Abstract
Polycomb Group (PcG) and Trithorax Group (TrxG) proteins are key epigenetic regulators of global transcription programs. Their antagonistic chromatin-modifying activities modulate the expression of many genes and affect many biological processes. Here we report that heterozygous mutations in two core subunits of Polycomb Repressive Complex 2 (PRC2), the histone H3 lysine 27 (H3K27)-specific methyltransferase E(Z) and its partner, the H3 binding protein ESC, increase longevity and reduce adult levels of trimethylated H3K27 (H3K27me3). Mutations in trithorax (trx), a well known antagonist of Polycomb silencing, elevate the H3K27me3 level of E(z) mutants and suppress their increased longevity. Like many long-lived mutants, E(z) and esc mutants exhibit increased resistance to oxidative stress and starvation, and these phenotypes are also suppressed by trx mutations. This suppression strongly suggests that both the longevity and stress resistance phenotypes of PRC2 mutants are specifically due to their reduced levels of H3K27me3 and the consequent perturbation of Polycomb silencing. Consistent with this, long-lived E(z) mutants exhibit derepression of Abd-B, a well-characterized direct target of Polycomb silencing, and Odc1, a putative direct target implicated in stress resistance. These findings establish a role for PRC2 and TRX in the modulation of organismal longevity and stress resistance and indicate that moderate perturbation of Polycomb silencing can increase longevity.
Keywords: aging, epigenetics, histone methyltransferase, Polycomb silencing
Trimethylation of Histone H3 lysine 27 (H3K27me3) is essential for the establishment and maintenance of Polycomb silencing and is carried out by Polycomb Repressive Complex 2 (PRC2), one of several PcG protein complexes that collaborate to implement Polycomb silencing (1). PRC2 contains the PcG proteins E(Z), SU(Z)12, ESC (or ESCL) and PCL (1). E(Z) is the catalytic subunit of PRC2 and its H3K27 trimethylation activity requires the other PRC2 subunits (2–6). Recent genome-wide mapping of sites on chromatin that contain H3K27me3 and are bound by PcG proteins have identified hundreds of putative Polycomb target genes in Drosophila and several thousand in human cells (7–10). In addition to their well-established role in maintaining cell identities, PcG proteins have been implicated in many processes relevant to aging, including stem cell pluripotency and self-renewal, regulation of cell cycle exit, regeneration and wound healing, tumorigenesis, and DNA repair, suggesting that PcG proteins may also modulate organismal life span (1, 11–16).
Studies in cultured mammalian cells have implicated PcG proteins in the regulation of cellular senescence (17–19). However, the relationship between cellular senescence and organismal aging remains uncertain (20, 21). Here we examine the role of PRC2 in the modulation of organismal life span. Using multiple alleles of several core subunits of PRC2, focusing on the subunits E(z) and esc, we provide genetic evidence that heterozygous mutations in these genes increase organismal longevity. Similar to other long-lived mutants, heterozygous E(z) and esc mutants exhibit increased resistance to oxidative stress and starvation. Furthermore, we provide evidence that the increased longevity and stress resistance are due to moderately reduced H3K27me3 and a consequent deficit in Polycomb silencing.
Results
To determine whether PRC2 modulates life span, we first examined several independently isolated mutant alleles of E(z), which encode the catalytic subunit of PRC2. Flies heterozygous for the protein null E(z)63 or the catalytically inactive E(z)731 mutation that were progeny of an out-cross to an Oregon-R (O-R) wild-type strain exhibited a substantially greater median life span than the O-R control (71% and 76%, respectively) (Fig. 1 A and B). When derived from an out-cross to a longer-lived Canton-S (C-S) wild-type strain, the median life span of E(z)63 heterozygotes was 33% longer than the C-S control (Fig. 2A).
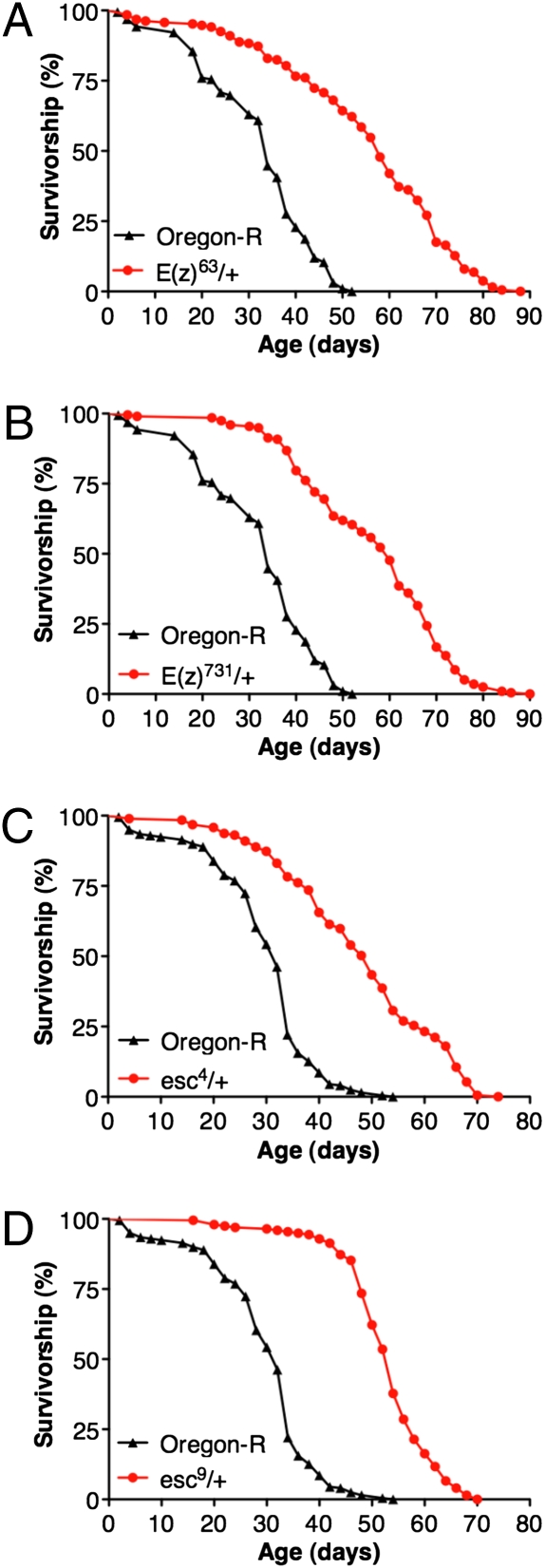
Fig. 1.
