Abstract
NF-κB, a central coordinator of immune and inflammatory responses, must be tightly regulated. We describe a NF-κB regulatory pathway that is driven by reversible lysine methylation of the p65 subunit, carried out by a lysine methylase, the nuclear receptor–binding SET domain-containing protein 1 (NSD1), and a lysine demethylase, F-box and leucine-rich repeat protein 11 (FBXL11). Overexpression of FBXL11 inhibits NF-κB activity, and a high level of NSD1 activates NF-κB and reverses the inhibitory effect of FBXL11, whereas reduced expression of NSD1 decreases NF-κB activation. The targets are K218 and K221 of p65, which are methylated in cells with activated NF-κB. Overexpression of FBXL11 slowed the growth of HT29 cancer cells, whereas shRNA-mediated knockdown had the opposite effect, and these phenotypes were dependent on K218/K221 methylation. In mouse embryo fibroblasts, the activation of most p65-dependent genes relied on K218/K221 methylation. Importantly, expression of the FBXL11 gene is driven by NF-κB, revealing a negative regulatory feedback loop. We conclude that reversible lysine methylation of NF-κB is an important element in the complex regulation of this key transcription factor.
Keywords: demethylase, histone, mass spectrometry, methylase, transcription factor
Members of the NF-κB family are central coordinators of innate and adaptive immune responses. Of the five family members in mammals [RelA (p65), RelB, c-Rel, NF-κB1 (p50), and NF-κB2 (p52)], the p65/p50 heterodimer functions most often in the “classical” signaling pathway (1). Constitutive NF-κB activation is frequently associated with a number of pathological conditions, including inflammation and cancer, and is well known to be involved in tumor angiogenesis and invasiveness (2). Loss of the normal regulation of NF-κB is a major contributor to the deregulated growth, resistance to apoptosis, and propensity to metastasize observed in many different cancers (3). Our data, showing NF-κB activation in many cancer-derived cell lines (4, 5), further support the strong correlation between constitutive NF-κB activation and cancer. Thus, understanding the complexity of NF-κB–dependent signaling is an important aspect of dealing with these diseases.
NF-κB has a variety of functions and is regulated by hundreds of different stimuli in different cell types (6). Specialized control of NF-κB activity is critical for achieving its normal transient activation in response to stress. Regardless of the stimulus, all pathways leading to NF-κB activation depend on inducing posttranslational modifications of the IκB kinase, IκB proteins, or NF-κB subunits. These regulatory modifications, including phosphorylation, ubiquitination, acetylation, sumolation, and nitrosylation, vary depending on the specific stimulus and context (7).
We have developed a lentiviral validation–based insertional (VBIM) method to create mutant cell lines in which the overexpression of specific cellular proteins, driven by inserted CMV promoters, is responsible for altered phenotypes. We used this method to identify the F-box and leucine-rich repeat protein 11 (FBXL11) as a potent negative regulator of NF-κB and to show that its demethylase activity was required for this function (8). FBXL11, previously shown to demethylate histone H3K36, contains an F-box, a JmjC domain, a CxxC zinc finger, a PHD domain, and three leucine-rich repeat elements (9). The point mutation H212A, in the JmjC domain, abolishes both H3K36 demethylase activity (9) and the ability of FBXL11 to inhibit NF-κB function (8). Here, we show that FBXL11 and its partner H3K36 methylase NSD1 function as an enzyme pair to regulate NF-κB through reversible lysine methylation of K218 and K221 of the p65 subunit of NF-κB, with important physiological consequences.
Results
FBXL11 and NSD1 Have Opposite Effects on NF-κB; Both Bind to p65 Subunit After Activation of NF-κB.
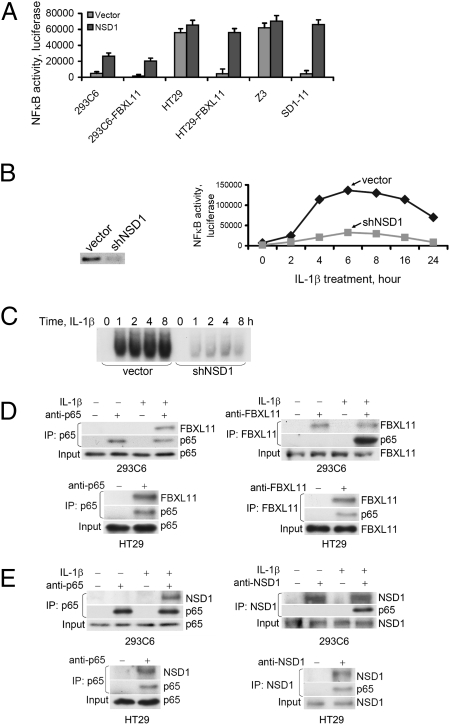
We tested whether NSD1, the known methylase for H3K36, also affects NF-κB activity, using human 293C6 and colon cancer HT29 cells, with either normal or greatly increased levels of FBXL11, and also Z3 cells, a 293C6-derived cell line with constitutive NF-κB activity (10), together with the SD1-11 mutant of Z3 cells in which FBXL11 is overexpressed (8). A κB-luciferase reporter was transfected transiently into the cells together with NSD1 and, 48 h later, luciferase assays were carried out, with β-galactosidase as an internal control. NF-κB activity was downregulated in all cells expressing high levels of FBXL11 (Fig. 1A), consistent with earlier results (8). Furthermore, overexpressed NSD1 activated NF-κB and reversed the inhibition caused by the overexpression of FBXL11 (Fig. 1A). Using shRNA to knock down the expression of NSD1 (Fig. 1B, Left) decreased both NF-κB activity (Fig. 1B, Right) and its ability to bind to DNA (Fig. 1C) in 293C6 cells treated with IL-1β. Therefore, the NSD1/FBXL11 pair of methylase/demethylase enzymes reversibly regulates NF-κB activity.
