Abstract
Both p53 and the Wnt signaling pathway play important roles in regulating the differentiation of mouse embryonic stem cells (mESCs). However, it is not known whether they directly and/or functionally crosstalk in mESCs. Here we report a surprising antidifferentiation function of p53 in mESCs through directly regulating the Wnt signaling pathway. A chromatin-immunoprecipitation-based microarray (ChIP-chip) and gene expression microarray assays reveal that the Wnt signaling pathway is significantly (P value, 0.000048) overrepresented in p53-regulated genes in mESCs. The expression of five Wnt ligand genes is robustly induced by various genotoxic and nongenotoxic insults in a p53-dependent manner. Moreover, the induction of these Wnt genes is greatly attenuated in mouse embryonic fibroblast (MEF) cells and ESC-derived neural stem/progenitor cells, suggesting that the induction is mESC specific. It is established that the activation of the Wnt signaling pathway inhibits the differentiation of mESCs. Consistent with this notion, we detected an antidifferentiation activity from the conditioned medium (CM) collected from UV (UV)-treated mESCs. This antidifferentiation activity can be lowered by either the addition of Wnt antagonists into the CM or the reduction of p53 levels in UV-treated mESCs. Therefore, reminiscent of its dual functions on death and survival in somatic cells, p53 appears to regulate both prodifferentiation and antidifferentiation programs in mESCs. Our findings uncover a direct and functional connection between p53 and the Wnt signaling pathway, and expand the catalog of p53 regulated genes in mESCs.
Keywords: transcription, epigenetic, modifications, DNA binding, neural stem cells
Tumor suppressor p53 is a sequence-specific transcription factor and critical for maintaining the genomic stability of an organism (1). Without stresses, the protein levels of p53 in cells are low and the majority of p53 remains in cytoplasm. Upon various stresses, the half-life of p53 increases from several minutes to hours. p53 then translocates into the nuclei and activates its target genes. Depending on cell and stress types, p53 can elicit different biological outcomes, such as cell cycle arrest, apoptosis, and senescence (2). The roles of p53 in somatic cells have been extensively studied. However, our knowledge about its roles in embryonic stem cells remains limited.
Embryonic stem cells (ESCs) are derived from inner cell mass of blastocysts and can develop into three germ layers of an embryo (3–5). Therefore, they hold great potential in tissue regeneration therapy. ESCs have an internal gene expression program that is largely governed by pluripotent factors, such as Nanog and Oct4 (6, 7). Several external signaling pathways also connect to this internal circuitry. For example, leukemia inhibitory factor (LIF)/gp130/STAT3 signaling pathway supports unlimited self-renewal of mouse ESCs (mESCs) (8). Recently, the Wnt signaling pathway has been linked to the self-renewal of ESCs (9, 10). Apart from the LIF/gp130/STAT3 signaling pathway, the activation of the Wnt signaling pathway alone does not sustain the long-term self-renewal of mESCs but only temporarily inhibits the differentiation of mESCs (11, 12).
Because of the ability to differentiate into many cell types, ESCs must have developed a mechanism to cope with various stresses, in particular, DNA damage insults, to avoid passing the mutation to their progeny cells. Indeed, it is known that ESCs have a lower mutation rate than somatic cells by two orders of magnitude (13). The failure of this stress-defense system in ESCs may cause various developmental abnormalities and cancers. As the “guardian of the genome,” p53 participates in the stress-defense program of ESCs. Compared to their differentiated counterparts, ESCs have two unique features in response to DNA damage insults, both of which are mediated by p53. First, ESCs are more sensitive to DNA damage agents than somatic cells (14, 15). Both p53-dependent and -independent pathways are involved in the rapid apoptosis of mESCs (16–18). Second, a p53-dependent repression of Nanog expression correlates with the differentiation of mESCs that are exposed to DNA damage (17, 19). In human ESCs (hESCs), p53 may also decrease the expression of Oct4 in response to genotoxic stresses (17). Regardless of the mechanism, p53-driven differentiation in ESCs may represent another means to clear the ESCs with DNA damage. Together, these two unique features of ESCs ensure ESCs to maintain the genomic stability of the whole population.
Despite these findings, our knowledge about whether p53 has additional roles in embryonic stem cells and how it functionally interacts with other signaling pathways is limited. In the current study, we identified p53 target genes in mESCs using a combination of ChIP-chip and gene expression microarray assays. Surprisingly, we identified the Wnt signaling pathway as one of the major targets of p53 in mESCs. The most notable observation in our study is that several Wnt ligand genes are highly induced by genotoxic and nongenotoxic stresses in a p53-dependent manner. Using a conditioned medium approach, we found that ultraviolet (UV)-treated mESCs secrete an antidifferentiation activity, which is dependent on the Wnt signaling and p53. Collectively, our results revealed a unique connection between p53 and the Wnt signaling in mESCs.
Results
Identification of p53 Binding Sites in mESC Using ChIP-Chip Assay.
Pluripotent mESCs express abundant p53 (18, 19). However, its precise roles in mESCs are not fully appreciated. To explore the potential functions of p53 in mESCs in a global and unbiased manner, we set out to map the genomewide binding sites of p53 using a ChIP-chip assay with an Agilent mouse promoter microarray. This microarray platform was designed to contain 60-bp DNA probes to cover from −5.5 kb upstream to +2.5 kb downstream of the transcriptional start sites (TSSs) of about 17,000 well-defined mouse transcripts. Adriamycin (also called doxorubicin), a DNA damage agent, was used to treat mESCs (R1E cells) for 8 h (Fig. 1A) (20, 21). Untreated cells served as a control.