Heterozygous mutations in E(z) and esc increase longevity. (A) E(z)63/+ live 71% longer (median life span) than Oregon-R (O-R) controls. (B) E(z)731/+ mutants live 76% longer than O-R (P < 0.001; O-R n = 192; E(z)63/+ n = 188; E(z)731/+ n = 197). (C) esc4/+ mutants live 47% longer than O-R. (D) esc9/+ mutants live 59% longer than O-R (P < 0.001; O-R n = 199; esc4/+ n = 189; esc9/+ n = 196). Life span assays in A and B were conducted in parallel, as were C and D. Assays were conducted at 25 °C under constant humidity (approximately 50%). Reported P values are from Mantel-Cox log-rank statistical analysis.
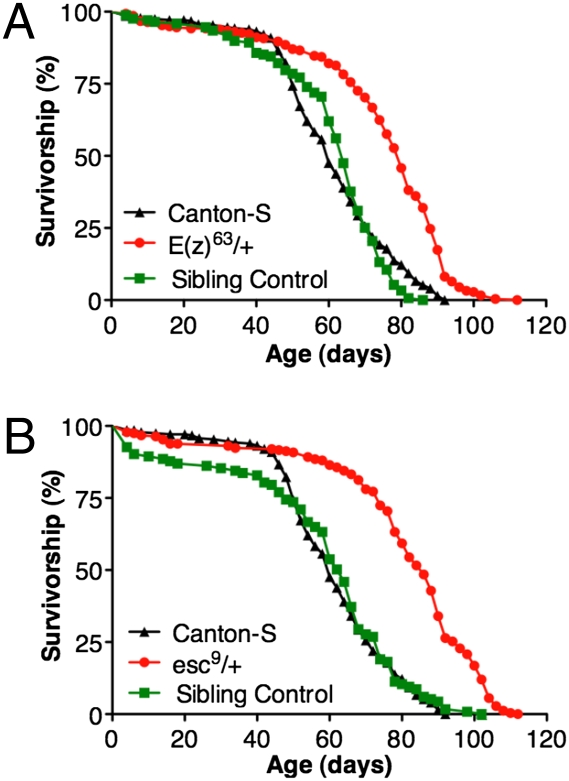
Fig. 2.
Heterozygous mutations in E(z) and esc increase longevity when out-crossed to a longer-lived C-S wild-type strain. (A) E(z)63/+ mutants live 33% longer than controls (P < 0.001; C-S n = 280; E(z)63/+ n = 278; Sibling control, n = 168). No difference in life span was observed between the C-S and E(z)63 sibling controls. (B) esc9/+ mutants live 43% longer than C-S controls (P < 0.001; C-S n = 280; esc9/+ n = 276; Sibling control n = 123). No difference in life span was observed between the C-S and esc9 sibling controls. Life span assays in A and B were conducted in parallel at 25 °C under constant humidity (approximately 50%). Sibling controls are genetically matched to experimental genotypes except for the chromosome containing the mutant allele. Reported P values are from Mantel-Cox log-rank statistical analysis.
The life spans of two heterozygous esc mutants were also determined. ESC binds directly to histone H3 and is required for H3K27 trimethylation by E(Z) (5). Males heterozygous for the null esc4 or the dominant negative esc9 mutation that were progeny of an out-cross to an O-R wild-type strain had median life spans that were, respectively, 47% and 60% longer than the O-R control (Fig. 1 C and D). When derived from an out-cross to a longer-lived C-S wild-type strain, heterozygous esc9 flies had a median life span that was 43% longer than the C-S control (Fig. 2B). The similar effects on longevity observed using four different mutations in both E(z) and esc out-crossed to two different wild-type strains strongly suggests that their increased longevity is due to the mutations in E(z) and esc and not other factors present in the genetic background. Initial results indicate that mutations in other PRC2 subunits including Su(z)12 and escl also cause a significant extension of life span when out-crossed to O-R (Fig. S1).
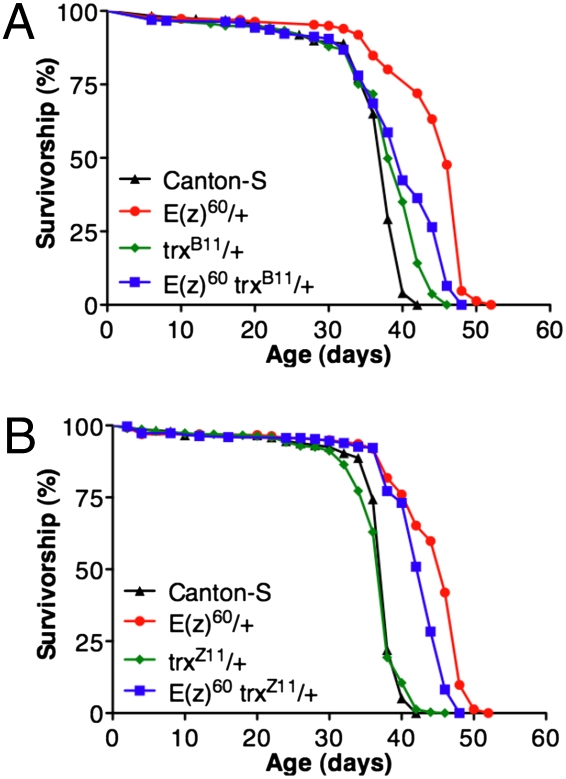
To determine whether disruption of Polycomb silencing is indeed the cause of the increased longevity, we tested whether mutations in trx could suppress the extended life span phenotype of E(z) mutants. TRX and other Trithorax Group (TrxG) proteins antagonize Polycomb silencing and are required for robust maintenance of active transcriptional states of Polycomb target genes (22–25). TRX itself is a histone methyltransferase that specifically trimethylates histone H3 on lysine 4 (H3K4me3), a modification associated with promoter regions of active genes (26–30). TRX also interacts with the histone acetyltransferase CBP and is required for CBP-mediated acetylation of H3K27, which directly blocks H3K27 trimethylation by E(Z), because the two modifications are mutually exclusive (31). Accordingly, mutations in the trx gene suppress the silencing defects of PcG mutants and would be expected to suppress the increased longevity phenotype of E(z) mutants if it were also due to compromised Polycomb silencing. As shown in Fig. 3, the null trxB11 mutation partially suppresses the long life span phenotype of E(z)60 mutants, eliminating 75% of the increase in E(z)60 longevity (Fig. 3A). The weaker trxZ11 allele also significantly shortened E(z)60 life span, but to a lesser extent, eliminating just 25% of the increase in its longevity, consistent with the hypomorphic nature of trxZ11 (Fig. 3B). These results suggest that the increased longevity of heterozygous PRC2 mutants is most likely due to reduced H3K27 trimethylation and consequent perturbation of Polycomb silencing.
