Abstract
Transacting siRNA (tasiRNA) biogenesis in Arabidopsis is initiated by microRNA (miRNA) –guided cleavage of primary transcripts. In the case of TAS3 tasiRNA formation, ARGONAUTE7 (AGO7)–miR390 complexes interact with primary transcripts at two sites, resulting in recruitment of RNA-DEPENDENT RNA POLYMERASE6 for dsRNA biosynthesis. An extensive screen for Arabidopsis mutants with specific defects in TAS3 tasiRNA biogenesis or function was done. This yielded numerous ago7 mutants, one dcl4 mutant, and two mutants that accumulated low levels of miR390. A direct genome sequencing-based approach to both map and rapidly identify one of the latter mutant alleles was developed. This revealed a G-to-A point mutation (mir390a-1) that was calculated to stabilize a relatively nonpaired region near the base of the MIR390a foldback, resulting in misprocessing of the miR390/miR390* duplex and subsequent reduced TAS3 tasiRNA levels. Directed substitutions, as well as analysis of variation at paralogous miR390-generating loci (MIR390a and MIR390b), indicated that base pair properties and nucleotide identity within a region 4–6 bases below the miR390/miR390* duplex region contributed to the efficiency and accuracy of precursor processing.
Keywords: high-throughput sequencing, miRNA, trans-acting siRNA
Small RNAs, including microRNA (miRNA), several classes of endogenous small interfering RNA (siRNA), and Piwi-associated RNA (piRNA), direct silencing activities that shape transcriptomes and proteomes of eukaryotic organisms. miRNAs arise from transcripts containing self-complementary foldback structures that are initially processed to form 21–22nt miRNA/miRNA* duplexes. In animals, primary transcripts with miRNA foldbacks (pri-miRNA) are processed first by the Microprocessor complex, which contains the RNase III-type protein Drosha and its cofactor Pasha (also known as DGCR8 in humans), then by Dicer, with partners that include the dsRNA-binding domain protein Loquacious (1). Plants orchestrate both pri-miRNA and pre-miRNA processing with the same (or very similar) complex, which includes the RNase-III like enzyme DICER-LIKE1 (DCL1) as the catalytic component (2–5). DCL1 interacts with the dsRNA binding protein HYPONASTIC LEAVES1 (HYL1) and the zinc-finger protein SERRATE (SE), both of which promote efficient and accurate miRNA biogenesis (2, 6–8).
The transacting siRNA (tasiRNA) class represents a specialized type of amplification-dependent siRNA (9, 10). Primary tasiRNA-generating transcripts are first processed by miRNA-guided cleavage (11, 12). Either the 3′ (TAS1, TAS2 and TAS4 families) or 5′ (TAS3 family) cleavage product is stabilized and converted to dsRNA by RNA-DEPENDENT RNA POLYMERASE6 (RDR6) (9, 10, 12, 13). Phased, 21-nt siRNAs are generated in register with the miRNA-guided cleavage site through sequential processing by DCL4. Routing of TAS3 precursor RNA requires two miRNA-guided events, both of which involve AGO7-miR390 complexes (14, 15). Interaction of AGO7-miR390 at a 3′ proximal target site results in primary transcript cleavage, and sets the register for phased siRNA generation. The 3′ cleavage function of AGO7-miR390 is generic, as any of several heterologous miRNA working through AGO1 can substitute for AGO7-miR390 (15). A second miR390 target site at a 5′-proximal position in the processed precursor interacts with AGO7-miR390 in a noncleavage mode (14, 16).
Here, we identify several mutants with defects in TAS3 tasiRNA biogenesis, including those with defects in the MIR390a-derived foldback, revealing a key role for structures near the base of the foldback for efficient and accurate miR390 processing.
Results and Discussion
Screen for TAS3-Based syn-tasiRNA–Deficient Mutants.
