5′-TAAGAGCAAAGTCTCCCTAG-3′). In addition, to confirm the disruption of the gene encoding ROP16, we analyzed total RNA from WT and ROP16-deficient parasites by Northern blotting using a DNA probe, which was generated by PCR (using primers 5′-TCTGGAAGAAGTTCAGCAGC-3′ and 5′-CTGTACGGCACCTTCGCTGC-3′).
Mammalian expression plasmids.
Luciferase reporters containing the Stat3-dependent promoter constructs (gifts from T. Hirano, Osaka University, Suita, Osaka, Japan) have been previously described (Nakajima et al., 1996; Ichiba et al., 1998). Fragments of type I and type II ROP16 lacking the N-terminal signal peptide (amplified using the primer 5′-GAATTCACCATGCGATACATGTCGTTTGAG-3′ and the common primer 5′-CTCGAGCATCCGATGTGAAAGAAAGTTCGGT-3′) and the series of type I ROP16 fragments (amplified using the following primers: Δ60, 5′-GAATTCACCATGGGGTCTCCTACGGCAGGGCAACCT-3′; Δ140, 5′-GAATTCACCATGGGACCGGGAGGATGGTTTCCAACA-3′; Δ220, 5′-GAATTCACCATGAATCCTCTTTTTCCTGGTCAGAGC-3′; Δ300, 5′-GAATTCACCATGCAGCTGAAAGCAGCTGCCGCACAG-3′; and ΔN, 5′-GAATTCACCATGCAACCTCCAGGAGCGGTGGAG-3′) and the common primer were obtained by PCR using genomic DNA from RH strain (type I) or ME49 (type II) and were ligated into the EcoRI and XhoI sites of pcDNA (Invitrogen) for expression of HA- or Flag-tagged proteins. Flag-tagged full-length ROP18 was obtained by PCR using the primers 5′-GGATCCACCACCATGGGTTTAGCGACTCTT-3′ and 5′-GCGGCCGCTACTTGTCATCGTCGTCCTTGTAGTCTTCTGTGTGGAGATGT-3′ and genomic DNA of RH strain as template and were ligated into the BamHI and NotI sites of pcDNA vector. A series of type I ROP16 mutants containing point mutations were generated using the following primers: G424E, 5′-GAACTGGAGGCGGAAATTTCCTCAGCT-3′ and 5′-AGCTGAGGAAATTTCCGCCTCCAGTTC-3′; V457A, 5′-CTTACAGAGACTGCGAGCCAATACGGT-3′ and 5′-ACCGTATTGGCTCGCAGTCTCTGTAAG-3′; S458E, 5′-TACAGAGACTGTGGAGCAATACGGTCTGC-3′ and 5′-GCAGACCGTATTGCTCCACAGTCTCTGTA-3′; F486V, 5′-AGGTTCTCGACTCGTTGTCGTGTCCAA-3′ and 5′-TTGGACACGACAACGAGTCGAGAACCT-3′; A502P, 5′-AATTGATGGCTCCCCATTGAACAGTCT-3′ and 5′-AGACTGTTCAATGGGGAGCCATCAATT-3′; L503S, 5′-GATGGCTCCGCATCGAACAGTCTAGTC-3′ and 5′-GACTAGACTGTTCGATGCGGAGCCATC-3′; L525I, 5′-AAGGGAAGCAATTATTGCATTGGCCAA-3′ and 5′-TTGGCCAATGCAATAATTGCTTCCCTT-3′; kinase inactive mutant D539A, 5′-GATTCGCGCATGGAGCTGTTAAATTGAACA-3′ and 5′-TTGTTCAATTTAACAGCTCCATGCGCGAAT-3′; NLS mutant, 5′-GTTGCTGCCCGTATGCGGATGAGAATGCAACCTCCAGGAGC-3′ and 5′-GCTCCTGGAGGTTGCATTCTCATCCGCAGACGGGCAGCAAC-3′; and S503L type II ROP16, 5′-TGATGGCTCCCCATTGAACAGTCTAGTCC-3′ and 5′-GGACTAGACTGTTCAATGGGGAGCCATCA-3′. Expression plasmids were generated using a site-directed mutagenesis kit (Stratagene). The sequences of all constructs were confirmed with an ABI PRISM Genetic Analyzer (Applied Biosystems). Flag-tagged Stat3 expression vector (a gift from S. Akira, Osaka University, Suita, Osaka, Japan) was described previously (Minami et al., 1996).
