Abstract
In diverse eukaryotic organisms, Dicer-processed, virus-derived small interfering RNAs direct antiviral immunity by RNA silencing or RNA interference. Here we show that in addition to core dicing and slicing components of RNAi, the RNAi-mediated viral immunity in Arabidopsis thaliana requires host RNA-directed RNA polymerase (RDR) 1 or RDR6 to produce viral secondary siRNAs following viral RNA replication-triggered biogenesis of primary siRNAs. We found that the two antiviral RDRs exhibited specificity in targeting the tripartite positive-strand RNA genome of cucumber mosaic virus (CMV). RDR1 preferentially amplified the 5′-terminal siRNAs of each of the three viral genomic RNAs, whereas an increased production of siRNAs targeting the 3′ half of RNA3 detected in rdr1 mutant plants appeared to be RDR6-dependent. However, siRNAs derived from a single-stranded 336-nucleotide satellite RNA of CMV were not amplified by either antiviral RDR, suggesting avoidance of the potent RDR-dependent silencing as a strategy for the molecular parasite of CMV to achieve preferential replication. Our work thus identifies a distinct mechanism for the amplification of immunity effectors, which together with the requirement for the biogenesis of endogenous siRNAs, may play a role in the emergence and expansion of eukaryotic RDRs.
Keywords: antiviral immunity, cucumber mosaic virus, RNA silencing, RNA-dependent RNA polymerase, secondary small interfering RNA
RNA silencing or RNA interference in fungi, nematodes and plants requires amplification of small interfering RNAs by eukaryotic RNA-directed RNA polymerases (RDRs) (1–3). Plant and fungal RDRs convert transcripts of target genes into dsRNA that is subsequently processed into secondary siRNAs by a Dicer or Dicer-like (DCL) nuclease. In contrast, Caenorhabditis elegans RDRs, such as RRF-1, may directly manufacture secondary siRNAs without dicing a dsRNA precursor (1–3). The genome of Arabidopsis thaliana encodes six RDRs that are grouped into four clusters (1, 2, 4), among which little is known about cluster III, consisting of RDRs 3a, 3b, and 3c. RDR2 and RDR6 are both required for the short-distance spread of transgene silencing and for the perception, but not the production, of the long-distance mobile silencing signal (1, 5–9). RDR2 is also essential for the biogenesis of the DCL3-dependent 24-nucleotide (nt) repeat-associated siRNAs (rasiRNAs) derived from transposons, retroelements, and other elements, which are the most abundant endogenous small RNAs in A. thaliana (3). Similarly, RDR6 coupled with DCL4 or DCL1 is responsible for the biogenesis of transacting siRNAs (tasiRNAs) and natural antisense siRNAs (nat-siRNAs), which silence expression of their target genes like microRNAs (miRNAs) (1–3).
RNA silencing controls antiviral immunity in fungi, plants, and invertebrates by producing virus-derived siRNAs to be loaded in an Argonaute protein for antiviral silencing (10–12). In A. thaliana, DCL4 and DCL2 produce viral siRNAs against distinct positive (+)-strand RNA viruses in a hierarchical and redundant manner (13–16). RDR1 and RDR6 of plants and RRF-1 of C. elegans have been implicated in antiviral silencing because RDR-defective mutants exhibit enhanced susceptibility to some of the RNA viruses examined (5, 13, 17–25). However, although two recent studies detected RDR-dependent biogenesis of viral siRNAs in A. thaliana, neither observed a consistent effect of secondary siRNAs on antiviral silencing (13, 18). For example, a modestly reduced viral siRNA production observed in rdr1 mutant plants was not associated with an expected increase in virus accumulation (13). Similarly, although virus accumulated to higher levels in both rdr2 rdr6 and rdr1 rdr2 rdr6 plants than in WT plants, a markedly reduced accumulation of viral siRNAs was detected only in rdr1 rdr2 rdr6 plants (18). Thus, it remains to be established if host RDRs regulate virus resistance either directly by amplification of viral siRNAs or indirectly by the activity of the endogenous RDR-dependent siRNAs of the host. In this regard, it should be pointed out that silencing against RNA viruses does not have to involve a host RDR. RNA viruses encode their own RNA-dependent RNA polymerase that synthesizes dsRNA replicative intermediates during viral RNA replication, which are sufficient to induce antiviral silencing in Drosophila melanogaster, an organism that neither encodes an RDR ortholog nor produces secondary siRNAs (26).
Plant and animal viruses encode essential pathogenesis factors to act as viral suppressors of RNA silencing (VSRs) (27). Recent studies have shown that use of VSR-deficient mutant viruses facilitates mapping the genetic requirements of the RNAi-mediated viral immunity (13, 14, 28, 29). Removal of VSRs enhances virus sensitivity to the immunity and thus is essential to reveal some of the induced antiviral silencing events that would otherwise be undetectable in hosts infected by a wild-type virus. In this study, we investigated the role of A. thaliana RDRs in the RNAi-mediated viral immunity by using a mutant of cucumber mosaic virus (CMV) that does not express the VSR protein 2b. CMV contains three positive-strand genomic RNAs and the 2b protein encoded by RNA2 is essential for infection by suppressing antiviral silencing initiated by either DCL4 or DCL2 (13). Our results demonstrate an essential role for the amplification of viral siRNAs by either RDR1 or RDR6 in antiviral silencing. Further analyses, including Illumina sequencing of more than 3.5 million viral siRNAs, indicated target specificity of the two antiviral RDRs. The possibility that emergence and expansion of eukaryotic RDRs represent an evolutionary adaptation to virus infection is discussed.
Results
An Essential Role for the Host RDR Function in Antiviral Silencing.