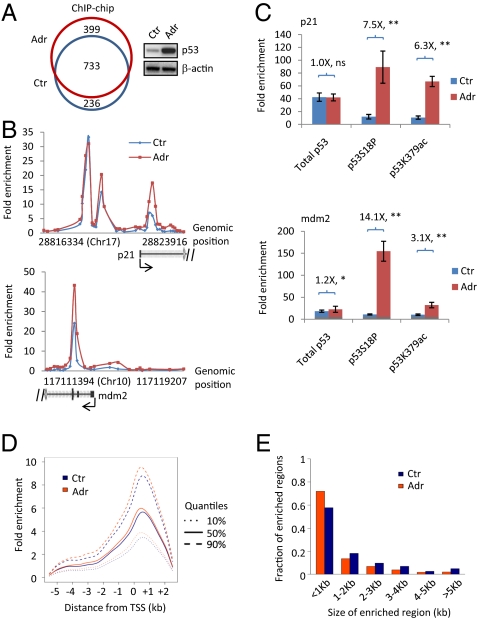
Fig. 1.
ChIP-chip assay to identify p53 bound genes in mESCs. (A) (Left) A Venn diagram of p53 bound genes in mESCs untreated (Ctr) and treated with adriamycin (Adr) for 8 h; (Right) Western blot analyses of p53 and β-actin. (B) Examples of p53 bound genes from ChIP-chip. (C) ChIP assay to measure the binding of p53 on p21 and mdm2 genes using antibodies targeting total p53, p53S18P, and p53K379ac. Fold enrichment was calculated as ratio of specific antibody signal versus nonspecific IgG. Numbers shown are ratios of fold enrichment with adriamycin treatment to untreated condition. **P < 0.01; *P < 0.05; ns, not significant. (D) Distribution of selected quantiles of fold enrichment over the distance between bound probes and the closest transcription start site. Probes within the −5.5 kb to +2.5 kb genomic region for all enriched genes were used to estimate the densities. (E) Frequencies of the size of p53 bound regions identified on the promoter microarrays.
We detected 1,132 genes bound by p53 in mESCs (R1E cells) treated with adriamycin [Fig. 1A, Fig. S1A, and Table S1]. Interestingly, we also identified 969 genes bound by p53 in untreated mESCs (Fig. 1A, Fig. S1A, and Table S2). About half (52.7 and 51.8% for untreated and treated, respectively) of the p53 bound regions located inside the coding region of the genes (Fig. S1B). The majority (65%, 733 out of 1,132) of the genes bound by p53 with DNA damage overlapped with that without DNA damage, indicating that p53 preoccupies many of its target genes before adriamycin treatment. Indeed, p53 binds to its two well-characterized target genes, p21 and mdm2, before and after adriamycin treatment (Fig. 1B). To assess whether the promoter bound p53 before adriamycin treatment is activated, we performed classical ChIP assay using antibodies specifically recognizing total p53, p53 serine 18 (serine 15 in human) phosphorylation (p53S18P), and p53 lysine 379 (lysine 382 in human) acetylation (p53K379ac). p53S18P and p53K379ac are two well-established posttranslational modifications that correlate with the activation of p53 (22–24). Although adriamycin did not increase the promoter occupancy of total p53 on the p21 gene, it greatly enhanced p53S18P and p53K379ac signals by 7.5- and 6.3-fold, respectively (Fig. 1C, Upper). Therefore, the ratios of p53S18P/total p53 and p53K379ac/total p53 increased 7.6- and 6.4-fold, respectively (Fig. S1C, Upper). A similar trend was also observed on the mdm2 gene (Fig. 1C and Fig. S1C, Lower). These results demonstrate that the main outcome of adriamycin treatment is to increase the portion of activated p53, but not the total p53, on its target genes. Thus, p53 is “poised” for activation on its target genes, presumably to facilitate a fast response in mESCs upon stresses.
The distribution of the distance between the transcription start sites (TSSs) and bound probes showed that the average binding peak of p53 in mESCs was 0.5 kb downstream of TSS (Fig. 1D). Surprisingly, adriamycin only had a minimal effect on the average fold enrichment of p53 on its bound genes (Fig. 1D) despite enhancing the total p53 levels fourfold in cells (Fig. 1A). These results suggest that the recruitment of p53 to many of its target genes is saturated before adriamycin treatment, which is consistent with the observations of p53 binding on p21 and mdm2 genes (Fig. 1 B and C). The sizes of the majority (∼60–70%) of p53 binding regions were less than 1 kb. Adriamycin treatment did not change the size distribution of p53 binding sites (Fig. 1E). Overall, our results provide a genomewide view of p53 on its target genes in mESCs.
The Wnt Signaling Pathway Is Enriched in p53-Regulated Genes in mESCs.
To search for the signaling pathway(s) that was (were) enriched in p53 target genes, we first determined the genes that are bound by p53 and differentially altered (corrected P value <0.05) after adriamycin treatment. Combining the ChIP-chip and gene expression microarray studies, we identified 573 genes that contain p53 binding site(s) and whose expression is significantly changed after adriamycin treatment (Fig. 2A, Fig. S1D, and Table S3). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (25) was then performed using this gene set (Table S3). This analysis revealed that the top four enriched signaling pathways were p53 signaling (P value, 3.12 × 10−8), basal cell carcinoma (P value, 2.94 × 10−5), Wnt signaling (P value, 4.8 × 10−5), and colorectal cancer signaling (P value, 1.06 × 10−3) (Table 1 and Fig. 2 B and C). The gene names in the basal cell carcinoma and colorectal cancer signaling heavily overlapped with those in the Wnt signaling pathway (Table 1), suggesting that the Wnt signaling pathway is one of the major targets of p53 in mESCs. To our knowledge, this report uniquely establishes a direct connection between p53 and the Wnt signaling pathway in mESCs using high throughput approaches.
