Abstract
Loss-of-function mutations in the genes associated with primary microcephaly (MCPH) reduce human brain size by about two-thirds, without producing gross abnormalities in brain organization or physiology and leaving other organs largely unaffected [Woods CG, et al. (2005) Am J Hum Genet 76:717–728]. There is also evidence suggesting that MCPH genes have evolved rapidly in primates and humans and have been subjected to selection in recent human evolution [Vallender EJ, et al. (2008) Trends Neurosci 31:637–644]. Here, we show that common variants of MCPH genes account for some of the common variation in brain structure in humans, independently of disease status. We investigated the correlations of SNPs from four MCPH genes with brain morphometry phenotypes obtained with MRI. We found significant, sex-specific associations between common, nonexonic, SNPs of the genes CDK5RAP2, MCPH1, and ASPM, with brain volume or cortical surface area in an ethnically homogenous Norwegian discovery sample (n = 287), including patients with mental illness. The most strongly associated SNP findings were replicated in an independent North American sample (n = 656), which included patients with dementia. These results are consistent with the view that common variation in brain structure is associated with genetic variants located in nonexonic, presumably regulatory, regions.
Keywords: brain morphology, cortical area, MRI, SNP, Imaging genetics
The human species is distinguished by the enormous size of its brain relative to its body size. In the primate lineage leading to humans, brain size and cerebral cortex surface area have increased dramatically (1). It is plausible that the genes driving the evolutionary expansion of the brain also determine to some extent differences in brain morphology among humans today. Despite the high heritability of brain morphology (2), the genetic mechanisms underlying normal variation in these phenotypes remain largely unknown. In many genes, loss-of-function mutations have been shown to have a profound effect on brain structure (3). Common variants of such genes could have less obvious, yet detectable, effects in healthy subjects, as well as in patients with psychiatric and neurological disorders.
Here we investigate four candidate genes selected because of their association with congenital primary recessive microcephaly (MCPH) (3): Microcephalin (MCPH1) (4), CDK5RAP2 (MCPH3) (5), ASPM (MCPH5) (6), and CENPJ (MCPH6) (7). Loss-of-function mutations and deletions in MCPH genes render the brain about one-third the normal size and result in a marked reduction in cerebral cortical area, without gross changes in basic physiology or structural plan (8). Animal studies have shown that these genes are expressed in the neuroepithelium in utero and influence proliferation of neuroblasts in the ventricular zone during cortical development (3). These genes were also selected on the basis of their hypothesized evolutionary significance (9–11). The molecular evolution of ASPM has been linked with major changes in relative cerebral cortex size across primates (12). There is also evidence of selective pressure on two haplotypes within MCHP1 and ASPM in recent human evolution, and the evolutionary rates of all four genes are higher in primates than in other mammals (9).
Previous attempts to link common gene variants in the MCPH family to normal variation in human brain morphology have been generally unsuccessful (13, 14). However, these studies only considered small numbers of exonic SNPs, which may not be in linkage disequilibrium (LD) with causative variants. Indeed, a causative variant is likely to be regulatory, rather than coding, and as such may be located in a region far upstream or downstream of the exon structure (15). In addition, most of these studies did not investigate sex-specific effects. Such effects might be expected, given the recent finding of an association between a MCPH1 SNP and head circumference in males only (16), and given the reported influence of the regulatory, X-linked MECP2 gene on MCPH gene expression (17). Previous studies also used rather crude phenotypes. Recent primate studies indicate that MCPH genes have specific effects on cortical structure, rather than overall brain or head size (8, 12).
The approach of the current study was to use microarray technology to genotype SNPs associated with the four MCPH genes, including upstream and downstream regions, and investigate associations with brain morphology phenotypes derived from MRI scans in an ethnically homogenous Norwegian single-site discovery sample (from the Thematic Organized Psychosis or TOP study, see Methods). In addition to the tag SNPs genotyped using microarray technology, two candidate exonic SNPs (rs930557, MCPH1; and rs41310927, ASPM) were genotyped using traditional methods (TaqMan assay). The significant findings from the discovery sample were then tested for replication in an ethnically heterogeneous North American multisite replication sample [from the Alzheimer's Disease Neuroimaging Initiative (ADNI) study, see Methods]. To reduce the number of statistical comparisons, we focused on four summary measures of brain morphometry: total brain volume, intracranial volume, total cortical surface area, and mean cortical thickness.
