Abstract
Interkingdom signaling is established in the gastrointestinal tract in that human hormones trigger responses in bacteria; here, we show that the corollary is true, that a specific bacterial signal, indole, is recognized as a beneficial signal in intestinal epithelial cells. Our prior work has shown that indole, secreted by commensal Escherichia coli and detected in human feces, reduces pathogenic E. coli chemotaxis, motility, and attachment to epithelial cells. However, the effect of indole on intestinal epithelial cells is not known. Because intestinal epithelial cells are likely to be exposed continuously to indole, we hypothesized that indole may be beneficial for these cells, and investigated changes in gene expression with the human enterocyte cell line HCT-8 upon exposure to indole. Exposure to physiologically relevant amounts of indole increased expression of genes involved in strengthening the mucosal barrier and mucin production, which were consistent with an increase in the transepithelial resistance of HCT-8 cells. Indole also decreased TNF-α-mediated activation of NF-κB, expression of the proinflammatory chemokine IL-8, and the attachment of pathogenic E. coli to HCT-8 cells, as well as increased expression of the antiinflammatory cytokine IL-10. The changes in transepithelial resistance and NF-κB activation were specific to indole: other indole-like molecules did not elicit a similar response. Our results are similar to those observed with probiotic strains and suggest that indole could be important in the intestinal epithelial cells response to gastrointestinal tract pathogens.
Keywords: host–pathogen interactions, interkingdom signaling, probiotics
The human gastrointestinal (GI) tract is rich in a diverse range of signaling molecules. A wide range of bacterial signals (e.g., autoinducer-2, autoinducer-3, and indole) (1) are produced by the ~1014 nonpathogenic commensal bacteria that coexist with host cells in the GI tract, and neuroendocrine hormones (e.g., norepinephrine and dopamine) are also synthesized in situ in the GI tract via the enteric nervous system. The close proximity of bacteria and the host cells in the GI tract, as well as the high local concentrations of the signals they secrete, has led to a signal-centric paradigm wherein GI tract signals are considered to be important mediators of homeostasis and infections through intrakingdom (i.e., recognition of bacterial signals by other bacteria) and/or interkingdom (i.e., recognition of host signals by bacteria and vice versa) signaling and communication.
Interkingdom recognition of hormones by pathogens was first demonstrated by Lyte et al. (2). Subsequently, Sperandio et al. (3) showed that enterohemorrhagic Escherichia coli (EHEC) virulence is increased upon exposure to epinephrine and norepinephrine, and that epinephrine binds and signals through the QseC receptor (4). Work from our group (5) has also shown that epinephrine and norepinephrine increase EHEC chemotaxis, motility, adherence to epithelial cells, and virulence gene expression. Although these studies provide evidence for the recognition of human hormones by pathogens, the converse (i.e., recognition of bacterial specific signaling molecules by intestinal epithelial cells and their role in GI tract function) has not been fully investigated. Although several studies have shown that culture supernatants from probiotic strains attenuate pathogen infection (6, 7), modulate inflammation in dendritic cells (8) and Caco-2 cells (9), and increase intestinal epithelial cell barrier function (10), the specific molecules contributing to these responses and the mechanisms involved are not known. In fact, most individual bacterial signals have been shown to be deleterious to host cells; for example, the Pseudomonas aeruginosa quorum-sensing signaling molecule N-(3-oxododecanoyl)-L-homoserine lactone disrupts epithelial barrier integrity in Caco-2 cells (11), increases the expression of proinflammatory cytokines in murine fibroblasts and human lung epithelial cells (12), and disrupts nuclear factor kappa B (NF-κB) signaling to promote persistence of a local P. aeruginosa infection (13).
Indole is produced in E. coli from L-tryptophan via tryptophanase (encoded by tnaA) (14). Prior work from our laboratory has shown that indole is an extracellular signal, represses E. coli K-12 biofilm formation through the sensor of quorum sensing signals, SdiA (14), is more effective in repressing E. coli K-12 biofilm at low temperatures (i.e., 25°C and 30°C) (15), and exerts the opposite effect as epinephrine and norepinephrine on EHEC chemotaxis, motility, adherence to epithelial cells, and virulence gene expression, at 37°C (5).
Because commensal E. coli produce as much as 600 μM indole in suspension cultures (16) and indole has been detected in human feces at comparable concentrations (~250–1,100 μM) (17, 18), it is likely that intestinal epithelial cells are continually exposed to high concentrations of indole. Therefore, we hypothesized that indole is recognized as an interkingdom signal in intestinal epithelial cells. Measurements of changes in gene expression in HCT-8 intestinal epithelial cells and phenotypic measurements of transepithelial resistance, NF-κB activation, IL-8 and IL-10 secretion, and EHEC attachment, show that indole modulates expression of proinflammatory genes, increases expression of antiinflammatory genes, strengthens epithelial cell barrier properties, and decreases pathogen colonization. Our data strongly suggest that indole is a beneficial interkingdom signal for intestinal epithelial cells.
Results
HCT-8 Intestinal Epithelial Cell Transcriptome Profiling on Exposure to Indole.