The 2b-deletion mutant of CMV used in this study, CMVf-Δ2b, was from the Fny strain of CMV, a subgroup I strain of CMV (30, 31). Unlike the Q strain used previously (13), Fny-CMV induces clearly visible disease symptoms (Fig. 1) in WT A. thaliana plants. CMVf-Δ2b caused no visible disease symptom (see Fig. 1) and accumulated to low levels in WT A. thaliana plants, but became as virulent as wt Fny-CMV in mutant plants defective for both DCL4 and DCL2. Thus, removal of the VSR of CMV rendered the mutant virus readily silenced in A. thaliana by the DCL4/DCL2-initiated RNA silencing immunity, similar to other VSR-deficient viruses previously described (13, 14).
Fig. 1.
CMVf-Δ2b caused diseases in rdr1/2 and rdr1/2/6 mutants, but not in WT, rdr1, rdr6, rdr1/2, and rdr2/6 mutants. Seedlings were photographed 3 weeks after inoculation with purified virions (20 μg/mL) of CMV or CMVf-Δ2b. Mutants rdr2, rdr1 rdr2, and rdr2 rdr6 were identical to WT and were not shown.
We compared the responses of the WT and the three RDR mutant plants (rdr1, rdr2, and rdr6) previously characterized (32, 33) and their double and triple mutants following inoculation with CMVf-Δ2b (see Fig. 1). Purified virions were used as the inoculum because virus inoculation by Agrobacterium infiltration triggers RDR6-dependent RNA silencing (34, 35) that may interfere with antiviral silencing. We found that the three single rdr mutants, as well as two double mutants, rdr1 rdr2 and rdr2 rdr6, showed no visible pathological changes with or without inoculation by CMVf-Δ2b, and thus were similar to WT plants. However, the two mutant combinations containing both rdr1-1 and rdr6-15 loss-of-function alleles, rdr1 rdr6 and rdr1 rdr2 rdr6, developed disease symptoms, including leaf deformation and severe stunting 3 weeks after inoculation with CMVf-Δ2b, but not with buffer alone (mock).
Following inoculation, the virus initially replicated and spread locally in the inoculated leaf (local infection) before moving out to systemically infect other parts and newly emerging tissues of the plant (systemic infection). As expected from previous studies (13, 31, 36), Northern blot hybridizations confirmed that CMVf-Δ2b locally and systemically infected all of the WT and mutant plants examined (Fig. S1 and Fig. 2A). Presence of the rdr1-1 allele both alone and in combination with rdr2-1 was reproducibly associated with a modestly enhanced (greater than twofold increase) virus accumulation in the systemically infected leaves (see Fig. 2A, compare Lanes 3 and 6 with Lanes 2, 4, 5 and 8; Fig. S2A). However, a markedly increased accumulation (>10-fold increase) of CMVf-Δ2b was detected only in the systemically infected leaves of rdr1 rdr6 and rdr1 rdr2 rdr6 plants, which carried both rdr1-1 and rdr6-15 alleles and developed disease symptoms (see Fig. 2A, compare Lanes 7 and 9 with Lanes 1–6 and 8; Fig. S2A). In contrast, presence of the rdr6-15 allele both alone and in combination with rdr2-1 had little effect (less than a onefold difference) on the accumulation of CMVf-Δ2b (see Fig. 2A). Compared to rdr1 rdr6 and rdr1 rdr2 rdr6 plants, however, CMVf-Δ2b consistently accumulated to higher levels in dcl2 dcl4 and dcl2 dcl3 dcl4 plants (see Fig. 2A, Lanes 11 and 13), which are completely defective in antiviral silencing against RNA viruses (13–16).
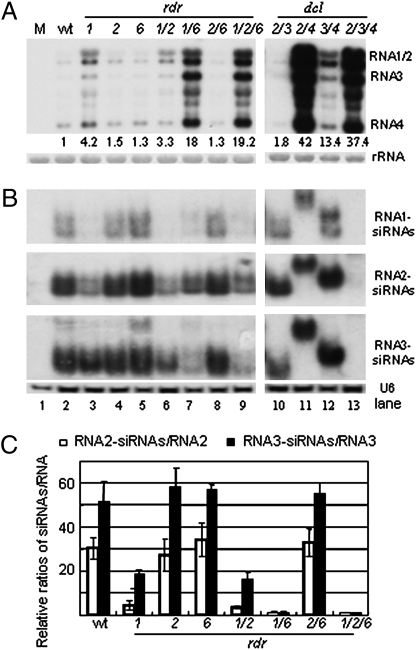
Fig. 2.
Accumulation of (A) viral genomic/subgenomic RNAs and (B) viral siRNAs derived from viral genomic RNAs 1 to 3 in the WT, rdr, and dcl mutant plants 2 weeks after inoculation with CMVf-Δ2b virions. The values below (A) referred to the relative hybridization signal intensity of genomic RNAs 1 to 3 measured for each sample with the accumulation level in WT plants set as 1; 25S rRNA and U6 RNA were used as loading controls for the high and low molecular weight RNA, respectively. RNA samples from WT plants mock inoculated with buffer were loaded in lane 1 (M). Note that lanes 1 to 13 were from the same membrane with the same exposure time. (C) The ratios of the relative accumulation levels for viral siRNAs versus their corresponding viral genomic RNAs 2 and 3 measured for three independent experiments. The ratios for both RNA2-siRNAs/RNA2 and RNA3-siRNAs/RNA3 detected in rdr1 rdr2 rdr6 plants were set as 1.