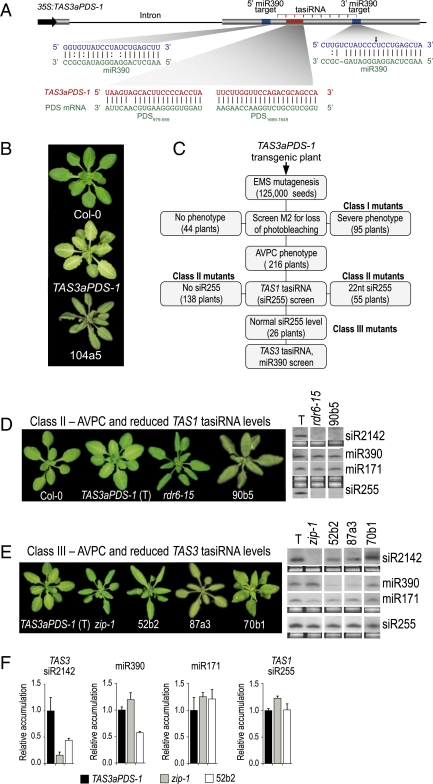
TAS3a-based synthetic (syn)-tasiRNAs with complementarity to the PDS mRNA provide a visual readout for tasiRNA activity in transgenic Arabidopsis (15). The 35S:TAS3aPDS-1 construct yields tandem syn-tasiRNAs from the 5′ D7[+] and 5′ D8[+] positions in place of siRNA2141 and siRNA2142, also known as tasi-ARFs (11, 12). These repress mRNAs encoding several AUXIN RESPONSE FACTORS, including ARF3 and ARF4, regulation of which is essential for proper developmental timing and lateral organ development (15, 17, 18) (Fig. 1A). In wild-type (Col-0) plants expressing 35S:TAS3aPDS-1, photobleaching emanates from the midrib and major veins, with the phenotype most prominent when viewed from the adaxial side of leaves (Fig. 1B) (15). Syn-tasiRNA accumulation and photobleaching are suppressed in plants containing loss-of-function rdr6-15, dcl4-2 and zip-1 (AGO7-defective) mutations (15).
Fig. 1.
Syn-tasiRNA strategy, mutant screen, and characterization of class II and III mutants (A) Organization of syn-tasiRNA construct 35S:TAS3aPDS-1. The miR390/miR390 target and syn-tasiRNA/PDS target sequences are shown in the expanded diagrams. (B) Photobleached phenotype of 35S:TAS3aPDS-1-transformed Col-0 and a class II mutant (104a5) recovered from the screen are shown next to nontransformed Col-0 (wt). (C) Flowchart of the screen for TAS3 tasiRNA-defective mutants using 35S:TAS3aPDS-1-transformed plants. (D and E) Representative images and select small RNA blot profiles from parental 35S:TAS3aPDS-1 transformed Col-0 (T) plants, reference mutants (rdr6-15 and zip1), and class II and class III mutants. Small RNA data using each radiolabeled probe in each panel were from the same blot. All small RNA, except for TAS3 siR2142 in the 70b1 mutant, comigrated with the 21-nt standard. (F) Mean (n = 3) relative level ± SD of TAS3 siR2142, miR390, miR171, and TAS1 siR255 (TAS3aPDS-1 = 1.0) in several mutants.
A screen for mutants with TAS3 tasiRNA specific defects was done using the syn-tasiRNA line. Besides loss of photobleaching, mutants with TAS3-specific defects were predicted to have 1) low or no syn-tasiRNA and endogenous tasi-ARF (siRNA2142), 2) normal levels of TAS1 tasiRNA (siR255), 3) normal levels of miRNA, such as miR171, that do not function in the TAS3 pathway, and 4) an accelerated vegetative phase change (AVPC) phenotype, which is associated with loss of TAS3 tasiRNA (15, 17–20). TAS3 pathway-specific mutants were not expected to have severe developmental defects, as would be expected for general loss-of-miRNA function mutants (21, 22). The AVPC phenotype is characterized by downward-curled rosette leaves, giving the appearance of a narrow leaf phenotype, and early development of abaxial trichomes (19). In all, 200 pools of seedlings from the M2 generation were screened. A total of 355 hygromycin-resistant (transgene-containing) individuals with a reduced-photobleaching phenotype were recovered (Fig. 1C).