Changes in intestinal epithelial cell gene expression upon exposure to 1 mM indole for 4 h and 24 h were determined by using whole-transcriptome profiling. Genes exhibiting a statistically significant change (P < 0.01) in expression relative to untreated controls were used to populate the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways with the Pathway Express software (19). Exposure to indole for 4 h resulted in the differential expression of 523 genes, of which 476 genes were induced and 47 genes were repressed. Importantly, no single pathway or ontology classification was dominant in the differentially expressed genes. On the other hand, ~8% (4,102 genes; 3,083 annotated) of the HCT-8 transcriptome was differentially expressed after a 24-h exposure to indole. Although several pathways were populated by this approach (e.g., metabolism, transport, protein modification, etc.) [Fig. S1], we limit our discussion to specific pathways—epithelial cell structure and organization, inflammation, and infection—that are related to pathogen colonization in the GI tract (Table S1). The entire list of differentially expressed genes, with fold-changes in expression, is included as Table S2.
Changes in Epithelial Cell Barrier Properties and Colonization.
Indole significantly induced several genes involved in the maintenance of epithelial cell structure and function. These included genes responsible for tight junction organization, actin cytoskeleton, mucin production, and adherens junction, which together suggest strengthening of epithelial cell barrier properties. Coordinated induction of seven claudin genes (Table S2) suggested that a 24-h exposure to indole increases paracellular resistance (Fig. 1A). The decrease in the expression of the pore-forming cldn2, which increases paracellular cation permeability (20), also further supports the theory that indole mediates an increase in paracellular resistance. Tight junction proteins TJP1, TJP3, and TJP4, which are downstream of claudin-mediated tight junction regulation (Fig. 1A), and gap junction proteins GJE1, GJB3, GJB4, and GJA8 were also induced by indole. Exposure to indole also induced the expression of 50 cytoskeleton genes (e.g., actinin, cingulin, and syntrophin), and the coordinated induction of several cytoskeleton gene families suggest fortification of the actin cytoskeleton upon exposure to indole. Similarly, genes involved in the production of mucins (muc1, muc3, muc13, muc20, and mucin and cadherin-like transcript variant 4), high-molecular-weight glycoproteins secreted by intestinal epithelial cells (21) and integral to epithelial cell defense, were also induced in expression upon exposure to indole. Together, these transcriptome changes suggest that indole promotes intestinal epithelial cell barrier function and increases resistance to pathogen colonization.
Fig. 1.
Changes in tight junction proteins and TER. (A) Pathways containing differentially expressed genes involved in tight junction formation were adapted from Pathway Express using KEGG classifications. Red, up-regulation; blue, down-regulation; gray, no change in expression; arrow, molecular interaction leading to activation; blunt line without an arrowhead, molecular interaction leading to inhibition. The pathway scheme shown is based on microarray data and does not include posttranscriptional regulation. (B) Changes in the TER of polarized HCT-8 cells exposed to either solvent (filled circles) or indole (open circles) for 24 h. Data represent mean ± SD from seven measurements at each time point and two independent experiments. (C) Effect of indole pretreatment on adherence of EHEC to HCT-8 cells in the presence of 500 μM norepinephrine. Data represent mean ± SD from three wells per experiment and three independent experiments. *, statistical significance, using Student's t test, at P < 0.05; **, significance at P < 0.005.
The transepithelial resistance (TER) of epithelial cells after indole exposure was measured to investigate the effect of indole on tight junction integrity and cell permeability. Fig. 1B shows that indole increased the TER of HCT-8 cells within 4 h of exposure and by 1.6-fold after 24 h. Because an increase in mucin production has been previously shown to attenuate pathogen colonization (21), we also investigated the effect of indole exposure on the colonization of HCT-8 cells by EHEC. Pretreatment of HCT-8 cells with 1 mM indole for 24 h decreased the norepinephrine-mediated EHEC colonization (5) by 1.4-fold (Fig. 1C). These phenotypic results are consistent with changes in gene expression and suggest that exposure to physiologically relevant concentrations of indole strengthens intestinal epithelial cell barrier properties and increases resistance to pathogen colonization.
Changes in Toll-Like Receptor Signaling.
Exposure to 1 mM indole led to an increase in the expression of two Toll-like receptor (TLR) genes—TLR-3 and TLR-9. TLRs respond to bacterial ligands and activate the host immune system; however, TLR-2, TLR-3, and TLR-9 have been reported to modulate intestinal inflammation and maintain homeostasis (22). More importantly, TLRs that play a significant role in responding to pathogens (e.g., TLR2 and TLR4) were not altered in expression. Additionally, genes involved in the modulation of TLR signaling—TOLLIP, SIGIRR, and SOCS5—were also induced by indole. Similarly, genes involved in the phosphoinositide-3-kinase (PI3K) pathway, which modulates TLR signaling (Fig. 2A) and leads to antiinflammatory cytokine IL-10 induction (8), were also induced by indole. Three genes involved in PI3K signaling (PIK3CA, PIK3CB, and PIK3CD) were induced, as was the transcription factor AKT1 that is activated by signaling through the PI3K pathway. Downstream target genes of AKT1, including the cAMP response element-binding protein and IL-10, were also induced upon addition of indole. Thus, our data strongly suggest that indole modulates TLR signaling in HCT-8 intestinal epithelial cells, both through up-regulation of antiinflammatory TLR, and through the PI3K-mediated down-regulation of inflammatory TLR signaling.
