Abstract
The dynamic behavior of proteins is critical for cellular homeostasis. However, analyzing dynamics of proteins and protein complexes in vivo has been difficult. Here we describe recombination-induced tag exchange (RITE), a genetic method that induces a permanent epitope-tag switch in the coding sequence after a hormone-induced activation of Cre recombinase. The time-controlled tag switch provides a unique ability to detect and separate old and new proteins in time and space, which opens up opportunities to investigate the dynamic behavior of proteins. We validated the technology by determining exchange of endogenous histones in chromatin by biochemical methods and by visualizing and quantifying replacement of old by new proteasomes in single cells by microscopy. RITE is widely applicable and allows probing spatiotemporal changes in protein properties by multiple methods.
Keywords: chromatin, histone, proteasome, protein dynamics, turnover
Proteins are dynamic molecules. Their abundance is controlled by synthesis and degradation and they can be subject to posttranslational processing, modification, and demodification. In addition, most proteins are very mobile and undergo interactions with multiple other protein partners (1–4). However, little is known about the dynamics of proteins within macromolecular complexes in vivo (2, 4). Studying time-dependent changes in physical properties of proteins or protein turnover requires methods to distinguish resident (old) proteins from new proteins. Current methods that do so are usually based on fluorescent reporters or differential chemical labeling. For example, fluorescence recovery after photo bleaching relies on exchange of the old bleached protein by nonbleached proteins (1, 3, 4). Alternative methods involve time-dependent changes in fluorescence, nonspecific pulse-chase labeling of proteins with labeled amino acids, or labeling with chemical dyes that specifically bind to short tags (5–7). Although suitable for detection of proteins by microscopy or mass spectrometry, a limitation of these methods is that they do not provide a handle for biochemical analysis of old and new proteins and their complexes. To solve this problem and to eliminate the requirement for chemical labels or UV light we developed recombination-induced tag exchange (RITE), a method in which a genetic epitope tag is switched by transient induction of a site-specific recombinase. As a consequence, old and newly synthesized proteins are differentially tagged, which enables monitoring of protein dynamics by multiple techniques, as illustrated here. In contrast to inducible expression strategies (8–12), differential tagging by a time-controlled site-specific protease (13), or the labeling methods described above, RITE allows parallel detection and purification of old and new proteins under physiological conditions and over long periods of time.
We used RITE to probe the stability of chromatin. Photobleaching experiments using histones tagged with fluorescent reporters suggest that chromatin is a static complex (14). However, recent work suggests that chromatin is more dynamic than previously anticipated (15). For example, ectopically induced histones can be incorporated into chromatin of nondividing yeast cells and gene activation of certain promoters is accompanied by transient loss of histones (8–12, 16). In metazoans, the histone H3 variant H3.3 can be assembled into chromatin by a replication-independent transcription-coupled process (17–19). We took advantage of RITE to determine whether endogenously expressed canonical histones undergo replication-independent exchange. RITE can also be used to visualize proteins by microscopy. To demonstrate this we applied RITE to the proteasome, a highly conserved and essential macromolecular complex critical for degradation of proteins by proteolysis (20). Using fluorescent RITE we could visualize the replacement of old by new proteasomes in the nucleus and cytoplasm of dividing cells.
Results
RITE Outline.
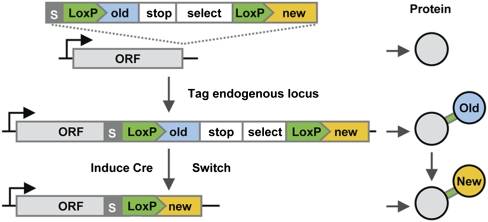
RITE can be applied by integration of a RITE cassette downstream of any gene of interest, resulting in a C-terminal tag situated between two LoxP sites with an orphan tag downstream. Upon a transient time-controlled activation of the site-specific Cre recombinase, recombination between the tandem LoxP sites results in exchange of the “old” tag by an orphan “new” tag in the coding sequence leading to an epitope-tag switch (Fig. 1). After switching, all newly synthesized mRNAs will encode for proteins containing the new epitope tag. The LoxP recombination sites are part of the coding sequence, which eliminates the need for introns and allows the tag cassette to be introduced directly at the 3′ end of any gene of interest to generate a switchable tag. As a consequence, the differentially tagged proteins are encoded by a single gene and under control of the endogenous promoter. Recombination can be induced using a constitutively expressed Cre recombinase fused to the human estrogen binding domain (EBD). This fusion protein is sequestered by heat-shock proteins and inactive (21). The nuclear activity of Cre-EBD can be rapidly activated by the addition of β-estradiol, which releases the fusion protein from heat-shock proteins (21). A major advantage of RITE is that the genetic switch is permanent. Therefore, after the switch both old and new proteins can be followed in the original cells and their descendants under any condition of interest. We applied this strategy in haploid yeast cells and integrated RITE cassettes by homologous recombination at endogenous gene loci.