Generation of transgenic parasites.
To complement the ROP16-deficient parasites, we generated an N-terminal signal peptide–containing ROP16 that was capable of being processed in the parasite by PCR using forward primer 5′-GAATTCCCTTTTTCGACAAAAATGAAAGTGACCACGAAAGGGCT-3′ and expressed a sag1-TgROP16-HA plasmid containing a pyrimethamine resistant gene cassette in the rop16 KO strain. We generated this vector by digesting the pyrimethamine resistance cassette of the p2854 plasmid with NotI and XhoI and ligated this into Klenow-treated pBluescript sag1-TgROP16-HA vector. A series of TgROP16-HA vectors were transfected into tachyzoites of the ROP16-deficient parasites. We selected for parasites stably expressing the complemented TgROP16-HA construct using 3-µM pyrimethamine (Sigma-Aldrich) selection and subjected to limiting dilution as described in the previous paragraph and previously (Donald et al., 1996).
Reagents.
Antiphosphorylated Stat3 Tyr705 or Ser727 was purchased from Cell Signaling Technology. Anti-actin, GST, Stat3, Toxoplasma p30 antigen, and HA probe were purchased from Santa Cruz Biotechnology, Inc. GST-Stat3 was purchased from SignalChem. Anti-Flag was obtained from Sigma-Aldrich. Kinase inhibitors (InSolution Casein Kinase II inhibitor I, PP2, SB202190, K-252a, PD153035, JAK inhibitor I, Y-27632, and GSK-3 inhibitor IX) were purchased from EMD.
Measurement of proinflammatory cytokine concentrations.
Peritoneal macrophages were cultured in 96-well plates (1 × 105 cells per well) with the indicated MOI of the indicated parasites for 24 h. Concentrations of IL-6 and IL-12 p40 in the culture supernatant were measured by ELISA according to manufacturer's instructions (eBioscience), as described previously (Yamamoto et al., 2004).
Luciferase reporter assay.
The reporter plasmids were transiently cotransfected into 293T cells with the control Renilla luciferase expression vectors using Lipofectamine 2000 reagent (Invitrogen). Luciferase activities of total cell lysates were measured using the Dual-Luciferase Reporter Assay System (Promega) as described previously (Yamamoto et al., 2006).
Western blot analysis and immunoprecipitation.
MEFs, 293T cells, and parasites were then lysed in a lysis buffer containing 0.1% Nonidet-P 40, 150 mM NaCl, 20 mM Tris-HCl, pH 7.5, and protease inhibitor cocktail (Roche). The cell lysates were separated by SDS-PAGE and transferred to PVDF membranes. For immunoprecipitation, cell lysates were precleared with Protein G–Sepharose (GE Healthcare) for 2 h and then incubated with Protein G–Sepharose containing 1.0 µg of the indicated antibodies for 12 h with rotation at 4°C. The immunoprecipitants were washed four times with lysis buffer, eluted by boiling with Laemmli sample buffer, and subjected to Western blot analysis using the indicated antibodies earlier in this paragraph and previously (Yamamoto et al., 2003).
In vitro kinase assay.