Fig. 3.
Mutations in the Polycomb silencing antagonist, trithorax, suppress the increased longevity of E(z) mutants. (A) E(z)60/+ mutants live 21% longer than C-S whereas E(z)60 trxB11/+ + double mutants live 5% longer than C-S. trxB11/+ mutants themselves show no difference in median life span compared to C-S (P < 0.001; C-S n = 298; E(z)60 n = 296; trxB11 n = 297; E(z)60 trxB11 n = 295). (B) E(z)60/+ mutants live 4.5% longer than E(z)60 trxZ11/+ double mutants. trxZ11/+ mutants show no difference in median life span compared to C-S (C-S n = 292; E(z)60 n = 296; trxZ11 n = 294; E(z)60 trxZ11 n = 293). Life span assays were conducted independently at 29 °C under constant humidity (approximately 50%). Reported P values are from Mantel-Cox log-rank statistical analysis.
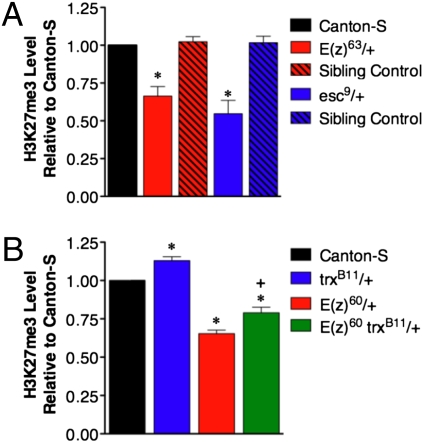
To assess the impact of the heterozygous PRC2 mutations on H3K27me3, we performed quantitative western analysis of whole adult male extracts, which revealed a 35% decrease in H3K27me3 levels in E(z)63 and E(z)60 heterozygotes and a 45% decrease in the even longer-lived esc9 heterozygotes (Fig. 4A). Conversely, the heterozygous trxB11 null mutation itself modestly but significantly increased H3K27me3 by 13% compared to C-S flies, and elevated the reduced H3K27me3 level of E(z)60 heterozygotes by 14% (Fig. 4B). The hypomorphic trxZ11 allele also promoted an increase in H3K27me3 in the E(z)60 trxZ11 double heterozygotes but to a lesser extent, increasing the H3K27me3 by just 7%, which is consistent with its more modest suppression of E(z)60 longevity (Fig. S2). The suppressing effect of two different trx mutations on both the longevity and reduced H3K27me3 level of E(z)60 mutants further suggests that the increased longevity of PRC2 subunit mutants is likely due to their reduced H3K27me3 levels and consequent perturbation of Polycomb silencing. This would imply that one or more Polycomb target genes can directly or indirectly modulate longevity (see below).
Fig. 4.
Long-lived E(z) and esc mutants have reduced levels of H3K27me3 that are partially restored by mutations in trx. (A) Quantitative Western blots from age-matched E(z)63 and esc9 adult male heterozygotes out-crossed to C-S. E(z)63/+ and esc9/+ mutants have, respectively, 35% less and 45% less H3K27me3 than their respective sibling controls and C-S controls. (B) Quantitative Western blots from age-matched E(z)60/+, trxB11/+, and E(z)60 trxB11/+ + adults out-crossed to C-S. E(z)60/+ mutants have 35% less H3K27me3 than C-S. trxB11/+ mutants have 13% more H3K27me3 than C-S. E(z)60 trxB11/+ + double mutants have 14% more H3K27me3 than E(z)60/+ mutants. The C-S control value was arbitrarily set to 1 in each experimental replicate. The normalized means from three independent replicates of each genotype are plotted with SEM (error bars). t-tests were performed to test for statistical significance. * indicates P ≤ 0.05 compared to C-S. + indicates P ≤ 0.05 compared to E(z)60/+.
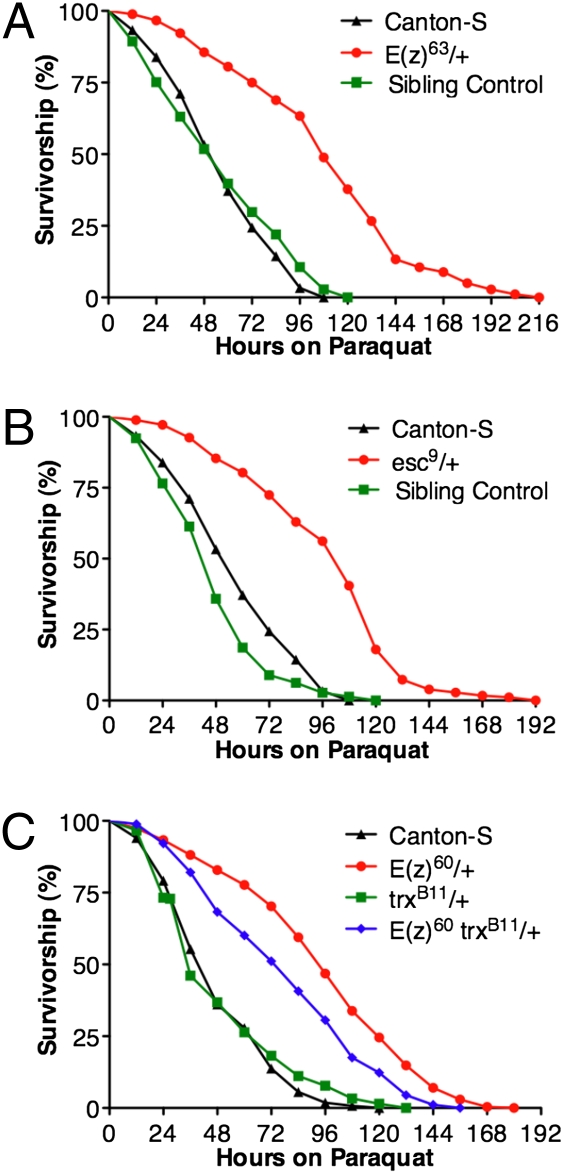
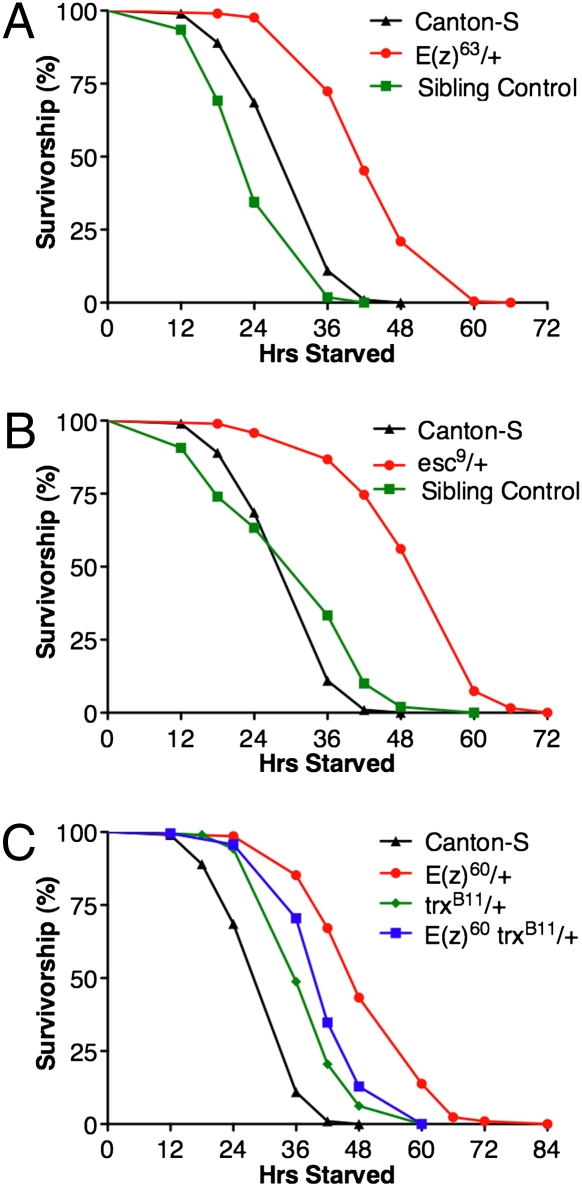
To begin to identify physiological characteristics of these mutants that may point to Polycomb target genes responsible for their increased longevity, we assayed their resistance to oxidative stress and starvation, two traits often correlated with increased longevity (32, 33). Genetic screens for mutations that confer resistance to multiple stresses have been shown to enrich for mutants with increased longevity (34). The E(z)63, E(z)60, and esc9 heterozygotes all display substantially greater resistance to acute oxidative stress (15 mM paraquat) and also have substantially greater resistance to starvation than C-S controls (Figs. 5 and 6). Both of these phenotypes of E(z)60 mutants are suppressed by the trxB11 mutation, suggesting that they are also due to impaired Polycomb silencing (Figs. 5C and 6C).
