Abstract
Hoxb13 is robustly transcribed in derivatives of posterior endoderm including the colon, rectum, and the prostate gland. Transcriptional activity in the prostate persists unabated under conditions of androgen deprivation and throughout the course of disease progression in a mouse prostate cancer model. To elucidate the molecular basis of prostate-restricted transcriptional activation of Hoxb13, a bacterial artificial chromosome (BAC)-based reporter gene deletion analysis was performed in transgenic mice. Two regions downstream of the Hoxb13 coding region were found to be required to support transcriptional activity in the prostate but were completely dispensable for expression in the colon and rectum. Bioinformatic analyses of one region identified a 37-bp element conserved in mammals. This element, which bears two potential binding sites for Forkhead class transcription factors, is occupied by FOXA1 in a human prostate cancer cell line. Precise replacement of this enhancer with an extended LoxP site in the context of a 218,555-bp BAC reporter nearly extinguished Hoxb13-mediated transcriptional activity in the mouse prostate. These data demonstrate that FOXA1 directly regulates HOXB13 in human prostate epithelial cells, and show that this prostate-specific regulatory mechanism is conserved in mice.
Keywords: androgen independent, recombineering, prostate-specific, Homeobox, Forkhead
Homeobox genes are major regulators of a diverse array of developmental processes (1). Within the past decade, a significant body of evidence has accumulated indicating that homeobox genes play important roles during prostate morphogenesis (2). Prominent among these are Nkx3.1, Hoxa10, and the group 13 paralogs Hoxa13, Hoxb13, and Hoxd13. Null mutations in these homeobox genes result in defects in branching morphogenesis and/or changes in gene expression that suggest altered identity of certain prostatic lobes (3–7).
During embryogenesis, Hoxb13 is robustly expressed in the posterior axial skeleton as well as the posterior endoderm that gives rise to the most distal regions of the gastrointestinal tract and the urogentital sinus (8). In adult mice, Hoxb13 expression is restricted to the prostate, descending colon, and rectum, which are all derived from posterior endoderm (9). Prostatic expression is highest in the ventral prostate lobe epithelium, with decreasing levels in the lateral, dorsal, and anterior lobe epithelia (9). An intriguing feature of Hoxb13 expression in the prostate is its persistence under androgen deprivation conditions (9). In marked contrast to virtually all other genes with prostate epithelial-restricted expression, castration does not diminish Hoxb13 mRNA accumulation, nor does it reduce expression of a reporter gene driven by Hoxb13 regulatory elements (10). This unusual expression pattern makes Hoxb13 an excellent paradigm to dissect regulatory mechanisms that can confer transcriptional activity in prostate epithelial cells under in vivo androgen depletion conditions. Understanding mechanisms that drive prostate-specific, androgen-independent gene expression is critical to support ongoing efforts to develop new strategies to treat lethal androgen-independent prostate cancer.
Reporter gene approaches in transgenic mice have been instrumental in the identification of prostate-restricted transcriptional control regions (11–18). However, these analyses have focused exclusively on androgen-stimulated genes. Recently, a large Hoxb13 reporter gene capable of recapitulating the androgen-independent, prostate-restricted transcriptional activity of the endogenous Hoxb13 gene has been reported (10). To shed light on the molecular mechanisms that mediate this gene expression pattern, we performed a detailed analysis of the regulatory regions that confer prostate specificity to Hoxb13. Using a combination of bioinformatic, biochemical, and transgenic analyses, we scanned a 218-kb genomic region near the Hoxb13 locus and functionally characterized a 37-bp enhancer module required for Hoxb13 transcriptional activity in the prostate. We further demonstrate that the Forkhead winged helix transcription factor FOXA1 binds this module in the LNCaP prostate cancer cell line. These data directly implicate FOXA proteins in the regulation of HOXB13 and provide new insights into the molecular basis of prostate-restricted gene expression.
Results
The B13/LZ218kb transgene, which contains a lacZ reporter gene under the control of mouse Hoxb13 regulatory elements within a 218-kb bacterial artificial chromosome (BAC) (Table 1; Fig. S1), was capable of robustly recapitulating the Hoxb13 expression pattern in the urogenital and gastrointestinal tracts (10). Reporter gene expression did not induce phenotypic changes.
Table 1.
Compendium of BAC and transgene data. The table indicates the number of independent transgenic strains surveyed, base pair sequence either upstream or downstream of Hoxb13, and intensity of X-gal staining in prostate or colon decreasing from (+++++) to (+), with (+/−) indicating highly variegated staining and (−) indicating no staining.