293T cells were transiently transfected with a total of 4.0 µg of either empty vector or the indicated plasmids (4 µg Flag-tagged ROP16WT or ROP16KD), using Lipofectamine 2000 as specified by the manufacturer (Invitrogen). Cells were harvested 48 h after transfection, lysed, and then immunoprecipitated with protein G–Sepharose together with 1.0 µg of anti-FLAG M2 mAb (Sigma-Aldrich) for 12 h by rotation. The beads were washed four times with lysis buffer, and another three times with kinase assay buffer (30 mM MOPS, pH 7.5, 50 mM NaCl, 10% glycerol, 10 mM MgCl2, and 10 mM MnCl2). The immunoprecipitates were incubated with 1 µg GST-Stat3 and 5 µM ATP (Wako Chemicals USA, Inc.) to detect specifically phospho-Stat3 Tyr705 at 30°C for 30 min in the absence (Fig. 5 B) or presence (Fig. 6 C) of DMSO or K-252a. Kinase reactions were stopped by addition of Laemmli sample buffer and were separated on a 5–20% poly acrylamide gradient gel. Gel was detained and subjected to Western blot analysis to detect phospho-Stat3, GST-tagged Stat3, and Flag-tagged ROP16WT or ROP16KD by anti–phospho-Stat3 Tyr705, anti-GST, and anti-Flag, respectively.
Microscopic analysis.
293T cells were transfected with 2 µg Flag-tagged ROP16 or ROP18 expression vectors. 48 h after transfection, the cells were fixed with a solution containing 4% PFA, permeabilized with 1% Triton-PBS, and stained with anti-STAT3 or anti-Flag, followed by treatment with the respective secondary antibody, Alexa Fluor 488–conjugated anti–mouse IgG (Invitrogen) or Alexa Fluor 594–conjugated anti–rabbit IgG antibody (Invitrogen). The immunostained cells were analyzed using a fluorescence microscope (IX71; Olympus). Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (Wako Chemicals USA, Inc.).
Structural modeling.
Models of the kinase domains of RHWT, RHL503S, ME49WT, and ME49S503L were constructed from ffas03 profile-profile alignments (http://ffas.burnham.org/ffas-cgi/cgi/ffas.pl) using the PDB0709 and SCOP175 profile databases. The top PDB0709 and SCOP175 alignments (corresponding to templates 3cokA and d1s9ja, respectively) were selected for each of the three sequences, resulting in a total of six structural models. Structural models were built using Spanner (http://sysimm.ifrec.osaka-u.ac.jp/cgi-bin/spanner), which employs a fragment assembly algorithm to produce a gapless alignment to the template. The structural models were then submitted to the SeSAW functional annotation server (http://sysimm.ifrec.osaka-u.ac.jp/SeSAW/). SeSAW uses a combined structure and sequence profile–profile similarity score (Standley et al., 2008) to rank their similarity to known structures. The model built on SCOP domain d1s9ja (dual-specificity mitogen-activated protein kinase kinase 1) resulted in significantly higher SeSAW scores than that built on PDB entry 3cokA (323.0 and 253.0, respectively) so the d1s9ja model was ultimately retained for further analysis. Residues 387–452 could not be modeled with as high a confidence as the rest of the structure as a result of a large insertion and were omitted from Fig. 6 and Fig. S7. The predicted active site cavity location was based on that of d1s9ja dual-specificity MAP kinase kinase 1. Electrostatic surfaces were prepared using the eF-surf server (http://ef-site.hgc.jp/eF-surf/) and eF-site (Kinoshita and Nakamura, 2004).
Online supplemental material.
Fig. S1 shows defective time-dependent Stat3 activation in cells infected with ROP16-deficeint parasites. Fig. S2 demonstrates cytoplasmic Stat3 localization in ROP18-transfected cells and activation of the Stat3-dependent promoter in cells transfected with NLS-mutated ROP16. Fig. S3 exhibits complementation of ROP16-deficient parasites by ROP16 mutants. Fig. S4 shows amino acid sequences of type I and type II ROP16 at position 373–547. Fig. S5 demonstrates time-dependent Stat3 activation in cells infected with ROP16-complemented parasites. Fig. S6 illustrates type I ROP16 deletion and point mutants used in this study. Fig. S7 shows in silico modeling of ROP16 variants and screening of kinase inhibitors. Table S1 provides a list of the 20 most similar kinases to the RHWT mode using the Spanner model built from SCOP domain d1s9jA. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091703/DC1.