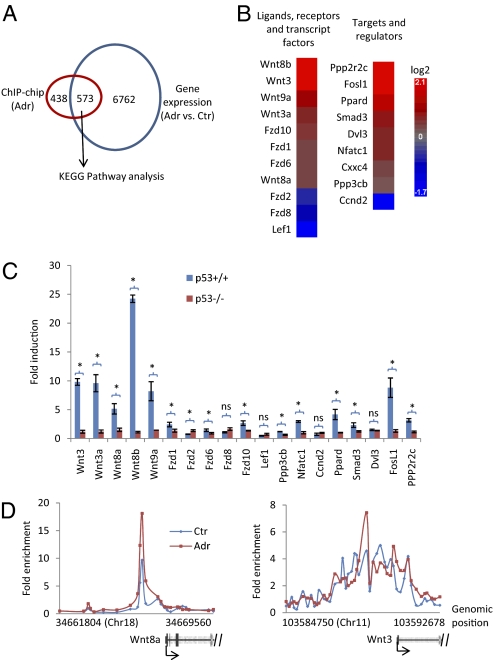
Fig. 2.
The Wnt signaling pathway is significantly enriched in p53 target. (A) A Venn diagram of genes bound by p53 with adriamcyin treatment from ChIP-chip and genes that were significantly altered in mESCs treated with adriamycin detected in gene expression microarray (false discovery rate corrected P value <0.05). Only genes that were represented by both ChIP-chip and gene expression microarray platforms were included in the analysis. (B) Heatmap of genes in the Wnt signaling pathway enriched in the overlapped gene list shown in A. (C) Real-time PCR result of the fold induction of enriched genes in the Wnt signaling pathway in p53+/+ and p53−/− mESCs treated with adriamycin for 8 h. Results are displayed as mean ± SEM; n = 3; *P < 0.05; ns, not significant. (D) Binding of p53 on the Wnt8a and Wnt3 genes from ChIP-chip.
Table 1.
Gene names of enriched pathways in the differentially altered p53-target genes in mESCs upon adriamycin treatment
| Term | P-value | Genes |
| p53 signaling pathway | 3.12E-08 | Gtse1, Puma, Sesn2, Ccng1, Ccnd2, Noxa, Lrdd, PIG8, Pirh2, Gadd45g, Trp73, Rrm2, Wig1, Bai1, p21 (Cdkn1a), mdm2, Fas |
| Basal cell carcinoma | 2.94E-05 | Wnt3, Wnt3a, Wnt8a, Wnt8b, Wnt9a, Fzd1, Fzd2, Fzd6, Fzd8, Fzd10, Lef1, Dvl3 |
| Wnt signaling pathway | 4.80E-05 | Wnt3, Wnt3a, Wnt8a, Wnt8b, Wnt9a, Fzd1, Fzd2, Fzd6, Fzd8, Fzd10, Lef1, Ppp3cb, Nfatc1, Ccnd2,Cxxc4, Ppp2r2c, Ppard, Smad3, Dvl3, Fosl1 |
| Colorectal cancer | 1.06E-03 | Fzd1, Fzd2, Fzd6, Fzd8, Fzd10, Lef1, Dvl3, Birc5, Smad3, Fosl1, Acvr1b, Pdgfra |
Gene set (Fig. 2A) was subject to KEGG pathway analysis to generate the gene list for each enriched pathway (P value < 0.005).
The major components in the Wnt signaling pathway include 19 secreted Wnt ligands, 10 transmembrane receptors and coreceptors, signal transducers (e.g., β-catenin and GSK3b), and Lef1/Tcf transcription complex. Secreted Wnt ligands bind to their receptor/coreceptor complex and elicit a series of downstream events, which eventually activates the Lef1/Tcf transcription complex. We identified 5 Wnt ligands, 5 receptors, one component of Lef1/Tcf complex, and nine putative regulators and downstream targets of the Wnt signaling as p53 target genes (Fig. 2B). Real-time PCR results confirmed that the gene expression changes of these identified genes in the Wnt signaling pathway are p53 dependent (Fig. 2C). Among these targets, the 5 Wnt ligands, Wnt3, Wnt3a, Wnt8a, Wnt8b, and Wnt9a, are highly induced (with fold induction larger than five) by p53 with adriamycin treatment.
The recruitment of p53 on the genes in the Wnt signaling can be arbitrarily grouped into two categories. The first category has relatively well-defined binding peaks, such as for the Wnt8a gene (Fig. 2D). The binding regions of p53 in the second category cover a large area of the corresponding genes without obvious peaks, as exemplified by that on the Wnt3 gene (Fig. 2D). Interestingly, the fold enrichment of p53 did not correlate with the fold induction of its target genes (compare Fig. 2C with 2D), suggesting that the fold induction is also influenced by other event(s) rather than by the DNA recruitment of p53 alone.
The Induction of the Wnt Genes by p53 Represents a General Stress Response of mESCs.