Fig. 1.
NSD1 activates NF-κB and reverses the inhibitory effect of FBXL11; FBXL11 and NSD1 bind to p65 upon NF-κB activation. (A) Luciferase assay of NF-κB activity. Expression of NSD1 activates NF-κB and reverses inhibition caused by increased expression of FBXL11 in different cells, including 293C6, HT29, and mutant Z3 cells, and corresponding cells in which FBXL11 is overexpressed. Results of triplicate luciferase assays are shown as means ± SD. (B) (Left) Western assay showing knockdown of expression of NSD1 in 293C6 cells with shRNA. (Right) Luciferase assay of NF-κB activity in 293C6 cells treated with IL-1β for different times, showing that reduced expression of NSD1 by shRNA decreased NF-κB activity. Results of triplicate luciferase assays are shown as means ± SD. (C) EMSA assay of NF-κB in 293C6 cells treated with IL-1β for different times, showing that reduced NSD1 expression by shRNA decreased NF-κB DNA binding activity. (D and E) p65 Coimmunoprecipitates with FBXL11 and NSD1; FBXL11 and NSD1 bind to p65 only when NF-κB is activated. Immunoprecipitation (IP) assays detect binding of endogenous FBXL11 or NSD1 to p65, either in 293C6 cells treated with IL-1β (Upper) or in HT29 cells with constitutive NF-κB activation (Lower).
Because the ability of NF-κB to bind to DNA is inhibited by FBXL11 (8), we reasoned that this enzyme was likely to affect NF-κB directly, rather than indirectly through the demethylation of H3K36. To test this possibility, we performed coimmunoprecipitation experiments with the endogenous proteins. Using either anti-p65 (Fig. 1D, Left) or anti-FBXL11 (Fig. 1D, Right), FBXL11 was pulled down together with p65, especially when NF-κB was strongly activated, either in response to IL-1β in 293C6 cells (Fig. 1D, Top), or constitutively in HT29 cells (Fig. 1D, Bottom). Therefore, it is likely that the direct demethylation of NF-κB by FBXL11 is responsible for inhibiting its activity. Furthermore, FBXL11 binds to NF-κB substantially only after it has been activated. In a similar series of experiments, we found that endogenous NSD1 can also be pulled down together with p65 in 293C6 cells treated with IL-1β (Fig. 1E, Top) or in HT29 cells with constitutive NF-κB (Fig. 1E, Bottom), but only after NF-κB has been activated.
p65 Subunit of Constitutively Activated NF-κB Is Methylated on K218 and K221 and Is Demethylated by FBXL11.
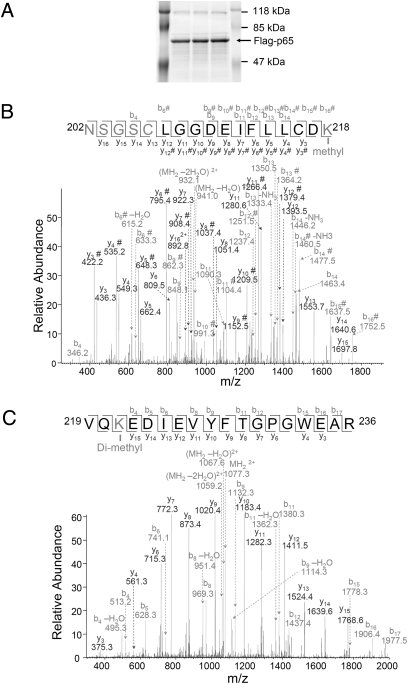
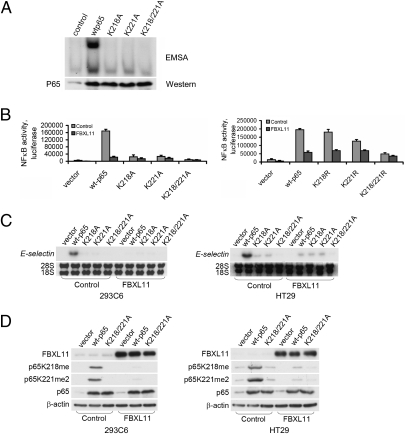
Analysis by mass spectrometry (MS) revealed that activated p65, purified from Z3 cells (Fig. 2A), is monomethylated on K218 (K218me, Fig. 2B) and dimethylated on K221 (K221me2, Fig. 2C). Experiments with 293C6 and HT29 cells yielded similar results. The mutant proteins K218A, K221A, or K218/221A were expressed transiently in 293C6 cells, together with a κB-dependent luciferase reporter, and NF-κB activity was analyzed by EMSA (Fig. 3A) or by use of a luciferase assay (Fig. 3B, Left). As is well known, the overexpression of wild-type p65 alone activated NF-κB (Fig. 3 A and B). The K218A and K221A mutations diminished NF-κB activation substantially (Fig. 3 A and B, Left) and the K218/221A double mutant yielded very little activation (Fig. 3B, Left). Similar experiments with the corresponding K-R mutants showed similar but less dramatic effects for the single mutants, but the K-R double mutant showed greatly reduced activity (Fig. 3B, Right). Experiments with HT29 cells and the K-A mutants gave similar results (Fig. S1). The inhibitory effect of FBXL11 on NF-κB activity was strongest with wild-type p65, and the K218A and K221A mutants were less susceptible to inhibition (Fig. S1). We also tested the expression of E-SELECTIN, a typical NF-κB target gene, in Northern assays (Fig. 3C). Wild-type p65 and the three K-A mutants were transfected transiently into 293C6 or HT29 cells, or into the same cells expressing high levels of FBXL11 and, 48 h later, samples were collected for Northern assay. The ability of overexpressed wild-type p65 to drive the expression of E-SELECTIN was greatly inhibited by high levels of FBXL11 (Fig. 3C). Furthermore, the K-A mutants had little ability to induce E-SELECTIN expression, and their small effect was only slightly inhibited in cells overexpressing FBXL11 (Fig. 3C). These data reveal that activated NF-κB is methylated on two functional lysine residues, K218 and K221, and, because single mutations of these residues significantly interfere with the inhibitory effect of FBXL11, that these two sites are likely to be important demethylation targets.