These analyses showed that the mutant virus replicated to high levels and caused severe disease symptoms in rdr1 rdr6 and rdr1 rdr2 rdr6 plants in which both RDR1 and RDR6 were not functional. Therefore, in addition to the core dicing and slicing components, A. thaliana requires an essential RDR function mediated by either RDR1 or RDR6 to enhance the potency of RNA silencing-mediated antiviral immunity. Notably, loss of RDR1 alone modestly, but significantly, increased the virus accumulation in the infected plants (P-values < 0.05), in contrast to removal of RDR6 and RDR2 both alone and together that had little effect on CMVf-Δ2b infection (see Fig. 2A and Figs. S1A and S2A). Moreover, we found that the increase of RNA3 levels in RDR-defective plants (rdr1 rdr6 and rdr1 rdr2 rdr6) from RDR-silencing plants (WT, rdr1, rdr2, rdr6, rdr1 rdr2, and rdr2 rdr6) was at least 10-fold higher than the increase of RNA2 levels (see Fig. 2A and Fig. S2A). Thus, RDR1 may not have the same function as RDR6 in antiviral silencing in targeting the genomic RNAs of CMV.
RDR-Dependent Antiviral Silencing Is Associated with Amplification of Virus-Derived siRNAs.
The contrasting effects of the single and double rdr1-1/rdr6-15 mutant alleles on the susceptibility of A. thaliana plants to CMVf-Δ2b provided a unique opportunity to determining if amplification of viral siRNAs plays a role in the RDR-dependent virus resistance. Thus, we next analyzed the accumulation of viral siRNAs corresponding to the three genomic RNAs of CMVf-Δ2b using the same samples analyzed in Fig. 2A. Compared to WT plants, a markedly decreased accumulation of viral siRNAs was observed in the plants containing both rdr1-1 and rdr6-15 alleles (Fig. 2B, compare Lane 2 with Lanes 7 and 9). The reduction was detected for siRNAs targeting all of the three genomic RNAs of CMVf-Δ2b and in both the inoculated leaves (see Fig. S1B) and systemically infected leaves (see Fig. 2B) of rdr1 rdr6 and rdr1 rdr2 rdr6 plants. It is striking to note that compared to WT plants, the levels of viral siRNAs were 1- to 2-fold lower in those symptomatic mutant plants even though CMVf-Δ2b replicated to 17- to 18-fold higher levels. Amplification of viral siRNAs were estimated by comparing the ratios of viral siRNAs/genomic RNAs between WT and mutant plants after the accumulation level of the viral genomic RNAs in WT plants and of viral siRNAs in rdr1 rdr2 rdr6 plants was both set as 1 (Fig. 2C). We found that the ratios of viral siRNAs/genomic RNAs in the symptomatic rdr1 rdr6 and rdr1 rdr2 rdr6 plants were at least 20-fold lower than that in the resistant WT plants (P-values < 0.05) (see Fig. 2C and Figs. S2 A and B), demonstrating that the RDR-dependent antiviral silencing in A. thaliana was associated with a substantial amount of RDR-mediated amplification of viral siRNAs. Participation of multiple RDRs in the biogenesis of viral siRNAs was also detected in a recent study (18), which, possibly because of the use of a VSR-expressing WT virus, did not find a consistent correlation between production of RDR-dependent viral siRNAs and antiviral silencing.
We found that consistent with previous studies (13–16), the silencing 21- and 22-nt viral siRNAs were undetectable in dcl4 dcl2 plants infected by CMVf-Δ2b under the same conditions (see Fig. 2B). This suggests that the RDR-dependent viral siRNAs were produced by the antiviral DCLs, similar to RDR6-dependent transgene siRNAs and tasiRNAs (2, 37), and that the low levels of viral siRNAs detected in rdr1 rdr6 and rdr1 rdr2 rdr6 plants were RDR-independent and represented primary viral siRNAs. Notably, CMVf-Δ2b accumulated to much lower levels in rdr1 rdr6 and rdr1 rdr2 rdr6 plants than in dcl2 dcl4 plants (see Fig. 2A). Thus, there was active antiviral silencing possibly mediated by the primary viral siRNAs in those RDR silencing-defective plants, indicating that antiviral RDRs are involved in the amplification, but not in the initiation, of antiviral silencing.
Further comparisons of the accumulation of viral siRNAs between WT and rdr mutants revealed distinct effects of rdr1-1 and rdr6-15 alleles on the amplification of viral siRNAs. The rdr6-15 allele, either alone or in combination with rdr2-1, had no obvious effect on the production of siRNAs corresponding to any of the three genomic RNAs (see Fig. 2). In contrast, and consistent with our early work (13), presence of the rdr1-1 allele, both alone and in combination with rdr2-1 and rdr6-15 alleles, markedly decreased the production of siRNAs targeting RNAs 1 and 2 in both the inoculated leaves (see Fig. S1) and systemically infected tissues (see Fig. 2). By comparison, the impact of the rdr1-1 allele either alone or in combination with rdr2-1 on the amplification of RNA3-specific siRNAs was much smaller than on the amplification of RNA1/2-specific siRNAs (see Fig. 2), and dramatically reduced amplification of RNA3-specific siRNAs was observed only when both rdr1-1 and rdr6-15 alleles were present. Subsequent analyses indicated that the three viral genomic RNAs were similarly targeted for siRNA amplification by RDR1 and an enhanced amplification of RNA3-specific siRNAs in rdr1 plants partially compensated the loss of RDR1-dependent siRNAs (see below).
Profiling Viral siRNAs Produced During Infection.