We were intrigued by the observation that p53 highly induces the expression of five Wnt ligand genes for two reasons. First, the fold induction of the Wnt genes by p53 is much higher than that of other identified genes in the Wnt signaling pathway (Fig. 2 B and C). Second, the Wnt signaling pathway plays an important role in regulating the self-renewal of mESCs (9, 10, 26–29). In particular, recombinant Wnt3a inhibits the differentiation of mESCs (11, 30). Paradoxically, p53 has been shown to possess a prodifferentiation role in mESCs (19). Therefore, the induction of Wnt ligand genes by p53 may represent a previously undiscovered antidifferentiation role of p53 in mESCs.
To further gain insights into the induction of the Wnt ligand genes by p53, we initially tested whether the induction of the five Wnt genes by p53 can be observed in other cell types. For this purpose, we treated mESC-derived neural progenitor cells (NPCs) and mouse embryonic fibroblast (MEF) cells with adriamycin for 8 h. The fold induction of the five Wnt genes was then measured by real-time PCR assay. For Wnt3a, Wnt8a, and Wnt8b genes, the induction was completely lost in NPCs and MEF cells (Fig. 3A). The induction of Wnt3 and Wnt9a was greatly attenuated. These results demonstrate that the induction of the Wnt ligands is tightly associated with mESCs and therefore suggest that the induction of the Wnt ligand genes by p53 plays an important role in mESCs. To explore the underlying mechanism(s) of this ESC-specific Wnt induction by p53, we performed classical ChIP assay in mESCs, NPCs, and MEFs using p53 antibody (Fig. S2A). The bindings of p53 on Wnt3, Wnt3a, Wnt8a, and Wnt8b genes in mESCs are significantly (P < 0.05) higher than those in NPCs and MEFs, strongly suggesting that the decrease of the bindings of p53 on Wnt ligand genes is one of the mechanisms of ESC-specific Wnt induction by p53. Because most of the previous studies on p53 were performed using either differentiated cells or cancer cell lines, it is not surprising that the induction of the Wnt ligand genes by p53, to our best knowledge, has not been detected.
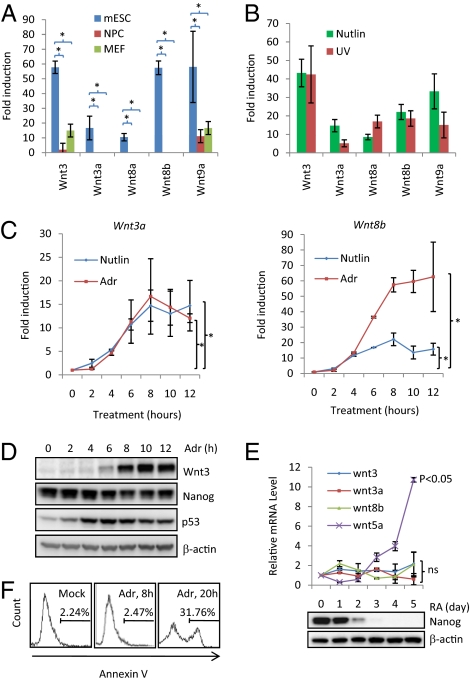
Fig. 3.
Characterization of the induction of the Wnt genes by p53. (A) Real-time PCR result of the fold induction of Wnt3, Wnt3a, Wnt8a, Wnt8b, and Wnt9a with adriamycin for 8 h in mESC (R1E), NPC (derived from R1E), and MEF cells. (B) Real-time PCR measurement of fold induction of Wnt3, Wnt3a, Wnt8a, Wnt8b, and Wnt9a with nutlin or UV in mESCs. (C) The kinetics of the induction of Wnt3a and Wnt8b upon nutlin or adriamycin treatment. (D) Western blot analysis to measure the protein levels of Wnt3, Nanog, p53, and β-actin. (E) (Upper) mRNA levels of Wnt3, Wnt3a, Wnt8b, and Wnt5a (control) during retinoic acid (RA)-induced differentiation of mESCs; (Lower) protein levels of Nanog and β-actin. Statistical analyses (comparing to day 0) were performed. (F) Annexin V staining to measure the apoptosis of R1E cells treated with adriamycin for 0, 8, and 20 h. All of the real-time PCR results are displayed as the mean ± SEM; n = 3; *P < 0.05; ns, not significant.
p53 acts as a general stress sensor (1, 2). Therefore, we examined whether other stresses besides adriamycin also induce the Wnt gene expression. mESCs were treated with nutlin, a nongenotoxic p53-specific inducer (31) and UV, another genotoxic insult. We found that both nutlin and UV treatment highly activate the expression of the five Wnt genes (Fig. 3B). Importantly, the induction of Wnt ligand genes by nutlin and UV are p53 dependent because the reduction of p53 by two short interference RNAs (siRNAs) decreased the fold induction (Fig. S2 B and C). Given that nutlin is a specific and nongenotoxic activator of p53, these results suggest that the induction of the Wnt genes is not limited to the genotoxic insults and more likely represent a general p53-mediated stress response.
The Induction of the Wnt Genes Is Not the Result of Nanog Repression and Apoptosis.
