Abstract
Changes in tooth shape have played a major role in vertebrate evolution with modification of dentition allowing an organism to adapt to new feeding strategies. The current view is that molar teeth evolved from simple conical teeth, similar to canines, by progressive addition of extra “cones” to form progressively complex multicuspid crowns. Mammalian incisors, however, are neither conical nor multicuspid, and their evolution is unclear. We show that hypomorphic mutation of a cell surface receptor, Lrp4, which modulates multiple signaling pathways, produces incisors with grooved enamel surfaces that exhibit the same molecular characteristics as the tips of molar cusps. Mice with a null mutation of Lrp4 develop extra cusps on molars and have incisors that exhibit clear molar-like cusp and root morphologies. Molecular analysis identifies misregulation of Shh and Bmp signaling in the mutant incisors and suggests an uncoupling of the processes of tooth shape determination and morphogenesis. Incisors thus possess a developmentally suppressed, cuspid crown-like morphogenesis program similar to that in molars that is revealed by loss of Lrp4 activity. Several mammalian species naturally possess multicuspid incisors, suggesting that mammals have the capacity to form multicuspid teeth regardless of location in the oral jaw. Localized loss of enamel may thus have been an intermediary step in the evolution of cusps, both of which use Lrp4-mediated signaling.
Keywords: cusp, Lrp4, tooth development, evo/devo, multicuspid crown
Vertebrates exhibit remarkable diversity in their dentitions, which is a feature of the importance of tooth shape in adaptation to new feeding strategies in evolution. Even quite closely related species of mammals can have different shapes of teeth and thus tooth development provides an excellent model for molecularly based evolutionary developmental biological studies (evo/devo). These tooth evolutional changes took place by the activation or inactivation of gene function, and thus evolutionary lost structures or gene activation/inactivation during evolution are occasionally retained as vestigial structures or latent gene activation/inactivation at embryonic stages.
The current view is that all mammalian teeth evolved from simple ancestral teeth with a conical shape not dissimilar to mammalian canines (1). Mammalian heterodont dentitions contain a variety of tooth shapes and most evo/devo studies have focused solely on the molar dentition, with cuspal morphology being used as the main comparative feature between specimens (1). A cusp is a pointed or rounded projection of the tooth that is composed of both enamel and dentin, and the general consensus is that multicuspid teeth (molariform) evolved from conical teeth by progressive addition of extra “cones” (1). Incisors however are a uniquely mammalian tooth type that are neither conical nor multicuspid and their evolutional process is not understood.
Among mammalian teeth, murine dentition has been used as a powerful tool for evo/devo studies because of the relative ease of gene manipulation. A major defining feature of Rodentia is the presence of continuously growing incisors. Most mammalian teeth consist of a clearly recognizable crown that consists of a thin coating of enamel covering a thicker layer of dentine, and roots that are composed only of dentine that is often surrounded by an external layer of a supporting tissue (e.g., periodontal ligament). Rodent incisors, however, have no obvious crown or roots but have two distinct surfaces: a labial surface of enamel-covered dentine and lingual surface of dentine only. It has been suggested that the labial side corresponds to the crown and the lingual side corresponds to the root (2, 3).
The low-density lipoprotein (LDL) receptor family is a large, evolutionarily conserved group of transmembrane proteins (4, 5). The LDL receptor was first identified as an endocytic receptor that transports the lipoprotein LDL into cells by receptor-mediated endocytosis. More recent findings have shown that LDL receptor family members can also function as direct signal transducers or modulators for a broad range of cellular signaling pathways (6–9).
We show here that rodent incisors possess a developmentally suppressed, cuspid crown–like morphogenesis program that is revealed by loss of Lrp4 activity. Lrp4 is thus responsible for maintaining the simple shape of incisors by suppression of cusp formation in development, a process that uncovers a likely route of mammalian incisor evolution.