| BAC Designation | Transgene Name | # of Strains | Upstream | Downstream | Prostate | Colon |
| RP23-335O22Hoxb13-lacZ | B13/LZ218kb | 6 | 158,454 | 60,101 | +++++ | +++++ |
| RP23-335O22Hoxb13-lacZ{Δ96kb-5′} | B13/LZΔ96kb5′ | 6 | 62,205 | 60,101 | +++++ | +++++ |
| RP23-335O22Hoxb13lacZ{Δ45kb-3′} | B13/LZΔ45kb3′ | 4 | 158,454 | 15,306 | + | +++++ |
| RP23-335O22Hoxb13-lacZ{Δ51kb-3′} | B13/LZΔ51kb3′ | 5 | 158,454 | 9,245 | − | +++++ |
| RP23-335O22Hoxb13-lacZ{R37bpLoxP} | B13/LZR37bpLoxP | 4 | 154,454 | 60,101 | +/− | +++++ |
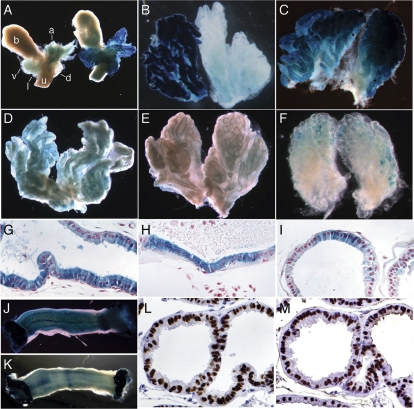
To localize prostate-specific regulatory elements contained within the 218,555-bp reporter construct, truncations were made in B13/LZ218kb by recombineering (19) (Table 1; Fig. S1). Deletion of 96 kb from the 5′ end (with respect to the transcriptional orientation of Hoxb13) of B13/LZ218kb (designated B13/LZΔ96kb5′) did not affect reporter gene expression in the prostate (Fig. 1A). Whole-mount X-gal analyses of six independent transgenic lines carrying the B13/LZΔ96kb5′ transgene showed strong, uniform reporter gene expression in the adult prostate that paralleled levels observed for the original B13/LZ218kb transgene (Fig. 1 A–C). The cellular distribution of reporter gene expression in B13/LZΔ96kb5′ transgenic prostates (Fig. 1G) was indistinguishable from that reported for the B13/LZ218kb transgene (10). In contrast, deletion of ∼45 kb from the 3′ end of B13/LZ218kb (designated B13/LZΔ45kb3′) resulted in a significant reduction in reporter gene activity (Fig. 1D). This construct retains 14 kb downstream of the Hoxb13 start of translation. Analyses performed in four independent lines carrying B13/LZΔ45kb3′ demonstrated that reporter gene expression was reduced by greater than 95% in neonatal anterior, dorsal, and lateral lobes, although a moderate level of expression was retained in the ventral lobes. In adult B13/LZΔ45kb3′ mice, reporter gene expression was no longer detectable above the level of endogenous β-galactosidase activity in the anterior, dorsal, and lateral lobes. Within the adult ventral lobe, staining did not appear uniform, giving rise to a variegated appearance; however, reporter gene expression remained restricted to epithelial cells (Fig. 1 D and H). These data demonstrate that enhancer elements required to support transcriptional activity of the Hoxb13 locus in the prostate gland reside between 15 and 60 kb downstream of the coding region. However, the B13/LZΔ45kb3′ transgene retained the ability to drive detectable reporter gene expression in the ventral lobe, albeit at a reduced level (Fig. 1D), indicating that cis elements that enable Hoxb13 expression in prostate epithelial cells lie elsewhere.
Fig. 1.
Reporter gene expression in B13/LZ transgenic mice. (A) Partial urogenital tract from a nontransgenic negative control FVB male (left) and a B13/LZΔ96kb5′ transgenic male (right) after X-gal staining. a, anterior prostate lobe; b, bladder; d, dorsal prostate lobe; l, lateral prostate lobe; u, urethra; v, ventral prostate lobe. (B) B13/LZ218kb transgenic ventral lobe (left) and nontransgenic FVB negative control (right). (C) B13/LZΔ96kb5′ transgenic ventral lobes. (D) B13/LZΔ45kb3′ ventral lobes. (E) B13/LZ51kb3′ ventral lobes. (F) B13/LZR37bpLoxP ventral prostate lobes. (G) Nuclear fast red (NFR)-stained section of a B13/LZΔ96kb5′ ventral lobe duct after whole-mount X-gal staining. (H) NFR-stained section of a ventral lobe duct from a B13/LZΔ96kb5′/ Δ45kb3′ male. The 3′ deletion in this construct is the same as that in B13/LZΔ45kb3′, and staining is indistinguishable from that in B13/LZΔ45kb3′ mice. (I) NFR-stained section of a ventral lobe duct from a B13/LZR37bpLoxP male. (J) B13/LZΔ96kb5′ colon segment. (K) B13/LZR37bpLoxP colon segment. (L) Immunohistochemical detection of Hoxb13 in the ventral prostate lobe. (M) Immunohistochemical detection of Foxa1 in the ventral prostate lobe.
In an attempt to locate elements important for supporting prostate-specific Hoxb13 transcriptional activity expression within the ∼15-kb downstream region that was retained in B13/LZΔ45kb3′, the VISTA browser was used to search for noncoding regions conserved between the mouse and human genomes (20). An ∼6-kb region located between 9 and 15 kb downstream of the Hoxb13 coding sequence showed clear evidence of conservation (Fig. 2A).
Fig. 2.
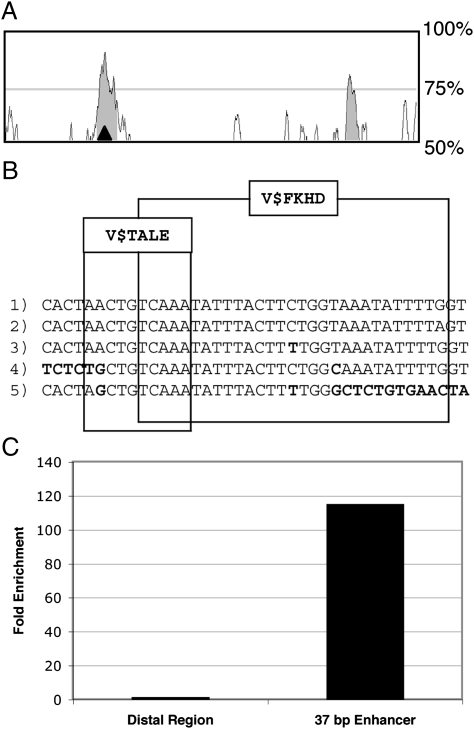
Analyses of the 37-bp conserved region. (A) VISTA Browser 2.0 plot of sequence conservation between the ∼9 kb and ∼15 kb region downstream of Hoxb13 aligned to the human genome. The calculation window is set at 50 bp, with the threshold set at 70% conservation identity over a 50-bp minimum conservation width and is indicated by shaded regions. y axis minimum and maximum are set at 50% and 100% conservation identity, respectively. The 37-bp conserved region is indicated by the black triangle in the shaded region. (B) The FKHD_TALE composite model defines the conserved 37-bp Hoxb13 enhancer. The Hoxb13 gene −15,000 and +30,000 region of rhesus monkey (3), rat (4), and dog (5) species was searched for homology to the mouse (1) and human (2) 37-bp sequences. The model was defined by taking into account sequence variations (bold characters) between the analyzed species. Vertebrate (V$) FKHD and TALE matrices of the FKHD_TALE model are marked by boxes. (C) Graph of the fold enrichment for human α-FOXA1 goat polyclonal antibody immunoprecipitated genomic regions over the goat IgG immunoprecipitated control. The quantitative PCR amplicons were a region containing the conserved 37-bp sequence and a region 2 kb distal to the conserved sequence.