Fig. 2.
Mass spectrometry (MS) shows that p65 is methylated on K218 and K221 on NF-κB activation. (A) GelCode blue-stained gel, showing that p65 is immunoprecipitated as a single strong band. The same samples were loaded into multiple lanes. (B) Analysis of tryptic peptides derived from p65 suggests that K218 is monomethylated. The pure single band was digested in the gel and samples were analyzed by LC-MS/MS. A mass shift of +14 was observed spanning peptide 202–218. Tandem MS analysis further suggested that K218 on the C-terminal side of the peptide is modified. Another +14 mass shift was also observed on the N-terminal side, suggesting an additional methylation within the N-terminal four residues. (C) Analysis of tryptic peptides derived from p65 suggests that K221 of p65 is dimethylated. A mass shift of +28 was observed spanning peptide 219–236. Tandem MS analysis further suggested that K221 on the N-terminal is dimethylated.
Fig. 3.
K218 and K221 are functional sites that are methylated on constitutive activation of NF-κB and demethylated on expression of FBXL11. (A) EMSA assay, showing that p65 mutant proteins K218A, K221A, and K218/221A differentially inhibited the ability of NF-κB to bind to DNA in 293C6 cells. (Lower) Western analysis showing that expression levels of the different p65 constructs are similar. (B) Mutations of K218, K221, and K218/221 of p65 differentially decreased NF-κB activity and the inhibitory effect of FBXL11 on NF-κB in 293C6 cells. (Left) K-A mutants. (Right) K-R mutants. Luciferase assay shows level of NF-κB activity after each mutant was transiently expressed for 48 h. Results of triplicate luciferase assays are shown as means ± SD. (C) Mutations K218A, K221A, and K218/221A of p65 differentially decreased expression of the typical NF-κB target gene E-SELECTIN and the inhibitory effect of FBXL11 on NF-κB in 293C6 cells (Left) and HT29 cells (Right). The different p65 constructs were transfected transiently into 293C6 or HT29 cells and, 48 h later, samples were collected for Northern analyses. (D) Western analyses, showing that p65 is monomethylated on K218 and dimethylated on K221 on constitutive activation of NF-κB, driven by the overexpression of wild-type p65, but not the K218/K221A mutant protein. The degree of methylation was dramatically decreased by the expression of FBXL11 in both 293C6 (Left) and HT29 cells (Right).
We raised rabbit polyclonal antibodies against p65 peptides containing K218me or K221me2 and confirmed their specificities by showing that they bound to the methylated peptides but not to the corresponding unmodified or acetylated peptides. Constructs encoding wild-type p65 or the K218/221A mutant protein were transfected transiently into 293C6 or HT29 cells or into corresponding cells in which FBXL11 is overexpressed; 48 h later, samples were collected for Western assay. p65 is methylated on K218 and K221 after the expression of wild-type p65 but not the K218/221A mutant (Fig. 3D). Furthermore, the methylation of these sites was dramatically decreased in cells overexpressing FBXL11, showing that the monomethylation of K218 and dimethylation of K221 occurs when NF-κB is constitutively activated and that the overexpression of FBXL11 decreases these modifications.
p65 Is Regulated by Reversible Lysine Methylation on K218 and K221 Through FBXL11 and NSD1 Enzyme Pair in Response to Cytokines.