Acknowledgments
We thank S. Hidaka and M. Yasuda for excellent secretarial assistance; Y. Magota for technical assistance; and members of Takeda's laboratory for discussions. We also thank Professors T. Hirano and S. Akira for supplying the plasmids.
This work was supported by grants from the Ministry of Education, Culture, Sports, Science and Technology, Takeda Science Foundation, Mishima Kaiun Memorial Foundation, Mochida Memorial Foundation for Medical and Pharmaceutical Research, Naito Foundation, Kowa Life Science Foundation, Ohyama health Foundation, The Japan Spina Bifida and Hydrocephalus Research Foundation, The Yasuda Medical Foundation, Kobayashi Foundation for Cancer Research, The Kato Memorial Trust for Nambyo Research, The Osaka Cancer Foundation, Showa Houkou Kai, Japan Leuckemia Research Fund, and Kudo Gakujutsu Zaidan.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- MEF
- mouse embryonic fibroblast
- MPA
- mycophenolic acid
- NLS
- nuclear localization signal
- TLR
- toll-like receptor
- YFP
- yellow fluorescent protein
References
- Ajzenberg D., Bañuls A.L., Su C., Dumètre A., Demar M., Carme B., Dardé M.L. 2004. Genetic diversity, clonality and sexuality in Toxoplasma gondii. Int. J. Parasitol. 34:1185–1196 10.1016/j.ijpara.2004.06.007 [DOI] [PubMed] [Google Scholar]
- Aliberti J., Valenzuela J.G., Carruthers V.B., Hieny S., Andersen J., Charest H., Reis e Sousa C., Fairlamb A., Ribeiro J.M., Sher A. 2003. Molecular mimicry of a CCR5 binding-domain in the microbial activation of dendritic cells. Nat. Immunol. 4:485–490 10.1038/ni915 [DOI] [PubMed] [Google Scholar]
- Boothroyd J.C., Dubremetz J.F. 2008. Kiss and spit: the dual roles of Toxoplasma rhoptries. Nat. Rev. Microbiol. 6:79–88 10.1038/nrmicro1800 [DOI] [PubMed] [Google Scholar]
- Bromberg J., Darnell J.E., Jr 2000. The role of STATs in transcriptional control and their impact on cellular function. Oncogene. 19:2468–2473 10.1038/sj.onc.1203476 [DOI] [PubMed] [Google Scholar]
- Butcher B.A., Kim L., Panopoulos A.D., Watowich S.S., Murray P.J., Denkers E.Y. 2005. IL-10-independent STAT3 activation by Toxoplasma gondii mediates suppression of IL-12 and TNF-α in host macrophages. J. Immunol. 174:3148–3152 [DOI] [PubMed] [Google Scholar]
- Crews C.M., Alessandrini A., Erikson R.L. 1992. The primary structure of MEK, a protein kinase that phosphorylates the ERK gene product. Science. 258:478–480 10.1126/science.1411546 [DOI] [PubMed] [Google Scholar]
- Del Rio L., Butcher B.A., Bennouna S., Hieny S., Sher A., Denkers E.Y. 2004. Toxoplasma gondii triggers myeloid differentiation factor 88-dependent IL-12 and chemokine ligand 2 (monocyte chemoattractant protein 1) responses using distinct parasite molecules and host receptors. J. Immunol. 172:6954–6960 [DOI] [PubMed] [Google Scholar]
- Denkers E.Y. 2003. From cells to signaling cascades: manipulation of innate immunity by Toxoplasma gondii. FEMS Immunol. Med. Microbiol. 39:193–203 10.1016/S0928-8244(03)00279-7 [DOI] [PubMed] [Google Scholar]
- Denkers E.Y., Kim L., Butcher B.A. 2003. In the belly of the beast: subversion of macrophage proinflammatory signalling cascades during Toxoplasma gondii infection. Cell. Microbiol. 5:75–83 10.1046/j.1462-5822.2003.00258.x [DOI] [PubMed] [Google Scholar]
