Abstract
Regulation of the homeostatic concentrations of specific sets of transcription factors is essential for correct programming of cell proliferation and differentiation. We have characterized the signal transduction pathways regulating the catabolisis of p45/NF-E2, a bZIP factor activating the erythroid and megakaryocytic gene transcription. Through use of different approaches including nano-scale proteomics, we show that activated-JNK, or Phospho-JNK (P-JNK), physically interacts with p45/NF-E2 and phosphorylates its Ser157 residue. This reaction leads to the poly-ubiquitination of p45/NF-E2 at one or more of six Lys residues, one of which being also a sumoylation site, and its degradation through the proteasome pathway. Significantly, this regulatory pathway of p45/NF-E2 by P-JNK exists only in uninduced murine erythroleukemia (MEL) cells but not in differentiated MEL cells in which JNK is inactivated on DMSO induction. Based on the above data and analysis of the chromatin-binding kinetics of p45/NF-E2 and the erythroid gene repressor Bach1 during the early phase of MEL differentiation, we suggest a model for the regulation of erythroid maturation. In the model, the posttranslational modifications and turnover of p45/NF-E2, as mediated by P-JNK, contribute to the control of its homeostatic concentration and consequently, its regulatory functions in the progression of erythroid differentiation and erythroid gene expression.
Keywords: erythroid differentiation, phosphorylation, proteasome degradation, ubiquitination, homeostatic
Regulation of erythroid differentiation is complex and occurs in multiple steps. Cytokines and nuclear hormones all play important roles in the maturation of erythroid cells. For example, it is known that signaling from the erythropoietin (Epo) receptor activates several pathways to promote cell proliferation, differentiation, and survival (1). Transcription factors such as GATA-1, EKLF, Bach1, and NF-E2 are also crucial for regulation of erythroid differentiation and maturation (2, 3). Among the many systems for studying the molecular mechanisms of erythroid differentiation is MEL (4–6). The MEL cells are derived from murine erythroleukemia by infection of the Friend virus, causing Epo-independent polyclonal expansion through a constitutive activation of the Epo receptor (7). During the early hours after exposure to differentiation-inducing agents such as DMSO or hexamethylene bisacetamide (HMBA), MEL cells undergo alterations that commit them to cessation of growth and to development of the characteristics of differentiation (8, 9).
Multiple kinases are involved during MEL differentiation. For example, the inhibition of PI3 kinase reduces the GATA-1 binding to its DNA targets and thus, prevents MEL differentiation. On one hand, the PKC δ, and possibly PKC ε and PKC ζ as well, also plays a role in the HMBA-mediated differentiation of MEL, although their downstream targets are not clear yet (10). On the other hand, the MAPK families including p38, JNK, and ERK have been shown to be essential for Epo-dependent cell growth (11, 12). This subfamily of the MAPK, including JNK and p38, could be activated by a number of growth factors such as Epo and stem cell factor (SCF), thus promoting the proliferation of immature erythroid cells as well as the survival of the hematopoietic cells (13). Also, a role for JNK in Epo-induced and stress-induced erythroid differentiation of MEL has been proposed (11, 14). However, the molecular basis for the role of JNK in the proliferation, differentiation, and survival in erythropoiesis is not clear.
The competitive chromatin-binding of two DNA-binding transcription factors to their cognate binding sequences seems to be one of the essential mechanisms of MEL differentiation. The regulatory regions of many erythroid genes [e.g., the locus-control region (LCRs) of the β and α globin gene families (6, 15–17)] consist of the Maf recognition element (MARE) that can be bound with a diverse set of leucine–zipper factors including p45, Bach1, Nrf1, and Nrf2, all of which could associate with the small Maf subunit as heterodimers (18–20). In undifferentiated MEL, the Bach1/sMaf heterodimer interacts with the MARE and thus, inhibits the expression of both erythroid-specific genes, including the globins and ubiquitous genes like the heme oxygenase 1. As suggested before (21, 22), the newly synthesized heme, during the induction of MEL to differentiate, binds to Bach1 and causes degradation of Bach1 through the ubiquitin–proteasome system (UPS). The degradation of Bach1 leads to the disruption of the corepressor complex(es) bound at the erythroid regulatory regions and recruitment of coactivators to these regions through binding of NF-E2, which consists of the erythroid-enriched bZIP factor p45 and the small Maf subunit. This displacement of Bach1 by p45 turns on the transcription of erythroid genes and allows MEL differentiation to proceed (21).
It seems that homeostatic control of the cellular concentrations of different factors is important for the erythroid differentiation process. The dynamic turnover of Bach1 during MEL differentiation leading to the exchange of DNA-binding between Bach1 and Maf and NF and E2, as described above, is one good example. Also, the concentrations of several protein kinases including phospho (P)-ERK and P-JNK decreased, and P-p38 increased on induction of MEL differentiation (23–26). The level of P-JNK in K562 cells also drastically dropped as early as 4 h after hemin induction (26). Finally, although it has been shown that the concentration of p45 protein is low in uninduced MEL cells and that it increases during erythroid differentiation, the regulation of turnover of p45 in erythroid cells before and after differentiation is not clear.