Results
Using a statistical model that estimates the genetic effect for males and females separately, while controlling for disease effects, we found in the discovery sample significant sex-specific associations between common variants of three MCPH genes and brain phenotypes after correction for multiple comparisons for each gene (Table 1). The majority of the significant SNPs affected total cortical area.
Table 1.
SNPs in the CDK5RAP2, MCPH1, and ASPM genes significantly associated with brain phenotypes
| Gene | SNP | Population | Phenotype | P-value | Effect size (±CI) | ||
| Nominal | Permuted | Corrected | |||||
| TOP sample | |||||||
| CDK5RAP2 | rs4836817 | Males | BV | 7 × 10−4 | 0.001 | 0.034 | −42 ± 24 cm3 |
| CDK5RAP2 | rs4836817 | Males | CA | 3 × 10−4 | 0.001 | 0.026 | −65 ± 35 cm2 |
| CDK5RAP2 | rs10818453 | Males | CA | 6 × 10−4 | 0.002 | 0.05 | −63 ± 35 cm2 |
| CDK5RAP2 | rs4836819 | Males | CA | 4 × 10−4 | 0.002 | 0.035 | −61 ± 34 cm2 |
| CDK5RAP2 | rs4836820 | Males | CA | 3 × 10−5 | 2 × 10−4 | 0.005 | −72 ± 33 cm2 |
| CDK5RAP2 | rs7859743 | Males | CA | 5 × 10−4 | 0.002 | 0.04 | −60 ± 33 cm2 |
| CDK5RAP2 | rs2297453* | Males | CA | 4 × 10−5 | 3 × 10−4 | 0.009 | −72 ± 34 cm2 |
| CDK5RAP2 | rs2282168 | Males | CA | 4 × 10−6 | 4 × 10−5 | 9 × 10−4 | −84 ± 35 cm2 |
| CDK5RAP2 | rs1888893 | Males | CA | 6 × 10−5 | 4 × 10−4 | 0.01 | −73 ± 35 cm2 |
| CDK5RAP2 | rs914592* | Males | CA | 5 × 10−6 | 5 × 10−5 | 0.001 | −83 ± 35 cm2 |
| CDK5RAP2 | rs914593 | Males | CA | 2 × 10−5 | 10−4 | 0.004 | −80 ± 36 cm2 |
| MCPH1 | rs2816514 | Females | BV | 2 × 10−4 | 7 × 10−5 | 0.019 | 44 ± 23 cm3 |
| MCPH1 | rs2816517 | Females | BV | 3 × 10−4 | 8 × 10−5 | 0.020 | 37 ± 20 cm3 |
| MCPH1 | rs11779303 | Females | CA | 2 × 10−4 | 4 × 10−5 | 0.009 | 62 ± 33 cm2 |
| MCPH1 | rs11779303 | Females | ICV | 2 × 10−4 | 3 × 10−5 | 0.006 | 61 ± 32 cm3 |
| ASPM | rs10922168 | Females | ICV | 0.030 | 0.014 | 0.037 | 42 ± 38 cm3 |
| ADNI sample | |||||||
| CDK5RAP2 | rs914592 | males | CA | 0.002 | 0.002 | — | −31 ± 11 cm2 |
| CDK5RAP2 | rs2297453 | males | CA | 0.005 | 0.007 | — | −26 ± 10 cm2 |
BV, brain volume; CA, cortical area; ICV, intracranial volume. The results from the TOP sample are presented in the upper section. Only SNPs with corrected P-values < 0.05 are shown (results for all four brain phenotypes are shown in Table S1). The results of the replication analysis in the ADNI sample are shown in the lower section (one-tailed t-test). Because these were planned comparisons, no correction for multiple comparisons was performed here. The effect sizes represent increase in volume and area per extra copy of the minor allele. Confidence Intervals (CI) are 95%, symmetrical (The genotypes for all SNPs in Table 1, including minor and major allele frequencies, are found in Table S2).
*SNPs genotyped on both Affymetrix and Illumina platforms.