Fig. 2.
Changes in Toll-like receptor, IL-8, and IL-10 signaling. (A) Pathways containing differentially expressed genes involved in tight junction formation were adapted from Pathway Express using KEGG classifications. Colors and symbols as in Fig. 1. The pathway scheme shown is based on microarray data and does not include posttranscriptional regulation. (B) Secretion of IL-8 in HCT-8 cells exposed to indole for 4 h or 24 h, in the presence or absence of 40 ng/mL TNF-α. Data represent mean ± SD from three independent experiments. (C) Intracellular staining and flow cytometry of IL-10 in HCT-8 cells exposed to indole for 24 h. Two-color dot plots of number of PE-stained cells and forward scatter for solvent control, indole-treated, and unstained cells are shown. Percentages in the boxed region indicate the proportion of HCT-8 cells producing IL-10 in one representative experiment. (D) Average increase in IL-10 expression data from three independent experiments. *, statistical significance, using Student’s t test, at P < 0.05; **, significance at P < 0.005.
Because TLR3 and TLR9 are not present on the cell membrane but are internal (i.e., on the endosomal membrane), we determined whether indole was internalized by HCT-8 cells. Measurement of extracellular indole (23) showed that indole was not significantly internalized by HCT-8 cells: ~90% of the indole added (1 mM) was still detected in the culture medium after 24 h.
Changes in Inflammatory Cytokine and Chemokine Gene Expression.
Exposure to indole for 24 h coordinately altered the expression of several cytokines in HCT-8 intestinal epithelial cells. Genes that were increased in expression upon exposure to 1 mM indole included the precursor to the proinflammatory cytokine IL-1 (pre-IL-1). In addition, the receptor for the proinflammatory cytokine IL-2, IL-4, IL-18 binding protein, IL-24, and IL-28 were all induced, suggesting that indole exposure leads to an increase in cytokine expression and signaling in HCT-8 intestinal epithelial cells. In addition to altering the expression of cytokines, indole also induced the expression of several chemokines (CCL25, CCL4, CCL16, CXCL3, XCL1, and CX3CL1) in HCT-8 cells.
However, not all changes in expression were proinflammatory: the expression of receptors for antiinflammatory cytokines (IL-10, IL-11) was also induced by indole. In addition, the expression of several other proinflammatory cytokines was decreased in HCT-8 cells upon exposure to indole. IL-8, an alarm cytokine that induces migration of neutrophils across the endothelium, induces basophil chemotaxis to the infection site, and enhances removal of microbes through neutrophil recruitment (24), was repressed (Fig. 2A) on exposure to indole. CXCL5 (or ENA-78), a chemokine that is structurally and functionally similar to IL-8 (25) and is highly expressed in intestinal epithelial cells during chronic inflammation (25), was also repressed. IL-12, which is one of the first cytokines to be released in response to detection of a pathogen through TLR signaling and is important in the initiation of the inflammatory response (26), was repressed. These changes in expression suggest that exposure to indole leads to coordinated changes in the expression of pro- and antiinflammatory cytokines and chemokines.
Changes in IL-8 expression upon 1 mM indole exposure were monitored by using ELISA to corroborate changes observed with microarrays (Fig. 2B). Indole decreased basal IL-8 production in HCT-8 cells at 4 h and 24 h by −1.9-fold and −1.8-fold, respectively. Additionally, indole also decreased TNF-α-induced IL-8 production by −1.8-fold, which is consistent with microarray data on the down-regulation of IL-8 mRNA. Intracellular staining and flow cytometry were used to also monitor expression of the antiinflammatory cytokine IL-10. Fig. 2 C and D show that indole increased IL-10 expression by 1.5-fold, which together with the increase in IL-10 receptor mRNA expression further suggests that indole increases antiinflammatory signaling as well.
Downstream mediators of cytokine signaling (i.e., signaling kinases) such as the mitogen-activated protein kinase (MAPK) pathway, which is central to the inflammatory signaling, were differentially expressed. Specifically, the expression of dual-specificity phosphatases (DUSPs)—Dusp1, Dusp3, Dusp10, Dusp16, Dusp18, and Dusp22, which function as inhibitors of MAPK signaling (27)—was increased upon indole exposure. However, because several MAPK genes were also up-regulated, further analysis of the phosphorylation state of different pathway components is required to fully understand the changes in MAPK signaling upon indole exposure. Indole also induced the expression of genes in the Jak/STAT signaling pathway, which is activated by proinflammatory molecules such as IFN-γ leading to increased microbial killing and cytokine production (28). Jak-1, -2, and -3 were induced along with STAT-1, -2, -3, -5, and -6 (Table S2). The IFN-stimulated transcription factor 3γ (ISGF3G), which is activated by STAT1 to regulate IFN-γ-stimulated genes, was also induced in expression upon exposure to indole, along with SOCS, a repressor of Jak-STAT signaling. Therefore, in addition to inducing changes in pro- and antiinflammatory mediators, exposure to indole also induced coordinated changes in the downstream signaling pathways used by these cytokines and chemokines.