Results and Discussion
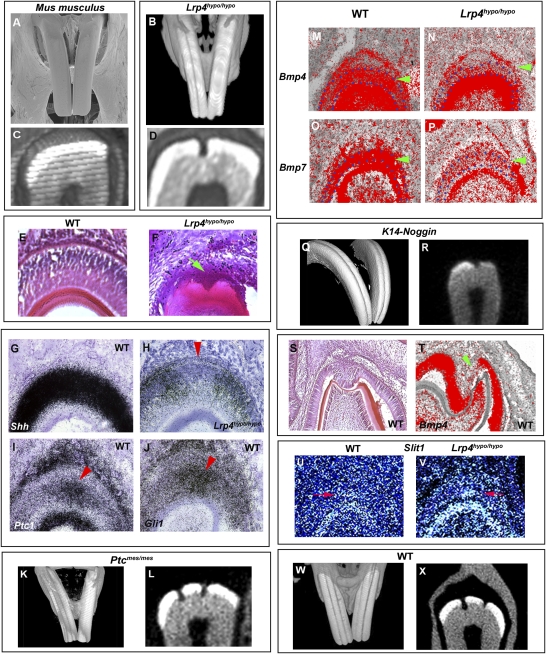
The incisors of laboratory mice (Mus musculus) have smooth enamel surfaces (Fig. 1A). Mice with a hypomorphic mutation in the LDL receptor 4 (Lrp4; also known as Megf7; Lrp4hypo/hypo) showed distinct grooved incisor labial surfaces (Fig. 1B). Cross section analysis of the grooved incisors of Lrp4hypo/hypo mice showed that the grooves were caused by a reduction of enamel on the labial surface (Fig. 1D).
Fig. 1.
Grooves in incisors of mutant mice. (A and B) Grooved incisors are found in Lrp4hypo/hypo (B), whereas there are no grooves in wild-type laboratory mice (Mus musculus; A). (C and D) Cross sections of incisors showed that grooves were caused by lack of enamel (D). (E and F) The first sign of grooves was at postnatal day 5 (P5) in Lrp4hypo/hypo mice (arrow in F). (G–J) Shh expression was downregulated at the presumptive groove region in Lrp4hypo/hypo mice at birth (arrowhead in H) whereas strong Ptc1 and Gli1 expression was observed in a similar region in wild-type (arrowhead in I and J). (M–P) Downregulation of Bmp4 (arrowhead in N) and Bmp7 (arrowhead in P) expression was observed in ameloblasts in Lrp4hypo/hypo mice at birth. (K, L, Q, and R) Ptcmes/mes (K and L) and K14-Noggin (Q and R) mice showed labial grooves that were caused by the lack of enamel. (S) Molar enamel–free zone in wild-type laboratory mice at P2. (T) Reduction in Bmp4 expression was observed at enamel-free zones at P2 (arrowhead). (U and V) Enamel-free zone marker gene, Slit1 was expressed at the presumptive groove region in Lrp4hypo/hypo mice at birth (arrow in V), whereas very faint Slit1 expression could be detected in similar regions in wild-type (arrow in U). (A–D, K, L, Q, and R) Images of incisors obtained from 3-month-old animals. (W and X) Incisors of 2-year-old wild-type laboratory mouse. Three-dimensional reconstructions (B, K, Q, and W) and cross-section (C, D, L, R, and X) based on micro-CT scans and SEM images (A) of maxillary incisors. Developing upper incisors (E–J, M–P, U, and V) and lower molars (S and T). Lrp4hypo/hypo (B, D, F, H, N, P, and V) and wild-type mice (A, C, E, G, I, J, M, O, and S–U). Ameloblasts are outline in blue (M–P).
Examination of the development of labial grooves in the mutants showed these to first appear shortly after birth; therefore we searched for molecular changes at this stage that might reveal a possible mechanism for the loss of enamel (Fig. 1F). The Shh pathway plays a critical role in ameloblast differentiation, the cells that co-ordinate enamel formation (10, 11). In frontal sections of wild-type laboratory mice, the Shh receptor Ptc1 was expressed in ameloblasts at the presumptive groove region, and Shh was expressed uniformly in ameloblasts (Fig. 1 G and I). Gli1 was also strongly expressed at the presumptive groove region (Fig. 1J). This indicates Shh activity in the ameloblasts of wild-type incisors at the position where the groove forms in the mutants. In the Lrp4hypo/hypo mutant, Shh expression in ameloblasts was generally downregulated, but a clear area of greatly reduced expression corresponded to the site of groove formation (Fig. 1H). An accompanying downregulation of Ptc1 and Gli1 expression was also observed at the presumptive groove region in Lrp4hypo/hypo mice (Fig. S1). To establish any causal link between loss of Shh activity and groove formation, we analyzed mice with mutations in the Shh pathway that survive after birth. The spontaneous mouse mutant, mesenchymal dysplasia, has an abnormal C terminus of the Ptc1 protein (Ptcmes/mes) that changes Shh activity (12–14). The maxillary incisors of these mice had grooves on their labial surfaces that were also caused by a lack of enamel (Fig. 1 K and L). This suggests that lowering of Shh activity in postnatal ameloblasts can lead to localized loss of enamel, in turn leading to the formation of labial grooves.