Because p53 promotes both the repression of the Nanog gene and apoptosis in mESCs (16, 19), we investigated whether the induction of the Wnt genes depends on apoptosis or the repression of Nanog. We first measured the kinetics of the induction of the Wnt genes upon adriamycin or nutlin treatment (Fig. 3C). The kinetics of Wnt induction by adriamycin and nutlin treatments was similar despite the fact that the maximum fold induction differed. The induction of the Wnt genes was detected as early as 4 h after adriamycin or nutlin treatment (Fig. 3C). As previously shown (19), the protein levels of Nanog decrease upon adriamycin treatment (Fig. 3D). The induction of Wnt genes is concomitant with the decrease of Nanog, which raises the possibility that the induction of Wnt is simply the result of Nanog repression. To test this possibility, we used retinoic acid treatment to induce the differentiation of mESCs, which in turn decreases the levels of Nanog (Fig. 3E, Lower). Although we observed a decrease of Nanog levels, we did not detect the induction of Wnt3, Wnt3a, and Wnt8b (Fig. 3E), suggesting that the induction of the Wnt genes could not simply be explained by the Nanog repression. To firmly dissociate the repression of Nanog and the induction of Wnt ligand genes, we used two siRNAs against Nanog to transiently reduce the levels of Nanog in mESCs. Cells then were treated with adriamycin for 8 h. Because of the mESC-specific nature of Wnt induction (Fig. 3A), we measured the induction of Wnt ligand genes 2 days after siRNA transfection (short term) with the hope that mESCs still maintained their stemness (Fig. S3A). Although the two siRNAs effectively reduced the levels of Nanog in mESCs, they did not significantly (P value >0.1) alter the fold induction of five Wnt ligand genes (Fig. S3B), strongly supporting the conclusion that the induction of Wnt ligand genes is not the result of Nanog repression. The apoptosis of mESCs was also measured after adriamycin treatment for 8 or 20 h. After 20-h treatment, about 31% of mESCs underwent apoptosis. Eight hours of adriamycin treatment did not cause detectable apoptosis in mESCs compared to untreated control (Fig. 3F). However, the induction of the Wnt genes already reached a plateau at the 8-h time point (Fig. 3C), demonstrating that the induction of the Wnt genes precedes apoptosis. Therefore, these results suggest that the induction of the Wnt genes is not the consequence of Nanog repression or apoptosis.
DNA-Damaged mESCs Secrete a Wnt- and p53-Dependent Antidifferentiation Activity.
Wnt ligands are secreted proteins and inhibit the differentiation of mESCs (9, 28). Because p53 induces the expression of the Wnt genes, we hypothesize that stressed mESCs secrete a Wnt- and p53-dependent antidifferentiation activity. To detect an antidifferentiation activity secreted from untreated (control) or UV-treated mESCs, conditioned media (CM) from mESCs untreated or treated with UV for 24 h were harvested (Fig. 4A). These media were then used to grow mESCs for 7 days in the absence of LIF, a cytokine that prevents mESCs from differentiation (Fig. 4A). The percentage of cells that have positive staining of Nanog or Oct4, two pluripotent markers for undifferentiated mESCs, served as an index of undifferentiated mESCs. In the absence of LIF, mESCs start to differentiate and the percentages of Nanog- and Oct4-positive cells decrease, as previously shown (8). If CM contains an antidifferentiation activity, it will increase the percentage of Nanog- or Oct4-positive cells. We chose to use UV treatment because it induces the expression of the Wnt genes (Fig. 3B) but does not contaminate the CM. Recombinant mouse Wnt3a (rWnt3a) was used as a positive control for the antidifferentiation activity.
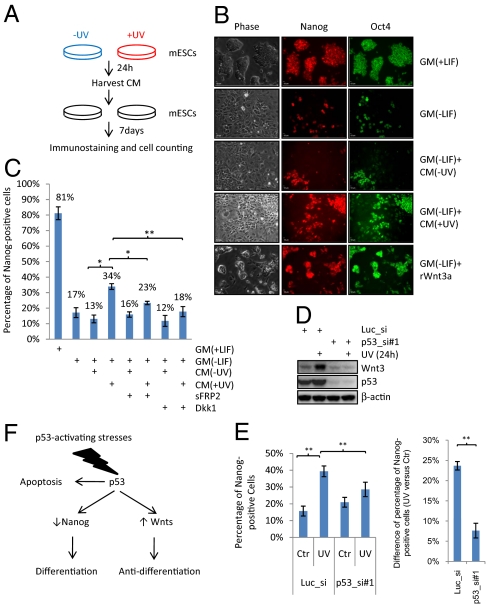
Fig. 4.
DNA-damaged mESC secrete a Wnt- and p53-dependent antidifferentiation activity. (A) Schematic representation of an assay to detect an antidifferentiation activity. CM, conditioned medium. (B) A representative illustration of Nanog and Oct4 immunostaining. GM, growth medium; LIF, 1,000 units/mL leukemia inhibitory factor; rWnt3a, 100 ng/mL recombinant mouse Wnt3a. The percentage of Nanog- or Oct4-positive cells was calculated from 10 random views and the results were shown in Fig. S4A. (C) Quantification of the percentage of Nanog-positive cells of mESCs grown in various combinations of GM and CM plus or minus sFRP2 or Dkk1 as indicated. (D) Western blot analyses of Wnt3, p53, and β-actin. (E) (Left) Quantification of the percentage of Nanog-positive cells of mESCs grown in CM collected from mESCs transfected with luciferase siRNA or p53 siRNA#1 (p53_si#1) untreated or treated with UV. (Right) The difference of the percentage of Nanog-positive cells between mESCs grown in CM from UV-treated mESCs and cells in CM from untreated mESCs was shown for luciferase and p53 siRNA transfections. (F) Summary of the roles of p53 in mESCs. All results are displayed as mean ± SEM; n = 3; *P < 0.05; **P < 0.01.