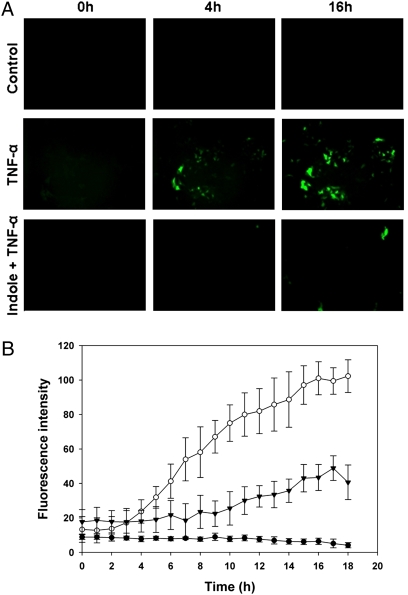
We also monitored the activation of the prototypical proinflammatory transcription factor NF-κB, which has been shown to mediate the inflammatory response both in vitro and in vivo (29, 30), using an HCT-8 GFP reporter cell line for NF-κB (31). Our data show that indole reduced the TNF-α-mediated increase in NF-κB activation in HCT-8 intestinal epithelial cells by 3-fold over 18 h (Fig. 3 A and B) and clearly indicate the ability of indole to attenuate TNF-α-mediated inflammation in HCT-8 cells.
Fig. 3.
Modulation of NF-κB activity by indole. (A) Effect of indole on NF-κB activation by TNF-α in HCT-8 NF-κB-GFP reporter cells and the resultant GFP expression. (B) Quantification of NF-κB activity in control cells (filled circles), cells treated with TNF-α to induce NF-κB activation (open circles), and cells treated with indole and TNF-α (filled inverted triangles). Data shown are mean ± SD from images obtained at four locations in three independent experiments.
Specificity of Indole in Attenuating NF-κB Activation and Promoting Epithelial Cell Barrier Properties.
We investigated the effect of five indole-like molecules (Fig. S2)—1H-indole-2,3-dione (isatin), 7-hydroxyindole (7-HI), 5-hydroxyindole (5-HI), 2-hydroxyindole (2-HI), and indole-3-acetic acid (I3AA)—for establishing the specificity of indole in attenuating NF-κB activation and promoting TER. Fig. 4A shows that only 5-HI significantly repressed NF-κB activity in a manner similar to that observed with indole (Fig. 3), whereas the effectiveness of the other molecules varied from having no effect or marginal (7-HI, I3AA, and 2-HI) to actually increasing NF-κB activation (isatin). However, 5-HI did not cause a significant change in the TER of HCT-8 cells after 24 h (Fig. 4B), and among the molecules tested, only 7-HI increased the TER to levels comparable with indole (Fig. 1B). Thus, although indole-like molecules were able to either attenuate NF-κB activation or increase TER at the concentrations tested, changes in both phenotypes were observed only with indole.
Fig. 4.
Changes in HCT-8 cell phenotypes with aromatic bicyclic molecules. (A) NF-κB activation in HCT-8 NF-κB-GFP reporter cells and the resultant GFP expression. Images shown are after 18 h of exposure to TNF-α and are representative of data from three independent experiments. (B) Changes in TER after exposure to indole-like molecules for 24 h. Data represent mean ± SD from three independent experiments. **, statistical significance, using Student's t test, at P < 0.005.
To further establish that the beneficial effects of indole were not caused by an increase in HCT-8 cell number or viability, we determined the change in HCT-8 cell density after exposure to indole for 24 h. Cells exposed to indole showed >99% viability after 24 h (live-cell density in control and indole-treated cells was 3.1 × 106 ± 0.6 × 106 and 3.0 × 106 ± 0.7 × 106 cells per mL, respectively), with a similar percentage of live cells for the solvent control. These data suggest that the beneficial effects of indole are not metabolic (i.e., because of the lack of an increase in cell numbers).
The whole-transcriptome data were substantiated with qRT-PCR where genes from inflammatory cytokines and signaling, and epithelial barrier function were profiled. ISGF3G was induced 4.1-fold in the microarrays and 5.1 ± 0.1-fold in qRT-PCR. Similar results were seen for CXCL5 (−5.2-fold in the arrays, −13.6 ± 0.6-fold in qRT-PCR), pre-IL-1 (5.6-fold in the arrays, 5.9 ± 0.2-fold in qRT-PCR), IL-8 (−1.8-fold in the arrays, −1.4 ± 0.1-fold in qRT-PCR), cldn3 (3.5-fold in the arrays, 4.9 ± 0.2-fold in qRT-PCR), TJP3 (5.7-fold in the arrays, 5.5 ± 0.2-fold in qRT-PCR), and muc13 (4.4-fold in the arrays, 2.3 ± 0.2-fold in qRT-PCR). Thus, the expression results from whole-transcriptome analysis were in good agreement with the qRT-PCR results.