To provide a genome view of viral siRNAs produced during infection, small RNA libraries were made from systemically infected tissues of two independent pools of WT, rdr1, and rdr1 rdr2 rdr6 plants inoculated with CMVf-Δ2b and sequenced with Solexa technology in Illumina, Inc and our campus core facility, respectively. We first verified the genotypes of the A. thaliana mutants used in this study by examining our small RNA libraries for tasiRNAs and rasiRNAs, which represent the endogenous products of RDR6 and RDR2, respectively (1–3). As expected, tasiRNAs and rasiRNAs were abundant in WT and rdr1 plants but both disappeared in rdr1 rdr2 rdr6 plants. Next, only small RNAs that are 100% identical or complementary to genomic RNAs of CMVf-Δ2b were considered because the inoculum was from a cloned virus. We found that the virus-derived siRNAs were 37.4%, 35.3%, and 16.3% of the total small RNA reads from WT, rdr1, and rdr1 rdr2 rdr6 plants, respectively. Thus, in spite of much higher levels of CMVf-Δ2b replication in rdr1 rdr2 rdr6 plants than in rdr1 and WT plants (see Fig. 2A), there was a twofold drop in the relative abundance of viral siRNAs in the library made from rdr1 rdr2 rdr6 plants as compared to the libraries from rdr1 and WT plants, which was consistent with the data presented in Fig. 2C and Figs. S2 A and B).
No preference for a specific nucleotide was observed for the 5′ termini of viral siRNAs. A great majority of the sequenced viral siRNAs were the 21-nt species in all of the libraries (72–86%) whereas 10 to 21% of viral siRNAs were the 22-nt species (Fig. S3A). Thus, although DCL4 clearly played a dominant role in antiviral silencing, there was active DCL2-dependent production of viral siRNAs in the presence of DCL4. Approximately equal ratios of viral siRNAs in all of the three libraries from WT and mutant plants were mapped respectively to the positive and negative strands of each of the three viral genomic RNAs (Fig. S3B). This result was similar to that found for several +RNA viruses, but was in contrast to several other +RNA viruses that produce significantly more positive-strand siRNAs than negative-strand siRNAs (11). Because +RNA viruses produce 10- to 100-fold excesses of positive- over negative-strand RNA (38), the absence of strand bias in virus-derived siRNAs produced in both wt and rdr1 rdr2 rdr6 plants indicates that the precursor of CMV-derived siRNAs was dsRNA whether or not there was RDR-dependent amplification of viral siRNAs.
We compared the relative abundances of siRNAs targeting the three genomic RNAs, which was not readily achieved by Northern blot analysis using different probes. In WT plants infected with CMVf-Δ2b, 100,581, 91,532, and 452,252 siRNAs (19- to 25-nt) were mapped to the viral RNAs 1, 2, and 3, respectively. Thus, the abundance of RNA3-specific siRNAs was more than twice the sum of RNA1- and RNA2-specific siRNAs. The density of siRNAs targeting RNA3 was at least fivefold higher than that of RNAs 1 and 2 (Fig. 3) because RNA3 is ≈1 and 0.5 kb shorter, respectively, than RNA1 and the 2b-deleted RNA2. The siRNA density of RNA3 was also much higher than that of either RNA1 or RNA2 in rdr1 rdr2 rdr6 plants (see Fig. 3). Thus, a higher replication rate of the shorter RNA3 may produce more abundant dsRNA replication intermediates for dicing, resulting in a higher density of primary siRNAs targeting RNA3 before further amplification by RDR.
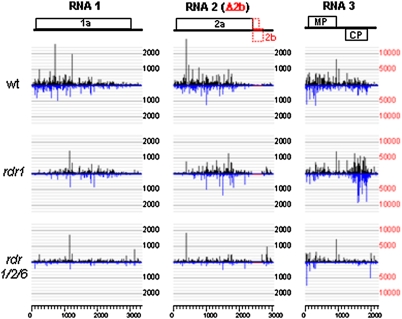
Fig. 3.
Mapping of viral siRNAs produced in infected plants. Reads of perfect match 21-nt viral siRNAs per million of the total sequenced small RNAs were plotted to the positive- (Top) and negative- (Bottom) strands of CMVf-Δ2b RNAs 1, 2 and 3 with a 20-nt window. Note that the scale of RNA3 is five times that of RNAs 1 and 2, and that no siRNAs were mapped to the deleted region of RNA2 shown in red. Because of 96% identity in the 240-nt 3′-untranslated regions among CMV RNAs 1, 2, and 3, siRNAs from this region were distributed to the individual genomic RNAs according to the overall siRNA density of each RNA. Genome organization of each viral genomic RNA is shown. CP, coat protein; MP, movement protein.
Although there were variations in the relative abundance of siRNAs targeting RNAs 1 and 2 between the duplicate libraries, RNA3-specific siRNAs was substantially and consistently reduced only in rdr1 rdr2 rdr6 plants, but not in rdr1 plants (Table S1). Because of the overwhelming abundance of RNA3-specific siRNAs produced during infection, there was little difference in the abundance of total viral siRNAs between WT and rdr1 plants (see Table S1). It is important to point out that some RNA3-specific siRNAs are expected to cross-target RNAs 1 and 2 for silencing because of 96% identity among the 240-nt 3′-untranslated regions of CMV RNAs (31).
Preferential Amplification of siRNAs Targeting Distinct Viral Genome Regions by RDR1 and RDR6.