Of the mutants, 95 had relatively strong or severe vegetative defects, and 44 had no visible morphological defects. Most of the severely affected plants (class I) were dwarfed, had serrated leaves, and resembled known miRNA-debilitated mutants, such as those with hen1, hyl1, se, or strong hypomorphic ago1 alleles (2, 8, 23) (Fig. S1). Among seven class I mutants analyzed, each had reduced levels of miR171 and TAS1 siR255, indicating that they were generally deficient in miRNA accumulation or activity (Fig. S1). Class I mutants were not analyzed further.
As exemplified by mutant 104a5, 216 mutants had an AVPC phenotype (Fig. 1 B and C). Approximately 64% (138) or 25% (55) of these mutants lacked the TAS1 tasiRNA siR255 altogether, or produced siR255-related small RNA that migrated during electrophoresis as a 22-nt RNA, respectively, indicating that they possessed general (TAS3 and TAS1) tasiRNA defects (Fig. 1 C and D). These 193 plants with general tasiRNA defects comprised class II mutants. Complementation analyses of a random sampling of 15 mutants, including 90b5 (Fig. 1D), revealed that the complete loss-of-siR255 subgroup within class II was dominated by rdr6 (10) and sgs3 (5) mutants (Table S1). Fourteen mutants produced 21-to-22 nt, size-shifted TAS1 siR255, and 14 of 14 of these possessed dcl4 defects based on complementation tests (Table S1). Loss of DCL4 is known to result in 22-nt size-shifted tasiRNA, because of the surrogate activity of DCL2 (24–26).
Only 12% (26) of plants possessed an AVPC phenotype, normal levels of 21-nt siR255, and normal levels of miR171 (Fig. 1C). Nearly all of these, which were designated as class III mutants, possessed low or undetectable levels of TAS3 siR2142. Among the class III mutants, complementation analysis revealed 23 independent ago7 mutants, 14 of which were subjected to ago7 allele sequencing. Most of the ago7 alleles contained substitutions affecting the PIWI domain, whereas single mutants with mid-domain or N-terminal domain substitutions were identified (Table S1). A TAS3-specific dcl4 mutant (70b1), in which TAS3 siR2142-related small RNA, but not TAS1 siR255, was shifted to 22 nt, was recovered, although minor reductions of both TAS1 and TAS3 tasiRNA were noted (Fig. 1E and Fig. S2). The 70b1 dcl4 allele contained a nonconserved Gly-to-Arg substitution affecting a region between the PAZ domain and first RNaseIII domain (Table S1).
Two recessive mutants, 52b2 and 87a3, could not be assigned to any of the complementation groups tested through crosses to zip1, rdr6-15, sgs3-11, and dcl4-2. These mutants had similar, moderate AVPC phenotypes (Fig. 1E). The 52b2 mutant accumulated significantly reduced levels siR2142 (44.9% compared with Col-0; P < 0.0028), but normal levels of miR171 and TAS1 siR255 (Fig. 1 E and F). Interestingly, both 52b2 and 87a3 had low levels of miR390 (Fig. 1E), with quantitative blot assays revealing a significant difference (P < 0.0001) between 52b2 and Col-0 plants (Fig. 1F).
Identification of mir390a-1 by Pooled Genome Sequencing.
In principle, direct genome sequencing of a mutant genome using high-throughput sequencing (HTS) technology can identify sites of mutation. However, each EMS-mutagenized genome can possess hundreds or thousands of changes in addition to the causal mutation. We developed a strategy for direct sequencing of a bulk segregant population of genomes for identification of the causative 52b2 mutation. A segregating F2 population from a cross between 52b2 (Col-0 background) and the polymorphic accession Ler was prepared, and 93 homozygous plants with both AVPC and low photobleaching phenotypes were identified. DNA from the 93 plants was pooled and subjected to high-throughput sequencing, which provided 221,000,000 36-base reads.