Discussion
The recognition of host cell signaling molecules by pathogens through interkingdom communication and its role in pathogenesis has been recently documented (3, 5). However, little is known about recognition of bacterial signals by host cells. To date, specific prokaryotic signals such as the P. aeruginosa quorum-sensing molecules have been shown to be deleterious to host cells as they help pathogen infections. Here, we show that the bacterial stationary-phase signal indole is recognized by intestinal epithelial cells and used to strengthen host cell-barrier properties and maintain controlled inflammation. This is supported by multiple lines of evidence showing that indole (i) increases the expression of genes involved in the formation of tight junctions and actin cytoskeleton, and increases the transepithelial resistance of polarized HCT-8 intestinal epithelial cells; (ii) attenuates IL-8 secretion and TNF-α-mediated NF-κB activation in HCT-8 cells while increasing IL-10 secretion; (iii) increases the resistance of HCT-8 cells to norepinephrine-mediated increase in EHEC colonization; and (iv) reduces the expression of proinflammatory cytokines and chemokines while inducing the expression of antiinflammatory cytokines. Therefore, we propose that indole can be a beneficial interkingdom signal that helps improve intestinal epithelial-cell function, maintain controlled inflammation, and increase resistance to pathogen colonization.
Several studies have shown that cell-free supernatants of probiotic bacterial cultures increase host epithelial-cell barrier properties and resistance to pathogen colonization. However, the identity of specific molecule(s) that contribute to this protective effect and the mechanisms involved are not known, because culture supernatants are complex mixtures comprised of media components, metabolites, and signals secreted by different bacterial species. The data presented here show that indole elicits a response similar to probiotic strains by strengthening host cell-barrier properties. Because indole is produced by several commensal bacteria (e.g., E. coli, Bacteroides ovatus, and Clostridium bifermentans) that colonize the human GI tract (32), it is intriguing to speculate that the beneficial effects of some probiotic strains are mediated, in part, through indole.
Although 4,102 genes (or 8% of the transcriptome) were differentially regulated by indole, it is worth noting that only 83 genes were regulated by 4.0-fold or higher. A moderate response to indole is to be expected, especially because indole is abundant in the human GI tract (~100 μM in human feces) (17), and is less likely to perturb the epithelial cell transcriptome like a pathogen. In this regard, our data are similar to that reported by Hasegawa et al. (27), who showed that two different species of commensal bacteria, Streptococcus gordonii and Fusobacterium nucleatum, perturbed the gingival epithelial cell transcriptome to a lesser extent than the pathogens Poryphyromonas gingivalis and Aggregatibacter actinomycetemcomitans, which elicited more widespread organism-specific changes in the transcriptome. Striking similarities are also evident between our data and changes in gene expression elicited by S. gordonii and F. nucleatum in gingival epithelial cells. For example, gene families that were differentially expressed in intestinal epithelial cells upon exposure to indole—tight junction proteins, actin cytoskeleton, cell cycle regulators, phosphatidylinositol signaling, MAP kinase signaling, TLR signaling, and Wnt signaling—were all altered in gingival epithelial cells as well when exposed to commensal bacteria.
In addition to the similarities with commensal response in gingival epithelial cells, significant differences are also observed between the indole response in intestinal epithelial cells and pathogen-mediated changes in expression in gingival epithelial cells. One such example is the activation of caspase-1 by pathogenic bacteria (e.g., lipopolysaccharide, Salmonella typhimurium type III secretion), which leads to increased secretion of proinflammatory cytokines such as IL-1β and the induction of apoptosis. Hasegawa et al. (27) reported that commensal bacteria did not activate caspase-1, thus indicating physiologic balance between the host and the organism. Similarly, no caspase-1 gene expression was observed in HCT-8 intestinal epithelial cells exposed to indole, which also suggests favorable recognition of indole by intestinal epithelial cells. Furthermore, the lack of activation of TLR signaling by indole, similar to that seen with oral commensal bacteria (27) and opposite to that of oral pathogens (30), also strongly suggests that indole is recognized as a beneficial signal and not detected as a pathogenic signal by intestinal epithelial cells.
IL-8 is an alarm chemokine that is highly induced by pathogens in a wide variety of cells, including human colon adenocarcinoma cells. For example, a 4- to 30-fold increase in IL-8 and ENA-78 (CXCL5) has been observed in HT-29 and Caco-2 intestinal epithelial cells upon exposure to Salmonella dublin (33). However, in our study, exposure to indole decreased the expression of ENA-78 and IL-8, which suggests that indole is not recognized as a pathogen signal by intestinal epithelial cells and strongly proposes an antiinflammatory role for indole in GI tract inflammation.
It has been proposed that bacteria (or molecules secreted by them) induce the expression of pro- and antiinflammatory cytokines (34), which function coordinately (i.e., as a network) to regulate the host response to commensal bacteria (34). Disruption of this regulated cytokine response leads to sustained inflammation, as has been shown with IL-10 knockout mice developing chronic enterocolitis even in the absence of pathogen colonization (35). In addition, this heightened level of alertness also enables host cells to respond rapidly to the presence of pathogens. Our data showing that indole up-regulates the expression of some proinflammatory cytokines, in addition to attenuating other indicators of inflammation, are consistent with this hypothesis, and suggest that indole-mediated communication is involved in the balance between pro- and antiinflammatory cytokine signaling in intestinal epithelial cells.