- Donald R.G., Carter D., Ullman B., Roos D.S. 1996. Insertional tagging, cloning, and expression of the Toxoplasma gondii hypoxanthine-xanthine-guanine phosphoribosyltransferase gene. Use as a selectable marker for stable transformation. J. Biol. Chem. 271:14010–14019 10.1074/jbc.271.24.14010 [DOI] [PubMed] [Google Scholar]
- Dubremetz J.F. 2007. Rhoptries are major players in Toxoplasma gondii invasion and host cell interaction. Cell. Microbiol. 9:841–848 10.1111/j.1462-5822.2007.00909.x [DOI] [PubMed] [Google Scholar]
- El Hajj H., Demey E., Poncet J., Lebrun M., Wu B., Galéotti N., Fourmaux M.N., Mercereau-Puijalon O., Vial H., Labesse G., Dubremetz J.F. 2006. The ROP2 family of Toxoplasma gondii rhoptry proteins: proteomic and genomic characterization and molecular modeling. Proteomics. 6:5773–5784 10.1002/pmic.200600187 [DOI] [PubMed] [Google Scholar]
- Gazzinelli R.T., Ropert C., Campos M.A. 2004. Role of the Toll/interleukin-1 receptor signaling pathway in host resistance and pathogenesis during infection with protozoan parasites. Immunol. Rev. 201:9–25 10.1111/j.0105-2896.2004.00174.x [DOI] [PubMed] [Google Scholar]
- Ichiba M., Nakajima K., Yamanaka Y., Kiuchi N., Hirano T. 1998. Autoregulation of the Stat3 gene through cooperation with a cAMP-responsive element-binding protein. J. Biol. Chem. 273:6132–6138 10.1074/jbc.273.11.6132 [DOI] [PubMed] [Google Scholar]
- Ihle J.N. 2001. The Stat family in cytokine signaling. Curr. Opin. Cell Biol. 13:211–217 10.1016/S0955-0674(00)00199-X [DOI] [PubMed] [Google Scholar]
- Joynson D.H., Wreghitt T.J., 2001. Toxoplasmosis: A Comprehensive Clinical Guide. Cambridge University Press, Cambridge, U.K: 395 pp [Google Scholar]
- Kim L., Butcher B.A., Lee C.W., Uematsu S., Akira S., Denkers E.Y. 2006. Toxoplasma gondii genotype determines MyD88-dependent signaling in infected macrophages. J. Immunol. 177:2584–2591 [DOI] [PubMed] [Google Scholar]
- Kinoshita K., Nakamura H. 2004. eF-site and PDBjViewer: database and viewer for protein functional sites. Bioinformatics. 20:1329–1330 10.1093/bioinformatics/bth073 [DOI] [PubMed] [Google Scholar]
- Kishimoto T. 2005. Interleukin-6: from basic science to medicine—40 years in immunology. Annu. Rev. Immunol. 23:1–21 10.1146/annurev.immunol.23.021704.115806 [DOI] [PubMed] [Google Scholar]
- Kruse M., Müller I.M., Müller W.E. 1997. Early evolution of metazoan serine/threonine and tyrosine kinases: identification of selected kinases in marine sponges. Mol. Biol. Evol. 14:1326–1334 [DOI] [PubMed] [Google Scholar]
- Mazumdar J., H Wilson E., Masek K., A Hunter C., Striepen B. 2006. Apicoplast fatty acid synthesis is essential for organelle biogenesis and parasite survival in Toxoplasma gondii. Proc. Natl. Acad. Sci. USA. 103:13192–13197 10.1073/pnas.0603391103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minami M., Inoue M., Wei S., Takeda K., Matsumoto M., Kishimoto T., Akira S. 1996. STAT3 activation is a critical step in gp130-mediated terminal differentiation and growth arrest of a myeloid cell line. Proc. Natl. Acad. Sci. USA. 93:3963–3966 10.1073/pnas.93.9.3963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montag D.T., Lotze M.T. 2006. Rapid flow cytometric measurement of cytokine-induced phosphorylation pathways [CIPP] in human peripheral blood leukocytes. Clin. Immunol. 121:215–226 10.1016/j.clim.2006.06.013 [DOI] [PubMed] [Google Scholar]