A pipeline, Mapping and Assembly with Short Sequences (MASS; Fig. 2A), was devised to map and assemble sequence data. Approximately 143,000 SNPs (27) were used to identify and quantify Col-0- and Ler-specific reads from repeat-filtered sequences. The ratios of summed Col-0 SNPs/summed Ler SNPs were calculated in 100,000 base windows (20,000 base scroll) across the Arabidopsis genome. A major peak of enriched Col-0 SNPs was identified on chromosome II (Fig. 2B). In addition, several minor peaks of Col-0-enriched SNPs were identified around pericentromeric regions. The basis for these minor peaks was not determined conclusively, although the peaks may reflect miscalled SNPs that do not exist in Ler. A 1.52-Mb region encompassing the major Col-0–enriched peak was assembled with the program Mapping and Assembly with Quality (MAQ) (28) using all high-quality sequencing reads, revealing five G-to-A or C-to-T changes that were consistent with EMS-induced mutation (Table S2). One mutation affected the sequence at the base of the foldback from MIR390a (Fig. 2 C and D). Portions of the 87a3 mutant genome were sequenced manually across the loci corresponding to the G-to-A or C-to-T positions in 52b2, revealing exactly the same mir390 mutation but wild-type Col-0 sequences at each of the other positions. Thus, 52b2 and 87a3 were independent mutants containing the same mir390a allele (mir390a-1). A genomic fragment containing wild-type MIR390a was introduced into 52b2 mutant plants. This restored photobleaching to the 52b2 mutant line and partially suppressed the leaf curling phenotype (Fig. 2E), confirming that the mir390a-1 mutation was causal.
Fig. 2.
High-throughput sequencing of the 52b2 mutant genome and identification of the causal mutation (A) Flowchart of sequence-based mapping and mutation identification using a bulk segregant population. (B) Scrolling window plot of ratios (Col-0/Ler) of total SNPs detected in the bulk segregant sequence dataset. (C) A 152-Mb interval spanning the major Col-0–enriched region of chromosome 2 is illustrated. Each nucleotide position that deviates from the reference genome position is indicated by an arrow. The complete or partial sequences of mapped reads from a 50-base segment (chromosome 2 16069100–16069149) from the MIR390a locus is shown in the expanded portion. (D) Restoration of photobleaching phenotype in 52b2 mutant plants by transformation with a wild-type MIR390a transgene. (E). Foldback sequence and predicted structure from wild-type MIR390a and mutant 52b2 mir390a-1 alleles. The position corresponding to the mutation is indicated by arrows. For comparative purposes, four foldback domains were assigned, as indicated by the brackets.
The sequencing-based approach that identified the mir390a-1 mutation should be broadly applicable to identification of other markerless (e.g., EMS-induced) mutations. The major benefit of the approach is the simultaneous mapping and sequencing at a genome-wide level. The ability to score all known polymorphisms in individuals from the mapping population affords tremendous marker density, and the MASS pipeline provides a straightforward route to identification of a small number of candidate genes within a relatively small interval of 1–2 Mb. Similar high-throughput sequencing-based approaches for identification of casual mutations were presented recently (29–31).
The mir390a-1 mutation affects position 94 (G94-to-A94 substitution) from the 5′ end of the predicted foldback. Using both mFOLD and RNAFold (32, 33), G94 in the wild-type sequence was predicted to be nonpaired, or to base pair with U12 with low probability, in the “C region” of the foldback below the miR390/miR390* segment (Fig. 2D). In mir390a-1, A94 was predicted to base pair with high probability to U12 (Fig. 2D). Due to the distance of the mutation away from the miR390/miR390* segment and the variability of this position between MIR390a and MIR390b (discussed below), this position may have been overlooked in a directed mutagenesis approach to identify precursor processing determinants.
Defective Processing of the mir390a-1 Foldback.
MIR390a, mir390a-1, and the paralogous MIR390b loci specify the identical miR390 sequence, but the foldbacks differ in sequence and predicted base-pair structure. The C region from the MIR390b foldback contains more predicted base-paired positions at and adjacent to C112, which occupies the spatially equivalent position as G94 in MIR390a (Fig. 3 A and B). The effects of the mir390a-1 A94 mutation, as well as the differences in the C region between MIR390a and MIR390b foldbacks, on miR390 biogenesis and TAS3 tasiRNA formation, were tested in a transient expression assay using Nicotiana benthamiana plants (34).
Fig. 3.