Fig. 3 showed the mapping of the positive- and negative-strand viral siRNAs sequenced from the infected plants to the top and bottom, respectively, of each of the three genomic RNAs of CMVf-Δ2b. Note that two different scales were used to accommodate the high abundance of RNA3-specific siRNAs. We found that in WT plants infected with CMVf-Δ2b, the density of siRNAs targeting the 5′ half of each of the three viral genomic RNAs was much higher than that of the 3′ half (see Fig. 3). However, siRNAs targeting the 5′-terminal regions of RNAs 1, 2, and 3 largely disappeared in rdr1 plants, but no major changes were observed for the density of siRNAs targeting the remaining regions between wt and rdr1 plants (see Fig. 3). For example, ≈30% of RNA1-specific siRNAs were mapped to the 1/5, 5′-termimal region of RNA1 in WT and rdr6 plants, but only 10% were mapped to the same region of RNA1 in rdr1 plants. The density of siRNAs targeting the 5′-terminal regions of RNAs 2 and 3 was also similarly reduced in rdr1 plants as compared to WT plants (see Fig. 3). These results indicate that RDR1 played a critical role in the amplification of siRNAs targeting the 5′-terminal regions of the three viral genomic RNAs.
We observed a marked increase in the density of siRNAs targeting the 3′ half of RNA3 in rdr1 plants compared to that in WT plants (see Fig. 3). More than 66% of the total RNA3-specific siRNAs were mapped to this region of RNA3 in rdr1 plants, whereas siRNAs targeting the same region of RNA3 in WT plants were reduced to ≈30% of the total RNA3-specific siRNAs (see Table S1). This region of RNA3 is identical in sequence to RNA4, a subgenomic RNA transcribed internally from the negative-strand RNA3 to act as mRNA of coat protein. Both the reduction of siRNAs targeting the 5′-terminal regions of the three viral genomic RNAs and the surge of siRNAs targeting the RNA4 region of RNA3 were reproducibly detected in rdr1 plants by sequencing two independently constructed small RNA libraries made from CMVf-Δ2b-infected wt and rdr1 plants, respectively (Fig. S4). In rdr1 rdr2 rdr6 plants, however, the density of siRNAs was uniformly reduced across the entire length of the three genomic RNAs and the surge of siRNAs targeting the RNA4 region of RNA3 observed in rdr1 plants was absent (see Fig. 3 and Fig. S4). Thus, whereas RDR1 preferentially amplifies the 5′-terminal viral siRNAs, siRNAs targeting the remaining regions of viral genomic RNAs as well as the RNA4 region of RNA3 in rdr1 plants may be amplified by RDR6.
To independently verify the observed distribution patterns of viral siRNAs, total 18- to 28-nt small RNAs were isolated from infected plants and labeled with 32P to probe three panels of DNA fragments synthesized by PCR representing the evenly divided regions of viral genomic RNAs 1, 2, and 3 (Fig. S5), as described previously (26, 39). This approach confirmed a higher density of siRNAs targeting RNA3 than those of RNAs 1 and 2 because the small RNA probes from the infected plants of all five different genotypes produced much stronger hybridization signals to RNA3 than RNAs 1 and 2 (see Fig. S5). The results also supported RDR1-dependent amplification of the 5′-terminal siRNAs, as strong siRNA signals for the 5′-terminal regions of genomic RNAs 1, 2, and 3 were visible in plants that are WT for RDR1, such as WT and rdr6 plants, but were absent in rdr1, rdr1 rdr6 and rdr1 rdr2 rdr6 plants that carry rdr1-1 allele (see Fig. S5). In rdr1 plants, siRNAs targeting the RNA4 region (the second fragment of RNA3 from the 3′-end) was the most abundant compared to the other regions of RNA3. This enhanced signal was RDR6-dependent because it was not visible in rdr1 rdr6 and rdr1 rdr2 rdr6 plants that contained an additional rdr6-15 allele (see Fig. S5). Importantly, no major difference was found for the distribution patterns of viral siRNAs produced in rdr1 rdr6 and rdr1 rdr2 rdr6 plants (see Fig. S5), supporting the genetic data that RDR2 may not play a critical role in the viral siRNA biogenesis.
Production of Satellite RNA-Derived siRNAs Is RDR-Independent.
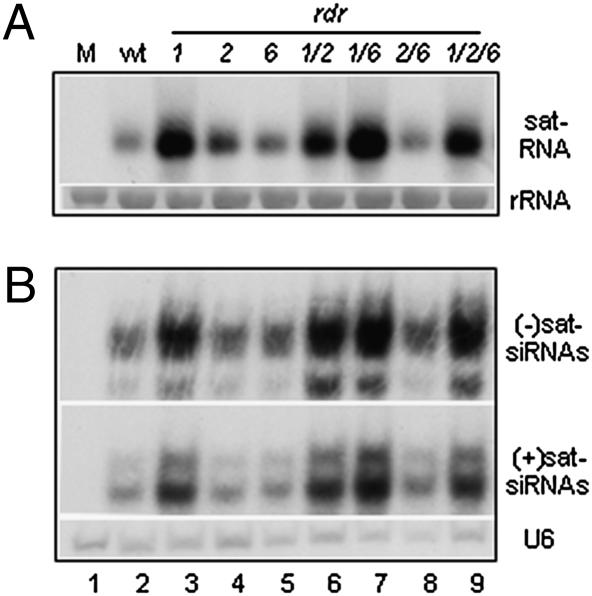
CMV infection is naturally associated with satellite RNAs (sat-RNAs), which depend on CMV for replication and transmission but share no sequence homology with the helper viral genome. The sat-RNA of CMV is a single-stranded noncoding RNA of 336-nt, and the same set of DCLs produce siRNAs from both sat-RNA and its helper virus (13, 15, 40). To determine if sat-RNA of CMV was a target of host RDRs, we inoculated the same panel of WT and mutant plants with both sat-RNA and CMVf-Δ2b because 2b is not required for sat-RNA replication (41). A modestly enhanced accumulation of sat-RNA was observed in all of the four mutant plants that contained the rdr1-1 allele (Fig. 4A), which was likely because of the slightly increased accumulation of its helper virus in these plants (Fig. S6B). However, presence of the rdr1-1 allele both alone and in combination with rdr6-15 or rdr2-1 was not associated with reduced accumulation of siRNAs derived from sat-RNA (Fig. 4B), unlike the helper viral siRNAs (Fig. S6D). In fact, a substantially increased accumulation of sat-siRNAs was observed in all of those plants that supported a higher level of sat-RNA replication, regardless of the presence of one or all of the three RDRs (see Fig. 4A and B, and Fig. S6B). As a result, the ratios of sat-siRNAs/sat-RNA differed less than half-fold between rdr1 rdr2 rdr6 plants and WT or various single/double rdr mutants with P-values >0.05 (Fig. S6E). These results indicate that production of sat-siRNAs positively correlated with the replication levels of the sat-RNA and that sat-RNA of CMV was not targeted for amplification by either of the host RDRs that amplify the helper viral siRNAs.