Fig. 1.
Outline of recombination-induced tag exchange (RITE). RITE cassettes contain two epitope tags (old and new), the first of which is in between two LoxP sites. Integration of a RITE cassette downstream of an ORF (ORF) results in a protein tagged with an old tag (blue). The old tag is preceded by an invariant flexible spacer (S) and a short peptide encoded by the LoxP sequence (LoxP) and is followed by a transcriptional terminator (stop) and a selectable marker (select). Upon induction of Cre recombinase, site-specific recombination between the tandem LoxP sites in the genome results in loss of the old tag and fusion of the ORF to the new tag. After the switch, newly synthesized proteins will contain the new tag (yellow), whereas existing proteins will contain the old tag. Old and new proteins are expressed from the same gene by the native promoter.
Application of RITE to Histone H3.
First RITE was applied to histone H3 to investigate the stability of histones within chromatin. One of the two histone H3 genes was tagged with a RITE cassette containing two small epitope tags, HA and T7 (H3-HA→T7) (Fig. 2A). The second histone H3 gene was deleted. As a consequence, in this strain all histone H3 proteins were tagged (Fig. 2A). Yeast cells expressing the tagged histones are viable (Fig. 2B). Because histone H3 is essential, this demonstrates that the tagged H3 proteins are functional. After addition of the hormone β-estradiol, which has no detectable effect on growth or transcription (22), most of the cells had undergone recombination within 2 h (Fig. 2C). To confirm that the genetic switch at the DNA level yields differentially tagged proteins, switched starved cells (see below) were released in fresh media and harvested at several time points after reentry into the cell cycle. Immunoblot analysis demonstrated replacement of old histone H3-HA protein by new H3-T7 in dividing cells (Fig. 2D). The replacement of one tagged protein by the other is in contrast to previously used “inducible-expression” strategies, which involve ectopic expression of a tagged (new) version of a protein by an inducible promoter in the presence of an endogenous copy. Because of ongoing synthesis of the endogenous gene copy, endogenous histones represent old as well as new proteins. As a consequence, the induced and endogenous proteins quickly reach a new steady state. Tagging a single endogenous gene with a RITE cassette eliminates this problem and allows simultaneous tracking of old and new proteins over many cell divisions.
Fig. 2.
Application of RITE to endogenous histone H3. (A) One of the two genes encoding histone H3 in yeast (HHT2) was tagged with a RITE cassette (H3-RITE) containing short epitope tags: HA (old) and T7 (new). The other gene encoding histone H3 (HHT1) was deleted. A Hygromycin resistance gene (Hygro) was used to select against illegitimate recombinants. The tag switch was under control of a constitutively expressed hormone-dependent Cre recombinase (Cre-EBD78). (B) Growth of wild-type and H3 RITE-tagged [before (HA) and after (T7) the switch] yeast cells spotted in a 10-fold dilution series. (C) The efficiency of recombination in the cell population was determined by Southern blot analysis of genomic DNA digested with HindIII (H) before (Pre) and after (Post) addition of the hormone β-estradiol. An invariant fragment was used as a control (Ctrl). (D) Detection of old (HA) and new (T7) histone H3 by quantitative immunoblot analysis of whole cell lysates of equal numbers of starved switched cells released into fresh media. The number of population doublings was calculated by staining the cells with N-hydroxysuccinimide-tetra-ethylrhodamine (NHS-TER) (SI Materials and Methods). (E) The percentage of old H3-HA plotted against the number of population doublings. The measured HA/T7 ratios of the blot in D were converted into H3-HA percentages by using standard curves of samples with known percentages of H3-HA and H3-T7 (SI Materials and Methods).
Immunodetection of Protein Turnover in Replicating and Nonreplicating Cells.