To determine whether the ∼6-kb region contained elements required for prostate expression of the reporter gene, the region downstream of Hoxb13 in B13/LZ218kb was reduced from 60,101 bp to 9,245 bp. This transgene was designated B13/LZΔ51kb3′. Whole-mount analyses of adult B13/LZΔ51kb3′ prostates revealed the complete absence of X-gal staining above background in all lobes (Fig. 1E, ventral lobe shown). Remarkably, lacZ expression in the colon remained robust and uniform at levels comparable to that of B13/LZ218kb colon (Fig. 1 J and K) (10). These data indicate that cis-regulatory elements in the region between +9,245 bp and +15,485 bp are required for Hoxb13 transcriptional activity in the ventral lobe. Furthermore, these data demonstrate that the cis-regulatory elements for prostatic expression are completely separable from the elements for colorectal expression.
Having identified a 6,420-bp sequence containing cis-regulatory elements required for ventral lobe expression located between +9,245 bp and +15,485 bp downstream of Hoxb13, bioinformatic analyses were used to search this region for conserved composite model matches (21). The FrameWorker module within the GEMS Launcher software (http://www.genomatix.de) (22) was employed to define common models of matching composite sites that are conserved in sequence, order, and spacing between the mouse 6,420-bp region of interest and the corresponding region of the human genome. FrameWorker defined a composite model within 37 bp (≈3 DNA helical turns) (23). The central element of this model consists of two Forkhead domain factor family (V$FKHD) FOXA class matrices (24, 25). The V$FKHD modules are in opposite orientations and separated by 12 bp, such that a complete DNA helical turn could position bound FOXA transcription factors to the same DNA helical surface. Furthermore, the position of an adjacent TALE (V$TALE) matrix match was also conserved within the 37-bp enhancer of mouse and human sequences (26–28).
Using the core DNA sequences of the mouse 37-bp enhancer defined by the composite model match, we searched the Hoxb13 loci (−15,000 to +30,000 bp) of rhesus monkey (Macaca mulatta), rat (Rattus norvegicus), dog (Canis lupus familiaris), and zebrafish (Danio rerio) vertebrate species by using the NCBI Blast Blt2seq program. The search identified DNA sequences highly similar to the mouse and human 37-bp enhancer in the rhesus monkey and rat genomes (Fig. 2B). In contrast, the search did not indicate similarity within the zebrafish Hoxb13A locus. FrameWorker analysis of the identified homologous sequences in human, mouse, rhesus monkey, rat, and dog confirmed the conservation of FKHD and TALE matrices. The presence of two Foxa protein binding sites in a region demonstrated to regulate urogenital sinus and prostate expression, coupled with similar reported expression patterns between Foxa1 and Hoxb13 in the urogenital sinus and prostate (29, 30), suggests that Foxa1 may directly regulate Hoxb13 expression.
To determine whether FOXA1 was recruited to the 37-bp conserved sequence in vivo, we employed the chromatin immunoprecipitation (ChIP) technique. The human prostate cancer cell line LNCaP was chosen, as it has high levels of FOXA1 protein (31) and also expresses HOXB13 (32). An anti-FOXA1 goat polyclonal antibody enriched the immunoprecipitated region of interest 115-fold over a nonspecific IgG immunoprecipitation control when detected by quantitative PCR. In a comparison that demonstrates antibody specificity and efficiency of chromatin shearing, a region 2 kb distal that is not predicted to be bound by FOXA1 was only enriched 1.8-fold over the IgG control (Fig. 2C). These data demonstrate that FOXA1 is bound to the 37-bp HOXB13 enhancer in human prostate epithelial cancer cells.
To compare the cellular distribution of Foxa1 and Hoxb13 in the mouse prostate gland, immunohistochemical analyses were performed on adjacent sections of adult mouse prostates. Both proteins were detected in a nearly uniform pattern in luminal epithelial cell nuclei (Fig. 1 L and M); however, Hoxb13 was also detected in a subset of basal cells in a manner reminiscent of the distribution of Nkx3.1 (16, 33).
To evaluate the role of the predicted 37-bp enhancer in vivo, the galK positive/negative selection-based BAC recombineering method (34) was employed to precisely replace the 37-bp enhancer contained in B13/LZ218kb with a 34-bp LoxP site and three additional base pairs. The LoxP sequence was chosen because the commonly used Cre recombination technique leaves a LoxP scar sequence in the genome which has not been reported to have effects on gene expression. The resulting recombinant BAC, RP23-335O22Hoxb13-lacZ{R37bpLoxP}, was sequenced across the recombination junctions to confirm correct replacement and subjected to restriction fragment length polymorphism analysis to verify BAC integrity. This transgene construct (B13/LZR37bpLoxP) was then used to generate transgenic mice for reporter gene analysis.