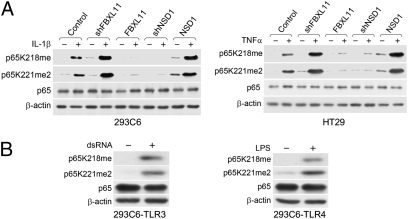
Using 293C6 or HT29 cells that either stably overexpress FBXL11 or in which the expression of this protein has been knocked down by shRNA (8), we observed that, after treatment with IL-1β or TNFα, K218me and K221me2 were formed in response to either cytokine, and that the induction was decreased by high levels and enhanced by low levels of FBXL11 (Fig. 4A). Furthermore, the methylation signal was greatly enhanced in cells overexpressing NSD1 and was reduced in cells in which an shRNA corresponding to NSD1 was used to knock down its expression (Fig. 4A). Collectively, these data confirm that FBXL11 and NSD1 constitute an enzyme pair that methylates and demethylates p65 on K218 and 221 in response to cytokine stimulation. In addition, in 293C6-TLR3 cells treated with double-stranded RNA (dsRNA) or in 293C6-TLR4 cells treated with lipopolysaccharide (LPS), the methylations of K218 and K221 were also induced (Fig. 4B), showing that these two residues are modified similarly in response to different NF-κB-activating stimuli.
Fig. 4.
K218 and K221 residues of p65 are methylated and demethylated by the NSD1-FBXL11 enzyme pair in response to cytokines; K218 and K221 are methylated in response to dsRNA or LPS. (A) Western analysis, showing that K218 and K221 of p65 are methylated and demethylated by NSD1 and FBXL11, respectively, in response to cytokines. 293C6 (Left) or HT29 cells (Right), and the corresponding FBXL11 or NSD1 cells, were treated with IL-1β or TNFα for 45 min before being collected for Western analyses. (B) K218 and K221 of p65 are methylated in response to either dsRNA in 293C6-TLR3 cells (Left) or LPS in 293C6-TLR4 cells (Right).
Methylation of K218 and K221 Plays an Important Role in Cell Proliferation, Colony Formation, and Gene Expression in Human Cancer HT29 Cells.
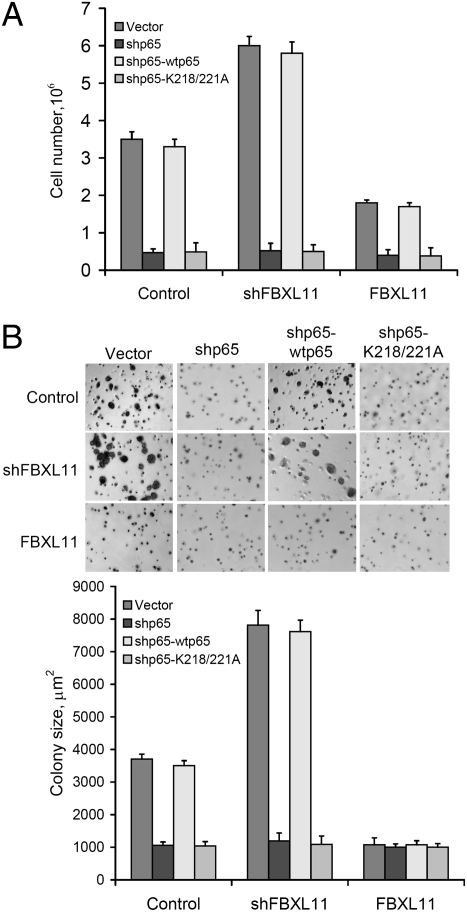
In HT29 cells, which have constitutively active NF-κB (4), knocking down FBXL11 markedly increased cell growth (Fig. 5A), whereas increased expression of FBXL11 decreased both cell growth and the sizes of colonies formed in soft agar (Fig. 5 A and B), indicating that FBXL11 is a potent suppressor of cell proliferation and colony formation in these cells. Furthermore, knocking down p65 expression with shRNA dramatically inhibited cell proliferation and colony formation in control HT29 cells or HT29 cells in which the FBXL11 level has been upregulated by overexpression or downregulated by shRNA, showing that the effect of FBXL11 on cell growth and colony formation is p65 dependent (Fig. 5 A and B). The phenotypes of cells in which shRNA to p65 is expressed is reversed by wild-type p65 but not the K218/221A mutant protein, again showing that the K218 and K221 sites are important for the function of p65 (Fig. 5 A and B). Analysis of global mRNA expression revealed that stable overexpression of FBXL11 in HT29 cells (8) greatly decreased the expression of some cytokine and chemokine genes and of some genes relevant to tumorigenesis (Table S1).
Fig. 5.