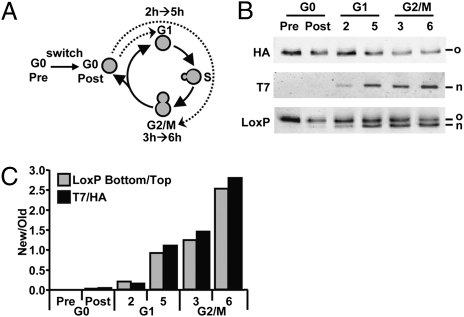
Quantification of the immunoblot shown in Fig. 2D showed that replacement of old H3-HA by new H3-T7 occurred at a rate faster than expected when only dilution due to replication is taken into account, suggesting histone turnover by replication-independent mechanisms (Fig. 2E). The fact that RITE introduces a permanent genetic switch after a transient signal allowed direct comparison of histone exchange in different cell cycle stages. To minimize new histone mRNA and protein expression during the recombination process the tag switch was performed in nutrient-starved cells, here referred to as G0 (Fig. 3A and Fig. S1). Switched H3-HA→T7 cells were released into fresh medium containing nocodazole to arrest the cells after passage through one S-phase in G2/M (Fig. 3A and Fig. S2). During S-phase, like the DNA, the amount of histones gets duplicated and incorporated into the chromatin. As expected, cells at the estimated start of the G2/M cell cycle block (t = 3 h) showed an approximately equal abundance of old H3-HA and new H3-T7 (Fig. 3 B and C). To investigate replication-independent histone exchange, the switched H3-HA→T7 cells were released into fresh media containing α-factor to arrest the cells in G1, to prevent passage through S-phase (Fig. 3A). New H3-T7 was detected at the start of the cell block (t = 2 h) and increased further during the next 3 h (t = 5 h). Moreover, the abundance of new histone H3-T7 after 5 h in G1 was similar to that of cells arrested in G2/M, which had undergone one round of genome duplication and therefore contain at least 50% new H3-T7 and 50% old H3-HA (Fig. 3 B and C). Thus, yeast cells that had been arrested in G1 for the duration of around three cell doubling times had replaced approximately half of the old H3-HA protein by new H3-T7 in the absence of DNA replication.
Fig. 3.
Global histone exchange determined by immunodetection. (A) Yeast strains were grown to saturation (here referred to as G0) in complete medium and recombination was induced overnight (switch) by addition of hormone (Fig. S1). Cells were released in fresh media and arrested in G1 (α-factor) or G2/M (nocodazole). Samples were taken at the estimated start of the arrest (2 h G1 and 3 h G2/M) and 3 h later. (B) Quantitative immunoblot analysis of old and new histone H3 in whole-cell lysates using antibodies against HA (old), T7 (new) or an antibody raised against the spacer-LoxP sequence (LoxP) recognizing old and new proteins simultaneously. (C) Relative H3-T7/H3-HA ratios (New/Old) were calculated on the basis of the ratio of the top band (H3-HA) and the bottom band (H3-T7) of the LoxP blot (absolute values) and the ratio of HA and T7 signals (arbitrary units).
Affinity Purification of Old and New Histones in Chromatin.
Because soluble histones represent a minor fraction of the total histone pool (23), these results suggested that the G1-arrested cells had incorporated new histone H3-T7 into chromatin. To address this question we took advantage of the possibility of using the epitope tags for affinity purification of chromatin fragments containing old and new histones. Following chromatin immunoprecipitation (ChIP) the ratio of new H3-T7 over old H3-HA was determined by real-time quantitative PCR (qPCR) for promoter regions of a set of genes with different transcriptional properties and for an intergenic region (Fig. 4A). Histone exchange in chromatin was already detectable in switched G0 cells before release. After supplementation of fresh medium containing α-factor, exchange increased in the transition to the G1 arrest and increased further during the arrest (2 and 5 h G1). Strikingly, 5 h after release into the G1 block, the replacement of old H3-HA by new H3-T7 was quantitatively similar at different loci to that of cells that had just duplicated their genome and histone content (3 h G2/M). This confirms that cells arrested in G1 had undergone rapid replication-independent exchange of chromatin-bound histones (Fig. 4A). However, histone exchange was not restricted to the G1 phase. Cells arrested in G2/M (from 3 h until 6 h) and even cells arrested by nutrient depletion (G0 pre until G0 post) accumulated new H3-T7 during the arrest, albeit slower (Figs. 3C and 4A). Identical results were obtained with a strain in which the old and new tags were swapped (H3-T7→HA; Fig. S3), showing that the characteristics of new histone deposition were not determined by the specific epitope tags. We conclude that replication-independent histone exchange is a common feature of arrested cells but the rate of exchange can vary between cell cycle phases.