Consistent with the fact that rWnt3a inhibits the differentiation of mESCs (9), mESCs grown in medium without LIF but containing rWnt3a have significantly higher (P value <0.01) Nanog- or Oct4-positive cells compared to those grown in medium without LIF (Fig. 4B and Fig. S5). Using this CM-based assay, we were able to detect a higher percentage of Nanog- (∼29%) or Oct4-positive (∼41%) cells in mESCs grown in CM collected from UV-treated mESCs (Fig. 4B and Fig. S4A) than mESCs grown in CM without UV treatment (17 and 29% for Nanog and Oct4, respectively). Results from alkaline phosphatase (AP) staining, a commonly used self-renewal assay, is consistent with that of Nanog and Oct4 staining (Fig. S4B). Therefore, these results demonstrate that UV-treated mESCs secrete an antidifferentiation activity.
We then addressed whether this antidifferentiation effect in CM from UV-treated mESCs was dependent on Wnt ligands. Because some Wnts have redundant effects (32), we used two general inhibitors of Wnt signaling, soluble frizzled-related protein 2 (sFRP2), and Dickoppf-1 (Dkk-1). sFRP2 prevents the binding of Wnts to their receptors, Frizzleds, and Dkk1 inhibits the function of coreceptor, LRP5/6 (33). The addition of sFRP2 or Dkk1 into the CM lowered the antidifferentiation activity (Fig. 4C), strongly suggesting that the secreted Wnts are, at least partially, responsible for the antidifferentiation activity in CM from UV-treated mESCs. It is noteworthy that our results do not rule out the possibility that other signaling pathways are also involved.
Because p53 activates the expression of the Wnt genes, the reduction of p53 levels in mESCs treated with UV should decrease the antidifferentiation activity in CM. To test this prediction, the levels of p53 in mESCs were ablated using siRNA and CM was collected from cells untreated or treated with UV (Fig. 4D). Indeed, mESCs grown in the CM from UV-treated p53 knockdown cells had a significantly lower (P value <0.01) percentage of Nanog-positive cells compared to mESCs grown in the CM from UV-treated mESCs transfected with luciferase siRNA (Fig. 4E, Left). In addition, the reduction of p53 levels decreased the effect of UV on increasing the antidifferentiation activity, as manifested by the percentage difference of Nanog-positive cells between with- and without-UV treatments (Fig. 4E, Right). Moreover, the decrease of the Nanog-positive cells correlates with the decrease of Wnt protein production (Fig. 4D). Using a second siRNA targeting p53, we observed similar results (Fig. S4 C and D). Of note, p53 may not be the only factor that mediates the UV-induced antidifferentiation activity because UV might have a p53-independent effect. Together, our results demonstrate that the CM from UV-treated mESCs contains an antidifferentiation activity, which depends at least partially on both Wnt ligands and p53.
CM from UV-Treated mESCs Promotes the Cell Proliferation and Inhibits the Differentiation into Smooth Muscle Lineage of mESCs.
To test whether CM from UV-treated mESCs has other effects on mESCs, several additional cellular assays were performed. We counted the cell numbers of mESCs grown in different combinations of growth medium (GM) and CM as indicated in Fig. 4B. The number of mESCs grown in CM from UV-treated mESCs for 7 day (5.3e + 7) is twice the number of those grown in CM without UV treatment (2.6e + 7) (Fig. S4E), suggesting that UV-treated CM either increases the cell proliferation and/or the cell survival of mESCs. Colony survival assay showed that UV-treated CM did not significantly (P > 0.1) affect the survival of mESCs (Fig. S4F), suggesting that the main role of UV-treated CM on the cell number is to promote the cell proliferation.
To address whether CM from UV-treated mESCs affects the differentiation of mESCs into various lineages, we carried out immunostaining using antibodies recognizing AFP (α-fetoprotein, liver, endoderm), SMA (smooth muscle α-actin, smooth muscle, mesoderm), and MAP2 (microtube-associated protein 2, neuron, ectoderm), three widely used lineage markers. We did not observe any staining signal using AFP and MAP2 antibodies. However, strong signal of SMA was observed (Fig. S5A). CM from UV-treated mESCs significantly (P < 0.05) decreased the percentage of SMA-positive cells (Fig. S5B), indicating that CM from UV-treated mESCs inhibits the differentiation of mESCs into smooth muscle lineage.
Discussion
To identify the unique pathway(s) that is regulated by p53 in mESCs, we used an unbiased genomewide approach: ChIP-chip assay combined with gene expression microarray followed by KEGG pathway analysis. The Wnt signaling pathway was identified as one of the major targets of p53 in mESCs. Of note, the linkage between p53 and the Wnt signaling has been reported in cancer cell lines (34). p53 activates the expression of SIAH-1, which is the E3 ligase of β-catenin (35, 36). However, this apparently is not the scenario in mESCs. In mESCs, p53 simultaneously regulates the expression of many genes in the Wnt signaling pathway (Fig. 2 A–C). Because p53 is regarded as an inducer of the differentiation of mESCs, it is surprising to observe that p53 induces the production of several Wnt ligands, antidifferentiation agents of mESCs. Our results support the notion that p53 has dual functions on the differentiation of mESCs. On the one hand, p53 represses the expression of Nanog, whose repression strictly correlates with the differentiation of mESCs. On the other hand, p53 induces the expression of several Wnt ligands to inhibit the differentiation of mESCs. The dual role of p53 in mESCs is reminiscent of its roles in somatic cells in controlling life and death, by activating the expression of prosurvival genes and apoptotic genes, respectively (37, 38). Therefore, p53 may act as a “decision maker” of mESCs as to whether to differentiate or not.