Fig. 5.
E(z) and esc mutants have increased resistance to oxidative stress that is partially suppressed by mutations in trx. Graphs depict survivorship during acute exposure to media containing 15 mM paraquat. (A) E(z)63/+ mutants have a significant increase in oxidative stress resistance compared to C-S and their sibling controls (P < 0.001; C-S n = 180; E(z)63/+ n = 180; Sibling control, n = 141). (B) esc9/+ mutants have a significant increase in oxidative stress resistance compared to C-S and their sibling controls (P < 0.001; C-S n = 180; esc9/+ n = 178; Sibling control, n = 145). (C) E(z)60/+ mutants have a similar resistance to oxidative stress as seen in E(z)63/+ mutants shown in A. E(z)60 trxB11/+ double mutants have significantly lower oxidative stress resistance than E(z)60/+ mutants alone (P < 0.001; C-S n = 269; E(z)60/+ n = 269; trxB11/+ n = 269; E(z)60 trxB11/+ + n = 268). Assays shown in A and B were conducted in parallel. Sibling controls are genetically matched to the experimental genotypes except for the chromosome carrying the mutant allele. Reported P values are from Mantel-Cox log-rank statistical analysis.
Fig. 6.
E(z) and esc mutants have increased resistance to starvation that is partially suppressed by mutations in trx. Graphs depict survivorship during acute starvation of adult males of each genotype. (A) E(z)63/+ mutants exhibit a significant increase in starvation resistance compared to C-S and their sibling controls (P < 0.001; C-S n = 210; E(z)63/+ n = 210; Sibling control, n = 107). (B) esc9/+ mutants exhibit a significant increase in starvation resistance compared to C-S and their sibling controls (P < 0.001; C-S n = 210; esc9/+ n = 189; Sibling control, n = 150). (C) E(z)60/+ mutants have significantly increased resistance to starvation, similar to the E(z)63/+ mutants shown in (A). E(z)60 trxB11/+ + double mutants have significantly lower oxidative stress resistance than E(z)60/+ mutants alone (P < 0.001; C-S n = 210; E(z)60/+ n = 210; trxB11/+ n = 209; E(z)60 trxB11/+ n = 210). Sibling controls are genetically matched to their experimental genotype except the chromosome carrying the mutant allele. Reported P values are from Mantel-Cox log-rank statistical analysis.
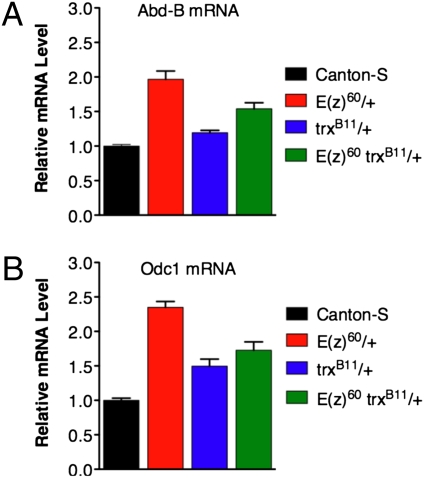
Are the reduced H3K27me3 levels of the long-lived mutants sufficient to compromise Polycomb silencing and alter expression of genes directly regulated by it? Although Polycomb silencing was originally thought to function as an on-off switch, it is now evident that it can also act like a “governor” to modulate the expression levels of some target genes without completely silencing them (10, 35). This suggests that moderate changes in H3K27me3 may lead to quantitative changes in expression of some Polycomb target genes. Even small changes in the levels of some PcG proteins can have biologically significant effects (36). To determine whether the reduced H3K27me3 levels observed are sufficient to lead to at least partial derepression of Polycomb target genes, we measured expression of the homeotic gene Abd-B, a well known target of Polycomb silencing, by quantitative real-time RT-PCR (qPCR). As shown in Fig. 7A, Abd-B expression in whole adult males is elevated in E(z)60 heterozygotes. This increase in Abd-B expression is partially suppressed by the trxB11 mutation in the E(z)60 trxB11 double heterozygotes, consistent with their modest increase in H3K27me3 (Fig. 4).