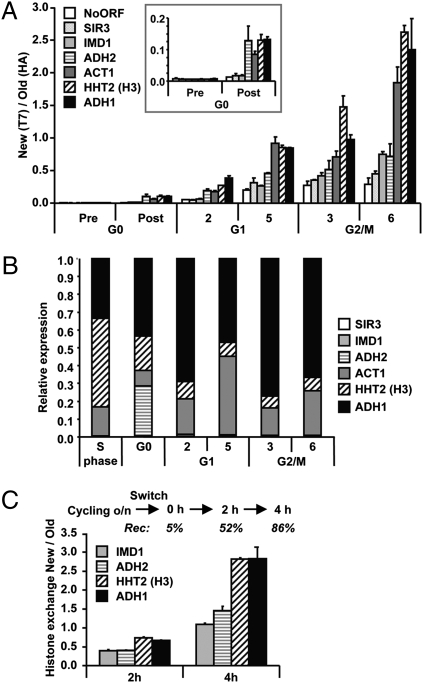
Fig. 4.
Replication-independent transcription-coupled histone turnover quantified by affinity purification. (A) Analysis of chromatin-bound histones by ChIP of HA (old) and T7 (new) histone H3 quantified by real-time quantitative PCR (qPCR). Histone exchange (ratio of new/old) was determined for promoters of the indicated genes and an intergenic region on chromosome V (NoORF). The genes are ranked by estimated transcription frequency in log phase, from low (open bars) to high (solid bars) frequency. The result shown is the average of two individual experiments (± SEM). The Inset is a zoom-in of the G0 time points. (B) Relative mRNA expression levels were determined by reverse-transcriptase qPCR (RT-qPCR). An S-phase sample of the same H3-RITE strain was used as a reference sample. A wild-type strain without a RITE tag showed very similar expression profiles (Fig. S4). (C) Histone turnover in H3-T7→HA cells without any arrest was determined by induction of Cre-recombinase in log-phase cells (OD660 = 0.25). The percentage of cells that had undergone recombination (Rec) is indicated for each time point (determined by a colony-plating assay). Histone replacement at promoters was determined by ChIP (HA/T7).
RITE allowed a direct and quantitative comparison between G1 and G2/M cells, which demonstrated that cells arrested in G1 replaced half of the old histones by new histones within 5 h by replication-independent mechanisms. Analysis of mRNA expression levels during the different phases of the cell cycle showed that the rate of histone exchange was coupled to the level of transcription at each time point or to previous transcription events (Fig. 4B and Fig. S4). Analysis of the inducible GAL1 promoter showed that induction of transcription caused an increase in histone exchange (Fig. S4), suggesting that transcription leads to histone exchange. In addition, transcription-coupled histone exchange also occurred in coding regions, at rates similar to the rates found at promoters (Fig. S5). Transcription-coupled histone exchange might be a specific property of arrested cells that cannot replace histones by replication-dependent mechanisms. To investigate this possibility, histone exchange in chromatin was determined in log-phase cells that had been grown for many generations without a growth arrest (Fig. 4C). In these cycling cells new histone H3-HA was also incorporated more efficiently in highly transcribed genes (Fig. 4C), suggesting that transcription-coupled histone exchange occurred on top of replication-dependent histone deposition. In addition, monitoring of old and new histones during successive cell divisions showed that transcription-coupled histone deposition was maintained during at least three cell divisions (Fig. S6). Thus, biochemical purification of old and new histones revealed that chromatin is a very dynamic macromolecular complex in dividing as well as nondividing cells and that transcription is a key determinant of chromatin instability.
Fluorescent RITE to Monitor Proteasome Replacement in Time and Space.