On one hand, the p45 was known to be modified by phosphorylation and sumoylation (27, 28), in particular, the cAMP-dependent PKA phosphorylates p45, but it had no effect on the DNA-binding or transactivation ability of NF-E2 (27). On the other hand, sumoylation of p45 enhances the transactivation ability of NF-E2, and this is modulated through the association between NF-E2 and the β-globin loci within the nuclear body PML oncogenic domains (POD) in erythroid cells including induced MEL (28). In contrast to sumoylation, very little, if any, is known about the processes of phosphorylation and ubiquitination of p45 in vivo and their biological roles in gene regulation and erythroid differentiation. As described below, we provide evidence that turnover of the p45 protein in undifferentiated MEL cells is regulated through the UPS as mediated by its phosphorylation by the P-JNK. On MEL differentiation, P-JNK is inactivated, and this is accompanied by stabilization of the p45 during the early phase of differentiation. We suggest a model of homeostatic control of the cellular concentrations of transcription factors p45 and Bach1, which is regulated by JNK, during erythroid differentiation.
Results
Phosphorylation of p45 Is Associated with Its Proteasome-Mediated Degradation.
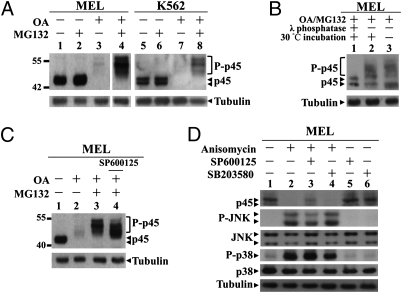
When MEL or the human K562 cells were treated for 2 h with 1 μM of Okadaic acid (OA), a Ser/Thr protein phosphatase inhibitor, disappearance of the p45 was observed (Fig. 1A, lanes 3 and 7). Furthermore, the proteasome inhibitor MG132 effectively overcame the OA-induced disappearance of p45 with slow-migrating species detected on the Western blots (WB) (Fig. 1A, lanes 4 and 8). These slow-migrating bands were phosphorylated p45 species, because incubation with λ phosphatase diminished the (OA+MG132)-induced band shift of p45 (Fig. 1B, lane 1). These results suggested that Ser/Thr phosphorylation of p45 caused the degradation of p45 through the proteasome (degradation) system.
Fig. 1.
Phosphorylation of p45 by P-JNK and its proteasome-mediated degradation in erythroid cells. (A) Phosphorylation-induced p45 degradation by the proteasome. MEL and K562 cells were treated with 1 μM OA with or without 60 μM MG132 for 2 h. The whole-cell extracts (WCE) were analyzed by WB with anti-p45 and anti-tubulin antibodies. (B) Dephosphorylation of p45 in vitro. WCE were prepared from MEL cells treated with (OA+MG132) for 2 h; 2 μg of the lysates were incubated at 30°C for 30 min with or without λ phosphatase and then analyzed by WB with anti-p45 Ab. (C) MEL cells were pretreated with 60 μM of SP600125 for 30 min before OA or (OA+MG132) treatment and then analyzed by WB. (D) Regulation of the p45 degradation by P-JNK. MEL cells were pretreated with 60 μM of SP600125 or 5 μM of SB203580 for 30 min before the incubation with 5 μg/mL of anisomycin for 1 h. The cell lysates were analyzed by WB as indicated.
JNK Mediates the Proteasomal Degradation of p45/NF-E2.
To find out the signaling pathways that contribute to the turnover of the p45 by the proteasome, we pretreated MEL cells with different kinase inhibitors followed by the OA or (OA+MG132) treatment. As seen, both staurosporine, a broad range Ser/Thr kinase inhibitor, and to lesser extent, the PKC inhibitor Calphostin C, inhibited the band shift of p45 (Fig. S1, compare lanes 5 and 11 to lanes 3 and 9).
To further identify the kinase(s) downstream of PKC that regulated the p45 degradation, different inhibitors and activators of ERK, p38, and JNK were used (Figs. 1 C and D and Fig. S1). Among the inhibitors tested, only the JNK inhibitor SP600125 affected the extent of phosphorylation of p45 (Fig. 1C, compare lanes 3 and 4).
In contrast, the JNK activator anisomycin caused a great increase of the levels of both P-JNK and P-p38 in MEL cells as well as a significant decrease of the amount of p45 (Fig. 1D, compare lanes 1 and 2). This decrease of p45 could be partially overcome by the JNK inhibitor SP600125 but not at all overcome by the p38 inhibitor SB203580 (Fig. 1D, compare lanes 2, 3 and 4). These data showed that P-JNK mediated the phosphorylation-dependent degradation of p45 in erythroid cells.
Active JNK Physically Interacts with p45 and Regulates the p45 Stability by Phosphorylating It at S157 and S346.
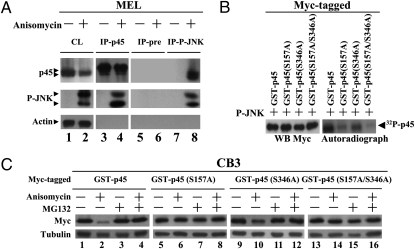
To examine how the active JNK triggered the proteasome degradation of p45, we first analyzed whether or not the endogenous p45 in MEL cells could interact with P-JNK. MEL cells were treated with anisomycin for 30 min, and the whole-cell lysates were subjected to immunoprecipitation (IP) with anti-p45 or anti-P-JNK Ab. As seen, p45 of anisomycin-treated MEL cells indeed interacted with P-JNK (Fig. 2A, lanes 4 and 8). Furthermore, transfection experiment in 293T cells showed that the domain of p45 interacting with P-JNK was located in the region of 1–83 aa (Fig. S2).