Fig. 7.
Gene expression changes in heterozygous E(z) mutants. (A and B) Moderate reductions in H3K27me3 in E(z)60 mutants derepress a known (A) and a putative (B) direct target of Polycomb Silencing. (A) Abd-B is derepressed approximately twofold in the E(z)60/+ mutants compared to C-S and is partially rerepressed compared to the E(z)60 trxB11/+ + double heterozygotes. (B) Odc1 is derepressed greater than twofold in the E(z)60/+ mutants compared to C-S and is partially rerepressed compared to the E(z)60 trxB11/+ + double heterozygotes. The normalized relative expression levels from three independent biological replicates of each genotype are plotted with SEM (error bars).
Genome-wide mapping of sites occupied by Polycomb proteins and H3K27me3 in S2 cells by chromatin immunoprecipitation combined with genome tiling arrays (ChIP-chip) has identified several hundred putative direct targets of Polycomb silencing (10, 37, 38). Genes with well-documented roles in modulating longevity are not among these direct targets (e.g., Sir2, Rpd3, foxo, InR, and others). However the histone deacetylases SIR2 and RPD3, which have been reported to modulate longevity in Drosophila, are also required for robust Polycomb silencing and associate with several larger PRC2 complexes (39–42). For this reason, we examined their protein levels after stringent RNAi knockdown of E(Z) in S2 cells and found them to be unchanged (Fig. S3). This indicates that changes in their levels are unlikely to play a role in the increased longevity or stress resistance of E(z) mutants and suggests that altered expression of genes not yet implicated in modulating life span may be responsible.
One candidate that might play a direct role in the increased longevity and stress resistance of E(z) mutants is Odc1, a putative direct target of Polycomb silencing identified by ChIP-chip (10). Odc1 encodes ornithine decarboxylase, an evolutionarily conserved enzyme that catalyzes the first, rate-limiting step in polyamine biosynthesis (43). Polyamines have been implicated in resistance to oxidative stress and were recently reported to increase longevity in Caenorhabditis elegans, Drosophila, and mice (44–47). Increased expression of ODC in mammalian cells protects them from apoptosis induced by oxidative damage and other stresses (48). As shown in Fig. 7B, Odc1 expression is elevated more than 2-fold in E(z)60 heterozygotes, and this increase is partially suppressed by the trxB11 mutation in the double heterozygotes. Further genetic analysis of Odc1 will be required to determine whether it plays a role in the increased longevity and stress resistance of PRC2 mutants. A comprehensive analysis of gene expression changes in the long-lived PRC2 mutant adults will also help identify additional candidates for future studies.
Discussion
The evidence presented here establishes a role for PRC2 and TRX in the modulation of life span and stress resistance. Using multiple alleles of several PRC2 subunits, we provide evidence that heterozygous mutations in the PRC2 subunits E(z) and esc extend life span and increase resistance to oxidative stress and starvation in Drosophila. Consistent with the enzymatic function of PRC2 in the methylation of H3K27, long-lived E(z) and esc mutants have reduced H3K27me3 levels. Furthermore, mutations in trx suppress the increased longevity and stress resistance phenotypes of E(z) mutants, while concomitantly increasing their reduced H3K27me3. The moderate reduction of H3K27me3 in long-lived E(z) mutants is sufficient to partially derepress some direct targets of Polycomb silencing, and this is also counteracted by mutations in trx. These results provide strong evidence that derepression of one or more Polycomb target genes is likely to be responsible for their increased longevity. Interestingly, E(z) was also recently identified as one of a number genes whose mRNA expression levels were significantly associated with variation in longevity in a large set of wild-derived inbred lines (49).
The counterbalancing effects of PRC2 and TRX on H3K27me3 levels suggest a simple model for their modulation of longevity (31). Although complete loss of PRC2 activity results in preadult lethality, moderately reducing H3K27me3 destabilizes Polycomb silencing sufficiently to cause partial derepression of some Polycomb target genes that can increase life span and stress resistance. Simultaneously reducing TRX and E(Z) exerts a compensatory effect, reestablishing more normal levels of H3K27me3 and Polycomb target gene expression. Based on this model, we expected that heterozygous trx mutations would decrease longevity. However, the modestly elevated H3K27me3 level (13%) of the heterozygous trxB11 null mutant may simply be insufficient to cause this effect in a wild-type background. It will be interesting to see whether increased TRX levels, which decrease H3K27me3 levels (much like PRC2 mutants) by elevating CBP-mediated H3K27 acetylation, will promote increased longevity, as our model would predict.
The evolutionary conservation of PRC2 components in metazoans and their conserved function in epigenetic silencing raises the possibility that they may play a conserved role in modulating life span in other organisms. Although histone methyltransferases have not been previously implicated in modulating organismal longevity, several other highly conserved chromatin-modifying enzymes have been. In addition to SIR2 and RPD3, the histone H3K4 demethylase LSD-1 has also recently been implicated in modulating longevity in C. elegans (50). Given the roles of these enzymes in the epigenetic maintenance of transcriptional states, it seems likely that additional chromatin modifying enzymes will be found to modulate longevity.
The most well-characterized targets of Polycomb silencing are the homeotic genes of the Bithorax and Antennapedia complexes. Although heterozygous PRC2 mutants exhibit no overt homeotic phenotypes, the elevated level of Abd-B expression in E(z) heterozygotes demonstrates that their moderately reduced H3K27me3 level is sufficient to partially derepress Polycomb target genes. Could modest derepression of one or more of the homeotic genes be responsible for the increased longevity? Given that they encode transcription factors, their potential for regulating expression of many other genes leaves this possibility open.