Whole-mount microscopic analyses of X-gal-stained tissue from B13/LZR37bpLoxP transgenic mice revealed that replacement of the 37-bp sequence within the context of the full-length 218,555-bp sequence resulted in a dramatic reduction of β-gal activity in the prostate (Fig. 1F) without altering cellular distribution (Fig. 1I), whereas strong staining in the colon was observed at comparable levels to those in mice carrying B13/LZΔ96kb5′ (Fig. 1K). The 37-bp enhancer replacement transgene phenocopied the loss of reporter gene activity observed in all lobes in B13/LZΔ45kb3′ transgenic mice, demonstrating that multiple downstream enhancers are required for robust Hoxb13 prostate-specific transcriptional activity. However, the 37-bp enhancer replacement alone also nearly eliminated reporter gene expression in the ventral lobe, indicating that this element plays a key role in mediating Hoxb13 transcriptional activity in the adult prostate.
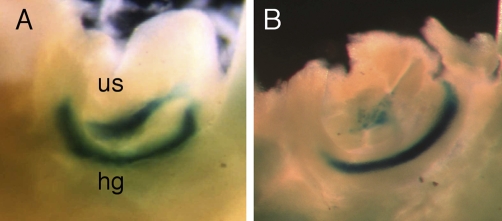
Both Foxa1 and Hoxb13 are expressed throughout the entire urogenital sinus epithelium before prostate development (29). Because two predicted Foxa binding sites are present in the 37-bp enhancer, reporter gene analyses of the B13/LZR37bpLoxP transgene were performed to determine the importance of the enhancer in the newly partitioned urogenital sinus of 12.5 days post coitum (dpc) embryos. Robust lacZ expression is found throughout the urogenital sinus in B13/LZ218kb transgenic embryos (Fig. 3A). A profound decrease in lacZ expression in the urogenital sinus was observed in B13/LZR37bpLoxP 12.5 dpc embryos; however, reporter gene expression in the hindgut remained strong (Fig. 3B). In contrast to the uniform reporter gene activity observed in the urogenital sinus of B13/LZ218kb transgenic embryos, expression in B13/LZR37bpLoxP embryos was limited to scattered cells within the central third of the urogenital sinus. These data indicate that replacement of the 37-bp module interferes with maintenance of reporter gene activity in the emerging urogenital sinus after septation of the cloaca.
Fig. 3.
Reporter gene expression in the urogenital sinus and hindgut of 12.5 dpc transgenic embryos. (A) B13/LZ218kb urogenital sinus (us) and hindgut (hg). (B) B13/LZR37bpLoxP urogenital sinus and hindgut.
We next sought to determine whether the 37-bp enhancer was sufficient to direct gene expression to the mouse prostatic epithelium. To this end, an 843-bp region of the Hoxb13 locus encompassing ∼500 bp upstream and 300 bp downstream of the enhancer was cloned upstream of the mouse hsp68 promoter linked to a lacZ reporter gene. Analysis using FrameWorker confirmed that the FKHD module was the only conserved element within this region. Four independent founder strains carrying this transgene were derived. Reporter gene analyses of male urogenital tissues did not detect β-gal activity in the prostate gland; however, one strain showed transgene expression in a few muscle fibers in the bladder, demonstrating that the construct was functional (Fig. S1C).
Discussion
Identifying genomic cis-regulatory regions that mediate tissue-restricted gene expression functionally underpins efforts to understand the molecular basis of animal development and homeostasis. In addition, these elements enable the development of tools to manipulate gene expression in model systems and, potentially, in a therapeutic setting in human disease patients. Discerning the location and function of distant elements that modulate complex temporal and spatial patterns of transcriptional activity is particularly challenging, and requires labor-intensive analyses in whole transgenic animals. The development of efficient methods to modify large DNA molecules with nucleotide precision combined with the availability of powerful informatic tools for genome analysis provides new opportunities to facilitate this arduous process. By applying these approaches to a canonical deletion analysis, we have been able to scan a large region of the Hoxb13 locus to functionally identify a 37-bp module that is necessary for prostate-restricted transcriptional activity.
Deletion analyses localized elements required to support robust transcriptional activity of Hoxb13 in the prostate within the region spanning +15 to +61 kb. Extending the deletion to encompass the region spanning +9 to +61 kb completely abolished transcriptional activity exclusively in the prostate, demonstrating that one or more key elements lie within a 6-kb sequence downstream of Hoxb13. Using comparative genomics of this ∼6-kb region between mice and humans, a conserved 37-bp region was recognized composed of two inversely orientated FKHD elements with preferential cognate sites for Foxa transcription factors. To enhance the specificity of this model, we examined the evolutionary conservation of this 37-bp sequence by searching the corresponding Hoxb13-containing regions of the rhesus monkey, rat, dog, and zebrafish genomes for sequence homology. Conserved sequences were found in all three mammalian genomes, but not in zebrafish (35). These data are consistent with the fact that the prostate gland is a mammalian innovation, and raise the possibility that this regulatory circuit evolved in concert with the prostate gland.
The location of the 37-bp enhancer 11.7 kb downstream of Hoxb13 and the dramatically reduced reporter gene expression in B13/LZR37bpLoxP transgenic prostates are consistent with predicted mechanisms of Foxa1-regulated gene activation. The transcription factor FOXA1 contributes to gene regulation by its ability to act as a pioneer factor that binds to nucleosomal DNA (36–41). In a recent report, it was demonstrated that FOXA1 cell-type-specific functions rely primarily on differential recruitment to chromatin predominantly at distant enhancers rather than at proximal promoters in the LNCaP and MCF-7 cell lines (42). This differential recruitment leads to cell-type-specific changes in chromatin structure and functional collaboration with lineage-specific transcription factors. The 37-bp enhancer may act to confer a chromatin structure to the region that is permissive for the binding of transcription factors that confer prostate-specific Hoxb13 expression by acting in concert with other elements in the region between +15 and +61 kb downstream. This interpretation is consistent with the fact that the 37-bp enhancer alone was not sufficient to drive expression of a heterologous promoter in the prostate. Loss of these 37 bp may result in a generally restrictive chromatin structure over a relatively broad region, but with some limited potential for coordinating transcriptional activity to confer the low level of variable X-gal staining observed.