BMP Signaling Has Been Shown to Induce Ameloblast Differentiation (15).
We found Bmp4 and Bmp7 expression to be specifically downregulated in ameloblasts of Lrp4hypo/hypo mutants at birth whereas expression was unaltered in odontoblasts (Fig. 1 N and P). Significantly, mice overexpressing the BMP antagonist, Noggin under the keratin 14 promoter (K14-Noggin) also showed grooves on the labial surface of maxillary incisors that were caused by loss of enamel (Fig. 1 Q and R). Thus, changes in both Shh and Bmp activity can lead to localized loss of enamel and groove formation.
The lack of enamel at the incisor grooves is reminiscent of the enamel-free zones located at the tip of the cusps of rodent molars that result from failure of complete ameloblast maturation (16–18). Before eruption of rodent molar teeth, the enamel-free zones are covered by ameloblasts, similar to those observed in the grooved incisors of Lrp4hypo/hypo mice (Fig. 1 F and S). To determine whether a conserved mechanism exists between lack of enamel on molar cusp tips and on grooved incisors, we compared the expression of genes known to be expressed during ameloblast differentiation in molars. Downregulation of Bmp4 expression, seen in the presumptive groove region of Lrp4hypo/hypo mice, was also observed in ameloblasts covering the enamel-free zones of wild-type molar teeth (Fig. 1T). A more specific marker of the enamel-free zone is expression of Slit1, which shows a localized patch of expression in ameloblasts at the tips of developing molar cusps (19, 20). Weak Slit1 expression was observed in odontoblasts of wild-type mouse incisors with a very small faint patch of expression at the groove location site at birth (Fig. 1U). In Lrp4hypo/hypo mice, Slit1 expression was increased in odontoblasts, and the patch of expression in ameloblasts at the groove location site was clearly visible (Fig. 1V). Thus the site of the formation of incisor labial grooves shares molecular characteristics with the molar enamel-free zone. Alteration of Shh or Bmp signaling pathways either directly (Ptcmes/mes, K14-Noggin) or indirectly via hypomorphic mutation of Lrp4 reveals the cryptic incisor enamel-free zone. The existence of a very weak patch of Slit1 expression in wild-type incisors at the position where a groove forms in the mutants, together with expression of Ptc1 and Gli1 in this same small region, all of which show changes in Lrp4hypo/hypo mice, suggests that this region of ameloblasts may be different from all other ameloblasts in the incisors. The obvious interpretation of these expression patterns is that ameloblasts in this region have compromised mineralization capacity. However, no obvious changes in enamel across the width of wild-type incisors have ever been reported. Based on the very weak patch of Slit1 expression observed in wild-type incisors (Fig. 1U), we reasoned that enamel might be susceptible in this area. We also reasoned that any defect in the ability of these cells to form enamel would be small and thus might only be evident in older mice. We thus analyzed the incisors of wild-type C57/BL6 mice that were 2 years old, and found very clear evidence of labial grooves as a result of a lack of enamel (Fig. 1 W and X). This suggests that the ameloblasts that coordinate enamel formation in the region of the groove become defective with age. Because ameloblasts in murine incisors are continually produced from the cervical loop stem cells, this may indicate age-related and location-specific defects in these cells (21).
Lrp4hypo/hypo mice often show supernumerary maxillary incisors (37%) whereas almost all of Lrp4 null mice exhibit supernumerary maxillary incisor tooth germs at birth (Fig. S2) (8). It has been shown that the extra incisor tooth germs grow from endogenous incisor tooth germs (22). However, grooved incisors were found in newborn Lrp4hypo/hypo mice that did not have the supernumerary maxillary incisors and were also found in wild-type old mice (Fig. S2). This excludes the possibility that the groove is formed by a failure of the separation of supernumerary incisor tooth germs from endogenous tooth germs.