PRC2 mutants exhibit increased resistance to oxidative stress and starvation. The elevated expression level of Odc1, a putative direct target of Polycomb silencing may contribute to this as it has been shown to mediate resistance to oxidative stress and a variety of other chemical and environmental stresses (44, 47, 48). Dietary supplementation with the polyamine spermidine was also recently shown to increase longevity in yeast, C. elegans, Drosophila, and mice, consistent with the possibility that Odc1 overexpression may contribute to the increased longevity of PRC2 mutants (46). Recent evidence suggests that other changes in metabolism and adult physiology might also contribute to the increased longevity of PRC2 mutants. YY1, the mammalian homolog of Drosophila PHO (a DNA-binding PcG protein involved in recruiting PRC2 to chromatin), directly regulates many genes required for mitochondrial oxidative metabolism (51–53). It will be interesting to determine whether transcriptional regulation of metabolic genes is a broader theme in the adult function of PcG proteins.
PRC2 and TRX play key roles in promoting epigenetically stable transcriptional states through their mutually antagonistic effects on H3K27me3 levels. Recent work has revealed a growing number of biological processes in which they play an important role. The results presented here now point to a role for these epigenetic transcriptional regulators in modulating life span.
Materials and Methods
Drosophila Strains.
O-R and C-S wild-type stocks were obtained from the Bloomington Stock Center, Bloomington, IN. E(z)63 (M1I) is a protein null mutation; E(z)731 (W638×) and E(z)60 produce catalytically dead truncated proteins (54–57). esc4 (Q113×) is a null allele resulting from an N-terminal truncation and esc9 (M236K) is a dominant negative allele (58, 59). trxB11 is a null allele resulting from an N-terminal truncation and trxZ11 (G3601S) is a hypomorphic mutation (60–63). The E(z)60 trxB11 and the E(z)60 trxZ11 recombinant flies lines were generated by Thomas Breen. escl1 is a homozygous viable, strong hypomorphic allele (Exelixis Inc.) (4, 64). The Su(z)124 allele is a null allele (65).
Life Span Assays.
Males carrying PcG mutations were out-crossed to wild-type O-R or C-S virgin females at 25 °C and approximately 50% relative humidity on media containing 2% yeast, 10% sucrose, 5% cornmeal, 0.6% agar (wt/vol). Newly eclosed adult males were collected and placed in vials (10-15 per vial). Media used for the life span assays contained 5% yeast, 10% sucrose, 5% cornmeal, 0.6% agar (wt/vol). Vials were kept at 25 °C or 29°C and approximately 50% relative humidity for the duration of the assays. Some life span assays as noted were conducted at 29 °C to reduce the time required to complete the assay. Increasing the temperature to 29 °C has been shown to accelerate life span assays without inducing a heat shock response (66). Flies were transferred to fresh food three times a week and dead flies were counted. Percent increases in life span are based on comparing the median survivals. Prism (GraphPad) was used for statistical analysis of life span data. Kaplan-Meyer survival curves display life span data and Mantel-Cox log-rank statistical analysis was used for testing statistical significance of the differences between the survivorship curves.
Quantitative Western Blots (H3K27me3 Levels).
Whole fly extracts were prepared from 7- to 10-day-old males collected under mild CO2 anesthesia and stored at −80 °C. Frozen flies were placed on ice and a volume (10 μL/mg flies) of PBS-T (0.1% Tween-20) containing protease inhibitors (Complete Mini Protease Inhibitor Mixture Tablet, Roche) was added. Flies were homogenized in 1.5 mL tubes using a pellet pestle (Kontes) and transferred to a Dounce homogenizer for further homogenization. Homogenates were filtered through a double layer of Miracloth (Calbiochem), mixed with an equal volume of 2× SDS sample buffer and stored at −80 °C. Before SDS/PAGE, samples were heated to 95 °C for 5–10 min and 5–10 μL was electrophoresed on 13% SDS/PAGE gels and transferred to 0.2 μm pore size nitrocellulose membrane. Antibodies and Quantitative Western blots were performed using an Odyssey Infrared Imager (LiCor) as previously described (31). Normalized means from three independent replicates of mutant and C-S (control) samples were compared by t tests. Data are displayed as the normalized means with SEM (error bars).
Oxidative Stress Resistance Assays.
Fifty to 100 7- to 10-day-old males (progeny of an out-cross to C-S) were placed in bottles without food or water for 4 h at 25 °C and approximately 50% relative humidity, then placed in vials (10-15 flies/vial) containing 3 mL of media [5% sucrose, 1.3% low-melt agar, and 15 mM paraquat (Sigma-Aldrich)]. Paraquat was added when media temperature reached 45 °C to prevent heat inactivation (67). Stress assays were conducted at 25 °C and approximately 50% relative humidity. Dead flies were scored every 12 h until all flies were dead. Nonmutant siblings and/or C-S flies served as controls. Data are displayed as survivorship. Each experiment was repeated at least two times. Results were pooled to generate the survivorship curves. Prism (GraphPad) was used for statistical analysis of the pooled data. Kaplan-Meyer survival curves display life span data and Mantel-Cox log-rank statistical analysis was used for testing statistical significance between the survivorship curves.
Starvation Resistance Assays.
Seven- to 10-day-old males (progeny of an out-cross to C-S) were placed into vials (10-15 flies/vial) containing 3 mL of 1.3% low-melt agar with water. Flies were kept at 25 °C with approximately 50% relative humidity for the duration of the assay. Dead flies were scored three times a day until all flies were dead. Nonmutant siblings and/or C-S flies served as controls. Data are displayed as survivorship. Each experiment was repeated at least two times and results were pooled to generate survivorship curves. Prism (GraphPad) was used for statistical analysis of the pooled data. Kaplan-Meyer survival curves display life span data and Mantel-Cox log-rank statistical analysis was used for testing statistical significance between the survivorship curves.
Real-Time PCR Analysis.
Total RNA was isolated from populations of 7- to 10-day-old whole adult males using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was synthesized using a High-Capacity cDNA Archive Kit (Applied Biosystems). Real-time PCR reactions were performed on a 7300 Real-Time PCR System (Applied Biosystems) using Perfecta SYBR green supermix (Quanta Biosciences). Reactions were run in triplicate using three independent biological replicates of each sample. Primer pairs to Abd-B, Odc1, and α-tubulin (control) yielded single peaks in the dissociation curve (primer sequences available upon request). mRNA expression levels of Abd-B and Odc1 were determined relative to α-tubulin expression by relative quantification. Relative expression levels of each gene in each genotype were normalized to C-S and statistical analysis was performed using t tests.