Fig. 2.
Phosphorylation of Ser157 of p45 by active JNK. (A) Interaction of the endogenous p45 with P-JNK. WCE were prepared from MEL cells with or without anisomycin treatment (5 μg/mL for 30 min), IP with anti-p45, anti-P-JNK, or the preimmune serum, and then analyzed by WB with anti-p45, anti-P-JNK, and anti-actin antibodies. CL, WCE without IP. (B) In vitro phosphorylation of p45 by P-JNK. Myc-tagged GST-p45-derived proteins (as indicated) were incubated with the active JNK in the in vitro phosphorylation reactions and analyzed either by WB and autoradiography. (C) Effects of S157A and S346A mutations on P-JNK-induced degradation of p45. CB3 cell pools stably expressing the indicated Myc-tagged GST-p45 variant were treated with 60 μM MG132 for 1 h, 5 μg/mL anisomycin for 2 h, or a combination of both. The WCE were then analyzed by WB with anti-Myc and anti-tubulin antibodies.
We then performed in vitro phosphorylation assay to map the site(s) of phosphorylation of p45 by P-JNK. As seen in the left panel of Fig. S3A, Myc-tagged GST-p45 could be labeled with 32P from [32P]ATP after in vitro phosphorylation reaction with the recombinant active JNK. The phosphorylated Myc-tagged GST-p45 band on the gel was then excised and subjected to mass spectrometry analysis. The MS/MS and MS3 spectra (Fig. S3A, right panel) indicated that S157 (major) and S346 (minor) of p45 were phosphorylated by the P-JNK. Indeed, mutations of these sites, in particular S157, greatly decreased the 32P-labeling of p45 by the active JNK in vitro phosphorylation reaction (Fig. 2B). Whether or not p45 phosphorylation at S157 and/or S346 by P-JNK affected its stability was directly tested by analysis of stable transfectants of CB3, a p45-null derivative of MEL (29). CB3 cells were stably transfected with pEF-based plasmids expressing GST-p45-Myc, GST-p45(S157A)-Myc, GST-p45(S346A)-Myc, or GST-p45(S157A/S346A)-Myc. Pools from independent transfections were selected and then treated with anisomycin with or without the presence of MG132. As seen in Fig. 2C, the S→A mutation at S157 (lanes 5–8) and to lesser extent, the mutation of S346 (lanes 9–12), overcame the P-JNK-dependent, proteasome-mediated degradation of p45.
We also measured the stability of p45 by the cycloheximide (CHX) chase experiments. As seen in Fig. S3B, the S157A substitution but not S346A drastically increased the stability of p45. The data of Figs. 1 and 2 together showed that p45 could be phosphorylated at S157 and S346 in uninduced MEL cells through the JNK-signaling pathway. Furthermore, the phosphorylation of p45 at S157 was required for the proteasome-mediated degradation of the p45.
Proteasome Degradation of p45 Occurs Through Poly-Ubiquitination at Multiple Sites.
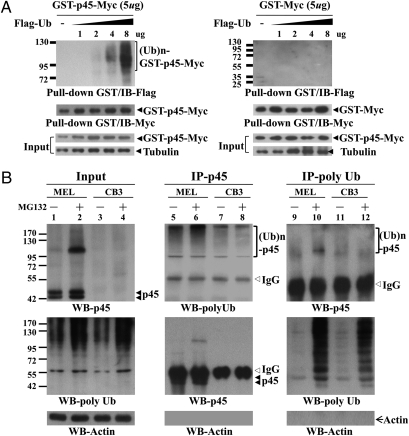
To examine the degradation of p45 by the UPS, we cotransfected either pEF-GST-Myc or pEF-GST-p45-Myc with different amounts of pEF-Flag-ubiquitin (Ub) into 293T cells. The whole-cells extracts were subjected to GST pull-down and WB analysis. As shown in the left panel of Fig. 3A, the Flag-Ub molecules were effectively conjugated to GST-p45-Myc to form poly-ubiquitinated species. The high molecular-weight bands resulted from poly-ubiquitination of p45, instead of GST or Myc of the GST-p45-Myc fusion polypeptide, because little poly-ubiquitination occurred on GST-Myc in transfected 293T cells (Fig. 3A, right panel). We then compared the stability of the endogenous p45 protein in MEL cells with or without MG132 treatment. As measured by the CHX chase experiments, p45 had a relatively short half-life (t1/2) of about 1–1.5 h (Fig. S4). As expected, inhibition of the proteasome activity maintained the level of p45 throughout the course of CHX treatment (Fig. S4, second panel). Furthermore, in contrast to CHX treatment alone, the MG132 cotreatment also led to a significant increase of discrete, high-molecular weight p45 bands, a pattern typical of poly-ubiquitinated proteins (Fig. S4, second panel). As expected, the p45-derived band patterns could not be observed in a parallel experiment using the p45-null cell line CB3 (Fig. S4, right two panels). Finally, the high molecular weight p45 species were indeed conjugated with Ub, as shown by Co-IP experiments (Fig. 3B) using both the anti-FK2, which selectively recognized ubiquitinated proteins but not free Ub, and anti-p45.