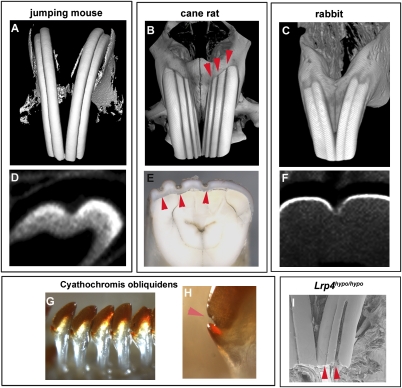
Several rodent species have been found to have grooved incisor labial surfaces. Species such as the meadow jumping mouse (Zapus hudsonius) and the cane rat (Thryonomys swinderianus) have one (Fig. 2A) to three (Fig. 2B) vertical grooves on the labial surfaces of their maxillary incisors, whereas other species such as grooved-toothed rats (Otomys tropicalis) display labial grooves in both maxillary and mandibular incisors (Tables S1–S3). Although rodents mostly have smooth incisors, we found grooved incisors in 60 rodents among 300 species investigated (Fig. S3 and Tables S1–S3). Labial grooves are also seen in species usually considered to be outside what are strictly considered as Rodentia, namely lagomorphs (picas, rabbits, and hares), which also possess lifelong continuously growing incisors (Fig. 2C). Cross-section analysis of the grooved incisors of several wild-type rodent species showed that the grooves were caused by a reduction of enamel on the labial surface (Fig. 2 D–F). Some fossil rodents also had grooved incisors, indicating that the labial grooves may have been lost in certain rodents, including Mus musculus, during evolution (Figs. S3 and S4) (23). In addition to rodent teeth, the enamel-free zone can also be observed in African cichlid fishes, suggesting that enamel-free zones are conserved structures in vertebrates (Fig. 2 G and H).
Fig. 2.
Grooved incisors in various wild-type rodents, rabbits, and fish. (A–C) Grooved incisors are found in jumping mice (Zapus hudsonius; A), cane rat (Thryonomys swinderianus; B), and rabbit (Oryctolagus cuniculus; C). (D–F) Cross-sections of incisors showed that grooves were caused by lack of enamel. Multiple grooves were found in cane rat and in Lrp4hypo/hypo mice (arrowheads in B, E, and I). Three-dimensional reconstructions (A–C) and cross-section (D and F) based on micro-CT scans, SEM images (I), and stereomicroscopic images (E) of maxillary incisors. (G and H) Enamel-free zone in cichlid fishes (Cyathochromis obliquidens; arrowhead in H). (A–F and I) All images of incisors were obtained from 3-month-old animals.
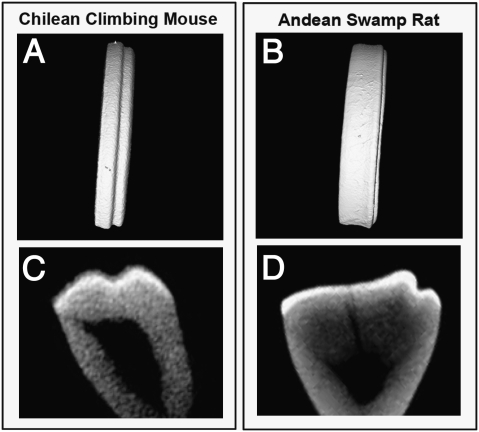
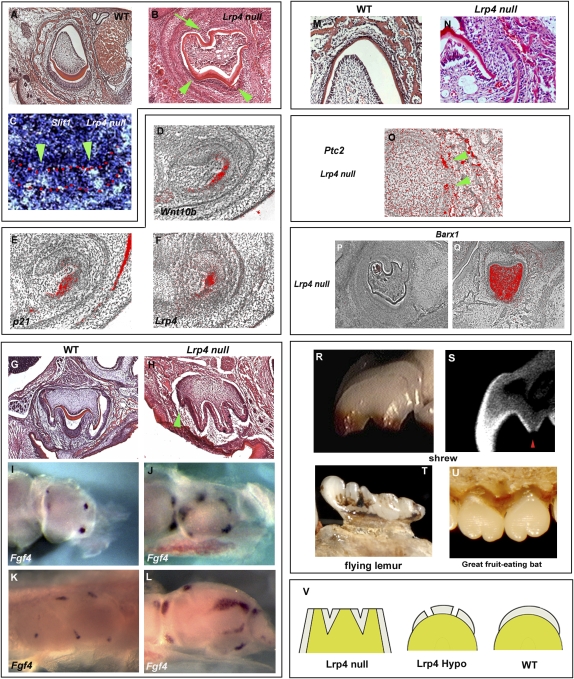
Some naturally occurring incisor grooves in several rodent species were, however, not found to be caused by loss of enamel but rather by folding of the enamel/dentin, similar to those observed in molar cusps (Fig. 3). When we examined the incisors of Lrp4 null mutant mice, we found a more severe phenotype than Lrp4hypo/hypo mice. Lrp4 null mutant incisors exhibited folded enamel and dentin on the labial side that was not observed in Lrp4hypo/hypo mice (Fig. 4B). Slit1, a marker of tertiary enamel knots as well as the enamel-free zone, was found in the tooth epithelium corresponding to the folded enamel/dentin in the mutant incisors (Fig. 4C) (19, 20). This suggests that the folded enamel/dentin represents a cusp-like structure. These mutant incisors are thus reminiscent of multicuspid crowns. Interestingly, multiple grooves (enamel-free zones) were also occasionally observed in Lrp4hypo/hypo and several rodent species (Fig. 2 B and I). In molars, cusp formation is initiated by a transient epithelial structure, the primary enamel knot (24, 25). To establish any link between Lrp4, primary enamel knots and cusp morphogenesis, we examined the expression of Lrp4, Wnt10b, and p21 during incisor development. The enamel knot marker genes, Wnt10b and p21 showed restricted coexpression with Lrp4 in incisor tooth epithelium (Fig. 4 D–F). Interestingly, Lrp4 expression was also observed in the primary enamel knot in the molars (8). These results suggest that the role of Lrp4 in incisors is probably similar to that in molars. Interestingly, Lrp4 null mice also had extra molar cusps, suggesting that Lrp4 might have a general role in suppressing cusp formation (Fig. 4 H, J, and L).