Effects of FBXL11 expression on cell proliferation and colony formation. (A) K218/221-dependent effect of FBXL11 on cell proliferation in colon cancer HT29 cells. High levels of FBXL11 decrease cell growth, whereas decreasing FBXL11 expression with shRNA increases cell growth in vector-transfected cells. However, knocking down p65 expression with an shRNA against its 5′-UTR greatly decreased the proliferation of HT29 cells and of cells expressing FBXL11 or shRNAs to FBXL11. The phenotype caused by shRNA to p65 can be reversed by expression of wild-type p65 but not the K218/221A mutant. (B) K218/221-dependent effect of FBXL11 on colony formation. (Upper) Images of colonies in a soft agar assay, showing that high levels of FBXL11 decrease sizes of colonies formed, whereas decreased expression of FBXL11 increases sizes of colonies in vector-transfected cells. However, knocking down p65 expression with shRNA against its 5′-UTR greatly decreases size of colonies formed by HT29 cells and cells expressing FBXL11 or shRNA to FBXL11. Phenotype caused by shRNA to p65 can be reversed by expression of wild-type p65 but not the K218/221A mutant protein. (Lower) Statistical analysis of the sizes of the colonies observed. Results of triplicate assays are shown as means ± SD.
p65-Dependent Gene Is Regulated by FBXL11 in Mouse Embryo Fibroblasts.
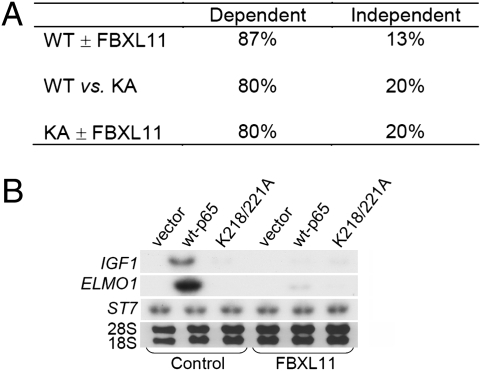
We expressed either wild-type p65 or the K218/221A double mutant protein in p65-null mouse embryo fibroblasts (MEFs). A global analysis of mRNA levels (Fig. 6A) revealed that, among the genes induced when NF-κB was activated by overexpressed wild-type p65 (“p65-dependent genes”), ∼87% were downregulated by FBXL11. It is interesting that quite a few of the corresponding proteins are involved in tumorigenesis (Table S2). Furthermore, ∼80% of these p65-dependent genes were not activated by the K218/221A mutant protein (Fig. 6A), showing that these residues play a key role in the expression of a substantial subset of the p65-dependent genes. Similarly, ∼80% of the KA-dependent genes were downregulated by FBXL11. The data strongly suggest that FBXL11 plays an important role in regulating the expression of some but not all p65-dependent genes and that the K218 and K221 sites are essential for this function. Interestingly, ∼13% of the p65-dependent genes were not downregulated by FBXL11, ∼20% of the p65-dependent genes were not affected by the K218/221A mutations, and ∼20% of the KA-dependent genes were FBXL11 independent (Fig. 6A), indicating the existence of other regulatory pathways. mRNAs from TNFα-treated MEFs expressing either wild-type p65 or the K218/221A mutant protein were also analyzed, yielding a similar pattern of expression, indicating that FBXL11 also plays an important role in regulating p65-dependent gene expression in response to cytokine-stimulated activation of NF-κB.
Fig. 6.
Regulation of p65-dependent gene expression by FBXL11 in MEFs. (A) Correlation between wild-type (WT) p65, K218/221A (KA) mutant p65, and FBXL11 on p65-dependent gene expression in p65-null MEFs. In the WT ± FBXL11 sample, “Dependent” represents genes that are downregulated by FBXL11. In the WT vs. 218/221 double KA sample, “Dependent” represents genes that are p65 dependent but not activated after expression of the KA mutant protein. In the KA ± FBXL11 sample, “Dependent” represents KA-affected genes that are also regulated by FBXL11. Constructs encoding wild-type or double KA mutant proteins were transfected transiently into p65-null MEFs and, 48 h later, samples were collected and RNAs were purified for analysis. Expression of wild-type p65 caused constitutive activation of NF-κB, leading to the induction of gene expression, which is designated as p65 dependent. The fold increase cutoff for analysis is 2. (B) Confirmation of the expression of some genes from the array analysis in MEFs. The same RNA samples were used for the Northern experiment. Probes for IGF1, ELMO1, and ST7 were used.
The list shown in Table S2 gives examples of the effect of high levels of FBXL11 on the expression of some p65-dependent genes in MEFs. Included are genes encoding cytokines and proteins relevant to tumorigenesis. Northern analyses confirmed the array results for several genes (Fig. 6B). For example, insulin-like growth factor 1 (IGF1) was induced upon the expression of wild-type p65, but not the K218/221A mutant protein. When FBXL11 was also overexpressed, the induction of IGF1 decreased significantly, showing that the induction of this gene depends upon K218 and K221 and is downregulated by FBXL11 (Fig. 6B). A similar result was obtained for the engulfment and cell motility 1 gene (ELMO1) (Fig. 6B). These two proteins play important roles in transformation, tumorigenesis, and metastasis (11, 12). We also show, as a control, that the p65-independent suppressor of tumorigenicity 7 gene (ST7) (13) is not induced upon expression of either wild-type p65 or the K218/221A mutant protein, and that its expression is not downregulated by FBXL11.