FOXA1 is an important regulator of prostate-specific expression. It has been demonstrated to interact with the androgen receptor on the promoters of PSA and Probasin and was essential for gene expression (14). Interestingly, in the absence of androgen stimulation, FOXA1 was still capable of binding the promoter regions but alone was unable to transactivate (14). FOXA1 may interact with a cofactor other than the androgen receptor on the 37-bp HOXB13 regulatory element to activate expression independent of androgen stimulation.
Interestingly, there are several parallels between Foxa1 and Hoxb13 expression and the consequences of their deletion in the mouse. Both Foxa1 and Hoxb13 are detected throughout the urogenital sinus epithelium at the beginning of prostate development by immunohistochemistry (10, 29). Both are expressed strongly in the prostate buds, and their expression continues through development and into adulthood (29). Foxa1 and Hoxb13 are expressed highest in the ventral lobes, at a lower level in the dorsal and lateral lobes, and weakest in the anterior lobes (9, 10, 29, 43). During prostate carcinogenesis in the human and mouse, studies suggest that expression of both genes is maintained (31, 44, 45). In the homozygous knockout of either Hoxb13 or Foxa1, defects are observed in luminal cell differentiation and secretory function, with a loss of some of the same major secretory products (6, 30).
Despite the similarities in Foxa1 and Hoxb13 expression and the in vivo evidence of FOXA1 binding to the 37-bp enhancer, it is possible that FOXA2 may also have the ability to bind to the enhancer and regulate HOXB13 expression. Their binding sites are highly conserved, and several prostate-specific promoters have been demonstrated to recruit either factor (46). Both Foxa1 and Foxa2 are expressed during the initial budding of the prostate (29) and may have redundant activities early in prostate development. In human and mouse prostate cancer, FOXA1 expression persists in early- and late-stage cancer, whereas FOXA2 is only detected in neuroendocrine small-cell carcinomas and in some high Gleason score adenocarcinomas (31). Interestingly, both factors have been demonstrated to interact with androgen receptor and, on prostate-specific promoters, FOXA2 was demonstrated to activate gene expression in an androgen-independent manner (46).
FOXA1 regulation of HOXB13 may also have important implications in breast cancer. FOXA1 has been demonstrated to be an important cofactor of the estrogen receptor in regulating gene networks in the breast (36). HOXB13 expression is repressed by estrogen signaling in human breast cancer cell lines and also in the neonatal estrogenized rat prostate (47, 48). However, HOXB13 is expressed in a subset of estrogen-receptor-positive breast cancer patients who are unresponsive to tamoxifen monotherapy (49). When estrogenic signaling is incapable of repressing HOXB13 expression during breast carcinogenesis, FOXA1 may invoke a mechanism similar to that used in the prostate to specifically activate HOXB13 expression. As deregulated expression of HOXB13 in breast cancer is associated with tumor aggressiveness, tamoxifen resistance, and poor prognosis, the mechanisms regulating HOXB13 expression in breast cancer warrant investigation (49).
The in vivo analyses of Hoxb13 regulatory elements reported here identify a 37-bp element distant from the coding region that is necessary but not sufficient to support robust transcriptional activity of Hoxb13 in the prostate. Additionally, we have demonstrated that FOXA1, a protein known to act as a pioneer factor in regulating gene expression, binds to a similar conserved region of the human genome, and is likely to enable transcription factors that confer prostate-specific expression of HOXB13. These factors may interact with FOXA1 directly to form a unique complex that activates HOXB13 expression in the absence of androgens, or may gain access to nearby cis-regulatory elements through FOXA1-mediated changes in chromatin structure. Elucidation of additional factors involved in HOXB13 expression and their mechanisms of action will further our understanding of prostate-specific gene regulation, and may have implications for understanding the emergence of androgen-independent prostate cancer.
Materials and Methods
Construction of Transgenes.
Construction of the B13/LZ218kb transgene (formerly designated RP23-335O22Hoxb13-lacZ) has been described (10). Truncations of B13/LZR37bpLoxP were generated using the standard DY380 recombineering protocol (19). The ampicillin-targeting cassettes to create truncations were generated by standard PCR amplifications from pTAMP (19), using the primers as follows: RP23-335O22Hoxb13-lacZ{Δ96-5′}, 5′-aatgggattcaccggcatgtccaaagaagttgagacagaaagtaaaattatcttagacgtcaggtggcac-3′ and 5′-acatcattttaaccatcttggcaccttctttctgcccacataatatatcactcacgttaagggattttggtc-3′; RP23-335O22Hoxb13lacZ{Δ45kb-3′}, 5′-tggccccgggtgcctttgccctttgattacattccgcgttgaggaggaactctagacgtcaggtggcac-3′ and 5′-acatttaacaggtataacaggagggtcaggagttaaaggggttttggcagctcacgttaagggattttggtc-3′; RP23-335O22Hoxb13-lacZ{Δ51kb-3′}, 5′-tggccccgggtgcctttgccctttgattacattccgcgttgaggaggaactctagacgtcaggtggcac-3′ and 5′-cggggaacccgggcgaagtgcgtctttcccagagatctgctgagagccgactcacgttaagggattttggtc-3′.
RP23-335O22Hoxb13-lacZ{R37bpLoxP} used the protocol as described (34), with the addition of 50 mg/mL kanamycin to the selection plates. Primers used for amplification of galK were 5′-tccctttgcctcccctcccCCTGTTGACAATTAATCATCGGCA-3′ and 5′-gctcaccccttgctagggTCAGCACTGTCCTGCTCCTT-3′; primers used for the replacement of the LoxP sequence were 5′-tccctttgcctcccctccctctccctctcctttcccgtctctatcataacttcgtataat-3′ and 5′-gctcaccccttgctagggtgttatctgtgctttgtagagaccgataacttcgtatagcat-3′.