Fig. 3.
Poly-ubiquitination of p45. (A) Poly-ubquitination of p45 in transfected 293T cells. The cells were cotransfected with pEF-GST-p45-Myc or pEF-GST-Myc with different amounts of pEF-Flag-Ub. At 45 h posttransfection, the cells were treated with 10 μM MG132 for 3 h. The WCE were then prepared, and the Myc-tagged GST-fusion proteins were isolated by GST pull-down assay. Then, they were analyzed by WB with anti-Flag and anti-Myc antibodies. (B) Poly-ubiquitination of the endogenous p45 in erythroid cell lines. The MEL and CB3 cells were treated with 20 μM MG132 for 3 h, and the WCE were prepared for analysis by IP. The IP samples were then analyzed by WB with uses of anti-p45, anti-FK2 (for poly-Ub), and anti-actin antibodies.
To identify the main ubiquitination sites, combined deletion and site-directed mutagenesis were first used. Constructs expressing different GST-fusion p45 fragments, including pEF-GST-p45(1-83)-Myc, pEF-GST-p45(78-206)-Myc, pEF-GST-p45(207-268)-Myc, pEF-GST-p45(269-297)-Myc, and pEF-GST- p45(298-373)-Myc, were individually cotransfected with pEF-Flag-Ub into 293T cells. The GST–fusion polypeptides in cell lysates were then purified by GST pull-down assay, and their conjugations by the Flag-Ub were analyzed by WB. The Lys residues modified by Flag-Ub were further mapped by site-directed mutagenesis, and their involvement in proteasome degradation was confirmed by transfection into K562 cells followed by (OA+MG132) treatment.
After the above initial mapping of the regions of p45 that could be poly-ubiquitinated in transfected 293T cells, we have identified six putative ubiquitinatable Lys residues including K108, K137, K215, K234, K241, and K368 (Fig. S5A, Top). To analyze the role of ubiquitination at these sites in p45 degradation, we generated six different so-called 5KR mutants of p45, each of which had one of the above Lys residues preserved whereas the other five substituted with Arg (R). These 5KR mutants were then individually expressed in K562 cells by transient transfection, treated or untreated with anisomycin, and analyzed by WB. As shown in the middle panels and bottom histogram of Fig. S5A, the wild-type p45 as well as the six 5KR mutants (Fig. S5A, panels 2–8) each exhibited a lowered steady-state level on anisomycin treatment. In interesting contrast, the JNK-mediated degradation was not observed for the p45 mutant with all of the six Lys mutated p45(6KR) (Fig. S5A, panel 9). Consistent with the data of Fig. S5, the WT p45 and the six 5KR mutants, but not p45(6KR), could all be poly-ubiquitinated, which is exemplified in Fig. S5B. These results together showed that the JNK-mediated degradation of p45 was through poly-ubiquitination at one or more of the Lys residues 108, 137, 215, 234, 241, and 368, although the contributions from K108 and K137 were not as big as the others. It is interesting to note here that K368 is also the sumoylation site of p45 (28).
P-JNK-Mediated Turnover and Differential Chromatin-Binding of p45 Protein Is Regulated During MEL Differentiation.
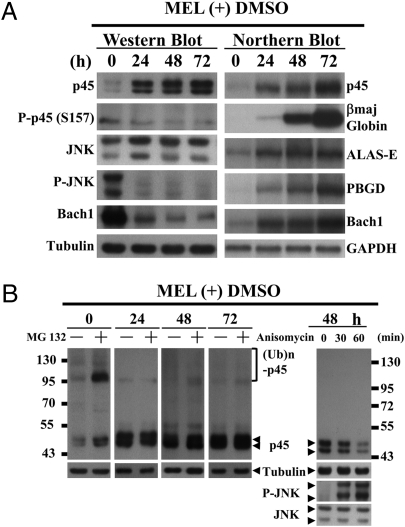
After the above studies, we have addressed the physiological role of P-JNK-mediated degradation of p45 by UPS. To examine this, we analyze the kinetics of changes of the amounts of p45, phosphorylated p45(S157) (P-p45(S157)), P-JNK, and Bach1 in MEL cells at the intervals of 24 h on differentiation (Fig. 4A Left). We also followed the 24-h interval kinetics of the changes of the mRNA levels of p45, βmaj globin, Bach1, 5-aminolevulinic acid synthase (ALAS-E), and porphobilinogen deaminase (PBGD), the latter two of which encoded enzymes required for the heme synthetic pathway (Fig. 4A Right).
Fig. 4.
Kinetics of p45 turnover during MEL differentiation. (A) Amount changes of specific proteins and RNAs during MEL cell differentiation. The levels of different proteins and RNAs in MEL cells without and with DMSO induction for 24–72 h were analyzed by WB with appropriate antibodies and analyses for the expression of p45, βmaj, ALAS-E, PBGD, Bach1, and GAPDH genes by Northern blotting. (B) DMSO-induced disappearance of poly-ubiquitination and proteasome-mediated degradation of p45 in MEL cells. MEL cells were treated with 20 μM MG132 for 1 h after DMSO induction for 24, 48, and 72 h, respectively, and then analyzed by WB with anti-p45 and anti-tubulin antibodies. The WCE in the right panel were prepared from MEL cells induced with DMSO for 48 h followed by treatment with anisomycin and then, analyzed by WB.