FBXL11 Gene Is Induced on NF-κB Activation.
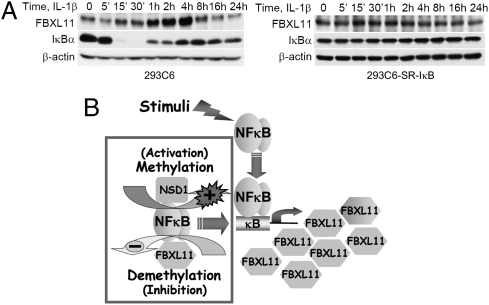
Importantly, the expression of endogenous FBXL11 was induced in a time-dependent manner in 293C6 cells treated with IL-1β, reaching a peak after 2–4 h (Fig. 7A, Left). IκBα was degraded and then resynthesized in parallel during this period. The induction of FBXL11 was blocked in 293C6SR-IκB cells, in which the dominant negative superrepressor of IκB is expressed (Fig. 7A, Right). This experiment shows that the FBXL11 gene is induced upon NF-κB activation (Fig. 7A, Left). Thus, similarly to IκB, FBXL11 is a negative regulator of NF-κB that is induced in response to NF-κB. Collectively, our data show that, in addition to previously known regulatory pathways, NF-κB is also regulated through methylation-demethylation of p65, catalyzed by the NSD1-FBXL11 enzyme pair, and that the FBXL11 protein participates in a negative regulatory feedback loop that further modulates NF-κB activity (Fig. 7 A and B).
Fig. 7.
FBXL11 gene is induced upon NF-κB activation. (A) Western analysis, showing that FBXL11 expression is induced in 293C6 cells on IL-1β treatment (Left) but is blocked in 293C6SR-κB cells, which express the dominant negative IκB superrepressor (Right). (B) Model. In addition to previously known regulatory pathways, NF-κB is regulated by the methylation and demethylation of p65, catalyzed by the NSD1-FBXL11 enzyme pair. FBXL11 gene is induced upon NF-κB activation, forming a negative regulatory loop.
Discussion
We demonstrated that, in response to an activating signal such as treatment with IL-1β or TNFα, or when NF-κB is constitutively activated, the p65 subunit is inducibly methylated and demethylated on lysine residues K218 and K221 by the chromatin-remodeling enzymes FBXL11 and NSD1, in ways that profoundly affect function. Recently, Huang et al. (14) showed that K370 of p53 is demethylated by LSD1, a histone H3K4 or H3K9 demethylase that downregulates p53 function. The dual function of LSD1 in regulating both histone H3 and p53 is strikingly similar to our finding that FBXL11 and NSD1 regulate both histone H3 and NF-κB. These two examples suggest that the regulation of transcription factors by histone methylases and demethylases may be a general phenomenon in mammalian cells, and probably in other biological systems as well.
Posttranslational modifications of NF-κB, reviewed recently (7, 15), include the acetylation of p65, which is inducibly modified at several sites, including K122, K123, K218, K221, K310, K314, and K315 under different conditions, with different effects on activity (16–18). In 293C6 cells, we observed the methylation of both K218 and K221 in response to activation of NF-κB. The p65 contains the Rel homology domain (amino acid residues 19–301), and two transactivation domains (amino acid residues 428–551) (7). The Rel homology domain contains sequences responsible for its dimerization, DNA binding, nuclear localization, and IκB binding. In the crystal structure, NF-κB dimers use loops instead of α-helices to mediate DNA contact. L1–L5 are the DNA-contacting loops of p65. Both K218 and K221 are located in loop L4 and are among the very few residues that directly contact the DNA backbone (19, 20). In addition, sequence analysis shows that these two sites are conserved from invertebrates, such as the sea squirt, to nonmammalian vertebrates, such as the zebra fish and frog, to many different mammals, including the mouse, dog, pig, horse, cow, and chimpanzee, indicating that these sites are essential for p65 function. Interestingly, K218 and K221 were previously reported to be acetylated in Cos7 cells and, upon the overexpression of the histone acetylases CBP/p300, also in 293T cells, leading to NF-κB activation (7, 21). Therefore, the same lysine residues can be methylated or acetylated, depending on the specific cellular context. Examples in which the same lysine residue can be either methylated or acetylated can be found elsewhere. Multiple residues of each core histone have been identified as modification sites and some of the same lysine side chains can be either methylated or acetylated in a cellular context–specific manner (22, 23). For p53, K372 is methylated by the H3K4 methylase SET7/9, enhancing the subsequent acetylation of the same residue and in turn stabilizing p53 (24). In addition, the previously identified acetylated sites K314/K315 that are modified by CBP/p300 (18) were also recently identified as substrates for monomethylation catalyzed by the histone H3K4 methylase SET9 (25). The ways in which lysine methylation and acetylation are controlled to finely regulate NF-κB activity in normal and tumor cells, and the mechanisms that control the timing, location, intensity, residue specificity, and type of modification, are important issues for future research.