Fig. 3.
Folding enamel/dentin in rodent incisors. Incisors with grooves caused by folded enamel/dentin in wild-type rodent species (Chilean climbing mouse; A and C) and Andean swamp rat; B and D). Three-dimensional reconstructions (A and B) and cross-section (C and D) based on micro-CT scans of maxillary incisors.
Fig. 4.
Cusp-like structure and molar-type roots in mammalian incisors. (A and B) Lrp4 null mutants showed folding of enamel/dentin on the labial sides of incisors at P1 (arrowheads in B). Molar root-like structures were also found at the lingual side of incisors in Lrp4 null mutants (arrow in B). (C) Slit1 a marker of tertiary enamel knot and the enamel-free zones, was found in the tooth epithelium corresponding to the folded enamel/dentin (arrowheads in C). Ameloblasts are outlined in red (C). (D–F) Expression of enamel knot marker genes (Wnt10b; D) and (p21; E), and Lrp4 expression (F) in lower incisors at E13.5 in wild type. (G and H) Extra cusps in upper molar of Lrp4 null mutant (arrowhead in H). (I–L) Extra Fgf4 expression domain was found in second lower molar (J) and first lower molars (L) of Lrp4 null mice. (M and N) Some incisors showed a gap at the lingual side in Lrp4 null mutants (N). (O) Ptc2 expression was observed at the gap (arrowheads). (P and Q) Barx1 expression in lower incisor (P) and lower molar (Q) of Lrp4 null mutant at P1. (R–U) Multicuspid incisors of shrews (Sorex arcticus; R and S), flying lemurs (Galeopterus variegates; T), and great fruit-eating bats (Artibeus lituratus; U). (S) Cross-section based on micro-CT scans of maxillary incisors, showing folded enamel/dentin with enamel-free zone (arrowhead). (V) Diagram of evolutionary mutant/modification series; incisor of Lrp4 null mutant (Left), Lrp4hypo/hypo (Center), and wild-type (Right) mice. (A–H and M–Q) Frontal sections. Wild-type mice (A, D–G, I, K, and M) and Lrp4 null mutant (B, C, H, J, L, and N–Q).
In addition to the labial side showing multiple cusp-like structures, the lingual surfaces of the Lrp4 null incisors were also very uneven, with protrusions producing a “corrugated” appearance (Fig. 4B). The lingual portion of the incisors also showed a discontinuity of epithelium that is not a normal feature of lingual incisor epithelium (Fig. 4 M and N). The apical edge of lingual epithelium histologically resembled an epithelial root sheath that is a unique structure found in developing molar tooth roots (Hertwig’s epithelial root sheath; 3, 26). Ptc2 expression, a marker of Hertwig’s epithelial root sheath in molars (14), was clearly identified in the apical edge of epithelium of the incisors of Lrp4 null mutants (Fig. 4O), suggesting that in the absence of Lrp4, epithelial cells on the lingual aspect of developing incisors were organized into root-forming structures usually seen in only molars. The existence of both a crown with multiple cusp-like structure and molar-type roots indicates that the Lrp4 null mutant incisors have undergone a transformation toward molars. To investigate this further, we examined the expression of molar mesenchyme marker Barx1 in the mutant incisors (27). Barx1 expression could not be detected in the incisors of Lrp4 null mice, suggesting that the mesenchyme of Lrp4 null mutant incisors has retained its incisor identity (Fig. 4 P and Q). The existence of cusp-like structures with enamel-free zones in the epithelium of Lrp4 null mutant incisors, together with the retention of incisor identity in the mesenchyme identifies an uncoupling of the processes of tooth shape determination (mesenchyme) and morphogenesis (epithelium). Interestingly, multicuspid incisors that are composed of folded enamel/dentin with enamel-free zones are naturally found in some mammalian species (Fig. 4 R–U). Furthermore, several extinct mammalian species also had multicuspid incisors (28–33).