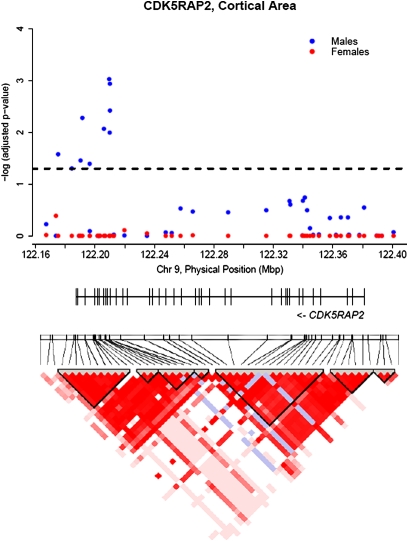
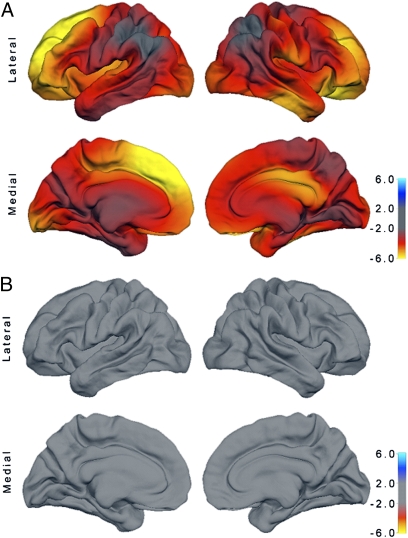
The locations of the significant SNPs within CDK5RAP2 affecting cortical area, are shown in Fig. 1. Three of the 10 significant SNPs (rs4836817, rs10818453, rs4836819) were located downstream, between 15 and 0.6 kb from the stop codon. The remaining seven significant SNPs were located in introns, and span across the last 7 of 38 exons in the CDK5RAP2 gene. However, all 10 SNPs were located within one LD block. There were no significant effects of CDK5RAP2 SNPs in females (see Fig. 1). Figure 2A shows the effect of the most significant SNP (rs2282168) in CDK5RAP2 on cortical areal expansion in males. These effects are bilaterally distributed and most prominent in the frontal cortex. There were no significant effects in females (Fig. 2B).
Fig. 1.
Negative log P-values corrected for multiple comparisons, for all SNPs in CDK5RAP2 for cortical area. The horizontal, dotted line marks the significance threshold (0.05). The bar below the figure marks the exon-intron structure of the gene. The LD map at the bottom was generated from the control group.
Fig. 2.
Association of CDK5RAP2 SNP rs2282168 with cortical area in (A) males and (B) females. The map shows the distribution of −log P-values (sign indicating direction of effect per copy of minor allele) across the reconstructed cortical surface. The corresponding maps for rs4836817, rs4836817, rs10818453, rs4836819, rs4836820, rs7859743, rs2297453, rs914592, rs1888893, and rs914593 were similar to that shown in this figure, which is probably because of the fact that all these polymorphisms are in strong LD. Fig. S1 shows the corresponding effect of MCPH1 SNP rs11779303 on cortical area in females.
The three significant markers within MCPH1 were located upstream, between 488 and 198 kb from the start codon, and only had effects in females. The significant SNP from ASPM was located in an intron, and affected only females. No significant effects on any brain phenotype were found for the two exonic candidate SNPs from MCPH1 and ASPM.
In line with recent guidelines for replicating genotype-phenotype associations (18), we subsequently tested the significant SNPs from the TOP study that were also genotyped on the Illumina platform for replication in the ADNI sample. Both of these SNPs, rs914592 and rs2297453, were associated with the CDK5RAP2 gene. Using the same brain phenotypes and the same sex-specific statistical model, while controlling for the effect of disease via regression modeling, we found a significant SNP dose-effect for rs914592, with a nominal P-value of 0.0035 on total cortical area in males and 0.55 in females. The other CDK5RAP2 SNP with a gene-wide significant effect on cortical area in the TOP sample, rs2297453, also showed significant male-specific effects on cortical area in the ADNI sample (P = 0.01 in males; P = 0.58 in females) (see Table 1). We did not find significant effects on other brain phenotypes, consistent with the results from the TOP sample.
Associations between the two replicated SNPs and schizophrenia and bipolar spectrum disorders in the TOP sample, and Alzheimer's Disease and mild cognitive impairment in the ADNI sample, were investigated using logistic regression. No significant associations were discovered for any of the diseases after multiple testing was accounted for.
Finally, to assess whether the current MCPH genes have been under positive selection, we considered three commonly investigated footprints of recent natural selection using standard statistics: population differentiation, as measured by fixation index, the presence of long haplotypes with high frequency (iHS), and low genetic diversity within a region (Tajima’s D).