Supplementary Material
Acknowledgments
We thank R. Jones, J. Müller, G. Struhl, Å, Rasmusson-Lestander, and the Bloomington Drosophila Stock Center for fly strains, Tom Breen for generating the E(z)60 trxB11 and E(z)60 trxZ11 recombinants, and members of the Harte Lab for technical assistance. This work was supported by a grant from the National Institutes of Health to P.J.H. (AG19981) and shared instrumentation grants (S10RR024536) and (AI36219) to P. Conrad and the Case/University Hospitals of Cleveland Center for AIDS Research for Western blot imaging equipment.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907739107/DCSupplemental.
References
- 1.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 2.Cao R, et al. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- 3.Cao R, Zhang Y. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED-EZH2 complex. Mol Cell. 2004;15:57–67. doi: 10.1016/j.molcel.2004.06.020. [DOI] [PubMed] [Google Scholar]
- 4.Kurzhals RL, Tie F, Stratton CA, Harte PJ. Drosophila ESC-like can substitute for ESC and becomes required for Polycomb silencing if ESC is absent. Dev Biol. 2008;313:293–306. doi: 10.1016/j.ydbio.2007.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tie F, Stratton CA, Kurzhals RL, Harte PJ. The N terminus of Drosophila ESC binds directly to histone H3 and is required for E(Z)-dependent trimethylation of H3 lysine 27. Mol Cell Biol. 2007;27:2014–2026. doi: 10.1128/MCB.01822-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nekrasov M, et al. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. EMBO J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyer LA, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- 8.Bracken AP, Dietrich N, Pasini D, Hansen KH, Helin K. Genome-wide mapping of Polycomb target genes unravels their roles in cell fate transitions. Genes Dev. 2006;20:1123–1136. doi: 10.1101/gad.381706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee TI, et al. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell. 2006;125:301–313. doi: 10.1016/j.cell.2006.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwartz YB, et al. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- 11.Holmes AM, Weedmark KA, Gloor GB. Mutations in the extra sex combs and Enhancer of Polycomb genes increase homologous recombination in somatic cells of Drosophila melanogaster. Genetics. 2006;172:2367–2377. doi: 10.1534/genetics.105.042473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simon JA, Lange CA. Roles of the EZH2 histone methyltransferase in cancer epigenetics. Mutat Res. 2008;647:21–29. doi: 10.1016/j.mrfmmm.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Oktaba K, et al. Dynamic regulation by polycomb group protein complexes controls pattern formation and the cell cycle in Drosophila. Dev Cell. 2008;15:877–889. doi: 10.1016/j.devcel.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaw T, Martin P. Epigenetic reprogramming during wound healing: Loss of polycomb-mediated silencing may enable upregulation of repair genes. EMBO Rep. 2009;10:881–886. doi: 10.1038/embor.2009.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee N, Maurange C, Ringrose L, Paro R. Suppression of Polycomb group proteins by JNK signalling induces transdetermination in Drosophila imaginal discs. Nature. 2005;438:234–237. doi: 10.1038/nature04120. [DOI] [PubMed] [Google Scholar]
- 17.Bracken AP, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 19.Gil J, Bernard D, Martínez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- 20.Pinkston JM, Garigan D, Hansen M, Kenyon C. Mutations that increase the life span of C. elegans inhibit tumor growth. Science. 2006;313:971–975. doi: 10.1126/science.1121908. [DOI] [PubMed] [Google Scholar]
- 21.Sedivy JM, Munoz-Najar UM, Jeyapalan JC, Campisi J. In: Cellular Senescence: A Link between Tumor Suppression and Organismal Aging? Molecular Biology of Aging. Guarente L, Partridge L, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. pp. 185–214. [Google Scholar]
- 22.Cavalli G, Paro R. Epigenetic inheritance of active chromatin after removal of the main transactivator. Science. 1999;286:955–958. doi: 10.1126/science.286.5441.955. [DOI] [PubMed] [Google Scholar]
- 23.Klymenko T, Müller J. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep. 2004;5:373–377. doi: 10.1038/sj.embor.7400111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poux S, Horard B, Sigrist CJ, Pirrotta V. The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development. 2002;129:2483–2493. doi: 10.1242/dev.129.10.2483. [DOI] [PubMed] [Google Scholar]
- 25.Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- 26.Smith ST, et al. Modulation of heat shock gene expression by the TAC1 chromatin-modifying complex. Nat Cell Biol. 2004;6:162–167. doi: 10.1038/ncb1088. [DOI] [PubMed] [Google Scholar]
- 27.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 28.Bernstein BE, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 29.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 30.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin YJ, Seroude L, Benzer S. Extended life-span and stress resistance in the Drosophila mutant methuselah. Science. 1998;282:943–946. doi: 10.1126/science.282.5390.943. [DOI] [PubMed] [Google Scholar]
- 33.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 34.Wang HD, Kazemi-Esfarjani P, Benzer S. Multiple-stress analysis for isolation of Drosophila longevity genes. Proc Natl Acad Sci USA. 2004;101:12610–12615. doi: 10.1073/pnas.0404648101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bloyer S, Cavalli G, Brock HW, Dura JM. Identification and characterization of polyhomeotic PREs and TREs. Dev Biol. 2003;261:426–442. doi: 10.1016/s0012-1606(03)00314-2. [DOI] [PubMed] [Google Scholar]
- 36.Klebes A, et al. Regulation of cellular plasticity in Drosophila imaginal disc cells by the Polycomb group, trithorax group and lama genes. Development. 2005;132:3753–3765. doi: 10.1242/dev.01927. [DOI] [PubMed] [Google Scholar]