As shown, the amounts of both the protein and mRNA of p45 were increased after DMSO induction as reported previously (30). Significantly, the increase of the level of p45 protein in MEL cells during differentiation was in parallel with the drastic decreases of both the level of p45 phosphorylated at S157 or P-p45(S157), and that of P-JNK (Fig. 4A, panels 2 and 4). This result provided further support to the scenario that the cellular level of the p45 protein was down-regulated by P-JNK-mediated phosphorylation and consequent poly-ubiquitination. Consistent with the above observations, WB showed that although the p45 protein level increased along the course of differentiation, the poly-ubiquitinated p45 species on the bolt decreased and were essentially undetectable after 24 h of differentiation (left 4 panels, Fig. 4B). Also, reactivation of P-JNK by anisomycin decreased the level of p45 in MEL cells induced by DMSO for 48 h, most likely through redirecting p45 into the UPS (Fig. 4B Right). As expected, this lowering of the p45 protein also caused decrease of the βmaj globin transcription (Fig. S6B). Significantly, CHX-chase experiments showed that the observations in Fig. 4 were accompanied with lengthening of the t1/2 of the p45 protein, by 0.5–1.5 h, in differentiated MEL cells (Fig. S6C).
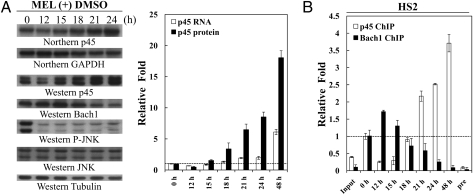
What is the functional consequence (s) of the stabilization of p45 protein during differentiation of the MEL cells? One likelihood was that the process ensured a rapid increase of the amount of p45 during the early phase of differentiation when the transcription of p45 gene was not fully activated yet. As shown in Fig. 5A, the p45 protein level increased rapidly, by eightfold, in between 12 and 24 h on DMSO induction, when compared with 0 h. During this time period, the level of the p45 mRNA was only increased to twofold of the 0-h level. Remarkably, this rapid increase of p45 protein was accompanied by the replacement of binding of Bach1 at the HS2 element by p45/NF-E2 as early as sometime between 18 and 21 h of DMSO induction (Fig. 5B). The data of Fig. 5 suggested that exchange of the p45/NF-E2 and Bach1-Maf bindings at different erythroid regulatory elements, including HS2, on erythroid differentiation were not only caused by the rapid degradation of Bach1 (21, 22) but that it was also facilitated by the rapid increase of p45 as the result of its stabilization on differentiation.
Fig. 5.
Kinetics of changes of the expression levels and chromatin binding of p45 and Bach1 during the early phase of MEL differentiation. (A) Changes of expression. The MEL cells were harvested at 12, 15, 18, 21, and 24 h after DMSO induction and analyzed by Northern blotting and WB. The relative levels of the mRNAs and proteins were calculated and compared in the histogram. (B) Changes of chromatin binding. ChIP assay was used to examine the differential bindings of p45 and Bach1 at the HS2 in MEL cells at differential time of DMSO induction. The signals were detected by quantitative RT-PCR and standardized to those of the input: the preimmune samples (pre) and the β-actin gene. The relative intensities of the PCR signals and folds of increases over the 0-h sample were then calculated.
Discussion
In this study, we have investigated the roles of the signal transduction pathways during erythroid differentiation of the MEL cells. Through combined uses of different kinase inhibitors and activators, inhibitor of the UPS, transient DNA cotransfection experiments, site-directed mutagenesis, and in vitro kinase reaction, we have showed that turnover of the p45 is mediated through the physical interaction between the proline-rich domain of p45 with the P-JNK that specifically phosphorylates p45 at S157 (Figs. 1–2 and Figs. S1–S3). This is followed by poly-ubiquitination of p45 at one or more of six specific Lys residues and its subsequent degradation by the UPS (Fig. 3 and Figs. S4 and S5). Significantly, this finding has provided important insight into the molecular/cellular mechanisms regulating the reciprocal changes of the cellular concentrations of the transcription factors NF-E2 and Bach1 (Figs. 4 and 5), which are known to be essential for the initiation and progression of MEL differentiation.
The regulation of the p45 stability is in great contrast to several other proteins, the stabilities of which are also known to be regulated by JNK, phosphorylation, and the UPS (31). For instance, in vitro experiments have suggested that inactive JNK, instead of P-JNK, targets c-jun for efficient poly-ubiquitination. Furthermore, phosphorylation of c-jun at S73 by P-JNK protects the factor from poly-ubiquitination and lengthens its t1/2 (32). In another case, the inactive JNK and MDM2, an E3 Ub ligase, can interact with p53 at G0/G1 phase and S/G2M phase, respectively. Both factors can trigger the poly-ubiquitination of p53 and its degradation by the 26S proteasome under nonstress conditions. After stress stimulation like UV, however, phosphorylation of the N-terminal domain of p53, by one or more kinases of the MEKK1-JNK pathway including P-JNK, induces a conformational change. This change dissociates p53 from MDM2 and thus, increases its t1/2 (33). It has also been shown that P-JNK phosphorylates Sp1 at T278 and T139 and this phosphorylation increase the Sp1 stability by repressing the Sp1 degradation through the proteasome-dependent pathway (34). Finally, the degradation of JunB by the proteasome has been shown to be initiated by P-JNK phosphorylation of the E3 ligase (Itch) instead of the JunB itself (35). Thus, the regulatory scenario of p45 is unique in that direct interaction of the factor with and its phosphorylation by P-JNK caused its poly-ubiquitination and proteasome degradation (Fig. S7). The contrast between p45 and the above-mentioned cases is likely caused by the different cellular environments and extracellular stimuli under which the stabilities of these factors are regulated.