Among the current associated markers, only rs10922168 in ASPM showed signs of population differentiation (between European and Asian populations), no SNPs showed extreme iHS scores, and Tajima’s D showed evidence of selection around SNPs within both ASPM and CDK5RAP2. In total, we found five SNPs for which the Tajima's D statistics associated with the surrounding 100-kb regions were in the lowest 5% quantile of this statistic’s empirical genome-wide distribution. All of this information and HapMap phase II allele frequencies are provided in (Table S3).
The University of California Santa Cruz (UCSC) genome browser was used for assessment of mammalian genomic conservation. Few of the significant SNPs are located in conserved regions in mammalian genomes. However, a region in intron 31 (chr9:123169718–123170071) (hg19), between the markers rs2282168 and rs1888893 (CDK5RAP2), is well conserved across mammalian genomes and is actually longer and more conserved than the flanking exons, suggesting functional importance.
Discussion
To our knowledge, the present findings are unique in their demonstration of an association between common variants of any of the MCPH genes and brain structure in humans. The replication of our initial findings for CDK5RAP2 in an independent sample represents a strong confirmation and implies that these results are highly unlikely to be chance findings. The present results revealed markedly different SNP effects between men and women in both samples, with CDK5RAP2 variations affecting only males. Sex-related differences in brain structure are believed to be determined mainly by the hormonal environment present during embryonic development (19). It is possible that the sex-related gene effects are caused by sex hormones influencing MCPH gene expression, or by interactions between MCPH genes and X-linked genes, such as MECP2 (17). In any case, this is a robust phenomenon across both samples, and could be of importance in understanding brain development and gender differentiation.
A striking aspect of the present findings is the specificity of the MCPH effects on cortical area, and not cortical thickness. Correspondingly, the increase in size of the cerebral cortex in the primate lineage leading to humans is coupled with large interspecies differences in cortical surface area, whereas the thickness of the neocortex is largely preserved across primates (20). In patients with microcephaly there is a reduction in brain size and cortical surface area, while cortical thickness is unaffected (21). All four MCPH genes are expressed in the neuroepithelium in utero and encode proteins that influence proliferation of neurons in the ventricular zone during cortical development (3). The ASPM, CDK5RAP2, and CENPJ proteins are important for the correct assembly of the mitotic spindle (6), and the MCPH1 protein regulates chromosome condensation during mitosis (4, 22). An increase in the number of symmetric cell-division cycles produces an increased neuronal progenitor pool, with a corresponding increase in the number of cortical columns, which leads to larger cortical surface area without substantial effect on cortical thickness (20). These data are consistent with our finding that MCPH genes are associated with cortical area and total brain volume, as opposed to cortical thickness. Moreover, cortical area and thickness may have independent cellular determinants (i.e., radial progenitor cells and intermediate progenitor cells, respectively) (23). It is possible that CDK5RAP2 is involved in regulating the formation of radial progenitor cells. Interestingly, studies in mice suggest that the primary expression of Cdk5rap2 during embryonic development is localized to the neuroepithelium in frontal cortex early in neurogenesis (24), consistent with the frontal distribution of cortical area effects observed in the present study.
It is an intriguing possibility that the same genes are involved in regulating both cellular proliferation during brain development and in brain enlargement during primate and human evolution. There is evidence for positive selection on MCPH1, ASPM, and CDK5RAP2 in the lineage leading to humans from a series of studies (for a review, see ref 25). CDK5RAP2 shows especially high rates of nonsynonymous substitutions in the human and chimpanzee terminal lineages (26). Our analyses of recent natural selection revealed evidence that some of our regions of interest, within ASPM and CDK5RAP2, have been under recent positive selection. Other authors have investigated other signals of selection in the microcephaly genes: Interspecies comparisons of primate mutation rate in synonymous and nonsynonymous substitution rates (Ka/Ks ratio) have revealed that MCPH1, ASPM, and CDK5RAP2 have undergone adaptive evolution in primates (4, 26). In addition, Mekel-Bobrov et al. (11) and Evans et al. (26) showed evidence of more recent selection, including extended haplotypes and regions of unusually high LD in MCPH1 and ASPM, although these results have been questioned by others (27). Two of our significant SNPs (rs2282168 and rs1888893) are located in an intron in CDK5RAP2 that has a large degree of conservation between mammals, which may suggest that this region is of importance for CDK5RAP2 regulation. Previous studies looking for genetic associations between polymorphisms in these genes and phenotypes derived from imaging have been unsuccessful (13, 14, 28). Our positive findings may justify further research into the relationship between genes with signs of recent selection and brain development.