Interestingly, the histone H3K36 methylases NSD1, NSD2, and NSD3 have been implicated in cancer (26–28). NSD2 and NSD3, which share 70–75% sequence identity with NSD1, contribute substantially less than NSD1 to tumorigenesis. Our discovery that NSD1 is capable of activating NF-κB provides a potential mechanism for how this protein might play a role in tumor formation, given that constitutive activation of NF-κB is a hallmark of most cancers.
An important issue for future research is where the transcription factors are when they are modified. Our study of NF-κB methylation has established that the p65 subunit is not significantly associated with histone-modifying enzymes FBXL11 or NSD1 until it becomes activated, suggesting that this event happens only after NF-κB is released from IκB and enters the nucleus. This interesting phenomenon is consistent with our previous data showing that FBXL11 regulates NF-κB activity downstream of the IKK complex and IκB (8). The data presented here for NF-κB lead to the hypothesis that reversible methylation of inducible transcription factors may occur, in concert with histone modifications, only when these factors enter the nucleus and bind to specific promoters where the local chromatin remodeling machinery is active. Confirmation and extension of this model requires more studies in the future.
Our gene array data in MEFs suggest that FBXL11 regulates most (∼87%), but not all, of the NF-κB–dependent genes. This finding further supports the notion summarized by Perkins and Gilmore (7) that the differential posttranslational modifications of NF-κB that occur in response to different signals, such as phosphorylation, acetylation, and the methylation reported here, may specify different promoter-specific responses to activated NF-κB, leading to signal-specific downstream effects that are responsible for the differences in NF-κB function that are found in response to different regulators, in different cell types, and in different specific contexts (6).
In addition, our MS data revealed monomethylation of at least one residue other than lysine (N202, S203, or S205; Fig. 2B), and we have not yet analyzed the methylation status of all of the lysine residues of p65 by MS. In addition to the methylation of K314/K315 discussed above (25), a recent report shows that K37 of p65 is also monomethylated by SET9 (29). Therefore, the possibility of additional methylation sites on p65 should be investigated in the future, as should the possibility that the p50 subunit may also be methylated.
Our observation that FBXL11, a histone H3K36 demethylase, is induced on NF-κB activation in response to cytokines, forming a feedback loop to help regulate NF-κB activity (Fig. 7A), connects well to the recent observations of De Santa et al. (30), who found that Jmjd3, a histone H3K27 demethylase, is also induced by NF-κB in macrophages in response to bacterial products and inflammatory cytokines, helping to control the downstream consequences of NF-κB activation in a feed-forward mechanism. Together, these two observations reveal the existence of an intimate link between histone H3 demethylases and the fine-tuned regulation of NF-κB.
Materials and Methods
Transfections and Luciferase Assays.
Constructs were transfected into cell lines by using the Lipofectamine and PLUS Reagents (Invitrogen). To establish stable pools, cells were cotransfected with a plasmid encoding a puromycin resistance gene, and selected in 1 μg/mL of puromycin 48 h later. For NF-κB luciferase assays, the κB-luciferase construct p5XIP10 κB (4), was transfected transiently into the cells and luciferase activity was assayed 48 h later. A β-galactosidase construct was cotransfected to normalize for transfection efficiency. Transfections and luciferase assays were carried out essentially as previously described (4).
Western and Northern Analyses.
Cells were cultured to ∼95% confluency, and samples were collected and assayed by the Western method as described by Lu et al. (4). Different antibodies were used to detect the target proteins. Northern analyses were carried out as described before (4). Cells cultured to about 80–90% confluency were transfected with different p65 constructs; 48 h later, samples were collected, total RNAs were purified, and 20 μg of each was loaded into each well. Human cDNA probe for E-selectin was made by reverse PCR from total RNA of 293C6 cells by using the SuperScript III First-Strand Synthesis System (Invitrogen). The cDNA fragments were cloned into pCR8/GW/TOPOTA (Invitrogen) and the fragments were sequenced, excised with EcoRI, and used as probes. For other Northern probes used with samples from MEFs, mouse mRNA was used to make the probes, following the procedure described above.
Liquid Chromatography–Tandem MS (LC-MS/MS) Analysis.
Analysis of the proteolytic digest was performed using a LTQ Orbitrap XL linear ion trap mass spectrometer (Thermo Fisher Scientific) coupled with Ultimate 3000 HPLC system (Dionex). The proteolytic digests were injected onto a reverse-phase C18 column (0.075 × 150 mm, Dionex) equilibrated with 0.1% formic acid/4% acetonitrile (vol/vol). A linear gradient of acetonitrile from 4% to 40% in water in the presence of 0.1% formic acid over a period of 45 min was used, at a flow rate of 300 nL/min. The spectra were acquired by data-dependent methods, consisting of a full scan (m/z 400–2,000) and then tandem MS on the five most abundant precursor ions. The previously selected precursor ions were scanned once during 30 s and then were excluded for 30 s. The obtained data were submitted to Mascot software (Matrix Science) to search for methylated, dimethylated, and trimethylated lysine residues in the p65 protein. The tandem mass spectra of the possibly modified peptides were further interpreted manually.