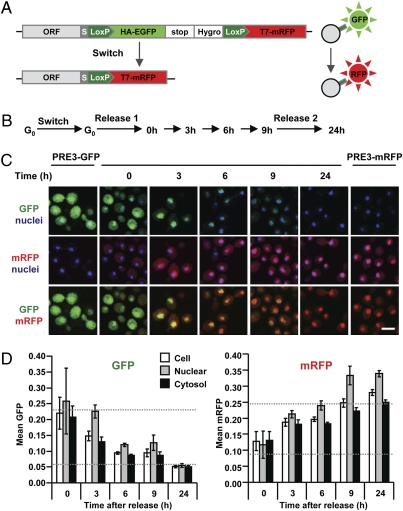
The methods discussed above probe protein dynamics in pools of cells. To visualize the behavior of old and new proteins in single cells, a fluorescent RITE cassette was constructed that switches from a green fluorescent protein (GFP) tag to a monomeric red fluorescent protein (mRFP) tag (Fig. 5A). To illustrate the use of fluorescent RITE, a constituent protein of another macromolecular complex, the proteasome, was tagged. Specifically, we constructed a yeast strain where the only endogenous PRE3 gene, encoding a catalytic β-subunit of the proteasome, was tagged with the GFP→mRFP RITE cassette. This strain has a normal growth rate, indicating that the RITE-tagged subunit is functional, because deletion or mutation of PRE3 is lethal. We note that yeast cells expressing H3-GFP→mRFP were inviable, indicating that not every protein can be safely tagged with the larger GFP→mRFP RITE cassette. To visualize the replacement of old by new proteasomes by microscopy, recombination was induced in G0 (Fig. 5B), during which very little proteasome synthesis occurs. Because the proteasome is a stable complex, many old proteasomes (Pre3-GFP) remain that are slowly replaced by new proteasomes (Pre3-mRFP) (Fig. 5C). When the cells were released in fresh media, the old proteasomes were more swiftly replaced by new proteasomes due to dilution during cell division (Fig. 5C). In yeast and mammalian cells the proteasome is present in both the nucleus and the cytosol (24). Quantification of GFP and mRFP signals showed that in the switched yeast cells, the appearance of new proteasome and loss of old proteasome followed similar kinetics in the two compartments (Fig. 5D). Thus fluorescent RITE enables visualization of replacement of old by new proteins in living cells in time and space during cell cycle arrests and during successive cell divisions.
Fig. 5.
Spatiotemporal analysis of old and new proteasomes by microscopy. (A) Schematic representation of fluorescent RITE. (B) PRE3-GFP→mRFP cells were grown to saturation (G0) and recombination was induced overnight (switch). Subsequently, cells were released in fresh media (release 1) and samples were taken at the indicated time points. Nine hours after the first release, cells were again supplemented with fresh media (release 2). Time points 3, 6, 9, and 24 h correspond to ≈0.3, 2, 3, and 8 cell divisions, respectively. (C) Representative confocal microscopy images of PRE3-GFP→mRFP grown as indicated in B and of control strains (PRE3-GFP and PRE3-mRFP). Hoechst was used as a nuclear counterstaining (blue). (Scale bar, 4 μm.) (D) The GFP and mRFP fluorescent intensities of micrographs from C were quantified and the value shown for each time point is an average of the mean fluorescence intensity in the nuclei, cytoplasm, and total surface of 400 cells (± SD). Dashed lines indicate GFP and mRFP signals in control cells expressing GFP or mRFP only (the bottom dashed lines indicate background levels).
Discussion
Here we show that RITE is a versatile method to study different parameters of protein dynamics such as protein turnover and exchange of subunits in macromolecular complexes. In contrast to other methods such as pulse-chase labeling, inducible expression, methods based on differential fluorescence, or TimeStamp (3, 5, 6, 13, 25), RITE provides the unique possibility to simultaneously monitor old and new proteins and to do so by multiple techniques. RITE has important additional advantages over existing technologies. It does not require addition of UV light, chemicals, or labels, circumventing the need for expensive ultrasensitive mass spectrometry technologies. Furthermore, because no heterologous inducible promoters are required to differentially express old and new proteins, tagged genes are regulated by their endogenous promoter and the switch can occur without perturbation under any condition of interest. Protein replacement of the stable proteasomes and histones could be assessed over long time periods in dividing and nondividing cells, indicating that RITE is suitable to study the dynamics of long-lived proteins, which are typically difficult to study with more traditional methods. RITE should also be applicable to shorter-lived proteins, however. Although it takes ∼2 h until the majority of the cells has switched, switched cells can already be detected as early as 15 min after activation of Cre. RITE may be less suitable for studies of very short-lived proteins.