All significant SNPs in the present study were located in nonexonic regions. The functional significance of these loci is not yet known, as is the case for a series of recent discoveries of gene variants related to human phenotypes (15, 29). However, given their location close to regulatory elements, it is possible that they are involved in gene regulation (30). These results therefore suggest that common variance in brain structure could be associated with differences in gene regulation rather than protein structure, consistent with recent findings in other complex human traits (15, 31).
Methods
Samples.
Discovery sample (TOP).
The discovery sample consisted of 287 ethnic-Norwegian subjects (age 35 ± 10.4 years), including healthy controls (n = 101; 41 males, 60 females) and subjects with severe mental disorders (n = 186; 94 males, 92 females), with structural MRI data and DNA from the Norwegian TOP study.
Replication sample (ADNI).
The significant findings from the discovery sample were tested for replication in the independent ADNI sample (www.adni.org) of 656 subjects, including healthy controls (n = 188; 102 males, 86 females), subjects with mild cognitive impairment (n = 322; 202 males, 120 females), and subjects with Alzheimer's disease (n = 146; 73 males, 73 females). A more detailed description of the subjects can be found in SI Methods.
Genotyping.
Discovery sample (TOP).
DNA was extracted from blood and genotyped by Expression Analysis (Durham, NC) with the Affymetrix 6.0 array. The SNPs associated with the specific microcephaly genes MCPH1, CDK5RAP2, ASPM, and CENPJ, as defined by Affymetrix, with a call rate of >93% and minor allele frequency of >0.05 were included in the analyses. This yielded 537 SNPs in MCPH1, 54 SNPs in CDK5RAP2, 8 SNPs in ASPM, and 12 SNPs in CENPJ. The two candidate exonic SNPs rs930557 (MCPH1) and rs41310927 (ASPM), were genotyped with the TaqMan assay (Applied Biosystems). Table S2 contains the genotypes for the significant snps.
Replication sample (ADNI).
DNA was extracted from blood and genotyped using the Illumina Human 610-Quad BeadChip assay for ADNI, by the Translational Genomics Research Institute (www.tgen.org). The SNPs with a significant finding in the TOP sample, and successfully assayed on the Illumina platform with the same quality control criteria, were included in the analyses of the ADNI sample.
Quality control genotypes.
The microarray genotype data went through standard quality control procedures for whole genome datasets. Subjects with low overall genotyping call rate (< 0.93) or potential gender mix-up, as defined by PLINK (32) (http://pngu.mgh.harvard.edu/purcell/plink/), were excluded from analysis.
Stratification analysis.
To detect genetic stratification within the Norwegian TOP sample, principal component analysis was performed with the EIGENSOFT smartpca tool (see SI Methods for details). No evidence was found for multiple genetic clusters, so no subjects in this group were removed. In the ADNI sample, Smartpca was used to identify 53 outlier subjects relative to a genetic cluster composed almost exclusively of subjects whose self-reported race was “White.” The self-reported race for the majority of the outliers was “Black/African American,” and for the remainder was “White,” “Asian,” or “Other.” The outliers were not included in subsequent analyses.
Population/evolutionary genetics.
The Web site Haplotter (http://hg-wen.uchicago.edu/selection/) was used to retrieve fixation index values between European and African, and between European and Asian populations, as well as iHS values for the European population, for the associated markers using HapMap phase II data (ref. 33) contains the definition of the iHS statistic, as well as a guide to how it should be interpreted). HapMap phase II allele frequencies were obtained through downloaded flat files. In addition, we extracted Tajima's D values for the 100-kb region centered on our candidate SNPs within a population of European descent (this information was first presented in ref. 34 and is available on the UCSC genome browser http://genome.ucsc.edu/). We considered the empirical genomewide distribution of this statistic; the position of our SNPs within this distribution is evidence as to whether selection has occurred. The UCSC genome browser was also used to investigate conserved regions in mammalian genomes.
Brain Imaging Protocol and Morphometric Parameters.
Discovery sample (TOP).