In addition to the prodifferentiation and antidifferentiation roles, p53 is also involved in the rapid apoptosis of mESCs (16). We observed about 30% of apoptotic cells after 20 h of adriamycin treatment (Fig. 3F). Almost no mESCs survived beyond 2 days after treatment, which agrees with previous reports that mESCs are sensitive to DNA damage insults (14, 15). The hypersensitivity of mESCs to DNA damage was proposed to be a mechanism for mESCs to quickly remove DNA damaged cells from the population, whereby maintaining the genomic stability of the whole population (39). It appears that the apoptotic program is dominant in mESCs in response to DNA damage, although the differentiation and antidifferentiation programs precede apoptosis (Fig. 3 C–F). Then, why does p53 induce Wnt production to inhibit differentiation and simultaneously repress Nanog expression and drive apoptosis in mESCs? We postulate a model to accommodate all of the observations (Fig. 4F). Upon stresses, mESCs use p53 to eradicate the cells with DNA damage burden by promoting their apoptosis and differentiation. This safeguard mechanism can ensure mESCs to maintain a population with low mutation rate. However, because of the hypersensitivity of mESCs to DNA damage and other stresses, mESCs could quickly lose their population. To maintain the stability of mESC populations, p53 induces the expression of Wnt ligands, which are secreted from stressed mESCs to inhibit the differentiation. Through this exquisite mechanism mediated by p53, mESCs can simultaneously maintain both genomic and population stability.
The induction of Wnt ligands by p53 is arguably ESC specific (Fig. 3A), highlighting the cell type-specific role of p53. This result also offers an explanation of why the induction of Wnt ligands by p53 has not been reported in the extensive studies on p53 in somatic cells. DNA sequences of the Wnt genes apparently do not explain this ESC specificity because mESCs, NPCs, and MEF cells have the same DNA sequences. Results from ChIP assay (Fig. S2A) indicate that one of the underlying mechanisms of ESC-specific induction of Wnt ligand genes is the differential binding of p53 in mESCs, NPCs, and MEFs. The fact that mESCs have more accessible chromatin than differentiated cells suggests certain epigenetic mechanism(s) might play a role (40). Indeed, epigenetic regulation has emerged as an important step to influence the binding of p53 to its target genes (38). The binding of p53 to the Wnt ligand genes is not typical because we did not find consensus p53 binding elements within the promoters and coding regions of the Wnt ligand genes using the Genomatix program. It is highly possible that the binding of p53 on the Wnt genes could be mediated by a mESC-specific protein. The tethering of p53 by other factors to its target genes has been shown in somatic cells (41). The underlying mechanism is currently under investigation in the laboratory.
Overactivation of the Wnt ligands is tumorigenic in certain types of somatic cells (26). Therefore, p53 could become “tumorigenic” if the induction of Wnt ligands by p53 is aberrantly inherited by the progeny cells of mESCs. The attenuation of the Wnt induction by p53 in more differentiated cells (Fig. 3A) could be one of the mechanisms to cancel the potential tumorigenic role of p53. Although the tumorigenic role of p53 has only been reported with mutant p53 (41, 42), recent progress of generating induced pluripotent stem (iPS) cells from MEFs and NPCs in vitro raises the possibility that wild-type p53 could switch from tumor suppressive to tumorigenic function under certain currently unknown conditions (43, 44). Our results merit further investigation of this possibility using an in vivo cancer model in the future.
Materials and Methods
Materials and methods include growth conditions for mESC, NPC, and MEF cells, ChIP-chip assay, genomewide location analysis, gene expression microarray and analysis, pathway analyses, preparation of conditioned medium and detection of the antidifferentiation activity, immunostaining, Western blotting, reverse transcription and real-time PCR, short interference RNA (siRNA) transfection, alkaline phosphatase (AP) staining, and colony survival assay. Details for each method are described in SI Text. High throughput data were submitted to GEO database (GSE16427 for ChIP-chip and GSE16428 for gene expression microarray).
Supplementary Material
Acknowledgments
We thank Bowen Cui for technical advice on ChIP-chip assay, and Glenn Merlino, Peter Blumberg, and Li Guo for critically reading the manuscript. This research was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, and Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909734107/DCSupplemental.