Foldback processing and TAS3 tasiRNA-initiation activity of mir390a-1 in transient assays. (A) Accumulation of miR390 and TAS3 syn-tasiRNA in N. benthamiana transient assays. One of three independent replicates is shown. Mean (n = 3) relative miR390 (blue) and TAS3a (red) syn-tasiRNA levels ± SD (lane 2 and lane 5 = 1.0) are shown. Syn-tasiRNA levels were measured only in assays containing 35S:TAS3aPDS-2 (lanes 5–8). U6 RNA is shown as a loading control. (B) Analysis of miR390 and miR390* sequences after transient expression of MIR390a, mir390a-1, and MIR390b. Proportions of reads containing specific sequences are plotted as stacked bars based on size (color coded), 5′ position and 3′ position, with end positions aligned to the respective sequences shown in the foldbacks. miR390 and related sequences are plotted upward, and miR390* sequences are plotted downward. (C) Shannon's entropy (H) for 5′ end (x axis), 3′ end (y axis) and size (gray scale) of small RNA populations shown in (B). High H values reflect high information content, which correlates with variability.
35S:MIR390a, 35S:mir390a-1, and 35S:MIR390b were expressed individually to analyze miR390 biogenesis and accumulation, or coexpressed with 35S:TAS3aPDS-2 (syn-tasiRNA) and 35S:HA-AGO7 to test for TAS3 tasiRNA initiation activity. Compared with 35S:MIR390a, 35S:mir390a-1 yielded miR390 at 28.3% (P < 3.02 × 10−5) or 28.6% (P < 0.002) when expressed individually or with the other TAS3 tasiRNA components, respectively (Fig. 3A), which was consistent with the low levels of TAS3 tasiRNA detected in the 52b2 mutant plants (Fig. 1 E and F). Interestingly, 35S:MIR390b also yielded low levels of miR390 when expressed individually (17.0%, P < 9.11 × 10−6) or with TAS3 tasiRNA components (19.8%, P < 0.0019)(Fig. 3A). In addition, the functional amounts of miR390, as reflected by the levels of TAS3-based syn-tasiRNA, were significantly lower using 35S:mir390a-1 (21.9%, P < 0.0072) and 35S:MIR390b (33.5%, P < 0.013), compared with using 35S:MIR390a (Fig. 3A). These data suggest that processing of the mir390a-1 and MIR390b foldbacks occurs inefficiently.
To analyze processing accuracy of MIR390a, mir390a-1, and MIR390b foldbacks, small RNA libraries from triplicate samples were subjected to high-throughput sequencing analysis after transient expression in N. benthamiana. Reads were first normalized based on library size and spike-in standards (35). Reads from within 29-nt windows, centered around the middle of the annotated miR390 or miR390* sequences, were analyzed for size, 5′ position, and 3′ position. The information content of each dataset was used to calculate Shannon's entropy (H) (36, 37), providing measures of small RNA uniformity or processing accuracy at both ends of each sequence (Fig. 3 B and C; SI Methods). MIR390a yielded predominantly 21-nt, canonical miR390 with highly uniform 5′ and 3′ ends, and moderately heterogeneous 20–21 nt miR390* sequences with uniform 5′ ends but with 3′ ends from two major positions (Fig. 3B, Fig. S3 and Table S3). In contrast, 35S:mir390a-1 yielded 5′-, 3′-, and size-heterogeneous miR390 and miR390* sequences, with only 45.6% ± 22.6% of miR390-related sequences containing accurately processed 5′ and 3′ ends (Fig. 3B and Fig. S3). This was reflected in high H values for each 35S:mir390a–derived miR390 and miR390* parameter (Fig. 3C). MIR390b yielded sequences with intermediate processing accuracy. Both ends of miR390, and the 5′ end of miR390*, exhibited more heterogeneity than the comparable ends of sequences from MIR390a (Fig. 3 B and C). Combined with the syn-tasiRNA biogenesis data (Fig. 3A), these experimental findings indicate that the mir390a-1 mutation affects both processing accuracy and efficiency, resulting in low levels of functional miR390. The findings also indicate that MIR390b possesses the properties of a low-efficiency mutant allele. Natural variation affecting foldback structure and miRNA biogenesis has been shown previously in plants and animals (38, 39).
Mutational Analysis of the MIR390a Foldback.