Fig. 4.
Sat-RNA derived siRNAs are RDR-independent. Accumulation of sat-RNA (A) and (+) and (−) siRNAs derived from sat-RNA (B) in the systemically infected leaves of WT and single, double, triple rdr mutant plants 3 weeks after inoculation with CMVf-Δ2b plus sat-RNA. 25S rRNA and U6 RNA were used as loading controls for the high and low molecular weight RNA, respectively. M, mock.
Discussion
Our findings demonstrate that in addition to the components involved in dicing and slicing, the RNA silencing antiviral defense in plants requires an essential RDR function to amplify viral siRNAs by either RDR1 or RDR6. We found that the VSR-deficient mutant of CMV became pathogenic and accumulated to high levels in A. thaliana plants when both RDR1 and RDR6 were not functional. Unlike RDR6, however, removal of RDR1 alone in rdr1 plants modestly enhanced virus accumulation in the infected plants. These phenotypes of the single and double knockout mutants of RDR1 and RDR6 in antiviral silencing are strikingly similar to those of dcl4/dcl2 single and double mutants: although the virulence of VSR-deficient virus mutants is restored only in dcl4 dcl2 double mutant plants, antiviral silencing is partially defective in dcl4 but not in dcl2 mutant plants (13–16). Importantly, analyses of viral siRNAs by both Northern blot hybridization and Illumina sequencing show that the RDR-dependent antiviral silencing is associated with more than 20-fold amplification of viral siRNAs. In contrast to dcl4 dcl2 plants that did not produce detectable silencing viral siRNAs, there was still accumulation of viral siRNAs at low levels in rdr1 rdr6 and rdr1 rdr2 rdr6 plants. These RDR-independent viral siRNAs, defined as primary viral siRNAs, were active in antiviral silencing because of reduced accumulation levels of CMVf-Δ2b in these RDR-defective plants as compared to dcl4 dcl2 plants that produce no detectable silencing viral siRNAs. Based on these and previous findings (13–16, 26, 37), we propose the following model for the induction of antiviral silencing by +RNA viruses in plants. After induction of antiviral silencing by the primary viral siRNAs processed from the dsRNA viral replicative intermediates (vRI-dsRNA), new viral dsRNA is synthesized by host RDR1 or RDR6, recognized by DCL4 or DCL2, and processed into the secondary viral siRNAs to direct more potent antiviral silencing. Viral dsRNA synthesized by both viral and host RNA polymerases as the target for dicing is also consistent with the demonstrated absence of strand bias in viral siRNAs produced in either RDR silencing or RDR silencing-defective plants.
Our results illustrate specificity of the two antiviral RDRs in the amplification of viral siRNAs from the three genomic RNAs of CMV. It has been previously reported that RDR1, but not RDR6, is induced by both salicylic acid treatment and virus infection (20, 21). We found that RDR1 preferentially amplifies siRNAs targeting the 5′-terminal region of each of the three viral genomic RNAs, whereas the remaining regions of the viral genomic RNAs are targeted by RDR6. The increased production of siRNAs targeting the RNA4 region of the genomic RNA3 observed in rdr1 plants also appears to be RDR6-dependent. The density of siRNAs targeting CMV RNA3 was higher than that of RNAs 1 and 2 in the infected plants and elimination of RDR-dependent silencing enhanced RNA3 levels at least 10-fold more than RNA2 levels. These findings indicate that the viral genomic RNA3 is more potently targeted for RDR-dependent silencing in the infected plants than RNAs 1 and 2. Preferential silencing of RNA3 by RDR6 may be critical for suppressing the pathogenicity of CMVf-Δ2b in rdr1 and rdr1 rdr2 mutant plants because the presence of the rdr1-1 allele was correlated with an enhanced RDR6-dependent production of siRNAs targeting the 3′ half of RNA3.
Unlike CMV, sat-RNA of CMV is not targeted for amplification by either of the two antiviral RDRs. This was an unexpected finding because both sat-RNA and CMV are single-stranded RNA with a m7G cap and 3′-OH replicated by the same viral replicase and targeted for the production of siRNAs by the same set of Dicer nucleases (15, 40). In this regard, sat-RNA of CMV is analogous to most of the mRNAs targeted by a miRNA because miRNA-guided cleavages of these mRNAs do not trigger RDR6-dependent production of ta-siRNAs (28). Similar to defective interfering RNAs and viroids, sat-RNAs contain extensive secondary structures and are poor targets of RNA silencing (39, 42, 43), which may play a role in sat-RNA resistance to RDR. Sat-RNAs are considered as molecular parasites of viruses because they compete for the replication machinery with the helper virus. Thus, avoiding RDR-dependent silencing that suppresses the helper viral replication represents a unique strategy of sat-RNAs to achieve preferential replication in the infected host, which may be critical for sat-RNAs to modulate helper-virus pathogenesis.