The evolutional processes of deriving complex heterodont mammalian dentitions from simple conical-shaped teeth has been much discussed in the last century (1, 34). Gaining cusps has been established as a major event in mammalian evolution. In East African cichlid fish, some species show tooth crown shape reversal where monocuspid teeth evolved from multicuspid teeth (35, 36). Interestingly, dolphins also possess a homodont conical tooth dentition whereas the primitive eutherian heterodont dentition included multicuspid teeth (1). Lrp4hypo/hypo mice show enamel-free zones in incisors and Lrp4 null mice exhibit a more severe phenotype of incisors with multiple cusp-like structures with enamel-free zones (Fig. 4V).
Lrp4 mutations reveal a developmentally suppressed program of molar-like epithelial changes in incisors. In some species this program is less suppressed, leading to incisors with obvious cuspid-like morphologies. Naturally occurring grooved incisors may represent a vestigial remnant of this suppressed molar program. Multiple signaling pathways are involved in cusp formation; in this context it is significant that Lrp4 is a known direct mediator of both Wnt and Bmp signaling and an indirect mediator of Shh (8, 9) and thus may have played a pivotal role in evolution of heterodontia (Fig. 4V). Lrp4 via its action on multiple signaling pathways including Shh, Bmp, and Wnt is thus central to a transition between a continuous enamel covering, grooved enamel, and folded enamel, all of which appear in the mammalian fossil record and extant species and raise the question of whether enamel grooves proceeded enamel folds in the evolution of multicuspid teeth.
Materials and Methods
Production and Analysis of Mutant Mice.
Lrp4hypo/hypo mice were produced as described by Johnson et al. (9). Lrp4 null mice were generated by deletion of the transcription start site and exon 1, which encodes the signal peptide and the initiating ATG. This strategy ensures that no residual functional protein can be generated. Ptcmes/mes were produced as described by Makino et al. (13). K14-Noggin were produced as described by Guha et al. (37).
In Situ Hybridization.
Whole-mount and radioactive section in situ hybridization was carried out using DIG labeled or 35S-UTP radiolabeled riboprobes (8) that were generated from mouse cDNA clones that were gifts from several laboratories: Fgf4 (G. R. Martin, University of California San Francisco), Ptc2 (A. Gritli-Linde, Göteborg University), and Shh (A. P. McMahon, Harvard University).
Scanning Electron Microscope Analysis.
Jaws were coated with gold and photographed using standard scanning electron microscopy.
Micro CT Analysis.
Heads of mice were scanned with Explore Locus SP (GE Preclinical Imaging) high-resolution micro-CT with a voxel dimension of 8 μm. Three-dimension reconstruction was performed by three structure analysis software, Microview (GE Preclinical Imaging).
Supplementary Material
Acknowledgments
We thank Robert Asher and Martyn T. Cobourne for critically reading the manuscript, Chris Healy for micro-CT analysis, and Tony Brain for SEM analysis. This work was supported by the Wellcome Trust and Medical Research Council. Support was also provided through a Research Councils UK Fellowship (A.O.); by grants from the National Institutes of Health (J.H., G.J.F., and J.T.S.); by a Wolfgang Paul Award from the Alexander-von-Humboldt Foundation (HL20948 and HL63762; J.H.); and by the QUENOTTES ANR program (C.C. and L.V.). Data in Tables S1, S2, and S3 were compiled from visits to the Natural History Museum of London, the Muséum National d’Histoire Naturelle of Paris, the Royal Museum for Central Africa of Tervuren, and the Museum of Vertebrate Zoology of Berkeley.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907236107/DCSupplemental.