Transgenic Mouse Generation and Genotyping.
BACs were linearized with PI-Sce I (New England Biolabs), and then purified by sucrose gradient centrifugation and dialyzed as described (50). The fragments were injected into FVB single-cell embryos, and transgenic mice were generated as described (51). Founders or transgenic offspring were identified by Southern blot analyses. Genomic DNA was isolated using the Gentra Puregene Mouse Tail kit (Qiagen). Ten micrograms of extracted DNA was digested with EcoRI and EcoRV, electrophoresed on a 0.8% agarose gel, and then transferred to a Nytran-N membrane (Whatman). The vector pLZKAN-Hoxb13 (10) was linearized with SacII, and then the SP6 universal primer was used to create a probe for Southern blot with the Ambion Strip-EZ PCR kit (Applied Biosystems). Blot hybridization was performed using Ambion ULTRAhyb solution (Applied Biosystems), was washed according to the manufacturer’s instructions, and exposed to X-ray film.
All animal procedures were approved by the University of Maryland, Baltimore County Institutional Animal Care and Use Committee.
Detection of β-Gal Activity.
Embryos or tissues were dissected and kept in cold PBS until fixed with 4% paraformaldehyde for 30–60 min. The staining was performed as described (50) until the desired intensity was achieved as compared to a nontransgenic negative control.
Chromatin Immunoprecipitation Assay.
This assay was adapted from Carroll et al. (36), with the following modifications: LNCaP cells were grown to 80% confluence, harvested, fixed with 37% formaldehyde (Sigma), and lysed. Cell lysates were sonicated with 8 pulses for 30 s each at power level 4 on a Branson Sonifier, model 450. Chromatin was evaluated for shearing efficiency on a 2.0% agarose gel, with optimal shearing consisting of a 200–1000 bp smear. Immunoprecipitation, reverse cross-linking, and DNA purification were performed as described (36), using an α-FOXA1 goat polyclonal antibody (Abcam, ab5089) and IgG (Jackson Immunoresearch, 005-000-003).
Quantitative PCR.
Quantitative PCR was performed using Sybr Green PCR Master Mix (Applied Biosystems) on a Bio-Rad iCycler IQ Real-time PCR machine. Primers were designed using Primer3 software (http://primer3.sourceforge.net) as follows: 37-bp enhancer, 5′-CCCACCCATCATCACTAACTG-3′ and 5′-GGATTGATTCACCCCTTCCT-3′; distal region, 5′-CCCAGGGACTCCTCTATTGTC-3′ and 5′-GTGATCTCAGCTGGGTGACTC-3′. The presence of a single amplicon was systemically verified by dissociation curve analysis. PCR amplifications for a region containing the 37-bp enhancer and a region 2 kb distal were conducted in quadruplicate and the calculated thresholds (CT) were averaged (Avg CT), removing outliers with >1 cycle difference. The nonspecific background, as detected by a negative control goat IgG immunoprecipitation, was used to derive (Avg CTIgG) for both regions. The change in cycle threshold for each region was normalized using the formula, dCT = (Avg CTα-FOXA1 − Avg CTIgG). The fold enrichment for both regions was calculated using the formula, fold enrichment = 2^ dCT.
Computational Analysis of Regulatory Elements.
The experimentally confirmed +9,245 bp to +15,485 bp DNA sequence of mouse Hoxb13 (chromosome 11, absolute positions: 96064905–96071145, NCBI build 36) was queried in the GenomeVISTA program to plot DNA conservation identity to the aligned human HOXB13 downstream sequences under conditions indicated in Fig. 2A (20). The experimentally confirmed +9,245 bp to +15,485 bp DNA sequence of mouse Hoxb13 (chromosome 11, absolute positions: 96064905–96071145, NCBI build 36) and the human HOXB13 downstream (chromosome 17, absolute positions 44144888–44148592, NCBI build 37) homologous sequences were searched for conserved composite models by FrameWorker 4.4.2 (2005) software with 1–10 bp distance between adjacent elements and allowing as many as five elements in the composite models. MatInspector Matrix Family Library Version 5.0 (February 2005) was used for the matrix family search. All vertebrate matrix families were queried with 0.75 core and “optimized” matrix similarity settings applying the “Urogenital Systems” cocitation filter (http://www.genomatix.de). To meet the DNA sequence length requirements of the software (<3000 bp), the actual input sequence lengths were kept under 3000 bp, assuring overlaps for exhaustive search. Only one composite model reached the maximum (five-element) complexity that was composed of vertebrate matrix TALE (strand +, matrix sim. 1.00) and FKHD (strands +/−, matrix sim. 0.91–0.98), and two minor matrices (V$) MYT1 (strand −, matrix sim. 0.93) and ETS (strand −, matrix sim. 0.85), respectively, reaching the maximum total FrameWork score of 1.00/1.00 (http://www.genomatix.de). However, a high degree of sequence conservation and the low number of input sequences may result in model overfitting, preventing the recognition of composite model matches in divergent species. Therefore, by using the 37-bp core sequences and 15-bp flanks of the mouse and human sequences, the Hoxb13 gene upstream −15,000 and downstream +30,000 sequences were searched in Macaca mulatta (rhesus monkey), Rattus norvegicus (rat), Canis lupus familiaris (dog), and Danio rerio (zebrafish) species for homology by using the NCBI Blast (Blt2seq) program. Homologous sequences of human, rhesus monkey, dog, mouse, and rat 37-bp region with 15-bp flanks were further analyzed by FrameWorker (5.4.3.3) for model definition using the Matrix Family Library 7.1, core/matrix similarity 0.75/optimized setting allowing 10-bp variance in distance between adjacent matrices. FrameWorker confirmed a single FKHD_TALE model [FKHD(−), matrix sim. 0.99, TALE(+), matrix sim. 1.00].