The molecular basis for the target specificity of RDR1 and RDR6 is unknown. Replication of the Flock house virus positive-strand RNA genome induces a strongly biased production of 5′-terminal siRNAs in D. melanogaster (26). In contrast, production of the abundant 5′-terminal viral siRNAs in A. thaliana is RDR1-dependent. Thus, the vRI-dsRNA made inside the replication complex may not be as readily accessible to the Dicer nucleases in plants as in insects so that host RDR-dependent amplification of viral siRNAs is necessary for an effective antiviral response in plants. In this regard, RDRs of plants, and perhaps of C. elegans as well, may have evolved not only for their roles in the biogenesis of endogenous siRNAs (2), but also to amplify viral siRNAs as a host adaptation to virus infection, thereby providing a distinct mechanism for the amplification of the immunity effector molecules.
Materials and Methods
Virus Infection and RNA Hybridizations.
A. thaliana mutants (in Columbia ecotype background) containing rdr1-1, rdr2-1, and rdr6-15 single loss-of-function alleles were characterized previously (32, 33), and used to generate double and triple mutants by genetic crosses. CMVf-Δ2b containing a 295-nt deletion in the 2b coding sequence of RNA2 was described previously (30, 31). CMVf-Δ2b with or without the sat-RNA of Q-CMV (41) was propagated in Nicotiana glutinosa plants and virions purified and inoculated to A. thaliana seedlings at the concentration of 20 μg/mL, as described (13). The inoculated leaves and systemically infected leaves were harvested from pools of 15 to 20 plants for RNA analyses by Northern blot hybridizations, as previously described (13). The probe to detect all CMV genomic and subgenomic RNAs corresponded to the 3′ terminal 240 nucleotides of Fny-CMV RNA2. Small RNAs were detected using mixtures of labeled DNA oligonucleotides corresponding to the three genomic RNAs and sat-RNA (13). Mapping of the distribution of siRNAs targeting different regions of the three genomic RNAs in Fig. S5 was as described (26) by using as probes total small RNAs harvested from virus-infected plants. All experiments were repeated at least three times with reproducible results.
Cloning, Sequencing, and Analysis of Viral siRNAs.
Small RNAs of 18- to 28-nt extracted from the systemically infected leaves 14 days after inoculation with CMVf-Δ2b were purified from 15% denatured polyacrylamide gel and cloned as described (44). Briefly, harvested small RNAs were directly ligated to the 3′-linker used for miRNA cloning (Integrated DNA Technology) by T4 RNA ligase 2Tr (New England Biolabs). The ligation products were purified and ligated to the 5′ Solexa linker (Illumina) by T4 RNA ligase I (New England Biolabs). The final ligation products were reverse transcribed using a primer targeting the 3′-linker and the cDNA pool was amplified by PCR and sent for sequencing by the Solexa technology (Illumina). Duplicate libraries were constructed from two independent RNA samples from WT, rdr1, and rdr1 rdr2 rdr6 plants and sequenced by the Solexa platform in Illumina, Inc, and the University of California–Riverside Core Facility, respectively. The analysis of the sequenced small RNAs was as described previously (26).
Supplementary Material
Acknowledgments
We thank Jim Carrington for the single rdr mutants. This work was supported by the National Research Initiative of the U.S. Department of Agriculture Cooperative State Research, Education, and Extension Service Grant 2007-35319-18325 (to S.D.) and National Institutes of Health Grant AI052447 (to S.D.). X.B.W. was supported in part by the China Scholarship Council.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904086107/DCSupplemental.
References
- 1.Voinnet O. Use, tolerance and avoidance of amplified RNA silencing by plants. Trends Plant Sci. 2008;13:317–328. doi: 10.1016/j.tplants.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 2.Poethig RS, et al. The function of RNAi in plant development. Cold Spring Harb Symp Quant Biol. 2006;71:165–170. doi: 10.1101/sqb.2006.71.030. [DOI] [PubMed] [Google Scholar]
- 3.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat Rev Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 4.Wassenegger M, Krczal G. Nomenclature and functions of RNA-directed RNA polymerases. Trends Plant Sci. 2006;11:142–151. doi: 10.1016/j.tplants.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 5.Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith LM, et al. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 8.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 9.Brosnan CA, et al. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mlotshwa S, Pruss GJ, Vance V. Small RNAs in viral infection and host defense. Trends Plant Sci. 2008;13:375–382. doi: 10.1016/j.tplants.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 11.Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–426. doi: 10.1016/j.cell.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang X, Segers GC, Sun Q, Deng F, Nuss DL. Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J Virol. 2008;82:2613–2619. doi: 10.1128/JVI.02324-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz-Pendon JA, Li F, Li WX, Ding SW. Suppression of antiviral silencing by cucumber mosaic virus 2b protein in Arabidopsis is associated with drastically reduced accumulation of three classes of viral small interfering RNAs. Plant Cell. 2007;19:2053–2063. doi: 10.1105/tpc.106.047449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deleris A, et al. Hierarchical action and inhibition of plant Dicer-like proteins in antiviral defense. Science. 2006;313:68–71. doi: 10.1126/science.1128214. [DOI] [PubMed] [Google Scholar]
- 15.Fusaro AF, et al. RNA interference-inducing hairpin RNAs in plants act through the viral defence pathway. EMBO Rep. 2006;7:1168–1175. doi: 10.1038/sj.embor.7400837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouché N, Lauressergues D, Gasciolli V, Vaucheret H. An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 2006;25:3347–3356. doi: 10.1038/sj.emboj.7601217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilkins C, et al. RNA interference is an antiviral defence mechanism in Caenorhabditis elegans. Nature. 2005;436:1044–1047. doi: 10.1038/nature03957. [DOI] [PubMed] [Google Scholar]