All participants underwent MRI scanning on a 1.5T Siemens Magnetom Sonata scanner (Siemens Medical Solutions) equipped with a standard head coil. After a conventional three-plane localizer, two sagittal T1-weighted magnetization prepared rapid-gradient echo (MPRAGE) volumes were acquired with the Siemens tfl3d1_ns pulse sequence (TE = 3.93 ms, TR =2,730 ms, TI = 1,000 ms, flip angle = 7°; FOV = 24 cm, voxel size= 1.33 × 0.94 × 1 mm3, number of partitions = 160) and subsequently averaged together to increase the signal-to-noise ratio. Acquisition parameters were optimized for increased gray/white matter image contrast.
Replication sample (ADNI).
All ADNI subjects were scanned using a scan protocol equivalent to that used for the TOP study. Briefly, the scan protocol included two sagittal, three-dimensional T1-weighted (MPRAGE-equivalent) acquisitions, with 1.3-mm isotropic resolution. The scans were performed on 1.5T Siemens, GE, and Philips scanners, at multiple sites across the United States and Canada (see http://adni-info.org for protocol parameters used for each scanner model).
MR image processing (common to discovery and replication sample).
The MR image files in DICOM format were transferred to the University of California San Diego MultiModal Imaging Laboratory (http://mmil.ucsd.edu) for morphometric analysis. Images were corrected for nonlinear warping caused by gradient coil nonlinearities, using tools developed through the Morphometry Biomedical Informatics Research Network (mBIRN). The two MPRAGE acquisitions were rigid-body registered to each other (motion corrected) and subsequently averaged together to increase the signal-to-noise ratio. The FreeSurfer 3.0.2 software package (http://freesurfer-software.org) was used to create a three-dimensional model of the cortical surface for cortical thickness and cortical surface area measurements (35–38). Based on this, we generated four summary measures: total brain volume, intracranial volume, total cortical surface area, and mean cortical thickness. A more detailed description of the MRI image processing can be found in SI Methods.
Statistical Model.
To account for sex-specific effects, we used a statistical model where the mean effect of SNP dose on the phenotype was allowed to differ for the two sexes. SNP-wise univariate General Linear Models were fit to each of the four morphometry phenotypes as the dependent variable, genotype as the independent variable, with age, sex, and the diagnosis categories as covariates. Genotype was modeled as minor allele SNP-dosage and was coded into different independent variables to account for SNP-dosage for each sex. A statistical significance test was performed on all SNPs, and the resulting P-values were subsequently corrected for multiple comparisons using a permutation procedure (39).
Genetic associations between the two replicated SNPs (rs914592 and rs2297453) and schizophrenia and bipolar spectrum disorders in the TOP sample, and Alzheimer's disease and mild cognitive impairment in the ADNI sample, were investigated using logistic regression. Two models were considered: an additive model on the log odds ratio in which the SNP dosage was the sole covariate, and another model in which the SNP dosage was divided by gender.
Correction for Multiple Comparisons in Analysis of Discovery Sample.
The statistical significance test was performed on all SNPs included from the TOP (discovery) sample, in total 611 SNPs from four genes, and the resulting P-values were subsequently corrected for multiple comparisons across all SNPs within the gene. Nonparametric P-values for each SNP-phenotype regression were calculated from the nominal P-values estimated by the model via permutation using 100,000 iterations. Specifically, composite genotypes were permuted among the male subjects and similarly for females, such that gene-wide LD structure was preserved. For each SNP, nominal P-values from the unshuffled data were converted to nonparametric P-values based on the proportion of nominal P-values calculated on permuted datasets, which were smaller than the nominal value on the unshuffled data. Subsequently, each nonparametric P-value was corrected to control the family-wise error rate stemming from multiple comparisons across all SNPs within the gene. First, the distribution of the minimum gene-wide, nonparametric, P-value (min-p) under the null hypothesis of no associations was estimated via permutation methods. Then the corrected, nonparametric P-value for each SNP/phenotype association was computed as the proportion of these min-ps, which were smaller than the corresponding uncorrected nonparametric P-value. These procedures are explained in more depth in Pantazis et al. (39).