As depicted in the cartoon model of Fig. S8, the regulation of the stability of p45 by P-JNK provides an essential link of the homeostatic program that regulates the MEL differentiation. On one hand, it is known that the JNK activity plays a crucial role in the proliferation of immature, primary erythroid cells as well as several erythroid cell lines, and it also has been found that it is not required for the survival or proliferation of certain erythroid cells like the CFU-es and proerythroblasts (36). On the other hand, it is known that p45 is a negative regulator of proliferation of erythroid cells including MEL (37). It is, thus, not surprising that erythroid cells like MEL has evolved to have such a pathway of P-JNK-mediated proteasome degradation of p45. In uninduced MEL, this pathway maintains the p45 at a relatively low level that is sufficient for the cell survival/growth but not enough to activate the erythroid genes (Fig. S8 Left). On DMSO induction of MEL to differentiate, the JNK is inactivated, and the diminishing of P-JNK prolongs the t1/2 of p45 and helps to elevate the amount of p45 during the early stage (<24 h) of MEL differentiation when the p45 gene transcription is not fully on yet. Previously, we have shown that p45 is sumoylated at the residue K368, one of the six sites of p45 poly-ubiquitination mapped in this study (Fig. S5). This sumoylation of p45 is required for NF-E2 to transactivate erythroid genes in erythroid cells, including induced MEL, because of enhanced DNA binding by the sumoylated p45/NF-E2 and its ability to bring in/maintain the active genes loci, such as the β globin, within the nuclear body POD (28). Thus, similar to other studies of the antagonistic effects of ubiquitination and sumoylation (38–40), K368 of p45 becomes accessible to sumoylation on inactivation of JNK in the induced MEL cells.
As a result of the above two scenarios, the effective concentration of active p45 at the early stage of differentiation becomes high enough to bind to and activate the promoters of a set of genes, including those encoding enzymes of the heme–synthesis pathway, such as ALAS-E (Fig. S8 Right). As shown previously by Zenke-Kawasaki et al. (41), after accumulation of heme to a threshold level, the Bach1 is degraded through the UPS. Indeed, our ChIP analysis of MEL cells during the early phase of DMSO-induced differentiation showed increased binding of p45 at the ALAS-E promoter (Fig. S9A). This was accompanied with the increase of the ALAS-E mRNA and a reciprocal decrease of the Bach1 protein during the early phase (12–21 h) of the MEL differentiation (Fig. S9B). At the mean time during the early to mid stage of MEL differentiation, an increasingly high level of the active p45 protein is assured through the continued inactivation of JNK and consequent lack of p45 degradation by the UPS, the accessibility of K368 for sumoylation of p45, and the further enhancement of the p45 gene transcription by p45 itself (42). Thus, this reciprocal change of the concentrations of p45 and Bach1 factors leads to the replacement of Bach1-Maf binding by that of p45/NF-E2 at the regulatory regions of different erythroid genes, including the HS2/β-LCR of the β globin locus (Fig. S8), as suggested previously (21, 22, 42). Our ChIP analysis indicates that this exchange of chromatin binding of Bach1 and p45 occurs as early as 12–21 h after MEL differentiation, and it is eventually completed at 48 h of differentiation (Fig. 5B and Fig. S9A).
In summary, we have uncovered and characterized in biochemical detail a cellular pathway, as initiated by P-JNK, that is important for the homeostatic control of the cellular concentration of the transcription factor p45 and consequently, of the other factors, including Bach1, during MEL differentiation. The study provides an evidence for P-JNK to function through physical interaction and phosphorylation of a target protein, thus triggering the turnover of the latter by the UPS. It will be interesting in the future to identify and characterize the upstream signals and molecular players that regulate JNK during erythroid differentiation.
Materials and Methods
Antibodies and Reagents.
Detailed sources of reagents and antibodies are described in SI Materials and Methods.
Cell Culture and Transfection.
For the culturing conditions of 293T, MEL, CB3, and K562 cells, see SI Materials and Methods. Erythroid differentiation of MEL cells was induced by adding 2% DMSO (Merck) to the media.
IP and WB.
The IP and WB procedures are as described in ref. 28.
In vitro dephosphorylation, phosphorylation, ubiquitination, and others protocols are described in detail in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Kazuhiko Igarashi for his gift of the Bach1 Ab, Dr. Zee-Fen Chang for the pMyc-Ub plasmid, and Dr. Ruey-Hwa Chen for her critical comments. We also thank A.Y. Chou and Y.J. Huang for their help. The critical comments from two anonymous reviewers are greatly appreciated. This research was supported by the National Health Research Institute Grant NHRI-EX98-9514SI, the National Science Council, and the Academia Sinica (AS), Taiwan, Republic of China. Y.-C.S. is an AS Distinguished Postdoctoral Scholar, and C.-K.J.S. is an AS Investigator Awardee.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0909153107/DCSupplemental.