- 18.Donaire L, et al. Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J Virol. 2008;82:5167–5177. doi: 10.1128/JVI.00272-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mourrain P, et al. Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell. 2000;101:533–542. doi: 10.1016/s0092-8674(00)80863-6. [DOI] [PubMed] [Google Scholar]
- 20.Xie Z, Fan B, Chen C, Chen Z. An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc Natl Acad Sci USA. 2001;98:6516–6521. doi: 10.1073/pnas.111440998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yu D, Fan B, MacFarlane SA, Chen Z. Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol Plant Microbe Interact. 2003;16:206–216. doi: 10.1094/MPMI.2003.16.3.206. [DOI] [PubMed] [Google Scholar]
- 22.Yang SJ, Carter SA, Cole AB, Cheng NH, Nelson RS. A natural variant of a host RNA-dependent RNA polymerase is associated with increased susceptibility to viruses by Nicotiana benthamiana. Proc Natl Acad Sci USA. 2004;101:6297–6302. doi: 10.1073/pnas.0304346101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qu F, et al. RDR6 has a broad-spectrum but temperature-dependent antiviral defense role in Nicotiana benthamiana. J Virol. 2005;79:15209–15217. doi: 10.1128/JVI.79.24.15209-15217.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu R, Yigit E, Li WX, Ding SW. An RIG-I-Like RNA helicase mediates antiviral RNAi downstream of viral siRNA biogenesis in Caenorhabditis elegans. PLoS Pathog. 2009;5:e1000286. doi: 10.1371/journal.ppat.1000286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schott DH, Cureton DK, Whelan SP, Hunter CP. An antiviral role for the RNA interference machinery in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2005;102:18420–18424. doi: 10.1073/pnas.0507123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aliyari R, et al. Mechanism of induction and suppression of antiviral immunity directed by virus-derived small RNAs in Drosophila. Cell Host Microbe. 2008;4:387–397. doi: 10.1016/j.chom.2008.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li F, Ding SW. Virus counterdefense: Diverse strategies for evading the RNA-silencing immunity. Annu Rev Microbiol. 2006;60:503–531. doi: 10.1146/annurev.micro.60.080805.142205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martín-Hernández AM, Baulcombe DC. Tobacco rattle virus 16-kilodalton protein encodes a suppressor of RNA silencing that allows transient viral entry in meristems. J Virol. 2008;82:4064–4071. doi: 10.1128/JVI.02438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li HW, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–1321. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 30.Ryabov EV, Fraser G, Mayo MA, Barker H, Taliansky M. Umbravirus gene expression helps potato leafroll virus to invade mesophyll tissues and to be transmitted mechanically between plants. Virology. 2001;286:363–372. doi: 10.1006/viro.2001.0982. [DOI] [PubMed] [Google Scholar]
- 31.Soards AJ, Murphy AM, Palukaitis P, Carr JP. Virulence and differential local and systemic spread of cucumber mosaic virus in tobacco are affected by the CMV 2b protein. Mol Plant Microbe Interact. 2002;15:647–653. doi: 10.1094/MPMI.2002.15.7.647. [DOI] [PubMed] [Google Scholar]
- 32.Xie Z, et al. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Allen E, et al. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 34.Johansen LK, Carrington JC. Silencing on the spot. Induction and suppression of RNA silencing in the Agrobacterium-mediated transient expression system. Plant Physiol. 2001;126:930–938. doi: 10.1104/pp.126.3.930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Voinnet O, Lederer C, Baulcombe DC. A viral movement protein prevents spread of the gene silencing signal in Nicotiana benthamiana. Cell. 2000;103:157–167. doi: 10.1016/s0092-8674(00)00095-7. [DOI] [PubMed] [Google Scholar]
- 36.Ding SW, Li WX, Symons RH. A novel naturally occurring hybrid gene encoded by a plant RNA virus facilitates long distance virus movement. EMBO J. 1995;14:5762–5772. doi: 10.1002/j.1460-2075.1995.tb00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ahlquist P, Noueiry AO, Lee WM, Kushner DB, Dye BT. Host factors in positive-strand RNA virus genome replication. J Virol. 2003;77:8181–8186. doi: 10.1128/JVI.77.15.8181-8186.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Szittya G, Molnár A, Silhavy D, Hornyik C, Burgyán J. Short defective interfering RNAs of tombusviruses are not targeted but trigger post-transcriptional gene silencing against their helper virus. Plant Cell. 2002;14:359–372. doi: 10.1105/tpc.010366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Du QS, et al. DCL4 targets Cucumber mosaic virus satellite RNA at novel secondary structures. J Virol. 2007;81:9142–9151. doi: 10.1128/JVI.02885-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding SW, et al. Efficient infection from cDNA clones of cucumber mosaic cucumovirus RNAs in a new plasmid vector. J Gen Virol. 1995;76:459–464. doi: 10.1099/0022-1317-76-2-459. [DOI] [PubMed] [Google Scholar]
- 42.Itaya A, et al. A structured viroid RNA serves as a substrate for dicer-like cleavage to produce biologically active small RNAs but is resistant to RNA-induced silencing complex-mediated degradation. J Virol. 2007;81:2980–2994. doi: 10.1128/JVI.02339-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang MB, et al. On the role of RNA silencing in the pathogenicity and evolution of viroids and viral satellites. Proc Natl Acad Sci USA. 2004;101:3275–3280. doi: 10.1073/pnas.0400104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mi S, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008;133:116–127. doi: 10.1016/j.cell.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.