The differential tagging of histone H3 showed that endogenously expressed canonical histones undergo turnover within chromatin in a transcription-dependent manner. Our results are in agreement with previous histone H3 turnover studies using time-controlled induced expression of a tagged ectopic histone copy in yeast (8–12, 16, 26). The direct comparison with replication-dependent assembly of new histones indicates that replication-independent histone exchange occurs at a high rate. This was unexpected when one considers the regulated expression of histones. We note that whereas H3 mRNA indeed peaks in S-phase when chromatin is duplicated, its expression is lower but still substantial outside of S-phase (Fig. 4B). This supports the idea that canonical histones are synthesized outside of S-phase for replication-independent histone exchange. Especially in starved cells, H3 mRNA is relatively abundant (Fig. 4B). The high rate of histone exchange suggests that posttranslational modifications in chromatin are continuously being erased in dividing and nondividing cells. Thus, replication-independent histone exchange might provide cycling and noncycling cells with a means to replace old histones that have acquired damage or that need to be epigenetically reset.
Using fluorescent RITE, replacement of old by new proteasomes in time and space was determined by microscopy. The amount of old proteasomes decreased at a very similar rate in the cytosolic and nuclear compartments, suggesting an even segregation during cell division and/or a fast reequilibration between proteasomes in both compartments. Likewise, the appearance of new proteasome in both compartments followed similar kinetics, indicating that the translocation of new proteasome subunits into the nucleus is a relatively fast phenomenon (Fig. 5D).
RITE is a widely applicable tool to dissect novel mechanisms and functions of protein dynamics. For example, RITE-tagged genes of interest and the Cre recombinase can be efficiently introduced into the collection of yeast deletion strains by one round of genetic crossing, which allows genomewide genetic screens for identification of factors involved in protein dynamics. RITE can also be applied to investigate whether new and aging proteins have different properties such as age-related posttranslational modifications or whether they show differential segregation between mother and daughter cells. Finally, although we have validated RITE in budding yeast, with minor modifications RITE technology may be adapted for use in higher eukaryotes. The RITE cassettes are universally applicable and conditional versions of Cre recombinase have already been developed for many cell systems or even whole organisms (27).
Materials and Methods
Yeast Strains and Growth Conditions.
Yeast strains and growth conditions are described in Table S1 and SI Materials and Methods. RITE cassettes contain an invariant short peptide spacer sequence (GGSGGS) that was found to be required for viability of strains carrying tagged histones. The spacer and ITSYNVCYTKLS peptide encoded by the LoxP DNA sequence are present in front of the epitope tags both before and after the switch. RITE cassettes were PCR amplified and targeted to the 3′ end of the endogenous genes by homologous recombination to tag the C terminus and ensure regulation by the endogenous promoter. The hormone-dependent Cre-EBD (Cre-EBD78) was described previously (22). A constitutively expressed copy was stably integrated in the yeast genome. For RITE experiments, yeast cells were grown overnight in YPD in the presence of Hygromycin B (200 μg/mL, Invitrogen). The cells were then diluted 1:10 into fresh YPD and incubated for 30–36 h. Recombination was induced by the addition of 1 μM β-estradiol (E-8875, Sigma-Aldrich). Subsequently, cells were diluted 1:25 in fresh YPD media to release the cells back into the cell cycle. Cells enter G1 arrest upon addition of 0.5 ng/μL of α-factor and G2/M arrest upon addition of 15 μg/mL Nocodazole (Sigma-Aldrich). Detailed protocols for ChIP, RT-PCR, immunoblot, Southern blot, FACS, and microscopy are described in SI Materials and Methods and Table S2.
Supplementary Material
Acknowledgments
We thank C. Logie for helpful suggestions; F. van Diepen, A. Pfauth, and L. Oomen for technical assistance; and G. Filion for statistical help. We thank members of the van Leeuwen lab and M. Fornerod for suggestions and critical reading of the manuscript. F.v.L. was supported by the European Union 6th framework program (Network of Excellence “The Epigenome” LSHG-CT-2004-503433) and by The Netherlands Organization for Scientific Research. D.L.L. was supported by postdoctoral fellowship PF-04-041-01-GMC from the American Cancer Society. V.M.B. was supported by a long-term European Molecular Biology Organization fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911164107/DCSupplemental.