Supplementary Material
Acknowledgments
We thank the study participants and the members of the Thematic Organized Psychosis study group involved in data collection, especially Drs. Jimmy Jensen, Per Nakstad, and Andres Server. We also thank Knut-Erik Gylder, Thomas Doug Bjella, Robin G. Jennings, Chris J. Pung, and Dr. Christine Fennema-Notestine. This work was supported by the Oslo University Hospital--Ullevål, Eastern Norway Health Authority (Grant 2004-123), and the Research Council of Norway (Grants167153/V50 and 163070/V50), and by Eli Lilly Inc. for parts of the genotyping costs of the TOP sample. This work was also supported in part by National Institutes of Health (NIH) Grant 1U54RR025204 (to A.H.J. and N.J.S.) and NIH Grant 1R01AG031224 (to A.M.D. and J.C.R.). The Alzheimer's Disease Neuroimaging Initiative (Principle Investigator Mike Weiner) is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Pfizer, Wyeth Research, Bristol-Myers Squibb, Eli Lilly, GlaxoSmithKline, Merck, AstraZeneca, Novartis Pharmaceuticals, Alzheimer's Association, Eisai Global Clinical Development, Elan, Forest Laboratories, and the Institute for the Study of Aging, with participation from the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institutes of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles.
Footnotes
Conflict of interest statement: A.M.D. is a founder and holds equity in CorTechs Labs and also serves on the Scientific Advisory Board. E.H. has equity interest in CorTechs Labs and also serves on its Board of Directors. The terms of this arrangement have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908454107/DCSupplemental.
References
- 1.Northcutt RG, Kaas JH. The emergence and evolution of mammalian neocortex. Trends Neurosci. 1995;18:373–379. doi: 10.1016/0166-2236(95)93932-n. [DOI] [PubMed] [Google Scholar]
- 2.Panizzon MS, et al. Distinct genetic influences on cortical surface area and cortical thickness. Cereb Cortex. 2009;19:2728–2735. doi: 10.1093/cercor/bhp026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Woods CG, Bond J, Enard W. Autosomal recessive primary microcephaly (MCPH): a review of clinical, molecular, and evolutionary findings. Am J Hum Genet. 2005;76:717–728. doi: 10.1086/429930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson AP, et al. Identification of microcephalin, a protein implicated in determining the size of the human brain. Am J Hum Genet. 2002;71(1):136–142. doi: 10.1086/341283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moynihan L, et al. A third novel locus for primary autosomal recessive microcephaly maps to chromosome 9q34. Am J Hum Genet. 2000;66:724–727. doi: 10.1086/302777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bond J, et al. ASPM is a major determinant of cerebral cortical size. Nat Genet. 2002;32:316–320. doi: 10.1038/ng995. [DOI] [PubMed] [Google Scholar]
- 7.Leal GF, et al. A novel locus for autosomal recessive primary microcephaly (MCPH6) maps to 13q12.2. J Med Genet. 2003;40:540–542. doi: 10.1136/jmg.40.7.540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ponting C, Jackson AP. Evolution of primary microcephaly genes and the enlargement of primate brains. Curr Opin Genet Dev. 2005;15:241–248. doi: 10.1016/j.gde.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 9.Vallender EJ, Mekel-Bobrov N, Lahn BT. Genetic basis of human brain evolution. Trends Neurosci. 2008;31:637–644. doi: 10.1016/j.tins.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Evans PD, et al. Microcephalin, a gene regulating brain size, continues to evolve adaptively in humans. Science. 2005;309:1717–1720. doi: 10.1126/science.1113722. [DOI] [PubMed] [Google Scholar]
- 11.Mekel-Bobrov N, et al. Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens. Science. 2005;309:1720–1722. doi: 10.1126/science.1116815. [DOI] [PubMed] [Google Scholar]
- 12.Ali F, Meier R. Positive selection in ASPM is correlated with cerebral cortex evolution across primates but not with whole-brain size. Mol Biol Evol. 2008;25:2247–2250. doi: 10.1093/molbev/msn184. [DOI] [PubMed] [Google Scholar]
- 13.Dobson-Stone C, et al. Investigation of MCPH1 G37995C and ASPM A44871G polymorphisms and brain size in a healthy cohort. Neuroimage. 2007;37:394–400. doi: 10.1016/j.neuroimage.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 14.Woods RP, et al. Normal variants of Microcephalin and ASPM do not account for brain size variability. Hum Mol Genet. 2006;15:2025–2029. doi: 10.1093/hmg/ddl126. [DOI] [PubMed] [Google Scholar]