The intestinal epithelial cell inflammatory response to pathogens is orchestrated, in part, through activation of NF-κB (29, 30), which is well established. Our data showing that indole reduced TNF-α-mediated NF-κB activation in intestinal epithelial cells support the antiinflammatory role for indole. Because commensal bacteria reduce the S. typhimurium-induced NF-κB activity (29) and inflammation in mice, it is intriguing to speculate that indole is a signal through which probiotics reduce intestinal inflammation.
The effect of indole on HCT-8 cells also extends beyond alterations in gene expression to changes in prototypical intestinal epithelial cell functions such as barrier integrity. First, the increase in tight junction protein expression and TER is significant, because an increase in tight junction resistance leads to decreased paracellular permeability and reduces the ability of pathogens to cross the epithelial cell barrier. Second, the induction of 50 cytoskeleton genes suggests increased resistance to pathogen colonization because pathogens like P. aeruginosa, Shigella flexneri, Listeria monocytogenes, and enteropathogenic E. coli disrupt epithelial cell morphology during infection by interfering with the actin cytoskeleton (36–38). Third, the increase in the expression of several mucin genes is also likely to decrease pathogen colonization: Kim et al. (21) recently reported that lactobacilli increased the production of mucin 2 (muc2) to inhibit EHEC attachment to epithelial cells in vitro. These three lines of evidence strongly support the hypothesis that indole elicits beneficial changes in intestinal epithelial cells.
The alteration of TER and the regulation of inflammation by indole has significant implications in the treatment of chronic intestinal inflammatory diseases such as Crohn’s disease, which is an autoimmune disease characterized by sustained inflammation and deterioration of epithelial cell barrier function. Currently no drug-based or surgical cure for this disease exists. Based on the data showing that indole elicits antiinflammatory changes in expression and increases epithelial cell barrier properties, we propose that indole could represent a new, yet safe (as it is naturally present in the human GI tract), modality for regulating intestinal inflammation and promoting epithelial cell function.
In summary, our in vitro studies strongly indicate the importance of indole in favorable interkingdom signaling interactions between the intestinal epithelial cells and commensal bacteria. Our results show coordinated control of inflammation in the presence of indole through the repression of several inflammatory cytokines and coordinated regulation of signaling pathways, along with fortification of cell structure through up-regulation of actin cytoskeleton and tight junction proteins. The recognition of indole as an interkingdom signal by human cells is also consistent with the recognition of the bacterial product indole-3-acetic acid, and provides an initial basis for understanding symbiotic interkingdom interactions at the molecular level, as well as insights into some GI tract diseases and pathogenic infections.
Materials and Methods
Details for all methods can be found in SI Materials and Methods.
HCT-8 Cell Culture.
The human colon-cancer cell line HCT-8 (ATCC), derived from enterocytes at the junction of the large and small bowel, is a polarizable cell line that has been used for investigating mechanisms underlying host-cell response to pathogens and inflammation (39, 40). Cells were maintained according to standard ATCC protocols.
RNA Isolation and Microarrays.
HCT-8 cells were exposed to 1 mM indole or solvent for 4 h or 24 h. RNA isolation, reverse transcription, cDNA labeling, hybridization of arrays, array scanning, and identification of genes exhibiting a statistically significant change in expression were performed as described in SI Materials and Methods.
NF-κB Activity Measurements.
HCT-8 cells transduced with a NF-κB-GFP reporter lentivirus (31) were exposed to nontoxic concentrations of indole or indole-like molecules for 4 h, followed by addition of 40 ng/mL TNF-α. Methods used for fluorescence microscopy and image analysis are described in SI Materials and Methods.
Flow Cytometry and Intracellular Cytokine Staining.
IL-10 expression in HCT-8 cells was determined by intracellular cytokine staining using a phycoerythrin-conjugated anti-human IL-10 antibody (eBioscience). Data (8,000 events per sample) were acquired on a BD FACSAria II cell sorter system (BD Biosciences) and analyzed by using FlowJo software (Tree Star).
In Vitro Colonization, Cell Viability Assay, Extracellular Indole Assay, Transepithelial Resistance, qRT-PCR, and IL-8 ELISA.
Assays were performed according to standard protocols, the details of which are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful for the HCT-8 cells provided by Dr. Guan Zhu (Texas A&M University), the NF-κB reporter lentivirus provided by Dr. Stelios Andreadis (State University of New York at Buffalo), the assistance with TER measurements provided by Dr. Sangeeta Khare and Manjunath Hegde, and the assistance with flow cytometry provided by Kristina Ryden and Jane Miller. This work was supported by funds from the National Science Foundation Grant CBET 0846453 (to A.J.) and National Institutes of Health Grant R01 EB003872 (to T.K.W.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE14379).
This article contains supporting information online at www.pnas.org/cgi/content/full/0906112107/DCSupplemental.