The G-to-A substitution in the mir390a-1 mutant could conceivably debilitate processing because of a change in foldback base pairing, loss of a base determinant, or both. Computational analysis of predicted foldback variants suggested that the mir390a-1 structure possessed a higher probability of base pairing between U12 and A94, compared with the probability of pairing between U12 and G94 in the wild-type foldback (Fig. 4A). This was reflected in a lower calculated entropy at both positions in the mir390a-1 foldback (Fig. 4A) (36, 37). The MIR390b predicted foldback, with even more extensive base pairing, yielded lower calculated positional entropies at nearly all bases in region C (Fig. 4A) (36, 37). Seven 35S:MIR390a mutants with substitutions at either position 94 and/or position 12 were constructed (Table S4). Including mir390a-1, the series resulted in foldbacks containing all possible single-base substitutions at both positions, and two combinations of dual-base substitutions (Fig. 4A). In addition, the sequences comprising MIR390a region C were substituted for the approximate equivalent sequences from MIR390b. Predicted foldback structures, positional entropies and miR390 biogenesis levels in a transient assay were determined.
Fig. 4.
Directed mutational analysis of MIR390a foldback (A) Predicted base pair structure of MIR390a and MIR390b foldbacks (Left). Enlarged region corresponds to the bracketed region for 11 mutant or variant foldbacks (Right). Positions 12 (U in MIR390a) and 84 (G in MIR390a) are indicated by arrows. Positional entropy values range from 0 (red) to 1.6 (purple) for all MIR390a-based foldbacks and 1.8 (purple) for MIR390b. Folding and Shannon's entropy values reflect the probability of variant base-pair states and were calculated using RNAfold. (B) Blot assays for miR390 derived from expression of 35S:MIR390a (lane 1), 35S:MIR390b (lane 2), 35S:mir390a-1 (lane 3), and each directed mutant construct (lanes 4–11) are shown, along with a negative control sample expressing empty vector (V). One of six independent replicates is shown. U6 RNA is shown as a loading control. (C) Mean relative miR390 levels ± SD (35S:MIR390a = 1.0).
Each substitution at position 94 (mir390a-1, mir390a-94U, and mir390a-94C) resulted in significantly (P < 0.003) lower miR390 levels compared with wild-type MIR390a, although the mir390a-94U and mir390a-94C defects were only modest (Fig. 4B). Unlike mir390a-1, mir390a-94U, and mir390a-94C mutations were not predicted to base pair with U12 (Fig. 4A). These data generally reinforce a role for G94, as either a single base-determinant or a high-entropy, weak–base-pair partner with U12. Among the position 12 substitutions, mir390-12C was significantly (P < 8.8 10−9) debilitated for miR390 biogenesis and was predicted to form a low-entropy base pair with G94. mir390a-12A retained both A12 and G94 in a predicted nonpaired configuration and yielded wild-type levels of miR390 (Fig. 4). These position 12 mutants lend support to the idea that a nonpaired or weakly paired G94 contributes to miR390 biogenesis. In contrast, mir390a-12G was predicted to adopt a fold involving low-entropy, highly base-paired 12G and G94 positions, but led to wild-type levels of miR390 (Fig. 4). However, the mir390a-12G local stem structure was predicted to include novel, high-entropy asymmetric bulges that differed from the comparable positions from MIR390a (Fig. 4A).
Among the double mutants, mir390a-12C94A contained the A94 mutation from mir390a-1 and a base-pair–disrupting change at position 12 (Fig. 4A). This mutant was highly debilitated for miR390 biogenesis, indicating that the mir390a-1 defect (A94) was not due solely to the increased base pair configuration between positions 12 and 94 (Fig. 4). Interestingly, the mir390a-12G94U mutant foldback, which contained the G and U positions from wild-type MIR390a reversed, yielded nearly wild-type levels of miR390 (Fig. 4).
Substitution of the base of the MIR390a stem with that from MIR390b led to significant (P < 6.73 10−6) debilitation of miR390 formation (Fig. 4 B and C). The mutant foldback region C was predicted to contain the same low-entropy, highly base-paired configuration as predicted for MIR390b foldback (Fig. 4A). However, the functional significance of the MIR390b locus remains unclear.