- Montoya J.G., Remington J.S. 2008. Management of Toxoplasma gondii infection during pregnancy. Clin. Infect. Dis. 47:554–566 10.1086/590149 [DOI] [PubMed] [Google Scholar]
- Mordue D.G., Sibley L.D. 2003. A novel population of Gr-1+-activated macrophages induced during acute toxoplasmosis. J. Leukoc. Biol. 74:1015–1025 10.1189/jlb.0403164 [DOI] [PubMed] [Google Scholar]
- Mun H.S., Aosai F., Norose K., Chen M., Piao L.X., Takeuchi O., Akira S., Ishikura H., Yano A. 2003. TLR2 as an essential molecule for protective immunity against Toxoplasma gondii infection. Int. Immunol. 15:1081–1087 10.1093/intimm/dxg108 [DOI] [PubMed] [Google Scholar]
- Nakajima K., Yamanaka Y., Nakae K., Kojima H., Ichiba M., Kiuchi N., Kitaoka T., Fukada T., Hibi M., Hirano T. 1996. A central role for Stat3 in IL-6-induced regulation of growth and differentiation in M1 leukemia cells. EMBO J. 15:3651–3658 [PMC free article] [PubMed] [Google Scholar]
- Ohren J.F., Chen H., Pavlovsky A., Whitehead C., Zhang E., Kuffa P., Yan C., McConnell P., Spessard C., Banotai C., et al. 2004. Structures of human MAP kinase kinase 1 (MEK1) and MEK2 describe novel noncompetitive kinase inhibition. Nat. Struct. Mol. Biol. 11:1192–1197 10.1038/nsmb859 [DOI] [PubMed] [Google Scholar]
- Plattner F., Yarovinsky F., Romero S., Didry D., Carlier M.F., Sher A., Soldati-Favre D. 2008. Toxoplasma profilin is essential for host cell invasion and TLR11-dependent induction of an interleukin-12 response. Cell Host Microbe. 3:77–87 10.1016/j.chom.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Qiu W., Wernimont A., Tang K., Taylor S., Lunin V., Schapira M., Fentress S., Hui R., Sibley L.D. 2009. Novel structural and regulatory features of rhoptry secretory kinases in Toxoplasma gondii. EMBO J. 28:969–979 10.1038/emboj.2009.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robben P.M., Mordue D.G., Truscott S.M., Takeda K., Akira S., Sibley L.D. 2004. Production of IL-12 by macrophages infected with Toxoplasma gondii depends on the parasite genotype. J. Immunol. 172:3686–3694 [DOI] [PubMed] [Google Scholar]
- Robben P.M., LaRegina M., Kuziel W.A., Sibley L.D. 2005. Recruitment of Gr-1+ monocytes is essential for control of acute toxoplasmosis. J. Exp. Med. 201:1761–1769 10.1084/jem.20050054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij J.P., Boyle J.P., Coller S., Taylor S., Sibley L.D., Brooke-Powell E.T., Ajioka J.W., Boothroyd J.C. 2006. Polymorphic secreted kinases are key virulence factors in toxoplasmosis. Science. 314:1780–1783 10.1126/science.1133690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeij J.P., Coller S., Boyle J.P., Jerome M.E., White M.W., Boothroyd J.C. 2007. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 445:324–327 10.1038/nature05395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanga C.A., Aliberti J., Jankovic D., Tilloy F., Bennouna S., Denkers E.Y., Medzhitov R., Sher A. 2002. Cutting edge: MyD88 is required for resistance to Toxoplasma gondii infection and regulates parasite-induced IL-12 production by dendritic cells. J. Immunol. 168:5997–6001 [DOI] [PubMed] [Google Scholar]