References
- 1.Berg RD. The indigenous gastrointestinal microflora. Trends Microbiol. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 2.Lyte M, Arulanandam BP, Frank CD. Production of Shiga-like toxins by Escherichia coli O157:H7 can be influenced by the neuroendocrine hormone norepinephrine. J Lab Clin Med. 1996;128:392–398. doi: 10.1016/s0022-2143(96)80011-4. [DOI] [PubMed] [Google Scholar]
- 3.Sperandio V, Torres AG, Jarvis B, Nataro JP, Kaper JB. Bacteria-host communication: the language of hormones. Proc Natl Acad Sci USA. 2003;100:8951–8956. doi: 10.1073/pnas.1537100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clarke MB, Hughes DT, Zhu C, Boedeker EC, Sperandio V. The QseC sensor kinase: a bacterial adrenergic receptor. Proc Natl Acad Sci USA. 2006;103:10420–10425. doi: 10.1073/pnas.0604343103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bansal T, et al. Differential effects of epinephrine, norepinephrine, and indole on Escherichia coli O157:H7 chemotaxis, colonization, and gene expression. Infect Immun. 2007;75:4597–4607. doi: 10.1128/IAI.00630-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medellin-Peña MJ, Wang H, Johnson R, Anand S, Griffiths MW. Probiotics affect virulence-related gene expression in Escherichia coli O157:H7. Appl Environ Microbiol. 2007;73:4259–4267. doi: 10.1128/AEM.00159-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reid G, Howard J, Gan BS. Can bacterial interference prevent infection? Trends Microbiol. 2001;9:424–428. doi: 10.1016/s0966-842x(01)02132-1. [DOI] [PubMed] [Google Scholar]
- 8.Hoarau C, et al. Supernatant from bifidobacterium differentially modulates transduction signaling pathways for biological functions of human dendritic cells. PLoS One. 2008;3:e2753. doi: 10.1371/journal.pone.0002753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kelly D, et al. Commensal anaerobic gut bacteria attenuate inflammation by regulating nuclear-cytoplasmic shuttling of PPAR-gamma and RelA. Nat Immunol. 2004;5:104–112. doi: 10.1038/ni1018. [DOI] [PubMed] [Google Scholar]
- 10.Ewaschuk JB, et al. Secreted bioactive factors from Bifidobacterium infantis enhance epithelial cell barrier function. Am J Physiol Gastrointest Liver Physiol. 2008;295:G1025–G1034. doi: 10.1152/ajpgi.90227.2008. [DOI] [PubMed] [Google Scholar]
- 11.Vikström E, Tafazoli F, Magnusson K-E. Pseudomonas aeruginosa quorum sensing molecule N-(3 oxododecanoyl)-l-homoserine lactone disrupts epithelial barrier integrity of Caco-2 cells. FEBS Lett. 2006;580:6921–6928. doi: 10.1016/j.febslet.2006.11.057. [DOI] [PubMed] [Google Scholar]
- 12.Jahoor A, et al. Peroxisome proliferator-activated receptors mediate host cell proinflammatory responses to Pseudomonas aeruginosa autoinducer. J Bacteriol. 2008;190:4408–4415. doi: 10.1128/JB.01444-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kravchenko VV, et al. Modulation of gene expression via disruption of NF-kappaB signaling by a bacterial small molecule. Science. 2008;321:259–263. doi: 10.1126/science.1156499. [DOI] [PubMed] [Google Scholar]
- 14.Lee J, Jayaraman A, Wood TK. Indole is an inter-species biofilm signal mediated by SdiA. BMC Microbiol. 2007;7:42. doi: 10.1186/1471-2180-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J, et al. Indole cell signaling occurs primarily at low temperatures in Escherichia coli. ISME J. 2008;2:1007–1023. doi: 10.1038/ismej.2008.54. [DOI] [PubMed] [Google Scholar]
- 16.Domka J, Lee J, Wood TK. YliH (BssR) and YceP (BssS) regulate Escherichia coli K-12 biofilm formation by influencing cell signaling. Appl Environ Microbiol. 2006;72:2449–2459. doi: 10.1128/AEM.72.4.2449-2459.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karlin DA, Mastromarino AJ, Jones RD, Stroehlein JR, Lorentz O. Fecal skatole and indole and breath methane and hydrogen in patients with large bowel polyps or cancer. J Cancer Res Clin Oncol. 1985;109:135–141. doi: 10.1007/BF00391888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuccato E, et al. Role of bile acids and metabolic activity of colonic bacteria in increased risk of colon cancer after cholecystectomy. Dig Dis Sci. 1993;38:514–519. doi: 10.1007/BF01316508. [DOI] [PubMed] [Google Scholar]
- 19.Draghici S, et al. A systems biology approach for pathway level analysis. Genome Res. 2007;17:1537–1545. doi: 10.1101/gr.6202607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krause G, et al. Structure and function of claudins. Biochim Biophys Acta - Biomem. 2008;1778(3):631–645. doi: 10.1016/j.bbamem.2007.10.018. [DOI] [PubMed] [Google Scholar]