- 37.Beisel C, et al. Comparing active and repressed expression states of genes controlled by the Polycomb/Trithorax group proteins. Proc Natl Acad Sci USA. 2007;104:16615–16620. doi: 10.1073/pnas.0701538104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nègre N, et al. Chromosomal distribution of PcG proteins during Drosophila development. PLoS Biol. 2006;4:e170. doi: 10.1371/journal.pbio.0040170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang YL, et al. Essential role of Drosophila Hdac1 in homeotic gene silencing. Proc Natl Acad Sci USA. 2001;98:9730–9735. doi: 10.1073/pnas.171325498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Furuyama T, Banerjee R, Breen TR, Harte PJ. SIR2 is required for polycomb silencing and is associated with an E(Z) histone methyltransferase complex. Curr Biol. 2004;14:1812–1821. doi: 10.1016/j.cub.2004.09.060. [DOI] [PubMed] [Google Scholar]
- 41.Kuzmichev A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tie F, Prasad-Sinha J, Birve A, Rasmuson-Lestander A, Harte PJ. A 1-megadalton ESC/E(Z) complex from Drosophila that contains polycomblike and RPD3. Mol Cell Biol. 2003;23:3352–3362. doi: 10.1128/MCB.23.9.3352-3362.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pegg AE. Regulation of ornithine decarboxylase. J Biol Chem. 2006;281:14529–14532. doi: 10.1074/jbc.R500031200. [DOI] [PubMed] [Google Scholar]
- 44.Zhou Y, Ma C, Karmouch J, Katbi HA, Liu XJ. Antiapoptotic role for ornithine decarboxylase during oocyte maturation. Mol Cell Biol. 2009;29:1786–1795. doi: 10.1128/MCB.01815-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soda K, Dobashi Y, Kano Y, Tsujinaka S, Konishi F. Polyamine-rich food decreases age-associated pathology and mortality in aged mice. Exp Gerontol. 2009;44:727–732. doi: 10.1016/j.exger.2009.08.013. [DOI] [PubMed] [Google Scholar]
- 46.Eisenberg T, et al. Induction of autophagy by spermidine promotes longevity. Nat Cell Biol. 2009;11:1305–1314. doi: 10.1038/ncb1975. [DOI] [PubMed] [Google Scholar]
- 47.Chattopadhyay MK, Tabor CW, Tabor H. Polyamine deficiency leads to accumulation of reactive oxygen species in a spe2Delta mutant of Saccharomyces cerevisiae. Yeast. 2006;23:751–761. doi: 10.1002/yea.1393. [DOI] [PubMed] [Google Scholar]
- 48.Park JK, et al. c-Myc exerts a protective function through ornithine decarboxylase against cellular insults. Mol Pharmacol. 2002;62:1400–1408. doi: 10.1124/mol.62.6.1400. [DOI] [PubMed] [Google Scholar]
- 49.Ayroles JF, et al. Systems genetics of complex traits in Drosophila melanogaster. Nat Genet. 2009;41:299–307. doi: 10.1038/ng.332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McColl G, et al. Pharmacogenetic analysis of lithium-induced delayed aging in Caenorhabditis elegans. J Biol Chem. 2008;283:350–357. doi: 10.1074/jbc.M705028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cunningham JT, et al. mTOR controls mitochondrial oxidative function through a YY1-PGC-1alpha transcriptional complex. Nature. 2007;450:736–740. doi: 10.1038/nature06322. [DOI] [PubMed] [Google Scholar]
- 52.Wang L, et al. Hierarchical recruitment of polycomb group silencing complexes. Mol Cell. 2004;14:637–646. doi: 10.1016/j.molcel.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 53.Wilkinson FH, Park K, Atchison ML. Polycomb recruitment to DNA in vivo by the YY1 REPO domain. Proc Natl Acad Sci USA. 2006;103:19296–19301. doi: 10.1073/pnas.0603564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Carrington EA, Jones RS. The Drosophila Enhancer of zeste gene encodes a chromosomal protein: examination of wild-type and mutant protein distribution. Development. 1996;122:4073–4083. doi: 10.1242/dev.122.12.4073. [DOI] [PubMed] [Google Scholar]
- 55.Jones RS, Gelbart WM. Genetic analysis of the enhancer of zeste locus and its role in gene regulation in Drosophila melanogaster. Genetics. 1990;126:185–199. doi: 10.1093/genetics/126.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones RS, Gelbart WM. The Drosophila Polycomb-group gene Enhancer of zeste contains a region with sequence similarity to trithorax. Mol Cell Biol. 1993;13:6357–6366. doi: 10.1128/mcb.13.10.6357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Müller J, et al. Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell. 2002;111:197–208. doi: 10.1016/s0092-8674(02)00976-5. [DOI] [PubMed] [Google Scholar]
- 58.Sathe SS, Harte PJ. The Drosophila extra sex combs protein contains WD motifs essential for its function as a repressor of homeotic genes. Mech Dev. 1995;52:77–87. doi: 10.1016/0925-4773(95)00392-e. [DOI] [PubMed] [Google Scholar]
- 59.Tie F, Furuyama T, Harte PJ. The Drosophila Polycomb Group proteins ESC and E(Z) bind directly to each other and co-localize at multiple chromosomal sites. Development. 1998;125:3483–3496. doi: 10.1242/dev.125.17.3483. [DOI] [PubMed] [Google Scholar]
- 60.Breen TR. Mutant alleles of the Drosophila trithorax gene produce common and unusual homeotic and other developmental phenotypes. Genetics. 1999;152:319–344. doi: 10.1093/genetics/152.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Breen TR, Chinwalla V, Harte PJ. Trithorax is required to maintain engrailed expression in a subset of engrailed-expressing cells. Mech Dev. 1995;52:89–98. doi: 10.1016/0925-4773(95)00393-f. [DOI] [PubMed] [Google Scholar]
- 62.Mazo AM, Huang DH, Mozer BA, Dawid IB. The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA. 1990;87:2112–2116. doi: 10.1073/pnas.87.6.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Srinivasan S, Dorighi KM, Tamkun JW. Drosophila Kismet regulates histone H3 lysine 27 methylation and early elongation by RNA polymerase II. PLoS Genet. 2008;4:e1000217. doi: 10.1371/journal.pgen.1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thibault ST, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 65.Birve A, et al. Su(z)12, a novel Drosophila Polycomb group gene that is conserved in vertebrates and plants. Development. 2001;128:3371–3379. doi: 10.1242/dev.128.17.3371. [DOI] [PubMed] [Google Scholar]
- 66.King V, Tower J. Aging-specific expression of Drosophila hsp22. Dev Biol. 1999;207:107–118. doi: 10.1006/dbio.1998.9147. [DOI] [PubMed] [Google Scholar]
- 67.Girardot F, Monnier V, Tricoire H. Genome wide analysis of common and specific stress responses in adult drosophila melanogaster. BMC Genomics. 2004;5:74. doi: 10.1186/1471-2164-5-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.