References
- 1.Rose KD. The Beginning of the Age of Mammals. Baltimore: Johns Hopkins University Press; 2006. [Google Scholar]
- 2.Tummers M, Yamashiro T, Thesleff I. Modulation of epithelial cell fate of the root in vitro. J Dent Res. 2007;86:1063–1067. doi: 10.1177/154405910708601108. [DOI] [PubMed] [Google Scholar]
- 3.Tummers M, Thesleff I. Observations on continuously growing roots of the sloth and the K14-Eda transgenic mice indicate that epithelial stem cells can give rise to both the ameloblast and root epithelium cell lineage creating distinct tooth patterns. Evol Dev. 2008;10:187–195. doi: 10.1111/j.1525-142X.2008.00226.x. [DOI] [PubMed] [Google Scholar]
- 4.Nykjaer A, Willnow TE. The low-density lipoprotein receptor gene family: A cellular Swiss army knife? Trends Cell Biol. 2002;12:273–280. doi: 10.1016/s0962-8924(02)02282-1. [DOI] [PubMed] [Google Scholar]
- 5.Herz J, Bock HH. Lipoprotein receptors in the nervous system. Annu Rev Biochem. 2002;71:405–434. doi: 10.1146/annurev.biochem.71.110601.135342. [DOI] [PubMed] [Google Scholar]
- 6.Johnson ML, Harnish K, Nusse R, Van Hul W. LRP5 and Wnt signaling: A union made for bone. J Bone Miner Res. 2004;19:1749–1757. doi: 10.1359/JBMR.040816. [DOI] [PubMed] [Google Scholar]
- 7.Gong Y, et al. Osteoporosis-Pseudoglioma Syndrome Collaborative Group. LDL receptor-related protein 5 (LRP5) affects bone accrual and eye development. Cell. 2001;107:513–523. doi: 10.1016/s0092-8674(01)00571-2. [DOI] [PubMed] [Google Scholar]
- 8.Ohazama A, et al. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One. 2008;3:e4092. doi: 10.1371/journal.pone.0004092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson EB, Hammer RE, Herz J. Abnormal development of the apical ectodermal ridge and polysyndactyly in Megf7-deficient mice. Hum Mol Genet. 2005;14:3523–3538. doi: 10.1093/hmg/ddi381. [DOI] [PubMed] [Google Scholar]
- 10.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–4785. doi: 10.1242/dev.127.22.4775. [DOI] [PubMed] [Google Scholar]
- 11.Gritli-Linde A, et al. Shh signaling within the dental epithelium is necessary for cell proliferation, growth and polarization. Development. 2002;129:5323–5337. doi: 10.1242/dev.00100. [DOI] [PubMed] [Google Scholar]
- 12.Sweet HO, Bronson RT, Donahue LR, Davisson MT. Mesenchymal dysplasia: A recessive mutation on chromosome 13 of the mouse. J Hered. 1996;87:87–95. doi: 10.1093/oxfordjournals.jhered.a022981. [DOI] [PubMed] [Google Scholar]
- 13.Makino S, Masuya H, Ishijima J, Yada Y, Shiroishi T. A spontaneous mouse mutation, mesenchymal dysplasia (mes), is caused by a deletion of the most C-terminal cytoplasmic domain of patched (ptc) Dev Biol. 2001;239:95–106. doi: 10.1006/dbio.2001.0419. [DOI] [PubMed] [Google Scholar]
- 14.Nakatomi M, Morita I, Eto K, Ota MS. Sonic hedgehog signaling is important in tooth root development. J Dent Res. 2006;85:427–431. doi: 10.1177/154405910608500506. [DOI] [PubMed] [Google Scholar]
- 15.Wang XP, et al. Follistatin regulates enamel patterning in mouse incisors by asymmetrically inhibiting BMP signaling and ameloblast differentiation. Dev Cell. 2004;7:719–730. doi: 10.1016/j.devcel.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 16.Cohn SA. Development of the molar teeth in the albino mouse. Am J Anat. 1957;101:295–319. doi: 10.1002/aja.1001010205. [DOI] [PubMed] [Google Scholar]
- 17.Gaunt WA. The development of enamel and dentine on the molars of the mouse, with an account of the enamel-free areas. Acta Anat (Basel) 1956;28:111–134. doi: 10.1159/000141138. [DOI] [PubMed] [Google Scholar]
- 18.Lyngstadaas SP, Møinichen CB, Risnes S. Crown morphology, enamel distribution, and enamel structure in mouse molars. Anat Rec. 1998;250:268–280. doi: 10.1002/(SICI)1097-0185(199803)250:3<268::AID-AR2>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 19.Løes S, Luukko K, Kvinnsland IH, Kettunen P. Slit1 is specifically expressed in the primary and secondary enamel knots during molar tooth cusp formation. Mech Dev. 2001;107:155–157. doi: 10.1016/s0925-4773(01)00454-3. [DOI] [PubMed] [Google Scholar]