- 15.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- 16.Wang JK, Li Y, Su B. A common SNP of MCPH1 is associated with cranial volume variation in Chinese population. Hum Mol Genet. 2008;17:1329–1335. doi: 10.1093/hmg/ddn021. [DOI] [PubMed] [Google Scholar]
- 17.Chahrour M, et al. MeCP2, a key contributor to neurological disease, activates and represses transcription. Science. 2008;320:1224–1229. doi: 10.1126/science.1153252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chanock SJ, et al. NCI-NHGRI Working Group on Replication in Association Studies. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 19.Negri-Cesi P, Colciago A, Celotti F, Motta M. Sexual differentiation of the brain: role of testosterone and its active metabolites. J Endocrinol Invest. 2004;27(6, Suppl):120–127. [PubMed] [Google Scholar]
- 20.Rakic P. A small step for the cell, a giant leap for mankind: a hypothesis of neocortical expansion during evolution. Trends Neurosci. 1995;18:383–388. doi: 10.1016/0166-2236(95)93934-p. [DOI] [PubMed] [Google Scholar]
- 21.Mochida GH, Walsh CA. Molecular genetics of human microcephaly. Curr Opin Neurol. 2001;14:151–156. doi: 10.1097/00019052-200104000-00003. [DOI] [PubMed] [Google Scholar]
- 22.Trimborn M, et al. Mutations in microcephalin cause aberrant regulation of chromosome condensation. Am J Hum Genet. 2004;75:261–266. doi: 10.1086/422855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontious A, Kowalczyk T, Englund C, Hevner RF. Role of intermediate progenitor cells in cerebral cortex development. Dev Neurosci. 2008;30(1–3):24–32. doi: 10.1159/000109848. [DOI] [PubMed] [Google Scholar]
- 24.Bond J, et al. A centrosomal mechanism involving CDK5RAP2 and CENPJ controls brain size. Nat Genet. 2005;37:353–355. doi: 10.1038/ng1539. [DOI] [PubMed] [Google Scholar]
- 25.Gilbert SL, Dobyns WB, Lahn BT. Genetic links between brain development and brain evolution. Nat Rev Genet. 2005;6:581–590. doi: 10.1038/nrg1634. [DOI] [PubMed] [Google Scholar]
- 26.Evans PD, Vallender EJ, Lahn BT. Molecular evolution of the brain size regulator genes CDK5RAP2 and CENPJ. Gene. 2006;375:75–79. doi: 10.1016/j.gene.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 27.Yu F, et al. Comment on “Ongoing adaptive evolution of ASPM, a brain size determinant in Homo sapiens.”. Science. 2007;316:370. doi: 10.1126/science.316.5823.370a. [DOI] [PubMed] [Google Scholar]
- 28.Rushton JP, Vernon PA, Bons TA. No evidence that polymorphisms of brain regulator genes Microcephalin and ASPM are associated with general mental ability, head circumference or altruism. Biol Lett. 2007;3:157–160. doi: 10.1098/rsbl.2006.0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. N Engl J Med. 2009;360:1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 30.Pastinen T, Hudson TJ. Cis-acting regulatory variation in the human genome. Science. 2004;306:647–650. doi: 10.1126/science.1101659. [DOI] [PubMed] [Google Scholar]
- 31.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carlson CS, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Res. 2005;15:1553–1565. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dale AM, Sereno MI. Improved localization of cortical activity by combining EEG and MEG with MRI cortical surface reconstruction—A linear-approach. J Cogn Neurosci. 1993;5:162–176. doi: 10.1162/jocn.1993.5.2.162. [DOI] [PubMed] [Google Scholar]
- 36.Dale AM, Fischl B, Sereno MI. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage. 1999;9:179–194. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- 37.Fischl B, Sereno MI, Dale AM. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage. 1999;9:195–207. doi: 10.1006/nimg.1998.0396. [DOI] [PubMed] [Google Scholar]
- 38.Fischl B, Dale AM. Measuring the thickness of the human cerebral cortex from magnetic resonance images. Proc Natl Acad Sci USA. 2000;97:11050–11055. doi: 10.1073/pnas.200033797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pantazis D, Nichols TE, Baillet S, Leahy RM. A comparison of random field theory and permutation methods for the statistical analysis of MEG data. Neuroimage. 2005;25:383–394. doi: 10.1016/j.neuroimage.2004.09.040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.