References
- 1.Gorski SA, Dundr M, Misteli T. The road much traveled: Trafficking in the cell nucleus. Curr Opin Cell Biol. 2006;18:284–290. doi: 10.1016/j.ceb.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 2.Russel D, et al. The structural dynamics of macromolecular processes. Curr Opin Cell Biol. 2009;21:97–108. doi: 10.1016/j.ceb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Reits EA, Neefjes JJ. From fixed to FRAP: Measuring protein mobility and activity in living cells. Nat Cell Biol. 2001;3:E145–E147. doi: 10.1038/35078615. [DOI] [PubMed] [Google Scholar]
- 4.D’Angelo MA, Hetzer MW. Structure, dynamics and function of nuclear pore complexes. Trends Cell Biol. 2008;18:456–466. doi: 10.1016/j.tcb.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adams SR, Tsien RY. Preparation of the membrane-permeant biarsenicals FlAsH-EDT2 and ReAsH-EDT2 for fluorescent labeling of tetracysteine-tagged proteins. Nat Protoc. 2008;3:1527–1534. doi: 10.1038/nprot.2008.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 7.Subach FV, et al. Monomeric fluorescent timers that change color from blue to red report on cellular trafficking. Nat Chem Biol. 2009;5:118–126. doi: 10.1038/nchembio.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schermer UJ, Korber P, Hörz W. Histones are incorporated in trans during reassembly of the yeast PHO5 promoter. Mol Cell. 2005;19:279–285. doi: 10.1016/j.molcel.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 9.Linger J, Tyler JK. Global replication-independent histone H4 exchange in budding yeast. Eukaryot Cell. 2006;5:1780–1787. doi: 10.1128/EC.00202-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 11.Rufiange A, Jacques PE, Bhat W, Robert F, Nourani A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol Cell. 2007;27:393–405. doi: 10.1016/j.molcel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 13.Lin MZ, Glenn JS, Tsien RY. A drug-controllable tag for visualizing newly synthesized proteins in cells and whole animals. Proc Natl Acad Sci USA. 2008;105:7744–7749. doi: 10.1073/pnas.0803060105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura H, Cook PR. Kinetics of core histones in living human cells: Little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol. 2001;153:1341–1353. doi: 10.1083/jcb.153.7.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat Rev Genet. 2008;9:15–26. doi: 10.1038/nrg2206. [DOI] [PubMed] [Google Scholar]
- 16.Kim HJ, et al. Histone chaperones regulate histone exchange during transcription. EMBO J. 2007;26:4467–4474. doi: 10.1038/sj.emboj.7601870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- 18.Wirbelauer C, Bell O, Schübeler D. Variant histone H3.3 is deposited at sites of nucleosomal displacement throughout transcribed genes while active histone modifications show a promoter-proximal bias. Genes Dev. 2005;19:1761–1766. doi: 10.1101/gad.347705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chow CM, et al. Variant histone H3.3 marks promoters of transcriptionally active genes during mammalian cell division. EMBO Rep. 2005;6:354–360. doi: 10.1038/sj.embor.7400366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Logie C, Stewart AF. Ligand-regulated site-specific recombination. Proc Natl Acad Sci USA. 1995;92:5940–5944. doi: 10.1073/pnas.92.13.5940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindstrom DL, Gottschling DE. The mother enrichment program: A genetic system for facile replicative life span analysis in Saccharomyces cerevisiae. Genetics. 2009;183:413–422. doi: 10.1534/genetics.109.106229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gunjan A, Verreault A. A Rad53 kinase-dependent surveillance mechanism that regulates histone protein levels in S. cerevisiae. Cell. 2003;115:537–549. doi: 10.1016/s0092-8674(03)00896-1. [DOI] [PubMed] [Google Scholar]
- 24.Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. EMBO J. 1997;16:6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yen HC, Xu Q, Chou DM, Zhao Z, Elledge SJ. Global protein stability profiling in mammalian cells. Science. 2008;322:918–923. doi: 10.1126/science.1160489. [DOI] [PubMed] [Google Scholar]
- 26.Choi ES, Shin JA, Kim HS, Jang YK. Dynamic regulation of replication independent deposition of histone H3 in fission yeast. Nucleic Acids Res. 2005;33:7102–7110. doi: 10.1093/nar/gki1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.