References
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 4.Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci USA. 1981;78:7634–7638. doi: 10.1073/pnas.78.12.7634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 6.Boyer LA, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loh YH, et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 8.Niwa H, Burdon T, Chambers I, Smith A. Self-renewal of pluripotent embryonic stem cells is mediated via activation of STAT3. Genes Dev. 1998;12:2048–2060. doi: 10.1101/gad.12.13.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- 10.Kielman MF, et al. Apc modulates embryonic stem-cell differentiation by controlling the dosage of beta-catenin signaling. Nat Genet. 2002;32:594–605. doi: 10.1038/ng1045. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa K, Nishinakamura R, Iwamatsu Y, Shimosato D, Niwa H. Synergistic action of Wnt and LIF in maintaining pluripotency of mouse ES cells. Biochem Biophys Res Commun. 2006;343:159–166. doi: 10.1016/j.bbrc.2006.02.127. [DOI] [PubMed] [Google Scholar]
- 12.Dravid G, et al. Defining the role of Wnt/beta-catenin signaling in the survival, proliferation, and self-renewal of human embryonic stem cells. Stem Cells. 2005;23:1489–1501. doi: 10.1634/stemcells.2005-0034. [DOI] [PubMed] [Google Scholar]
- 13.Cervantes RB, Stringer JR, Shao C, Tischfield JA, Stambrook PJ. Embryonic stem cells and somatic cells differ in mutation frequency and type. Proc Natl Acad Sci USA. 2002;99:3586–3590. doi: 10.1073/pnas.062527199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Waard H, et al. Cell type-specific hypersensitivity to oxidative damage in CSB and XPA mice. DNA Repair (Amst) 2003;2:13–25. doi: 10.1016/s1568-7864(02)00188-x. [DOI] [PubMed] [Google Scholar]
- 15.Van Sloun PP, et al. The role of nucleotide excision repair in protecting embryonic stem cells from genotoxic effects of UV-induced DNA damage. Nucleic Acids Res. 1999;27:3276–3282. doi: 10.1093/nar/27.16.3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Vries A, et al. Targeted point mutations of p53 lead to dominant-negative inhibition of wild-type p53 function. Proc Natl Acad Sci USA. 2002;99:2948–2953. doi: 10.1073/pnas.052713099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Qin H, et al. Regulation of apoptosis and differentiation by p53 in human embryonic stem cells. J Biol Chem. 2007;282:5842–5852. doi: 10.1074/jbc.M610464200. [DOI] [PubMed] [Google Scholar]
- 18.Aladjem MI, et al. ES cells do not activate p53-dependent stress responses and undergo p53-independent apoptosis in response to DNA damage. Curr Biol. 1998;8:145–155. doi: 10.1016/s0960-9822(98)70061-2. [DOI] [PubMed] [Google Scholar]
- 19.Lin T, et al. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat Cell Biol. 2005;7:165–171. doi: 10.1038/ncb1211. [DOI] [PubMed] [Google Scholar]
- 20.Huang J, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444:629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 22.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 23.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 24.Brooks CL, Gu W. Ubiquitination, phosphorylation and acetylation: The molecular basis for p53 regulation. Curr Opin Cell Biol. 2003;15:164–171. doi: 10.1016/s0955-0674(03)00003-6. [DOI] [PubMed] [Google Scholar]
- 25.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 26.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 27.Goessling W, et al. Genetic interaction of PGE2 and Wnt signaling regulates developmental specification of stem cells and regeneration. Cell. 2009;136:1136–1147. doi: 10.1016/j.cell.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cadigan KM, Nusse R. Wnt signaling: A common theme in animal development. Genes Dev. 1997;11:3286–3305. doi: 10.1101/gad.11.24.3286. [DOI] [PubMed] [Google Scholar]
- 29.Miyabayashi T, et al. Wnt/beta-catenin/CBP signaling maintains long-term murine embryonic stem cell pluripotency. Proc Natl Acad Sci USA. 2007;104:5668–5673. doi: 10.1073/pnas.0701331104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Singla DK, Schneider DJ, LeWinter MM, Sobel BE. wnt3a but not wnt11 supports self-renewal of embryonic stem cells. Biochem Biophys Res Commun. 2006;345:789–795. doi: 10.1016/j.bbrc.2006.04.125. [DOI] [PubMed] [Google Scholar]
- 31.Vassilev LT, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 32.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 33.Nusse R. Developmental biology. Making head or tail of Dickkopf. Nature. 2001;411:255–256. doi: 10.1038/35077199. [DOI] [PubMed] [Google Scholar]
- 34.Harris SL, Levine AJ. The p53 pathway: Positive and negative feedback loops. Oncogene. 2005;24:2899–2908. doi: 10.1038/sj.onc.1208615. [DOI] [PubMed] [Google Scholar]
- 35.Fiucci G, et al. Siah-1b is a direct transcriptional target of p53: identification of the functional p53 responsive element in the siah-1b promoter. Proc Natl Acad Sci USA. 2004;101:3510–3515. doi: 10.1073/pnas.0400177101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwai A, et al. Siah-1L, a novel transcript variant belonging to the human Siah family of proteins, regulates beta-catenin activity in a p53-dependent manner. Oncogene. 2004;23:7593–7600. doi: 10.1038/sj.onc.1208016. [DOI] [PubMed] [Google Scholar]
- 37.Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130:597–600. doi: 10.1016/j.cell.2007.08.005. [DOI] [PubMed] [Google Scholar]
- 38.Tanaka T, Ohkubo S, Tatsuno I, Prives C. hCAS/CSE1L associates with chromatin and regulates expression of select p53 target genes. Cell. 2007;130:638–650. doi: 10.1016/j.cell.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Hong Y, Stambrook PJ. Restoration of an absent G1 arrest and protection from apoptosis in embryonic stem cells after ionizing radiation. Proc Natl Acad Sci USA. 2004;101:14443–14448. doi: 10.1073/pnas.0401346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meshorer E, et al. Hyperdynamic plasticity of chromatin proteins in pluripotent embryonic stem cells. Dev Cell. 2006;10:105–116. doi: 10.1016/j.devcel.2005.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Di Agostino S, et al. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 42.Adorno M, et al. A Mutant-p53/Smad complex opposes p63 to empower TGFbeta-induced metastasis. Cell. 2009;137:87–98. doi: 10.1016/j.cell.2009.01.039. [DOI] [PubMed] [Google Scholar]
- 43.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 44.Kim JB, et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.