- Sinha P., Clements V.K., Ostrand-Rosenberg S. 2005. Interleukin-13-regulated M2 macrophages in combination with myeloid suppressor cells block immune surveillance against metastasis. Cancer Res. 65:11743–11751 10.1158/0008-5472.CAN-05-0045 [DOI] [PubMed] [Google Scholar]
- Standley D.M., Toh H., Nakamura H. 2008. Functional annotation by sequence-weighted structure alignments: statistical analysis and case studies from the Protein 3000 structural genomics project in Japan. Proteins. 72:1333–1351 10.1002/prot.22015 [DOI] [PubMed] [Google Scholar]
- Taylor S., Barragan A., Su C., Fux B., Fentress S.J., Tang K., Beatty W.L., Hajj H.E., Jerome M., Behnke M.S., et al. 2006. A secreted serine-threonine kinase determines virulence in the eukaryotic pathogen Toxoplasma gondii. Science. 314:1776–1780 10.1126/science.1133643 [DOI] [PubMed] [Google Scholar]
- Trinchieri G. 2003. Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol. 3:133–146 10.1038/nri1001 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. 2003. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 301:640–643 10.1126/science.1087262 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Yamazaki S., Uematsu S., Sato S., Hemmi H., Hoshino K., Kaisho T., Kuwata H., Takeuchi O., Takeshige K., et al. 2004. Regulation of Toll/IL-1-receptor-mediated gene expression by the inducible nuclear protein IkappaBzeta. Nature. 430:218–222 10.1038/nature02738 [DOI] [PubMed] [Google Scholar]
- Yamamoto M., Okamoto T., Takeda K., Sato S., Sanjo H., Uematsu S., Saitoh T., Yamamoto N., Sakurai H., Ishii K.J., et al. 2006. Key function for the Ubc13 E2 ubiquitin-conjugating enzyme in immune receptor signaling. Nat. Immunol. 7:962–970 10.1038/ni1367 [DOI] [PubMed] [Google Scholar]
- Yap G.S., Shaw M.H., Ling Y., Sher A. 2006. Genetic analysis of host resistance to intracellular pathogens: lessons from studies of Toxoplasma gondii infection. Microbes Infect. 8:1174–1178 10.1016/j.micinf.2005.10.031 [DOI] [PubMed] [Google Scholar]
- Yarovinsky F., Sher A. 2006. Toll-like receptor recognition of Toxoplasma gondii. Int. J. Parasitol. 36:255–259 10.1016/j.ijpara.2005.12.003 [DOI] [PubMed] [Google Scholar]
- Yarovinsky F., Zhang D., Andersen J.F., Bannenberg G.L., Serhan C.N., Hayden M.S., Hieny S., Sutterwala F.S., Flavell R.A., Ghosh S., Sher A. 2005. TLR11 activation of dendritic cells by a protozoan profilin-like protein. Science. 308:1626–1629 10.1126/science.1109893 [DOI] [PubMed] [Google Scholar]
- Zhao B., Lehr R., Smallwood A.M., Ho T.F., Maley K., Randall T., Head M.S., Koretke K.K., Schnackenberg C.G. 2007. Crystal structure of the kinase domain of serum and glucocorticoid-regulated kinase 1 in complex with AMP PNP. Protein Sci. 16:2761–2769 10.1110/ps.073161707 [DOI] [PMC free article] [PubMed] [Google Scholar]