References
- 1.Ingley E, Tilbrook PA, Klinken SP. New insights into the regulation of erythroid cells. IUBMB Life. 2004;56:177–184. doi: 10.1080/15216540410001703956. [DOI] [PubMed] [Google Scholar]
- 2.Andrews NC, Erdjument-Bromage H, Davidson MB, Tempst P, Orkin SH. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature. 1993;362:722–728. doi: 10.1038/362722a0. [DOI] [PubMed] [Google Scholar]
- 3.Koury MJ, Sawyer ST, Brandt SJ. New insights into erythropoiesis. Curr Opin Hematol. 2002;9:93–100. doi: 10.1097/00062752-200203000-00002. [DOI] [PubMed] [Google Scholar]
- 4.Curtis PJ. Globin mRNA in Friend cells: Its structure, function and synthesis. Biochim Biophys Acta. 1980;605:347–364. doi: 10.1016/0304-419x(80)90016-5. [DOI] [PubMed] [Google Scholar]
- 5.Andrews NC, Orkin SH. Transcriptional control of erythropoiesis. Curr Opin Hematol. 1994;1:119–124. [PubMed] [Google Scholar]
- 6.Bulger M, Sawado T, Schübeler D, Groudine M. ChIPs of the beta-globin locus: Unraveling gene regulation within an active domain. Curr Opin Genet Dev. 2002;12:170–177. doi: 10.1016/s0959-437x(02)00283-6. [DOI] [PubMed] [Google Scholar]
- 7.Marks PA, Richon VM, Kiyokawa H, Rifkind RA. Inducing differentiation of transformed cells with hybrid polar compounds: A cell cycle-dependent process. Proc Natl Acad Sci USA. 1994;91:10251–10254. doi: 10.1073/pnas.91.22.10251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Levy J, Terada M, Rifkind RA, Marks PA. Induction of erythroid differentiation by dimethylsulfoxide in cells infected with Friend virus: Relationship to the cell cycle. Proc Natl Acad Sci USA. 1975;72:28–32. doi: 10.1073/pnas.72.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fibach E, Reuben RC, Rifkind RA, Marks PA. Effect of hexamethylene bisacetamide on the commitment to differentiation of murine erythroleukemia cells. Cancer Res. 1977;37:440–444. [PubMed] [Google Scholar]
- 10.Leng L, et al. Differential modulation of protein kinase C isoforms in erythroleukemia during induced differentiation. Cancer Res. 1993;53:5554–5558. [PubMed] [Google Scholar]
- 11.Nagata Y, Takahashi N, Davis RJ, Todokoro K. Activation of p38 MAP kinase and JNK but not ERK is required for erythropoietin-induced erythroid differentiation. Blood. 1998;92:1859–1869. [PubMed] [Google Scholar]
- 12.Jacobs-Helber SM, Ryan JJ, Sawyer ST. JNK and p38 are activated by erythropoietin (EPO) but are not induced in apoptosis following EPO withdrawal in EPO-dependent HCD57 cells. Blood. 2000;96:933–940. [PubMed] [Google Scholar]
- 13.Arcasoy MO, Jiang X. Co-operative signaling mechanisms required for erythroid precursor expansion in response to erythropoietin and stem cell factor. Br J Haematol. 2005;130:121–129. doi: 10.1111/j.1365-2141.2005.05580.x. [DOI] [PubMed] [Google Scholar]
- 14.Nagata Y, Todokoro K. Requirement of activation of JNK and p38 for environmental stress-induced erythroid differentiation and apoptosis and of inhibition of ERK for apoptosis. Blood. 1999;94:853–863. [PubMed] [Google Scholar]
- 15.Grosveld F. Activation by locus control regions? Curr Opin Genet Dev. 1999;9:152–157. doi: 10.1016/S0959-437X(99)80023-9. [DOI] [PubMed] [Google Scholar]
- 16.Tanimoto K, Engel JD. In vivo modulation of human beta-globin gene switching. Trends Cardiovasc Med. 2000;10:15–19. doi: 10.1016/s1050-1738(00)00035-9. [DOI] [PubMed] [Google Scholar]
- 17.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blank V, Andrews NC. The Maf transcription factors: Regulators of differentiation. Trends Biochem Sci. 1997;22:437–441. doi: 10.1016/s0968-0004(97)01105-5. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi K. Function of NF-E2-related transcription factors. Seikagaku. 2000;72:257–268. [PubMed] [Google Scholar]
- 20.Motohashi H, O'Connor T, Katsuoka F, Engel JD, Yamamoto M. Integration and diversity of the regulatory network composed of Maf and CNC families of transcription factors. Gene. 2002;294:1–12. doi: 10.1016/s0378-1119(02)00788-6. [DOI] [PubMed] [Google Scholar]
- 21.Brand M, et al. Dynamic changes in transcription factor complexes during erythroid differentiation revealed by quantitative proteomics. Nat Struct Mol Biol. 2004;11:73–80. doi: 10.1038/nsmb713. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, et al. Heme regulates the dynamic exchange of Bach1 and NF-E2-related factors in the Maf transcription factor network. Proc Natl Acad Sci USA. 2004;101:1461–1466. doi: 10.1073/pnas.0308083100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Matsuzaki T, Aisaki K, Yamamura Y, Noda M, Ikawa Y. Induction of erythroid differentiation by inhibition of Ras/ERK pathway in a friend murine leukemia cell line. Oncogene. 2000;19:1500–1508. doi: 10.1038/sj.onc.1203461. [DOI] [PubMed] [Google Scholar]