Supplementary Material
Acknowledgments
This work was supported by grant DK59152 from the National Institutes of Health, and grant DAMD17-98-1-8477 from the Congressionally Directed Medical Research Program. The authors gratefully acknowledge Carlise R. Bethel for technical assistance with Foxa1 immunohistochemistry, Albert Zhou for assistance analyzing R37pb-hsp68-lacZ transgenic mice, and Sandra Tickle for animal care.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902001107/DCSupplemental.
References
- 1.Hombría JC, Lovegrove B. Beyond homeosis—HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 2.Thomson AA, Marker PC. Branching morphogenesis in the prostate gland and seminal vesicles. Differentiation. 2006;74:382–392. doi: 10.1111/j.1432-0436.2006.00101.x. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka M, et al. Nkx3.1, a murine homolog of Drosophila bagpipe, regulates epithelial ductal branching and proliferation of the prostate and palatine glands. Dev Dyn. 2000;219:248–260. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1054>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- 4.Podlasek CA, et al. Hoxa-10 deficient male mice exhibit abnormal development of the accessory sex organs. Dev Dyn. 1999;214:1–12. doi: 10.1002/(SICI)1097-0177(199901)214:1<1::AID-DVDY1>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Podlasek CA, Clemens JQ, Bushman W. Hoxa-13 gene mutation results in abnormal seminal vesicle and prostate development. J Urol. 1999;161:1655–1661. [PubMed] [Google Scholar]
- 6.Economides KD, Capecchi MR. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development. 2003;130:2061–2069. doi: 10.1242/dev.00432. [DOI] [PubMed] [Google Scholar]
- 7.Podlasek CA, Duboule D, Bushman W. Male accessory sex organ morphogenesis is altered by loss of function of Hoxd-13. Dev Dyn. 1997;208:454–465. doi: 10.1002/(SICI)1097-0177(199704)208:4<454::AID-AJA2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 8.Zeltser L, Desplan C, Heintz N. Hoxb-13: A new Hox gene in a distant region of the HOXB cluster maintains colinearity. Development. 1996;122:2475–2484. doi: 10.1242/dev.122.8.2475. [DOI] [PubMed] [Google Scholar]
- 9.Sreenath T, Orosz A, Fujita K, Bieberich CJ. Androgen-independent expression of hoxb-13 in the mouse prostate. Prostate. 1999;41:203–207. doi: 10.1002/(sici)1097-0045(19991101)41:3<203::aid-pros8>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 10.McMullin RP, Mutton LN, Bieberich CJ. Hoxb13 regulatory elements mediate transgene expression during prostate organogenesis and carcinogenesis. Dev Dyn. 2009;238:664–672. doi: 10.1002/dvdy.21870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleutjens KB, et al. A 6-kb promoter fragment mimics in transgenic mice the prostate-specific and androgen-regulated expression of the endogenous prostate-specific antigen gene in humans. Mol Endocrinol. 1997;11:1256–1265. doi: 10.1210/mend.11.9.9974. [DOI] [PubMed] [Google Scholar]
- 12.Cleutjens KB, et al. An androgen response element in a far upstream enhancer region is essential for high, androgen-regulated activity of the prostate-specific antigen promoter. Mol Endocrinol. 1997;11:148–161. doi: 10.1210/mend.11.2.9883. [DOI] [PubMed] [Google Scholar]
- 13.Cleutjens KB, van Eekelen CC, van der Korput HA, Brinkmann AO, Trapman J. Two androgen response regions cooperate in steroid hormone regulated activity of the prostate-specific antigen promoter. J Biol Chem. 1996;271:6379–6388. doi: 10.1074/jbc.271.11.6379. [DOI] [PubMed] [Google Scholar]
- 14.Gao N, et al. The role of hepatocyte nuclear factor-3 α (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol Endocrinol. 2003;17:1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- 15.Schuur ER, et al. Prostate-specific antigen expression is regulated by an upstream enhancer. J Biol Chem. 1996;271:7043–7051. doi: 10.1074/jbc.271.12.7043. [DOI] [PubMed] [Google Scholar]
- 16.Chen H, Mutton LN, Prins GS, Bieberich CJ. Distinct regulatory elements mediate the dynamic expression pattern of Nkx3.1. Dev Dyn. 2005;234:961–973. doi: 10.1002/dvdy.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenberg NM, et al. The rat probasin gene promoter directs hormonally and developmentally regulated expression of a heterologous gene specifically to the prostate in transgenic mice. Mol Endocrinol. 1994;8:230–239. doi: 10.1210/mend.8.2.8170479. [DOI] [PubMed] [Google Scholar]
- 18.Yan Y, et al. Large fragment of the probasin promoter targets high levels of transgene expression to the prostate of transgenic mice. Prostate. 1997;32:129–139. doi: 10.1002/(sici)1097-0045(19970701)32:2<129::aid-pros8>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 19.Lee EC, et al. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics. 2001;73:56–65. doi: 10.1006/geno.2000.6451. [DOI] [PubMed] [Google Scholar]
- 20.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: Computational tools for comparative genomics. Nucleic Acids Res. 2004;32(Web Server issue):W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda K, et al. Androgen receptor binding sites identified by a GREF_GATA model. J Mol Biol. 2005;353:763–771. doi: 10.1016/j.jmb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 22.Klingenhoff A, Frech K, Quandt K, Werner T. Functional promoter modules can be detected by formal models independent of overall nucleotide sequence similarity. Bioinformatics. 1999;15:180–186. doi: 10.1093/bioinformatics/15.3.180. [DOI] [PubMed] [Google Scholar]