- 20.Luukko K, et al. Identification of a novel putative signaling center, the tertiary enamel knot in the postnatal mouse molar tooth. Mech Dev. 2003;120:270–276. doi: 10.1016/s0925-4773(02)00458-6. [DOI] [PubMed] [Google Scholar]
- 21.Harada H, et al. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–120. doi: 10.1083/jcb.147.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munne PM, Tummers M, Järvinen E, Thesleff I, Jernvall J. Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development. 2009;136:393–402. doi: 10.1242/dev.025064. [DOI] [PubMed] [Google Scholar]
- 23.Ortiz PE, Pardinas UFJ, Steppan SU. A new fossil phyllotine (Rodentia: muroidae) from northwestern Argentina and relationships of the reithrodon group. J Mammal. 2000;81:37–51. [Google Scholar]
- 24.Tucker AS, Sharpe PT. Molecular genetics of tooth morphogenesis and patterning: The right shape in the right place. J Dent Res. 1999;78:826–834. doi: 10.1177/00220345990780040201. [DOI] [PubMed] [Google Scholar]
- 25.Jernvall J, Thesleff I. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. doi: 10.1016/s0925-4773(99)00322-6. [DOI] [PubMed] [Google Scholar]
- 26.Yokohama-Tamaki T, et al. Cessation of Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development. 2006;133:1359–1366. doi: 10.1242/dev.02307. [DOI] [PubMed] [Google Scholar]
- 27.Tucker AS, Matthews KL, Sharpe PT. Transformation of tooth type induced by inhibition of BMP signaling. Science. 1998;282:1136–1138. doi: 10.1126/science.282.5391.1136. [DOI] [PubMed] [Google Scholar]
- 28.McKenna MC. Primitive Paleocene and Eocene Apatemyidae (Mammalia, Insectivora) and the primate-insectivore boundary. Am Mus Novit. 1963;2160:1–39. [Google Scholar]
- 29.Gingerich PD, Rose KD. Studies on Paleocene and early Eocene Apatemyidae (Mammalia, Insectivora): Part I. Dentition of Clarkforkian Labidolemur kayi. Contrib Mus Paleontol Univ Mich. 1982;26:49–55. [Google Scholar]
- 30.Rose KD, Gingerich PD. A new insectivore from the Clarkforkian (Earliest Eocene) of Wyoming. J Mammal. 1987;68:17–27. [Google Scholar]
- 31.Butler PM. Phylogeny of the insectivores. In: Benton MJ, editor. Mammals (The Phylogeny and Classification of the Tetrapods) Vol 2. Oxford: Clarendon Press; 1988. pp. 117–141. [Google Scholar]
- 32.McKenna MC, Bell SK. In: Classification of Mammals Above the Species Level. McKenna MC, Bell SK, editors. New York: Columbia University Press; 1997. [Google Scholar]
- 33.Lofgren DL, Lillegraven JA, Clemens WA, Gingerich PD, Williamson TE. Paleocene biochronology: The Puercan through Clarkforkian land mammal ages. Late Cretaceous and Cenozoic Mammals of North America. In: Woodburne MO, editor. Biostratigraphy and Geochronology. New York: Columbia University Press; 2004. [Google Scholar]
- 34.Widdowson TW. The evolution of mammalian teeth. In: Widdowson TW, editor. Special or Dental Anatomy and Physiology and Dental Histology The Evolution of Mammalian Teeth. London: Staples Press; 1952. pp. 280–306. [Google Scholar]
- 35.Fraser GJ, Bloomquist RF, Streelman JT. A periodic pattern generator for dental diversity. BMC Biology. 2008;6:32. doi: 10.1186/1741-7007-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Streelman JT, Albertson RC. Evolution of novelty in the cichlid dentition. JEZ B: Mol Dev Evol. 2006;306B:216–226. doi: 10.1002/jez.b.21101. [DOI] [PubMed] [Google Scholar]
- 37.Guha U, et al. Target-derived BMP signaling limits sensory neuron number and the extent of peripheral innervation in vivo. Development. 2004;131:1175–1186. doi: 10.1242/dev.01013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.