- 24.Uddin S, Ah-Kang J, Ulaszek J, Mahmud D, Wickrema A. Differentiation stage-specific activation of p38 mitogen-activated protein kinase isoforms in primary human erythroid cells. Proc Natl Acad Sci USA. 2004;101:147–152. doi: 10.1073/pnas.0307075101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishigaki K, Hanson C, Thompson D, Yugawa T, Ruscetti S. Activation of the Jun N-terminal kinase pathway by friend spleen focus-forming virus and its role in the growth and survival of friend virus-induced erythroleukemia cells. J Virol. 2005;79:12752–12762. doi: 10.1128/JVI.79.20.12752-12762.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDevitt MA, et al. A critical role for the host mediator macrophage migration inhibitory factor in the pathogenesis of malarial anemia. J Exp Med. 2006;203:1185–1196. doi: 10.1084/jem.20052398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casteel D, Suhasini M, Gudi T, Naima R, Pilz RB. Regulation of the erythroid transcription factor NF-E2 by cyclic adenosine monophosphate-dependent protein kinase. Blood. 1998;91:3193–3201. [PubMed] [Google Scholar]
- 28.Shyu YC, et al. Sumoylation of p45/NF-E2: Nuclear positioning and transcriptional activation of the mammalian beta-like globin gene locus. Mol Cell Biol. 2005;25:10365–10378. doi: 10.1128/MCB.25.23.10365-10378.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu SJ, Rowan S, Bani MR, Ben-David Y. Retroviral integration within the Fli-2 locus results in inactivation of the erythroid transcription factor NF-E2 in Friend erythroleukemias: Evidence that NF-E2 is essential for globin expression. Proc Natl Acad Sci USA. 1994;91:8398–8402. doi: 10.1073/pnas.91.18.8398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Francastel C, Poindessous-Jazat V, Augery-Bourget Y, Robert-Lézénès J. NF-E2p18/mafK is required in DMSO-induced differentiation of Friend erythroleukemia cells by enhancing NF-E2 activity. Leukemia. 1997;11:273–280. doi: 10.1038/sj.leu.2400552. [DOI] [PubMed] [Google Scholar]
- 31.Bogoyevitch MA, Kobe B. Uses for JNK: The many and varied substrates of the c-Jun N-terminal kinases. Microbiol Mol Biol Rev. 2006;70:1061–1095. doi: 10.1128/MMBR.00025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fuchs SY, Dolan L, Davis RJ, Ronai Z. Phosphorylation-dependent targeting of c-Jun ubiquitination by Jun N-kinase. Oncogene. 1996;13:1531–1535. [PubMed] [Google Scholar]
- 33.Fuchs SY, Fried VA, Ronai Z. Stress-activated kinases regulate protein stability. Oncogene. 1998;17:1483–1490. doi: 10.1038/sj.onc.1202184. [DOI] [PubMed] [Google Scholar]
- 34.Chuang JY, et al. Phosphorylation by c-Jun NH2-terminal kinase 1 regulates the stability of transcription factor Sp1 during mitosis. Mol Biol Cell. 2008;19:1139–1151. doi: 10.1091/mbc.E07-09-0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu YC, Penninger J, Karin M. Immunity by ubiquitylation: A reversible process of modification. Nat Rev Immunol. 2005;5:941–952. doi: 10.1038/nri1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jacobs-Helber SM, Sawyer ST. Jun N-terminal kinase promotes proliferation of immature erythroid cells and erythropoietin-dependent cell lines. Blood. 2004;104:696–703. doi: 10.1182/blood-2003-05-1754. [DOI] [PubMed] [Google Scholar]
- 37.Li YJ, et al. p45(NFE2) is a negative regulator of erythroid proliferation which contributes to the progression of Friend virus-induced erythroleukemias. Mol Cell Biol. 2001;21:73–80. doi: 10.1128/MCB.21.1.73-80.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastushok L, Xiao W. DNA postreplication repair modulated by ubiquitination and sumoylation. Adv Protein Chem. 2004;69:279–306. doi: 10.1016/S0065-3233(04)69010-3. [DOI] [PubMed] [Google Scholar]
- 39.Steffan JS, et al. SUMO modification of Huntingtin and Huntington's disease pathology. Science. 2004;304:100–104. doi: 10.1126/science.1092194. [DOI] [PubMed] [Google Scholar]
- 40.Ulrich HD. Mutual interactions between the SUMO and ubiquitin systems: A plea of no contest. Trends Cell Biol. 2005;15:525–532. doi: 10.1016/j.tcb.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 41.Zenke-Kawasaki Y, et al. Heme induces ubiquitination and degradation of the transcription factor Bach1. Mol Cell Biol. 2007;27:6962–6971. doi: 10.1128/MCB.02415-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee TL, Wen SC, Hsiao WY, Shen CK, Shyu YU. Self-regulation of mouse p45/NF-E2 during murine erythroleukemia cell differentiation. Zool Studies. 2009;48:362–369. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.