- 23.Cartharius K, et al. MatInspector and beyond: Promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 24.Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kaufmann E, Müller D, Knöchel W. DNA recognition site analysis of Xenopus winged helix proteins. J Mol Biol. 1995;248:239–254. doi: 10.1016/s0022-2836(95)80047-6. [DOI] [PubMed] [Google Scholar]
- 26.Jiang Y, et al. A novel family of Cys-Cys, His-Cys zinc finger transcription factors expressed in developing nervous system and pituitary gland. J Biol Chem. 1996;271:10723–10730. doi: 10.1074/jbc.271.18.10723. [DOI] [PubMed] [Google Scholar]
- 27.Bertolino E, Reimund B, Wildt-Perinic D, Clerc RG. A novel homeobox protein which recognizes a TGT core and functionally interferes with a retinoid-responsive motif. J Biol Chem. 1995;270:31178–31188. doi: 10.1074/jbc.270.52.31178. [DOI] [PubMed] [Google Scholar]
- 28.Mao X, Miesfeldt S, Yang H, Leiden JM, Thompson CB. The FLI-1 and chimeric EWS-FLI-1 oncoproteins display similar DNA binding specificities. J Biol Chem. 1994;269:18216–18222. [PubMed] [Google Scholar]
- 29.Mirosevich J, Gao N, Matusik RJ. Expression of Foxa transcription factors in the developing and adult murine prostate. Prostate. 2005;62:339–352. doi: 10.1002/pros.20131. [DOI] [PubMed] [Google Scholar]
- 30.Gao N, et al. Forkhead box A1 regulates prostate ductal morphogenesis and promotes epithelial cell maturation. Development. 2005;132:3431–3443. doi: 10.1242/dev.01917. [DOI] [PubMed] [Google Scholar]
- 31.Mirosevich J, et al. Expression and role of Foxa proteins in prostate cancer. Prostate. 2006;66:1013–1028. doi: 10.1002/pros.20299. [DOI] [PubMed] [Google Scholar]
- 32.Xu LL, et al. Quantitative expression profile of androgen-regulated genes in prostate cancer cells and identification of prostate-specific genes. Int J Cancer. 2001;92:322–328. doi: 10.1002/ijc.1196. [DOI] [PubMed] [Google Scholar]
- 33.Wang X, et al. A luminal epithelial stem cell that is a cell of origin for prostate cancer. Nature. 2009;461:495–500. doi: 10.1038/nature08361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dieterich C, Rahmann S, Vingron M. Functional inference from non-random distributions of conserved predicted transcription factor binding sites. Bioinformatics. 2004;20(Suppl 1):i109–i115. doi: 10.1093/bioinformatics/bth908. [DOI] [PubMed] [Google Scholar]
- 36.Carroll JS, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 37.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–2526. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laganière J, et al. Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA. 2005;102:11651–11656. doi: 10.1073/pnas.0505575102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bossard P, Zaret KS. Repressive and restrictive mesodermal interactions with gut endoderm: Possible relation to Meckel’s Diverticulum. Development. 2000;127:4915–4923. doi: 10.1242/dev.127.22.4915. [DOI] [PubMed] [Google Scholar]
- 40.Gualdi R, et al. Hepatic specification of the gut endoderm in vitro: Cell signaling and transcriptional control. Genes Dev. 1996;10:1670–1682. doi: 10.1101/gad.10.13.1670. [DOI] [PubMed] [Google Scholar]
- 41.Cirillo LA, et al. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 42.Lupien M, et al. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–970. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berquin IM, Min Y, Wu R, Wu H, Chen YQ. Expression signature of the mouse prostate. J Biol Chem. 2005;280:36442–36451. doi: 10.1074/jbc.M504945200. [DOI] [PubMed] [Google Scholar]
- 44.Chiaverotti T, et al. Dissociation of epithelial and neuroendocrine carcinoma lineages in the transgenic adenocarcinoma of mouse prostate model of prostate cancer. Am J Pathol. 2008;172:236–246. doi: 10.2353/ajpath.2008.070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Edwards S, et al. Expression analysis onto microarrays of randomly selected cDNA clones highlights HOXB13 as a marker of human prostate cancer. Br J Cancer. 2005;92:376–381. doi: 10.1038/sj.bjc.6602261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, et al. Foxa1 and Foxa2 interact with the androgen receptor to regulate prostate and epididymal genes differentially. Ann N Y Acad Sci. 2005;1061:77–93. doi: 10.1196/annals.1336.009. [DOI] [PubMed] [Google Scholar]
- 47.Wang Z, et al. The prognostic biomarkers HOXB13, IL17BR, and CHDH are regulated by estrogen in breast cancer. Clin Cancer Res. 2007;13:6327–6334. doi: 10.1158/1078-0432.CCR-07-0310. [DOI] [PubMed] [Google Scholar]
- 48.Prins GS, et al. Influence of neonatal estrogens on rat prostate development. Reprod Fertil Dev. 2001;13:241–252. doi: 10.1071/rd00107. [DOI] [PubMed] [Google Scholar]
- 49.Ma XJ, et al. A two-gene expression ratio predicts clinical outcome in breast cancer patients treated with tamoxifen. Cancer Cell. 2004;5:607–616. doi: 10.1016/j.ccr.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 50.Bieberich CJ, Utset MF, Awgulewitsch A, Ruddle FH. Evidence for positive and negative regulation of the Hox-3.1 gene. Proc Natl Acad Sci USA. 1990;87:8462–8466. doi: 10.1073/pnas.87.21.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gordon JW, Ruddle FH. Gene transfer into mouse embryos: Production of transgenic mice by pronuclear injection. Methods Enzymol. 1983;101:411–433. doi: 10.1016/0076-6879(83)01031-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.