The relatively high diversity of sequences, sizes, and secondary structures of plant MIRNA foldbacks (40) means that processing determinants are not particularly obvious. Based on in vitro processing assays with MIR167b foldbacks, DCL1 is sufficient to catalyze ATP-dependent pri- and premiRNA transcript processing, although only a minority of such products possess accurate 5′ and 3′ ends (6). The dsRNA binding motifs of DCL1 may provide a basal function for foldback recognition. However, inclusion of both SE and HYL1 in these reactions increases the rate and accuracy of processing (6, 41). This may indicate that SE and HYL1 function as accessory factors that position DCL1 accurately on substrates through interaction with one or more foldback structural features. We propose that the inaccurate and inefficient processing of the mir390a-1 foldback is due to loss of interaction with key factors promoting miRNA biogenesis. In particular, it is attractive to consider G94 in a flexible, high-entropy context as a recognition determinant for HYL1 and/or SE. Both SE and HYL1 promote miR390 accumulation in vivo (23, 42). Importantly, the effects of the mir390a-1 mutation on foldback processing in transient assays are very similar to the effects of MIR167b foldback processing in the absence of SE and HYL1 in vitro (6). It seems unlikely, however, that foldback position G94 is the sole determinant for such interactions, as there is high sequence and structural diversity at this position among foldbacks from conserved MIRNA families. By analogy with the Drosha-Pasha/DGCR8 complex interacting with the base of animal foldbacks (43), features defining the junction between the base of the stem and the nonpaired region outside of the stem may also interact with the DCL1-HYL1-SE complex for positioning of the first set of cuts at the proximal end of the miRNA/miRNA* duplex. Indeed, Mateos et al. (44), Song et al. (45), and Werner et al. (46) revealed a key role for a single-stranded/base-paired stem junction ∼15 nucleotides from the miRNA/miRNA* duplex in accurate processing of many Arabidopsis miRNAs.
Methods
References for rdr6-15, dcl4-2, sgs3-11, hen1-1, hyl1-2, se-2, hst-15, dcl1-7, and zip-1 alleles were described (12). Detailed descriptions, protocols, and references for transgenic plant materials, RNA blot assays, transient expression assays in N. benthamiana, Arabidopsis mutagenesis and genetic screen, sequencing and analysis of small RNA populations, and the MASS pipeline are provided in SI Methods.
Supplementary Material
Acknowledgments
We thank Sarah Dvorak for assistance with the genetics screen and technical assistance. We also thank Detlef Weigel, Korbinian Schneeberger, and Richard Clark for helpful discussion, genomic sequencing test data sets, and SNP data; and we thank Nina Fedoroff, Mike Axtell, Detlef Weigel, and Javier Palatnik for sharing data and manuscripts prior to publication. This work was supported by grants from the National Science Foundation (MCB-0618433), National Institutes of Health (AI43288) and United States Department of Agriculture–National Research Initiative (2006-35301-17420).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0913203107/DCSupplemental.
References
- 1.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kurihara Y, Takashi Y, Watanabe Y. The interaction between DCL1 and HYL1 is important for efficient and precise processing of pri-miRNA in plant microRNA biogenesis. RNA. 2006;12:206–212. doi: 10.1261/rna.2146906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP. MicroRNAs in plants. Genes Dev. 2002;16:1616–1626. doi: 10.1101/gad.1004402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Golden TA, et al. SHORT INTEGUMENTS1/SUSPENSOR1/CARPEL FACTORY, a Dicer homolog, is a maternal effect gene required for embryo development in Arabidopsis. Plant Physiol. 2002;130:808–822. doi: 10.1104/pp.003491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park W, Li J, Song R, Messing J, Chen X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr Biol. 2002;12:1484–1495. doi: 10.1016/s0960-9822(02)01017-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dong Z, Han MH, Fedoroff N. The RNA-binding proteins HYL1 and SE promote accurate in vitro processing of pri-miRNA by DCL1. Proc Natl Acad Sci USA. 2008;105:9970–9975. doi: 10.1073/pnas.0803356105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiraguri A, et al. Specific interactions between Dicer-like proteins and HYL1/DRB-family dsRNA-binding proteins in Arabidopsis thaliana. Plant Mol Biol. 2005;57:173–188. doi: 10.1007/s11103-004-6853-5. [DOI] [PubMed] [Google Scholar]