Additional experimental details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health grants CA95851 (to G.R.S.) and CA60730 (to A.V.G.). We acknowledge Cleveland Clinic cores for synthetic peptides, antibodies, and microarray analyses. We also appreciate the support of the Center for Proteomics and Bioinformatics at Case Western Reserve University.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0912493107/DCSupplemental.
References
- 1.Karin M, Yamamoto Y, Wang QM. The IKK NF-κB system: A treasure trove for drug development. Nat Rev Drug Discov. 2004;3:17–26. doi: 10.1038/nrd1279. [DOI] [PubMed] [Google Scholar]
- 2.Karin M. Nuclear factor-κB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin AS. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-κB. J Clin Invest. 2001;107:241–246. doi: 10.1172/JCI11991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu T, Sathe SS, Swiatkowski SM, Hampole CV, Stark GR. Secretion of cytokines and growth factors as a general cause of constitutive NFκB activation in cancer. Oncogene. 2004;23:2138–2145. doi: 10.1038/sj.onc.1207332. [DOI] [PubMed] [Google Scholar]
- 5.Lu T, et al. Secreted transforming growth factor β2 activates NF-κB, blocks apoptosis, and is essential for the survival of some tumor cells. Proc Natl Acad Sci USA. 2004;101:7112–7117. doi: 10.1073/pnas.0402048101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perkins ND, Gilmore TD. Good cop, bad cop: The different faces of NF-κB. Cell Death Differ. 2006;13:759–772. doi: 10.1038/sj.cdd.4401838. [DOI] [PubMed] [Google Scholar]
- 7.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor κB pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 8.Lu T, et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFκB. Proc Natl Acad Sci USA. 2009;106:16339–16344. doi: 10.1073/pnas.0908560106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 10.Sathe SS, et al. Mutant human cells with constitutive activation of NF-κB. Proc Natl Acad Sci USA. 2004;101:192–197. doi: 10.1073/pnas.0306812101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ouban A, Muraca P, Yeatman T, Coppola D. Expression and distribution of insulin-like growth factor-1 receptor in human carcinomas. Hum Pathol. 2003;34:803–808. doi: 10.1016/s0046-8177(03)00291-0. [DOI] [PubMed] [Google Scholar]
- 12.Jarzynka MJ, et al. ELMO1 and Dock180, a bipartite Rac1 guanine nucleotide exchange factor, promote human glioma cell invasion. Cancer Res. 2007;67:7203–7211. doi: 10.1158/0008-5472.CAN-07-0473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hooi CF, et al. ST7-mediated suppression of tumorigenicity of prostate cancer cells is characterized by remodeling of the extracellular matrix. Oncogene. 2006;25:3924–3933. doi: 10.1038/sj.onc.1209418. [DOI] [PubMed] [Google Scholar]
- 14.Huang J, et al. p53 Is regulated by the lysine demethylase LSD1. Nature. 2007;449:105–108. doi: 10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 15.Neumann M, Naumann M. Beyond IκBs: Alternative regulation of NF-κB activity. FASEB J. 2007;21:2642–2654. doi: 10.1096/fj.06-7615rev. [DOI] [PubMed] [Google Scholar]
- 16.Chen LF, Greene WC. Shaping the nuclear action of NF-κB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 17.Quivy V, Van Lint C. Regulation at multiple levels of NF-κB-mediated transactivation by protein acetylation. Biochem Pharmacol. 2004;56:1221–1229. doi: 10.1016/j.bcp.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 18.Buerki C, et al. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-κB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 20.Chen FE, Ghosh G. Regulation of DNA binding by Rel/NF-κB transcription factors: Structural views. Oncogene. 1999;18:6845–6852. doi: 10.1038/sj.onc.1203224. [DOI] [PubMed] [Google Scholar]
- 21.Chen LF, Mu Y, Greene WC. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 2002;21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 23.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 24.Ivanov GS, et al. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol Cell Biol. 2007;27:6756–6769. doi: 10.1128/MCB.00460-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang XD, et al. Negative regulation of NF-κB action by Set9-mediated lysine methylation of the RelA subunit. EMBO J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rayasam GV, et al. NSD1 is essential for early post-implantation development and has a catalytically active SET domain. EMBO J. 2003;22:3153–3163. doi: 10.1093/emboj/cdg288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider R, Bannister AJ, Kouzarides T. Unsafe SETs: Histone lysine methyltransferases and cancer. Trends Biochem Sci. 2002;27:396–402. doi: 10.1016/s0968-0004(02)02141-2. [DOI] [PubMed] [Google Scholar]
- 28.Strahl BD, et al. Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol. 2002;22:1298–1306. doi: 10.1128/mcb.22.5.1298-1306.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ea CK, Baltimore D. Regulation of NF-κB activity through lysine monomethylation of p65. Proc Natl Acad Sci USA. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Santa F, et al. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.