- 21.Kim Y, Kim S-H, Whang K-Y, Kim Y-J, Oh S. Inhibition of Escherichia coli O157:H7 attachment by interactions between lactic acid bacteria and intestinal epithelial cells. J Microbiol Biotechnol. 2008;18:1278–1285. [PubMed] [Google Scholar]
- 22.Gribar SC, Anand RJ, Sodhi CP, Hackam DJ. The role of epithelial Toll-like receptor signaling in the pathogenesis of intestinal inflammation. J Leukoc Biol. 2008;83:493–498. doi: 10.1189/jlb.0607358. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Bansal T, Jayaraman A, Bentley WE, Wood TK. Enterohemorrhagic Escherichia coli biofilms are inhibited by 7-hydroxyindole and stimulated by isatin. Appl Environ Microbiol. 2007;73:4100–4109. doi: 10.1128/AEM.00360-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mukaida N. Pathophysiological roles of interleukin-8/CXCL8 in pulmonary diseases. Am J Physiol Lung Cell Mol Physiol. 2003;284:L566–L577. doi: 10.1152/ajplung.00233.2002. [DOI] [PubMed] [Google Scholar]
- 25.Walz A, Schmutz P, Mueller C, Schnyder-Candrian S. Regulation and function of the CXC chemokine ENA-78 in monocytes and its role in disease. J Leukoc Biol. 1997;62:604–611. doi: 10.1002/jlb.62.5.604. [DOI] [PubMed] [Google Scholar]
- 26.Zhang S, Wang Q. Factors determining the formation and release of bioactive IL-12: regulatory mechanisms for IL-12p70 synthesis and inhibition. Biochem Biophys Res Commun. 2008;372:509–512. doi: 10.1016/j.bbrc.2008.05.081. [DOI] [PubMed] [Google Scholar]
- 27.Hasegawa Y, et al. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect Immun. 2007;75:2540–2547. doi: 10.1128/IAI.01957-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu X, Chen J, Wang L, Ivashkiv LB. Crosstalk among Jak-STAT, Toll-like receptor, and ITAM-dependent pathways in macrophage activation. J Leukoc Biol. 2007;82:237–243. doi: 10.1189/jlb.1206763. [DOI] [PubMed] [Google Scholar]
- 29.O'Mahony C, et al. Commensal-induced regulatory T cells mediate protection against pathogen-stimulated NF-kappaB activation. PLoS Pathog. 2008;4:e1000112. doi: 10.1371/journal.ppat.1000112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chaouche-Drider N, et al. A commensal Helicobacter sp. of the rodent intestinal flora activates TLR2 and NOD1 responses in epithelial cells. PLoS One. 2009;4:e5396. doi: 10.1371/journal.pone.0005396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian J, Andreadis ST. Independent and high-level dual-gene expression in adult stem-progenitor cells from a single lentiviral vector. Gene Ther. 2009;16:874–884. doi: 10.1038/gt.2009.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smith EA, Macfarlane GT. Enumeration of human colonic bacteria producing phenolic and indolic compounds: effects of pH, carbohydrate availability and retention time on dissimilatory aromatic amino acid metabolism. J Appl Bacteriol. 1996;81:288–302. doi: 10.1111/j.1365-2672.1996.tb04331.x. [DOI] [PubMed] [Google Scholar]
- 33.Yang SK, Eckmann L, Panja A, Kagnoff MF. Differential and regulated expression of C-X-C, C-C, and C-chemokines by human colon epithelial cells. Gastroenterology. 1997;113:1214–1223. doi: 10.1053/gast.1997.v113.pm9322516. [DOI] [PubMed] [Google Scholar]
- 34.Henderson B, Poole S, Wilson M. Microbial/host interactions in health and disease: who controls the cytokine network? Immunopharmacology. 1996;35:1–21. doi: 10.1016/0162-3109(96)00144-0. [DOI] [PubMed] [Google Scholar]
- 35.Kühn R, Löhler J, Rennick D, Rajewsky K, Müller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 36.Cowell BA, Evans DJ, Fleiszig SMJ. Actin cytoskeleton disruption by ExoY and its effects on Pseudomonas aeruginosa invasion. FEMS Microbiol Lett. 2005;250:71–76. doi: 10.1016/j.femsle.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 37.Athman R, et al. Shigella flexneri infection is dependent on villin in the mouse intestine and in primary cultures of intestinal epithelial cells. Cell Microbiol. 2005;7:1109–1116. doi: 10.1111/j.1462-5822.2005.00535.x. [DOI] [PubMed] [Google Scholar]
- 38.Veiga E, et al. Invasive and adherent bacterial pathogens co-Opt host clathrin for infection. Cell Host Microbe. 2007;2:340–351. doi: 10.1016/j.chom.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leblond J, et al. Regulation of proteolysis by cytokines in the human intestinal epithelial cell line HCT-8: role of IFNgamma. Biochimie. 2006;88:759–765. doi: 10.1016/j.biochi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 40.Roy K, et al. Enterotoxigenic Escherichia coli EtpA mediates adhesion between flagella and host cells. Nature. 2009;457:594–598. doi: 10.1038/nature07568. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.