- 8.Yang L, Liu Z, Lu F, Dong A, Huang H. SERRATE is a novel nuclear regulator in primary microRNA processing in Arabidopsis. Plant J. 2006;47:841–850. doi: 10.1111/j.1365-313X.2006.02835.x. [DOI] [PubMed] [Google Scholar]
- 9.Vazquez F, et al. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 10.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- 15.Montgomery TA, et al. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 16.Howell MD, et al. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fahlgren N, et al. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Curr Biol. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- 18.Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Curr Biol. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- 19.Poethig RS. Phase change and the regulation of developmental timing in plants. Science. 2003;301:334–336. doi: 10.1126/science.1085328. [DOI] [PubMed] [Google Scholar]
- 20.Adenot X, et al. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Curr Biol. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 21.Clarke JH, Tack D, Findlay K, Van Montagu M, Van Lijsebettens M. The SERRATE locus controls the formation of the early juvenile leaves and phase length in Arabidopsis. Plant J. 1999;20:493–501. doi: 10.1046/j.1365-313x.1999.00623.x. [DOI] [PubMed] [Google Scholar]
- 22.Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–5243. doi: 10.1242/dev.126.23.5231. [DOI] [PubMed] [Google Scholar]
- 23.Montgomery TA, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 25.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 26.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Clark RM, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 28.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sarin S, Prabhu S, O'Meara MM, Pe'er I, Hobert O. Caenorhabditis elegans mutant allele identification by whole-genome sequencing. Nat Methods. 2008;5:865–867. doi: 10.1038/nmeth.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blumenstiel JP, et al. Identification of EMS-induced mutations in Drosophila melanogaster by whole-genome sequencing. Genetics. 2009;182:25–32. doi: 10.1534/genetics.109.101998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneeberger K, et al. SHOREmap: Simultaneous mapping and mutation identification by deep sequencing. Nat Methods. 2009;6:550–551. doi: 10.1038/nmeth0809-550. [DOI] [PubMed] [Google Scholar]
- 32.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053–2056. doi: 10.1126/science.1076311. [DOI] [PubMed] [Google Scholar]
- 35.Fahlgren N, et al. Computational and analytical framework for small RNA profiling by high-throughput sequencing. RNA. 2009;15:992–1002. doi: 10.1261/rna.1473809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shannon CE. A mathematical theory of communication. Bell Sys Tech J. 1948;27:379–423. 623–656. [Google Scholar]
- 37.Schneider TD. Information content of individual genetic sequences. J Theor Biol. 1997;189:427–441. doi: 10.1006/jtbi.1997.0540. [DOI] [PubMed] [Google Scholar]
- 38.de Meaux J, Hu JY, Tartler U, Goebel U. Structurally different alleles of the ath-MIR824 microRNA precursor are maintained at high frequency in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:8994–8999. doi: 10.1073/pnas.0803218105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun G, et al. SNPs in human miRNA genes affect biogenesis and function. RNA. 2009;15:1640–1651. doi: 10.1261/rna.1560209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Axtell MJ. Evolution of microRNAs and their targets: Are all microRNAs biologically relevant? Biochim Biophys Acta. 2008;1779:725–734. doi: 10.1016/j.bbagrm.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 41.Han MH, Goud S, Song L, Fedoroff N. The Arabidopsis double-stranded RNA-binding protein HYL1 plays a role in microRNA-mediated gene regulation. Proc Natl Acad Sci USA. 2004;101:1093–1098. doi: 10.1073/pnas.0307969100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chitwood DH, et al. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 44.Mateos JL, Bologna NG, Chorostecki U, Palatnik J. Identification of structural determinants for microRNA processing in plants by random mutagenesis of MIR172a precursor. Curr Biol. 2010 doi: 10.1016/j.cub.2009.10.072. in press. [DOI] [PubMed] [Google Scholar]
- 45.Song L, Axtell MJ, Fedoroff NV. RNA secondary structural determinants of miRNA precursor processing in Arabidopsis. Curr Biol. 2010 doi: 10.1016/j.cub.2009.10.076. in press. [DOI] [PubMed] [Google Scholar]
- 46.Werner S, Wollman H, Weigel D. Sequence determinants for accurate processing of miR172a in Arabidopsis thaliana. Curr Biol. 2010 doi: 10.1016/j.cub.2009.10.073. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.