Abstract
DNA flows between chromosomes and mobile elements, following rules that are poorly understood. This limited knowledge is partly explained by the limits of current approaches to study the structure and evolution of genetic diversity. Network analyses of 119,381 homologous DNA families, sampled from 111 cellular genomes and from 165,529 phage, plasmid, and environmental virome sequences, offer challenging insights. Our results support a disconnected yet highly structured network of genetic diversity, revealing the existence of multiple “genetic worlds.” These divides define multiple isolated groups of DNA vehicles drawing on distinct gene pools. Mathematical studies of the centralities of these worlds’ subnetworks demonstrate that plasmids, not viruses, were key vectors of genetic exchange between bacterial chromosomes, both recently and in the past. Furthermore, network methodology introduces new ways of quantifying current sampling of genetic diversity.
Keywords: evolution, lateral gene transfer, mobile genetic elements, phages, plasmids
Most of the genetic biodiversity found in cells is considered to be largely distributed among bacteria and archaea. The genetic diversity of eukaryotes appears comparatively smaller (1). Thus, any complete picture of the genetic evolution requires a deep study of the prokaryotic genomes. Two decades of investigations have demonstrated that the evolutionary processes leading to their extant genetic diversity were not simply tree-like (2), i.e., involving only the vertical transmission of genes in diverging lineages from ancestors to descendants. Rather, genetic material was also exchanged laterally between contemporary prokaryotes (3). In a still cell-centered view of molecular evolution, the main vectors of these genetic exchanges, presumably the phages (4) and the plasmids (5), undoubtedly encompassing substantial quantities of DNA molecules, were given the essential but secondary role of “gene carriers” or “gene weavers.”
Recently, metagenomics (i.e., environmental genomics) started enlarging this perspective by providing an unprecedented wealth of DNA molecules directly from nature (6). Biologists became able to simultaneously sequence many of the key players involved in the DNA flow of a given environment, offering a much more integrated view (5, 7). Consequently, the evolutionary analyses of many metagenomes focused more on the global genetic diversity (or global functional diversity) of an environment rather than on traditional issues of systematics (8, 9).
These 2 developments—the accumulation at an unprecedented pace of DNA from all sorts of DNA vehicles (e.g., cellular chromosomes, phages, plasmids), coupled to the recognition that a species tree model might be a poor descriptor of the evolution of genetic diversity—encouraged us to study the genetic diversity in fundamentally new ways. First, we considered the evolution of genetic diversity from a DNA-centered perspective, assuming that all types of DNA-carrying entities are “vehicles” through which DNA molecules flow, via mechanisms of lateral exchange, recombination, and vertical inheritance. Second, taking advantage of developments in network theory, we built a series of edge-weighted networks, displaying the evolution of all the genetic connections between all these DNA vehicles. Interestingly, these networks presented modules and centralities suggesting fundamental divides in genetic diversity, which we call “genetic worlds.” These worlds correspond to isolated clusters of vehicles—always of the same type—by which DNA is principally or exclusively shared, and in which molecular evolution seems to obey a particular mode and tempo. When studied from a cell-centered perspective, our networks greatly improve the current views of the processes and mechanisms responsible for the genetic diversity in various cellular lineages, and how they evolve.
Disconnected Structured Network
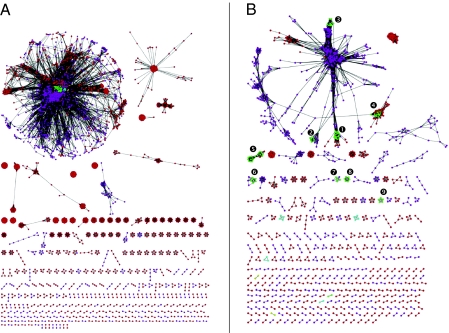
Giving up the preconception of a cell-centered perspective (5, 7) and of a tree-centered model (10) when exploring the structure and evolution of genetic diversity, we studied the DNA pools shared by 3 different types of DNA vehicles by using a single network. Each node in this global network (Fig. 1A) corresponds to an extant DNA vehicle, either the genome of a given plasmid, a given phage, or the chromosomal portion of a cellular genome [known as the organism’s “private pool” (5)]. These vehicles are connected in the network by an edge when they share at least one homologous DNA fragment of >300 bp, when the minimal reciprocal best BLAST score required to be considered homologous is 1e-20. Although this network is quite complex, it is nonetheless structured and informative. First, it contains multiple connected components of different sizes, corresponding to obvious clusters of vehicles that share a same DNA pool to the exclusion of other vehicles. Although most vehicles fell within a single connected component, 98.5% of this network connected components comprised only one type of vehicle, which indicates that their DNA pool circulates between vehicles of the same type. Preferential recombination between a given type of DNA vehicle and subsequent genetic drift could explain this discontinuity.
Fig. 1.
Network of shared DNA families among cellular, plasmid, and phage genomes. (A) Global network in which each node represents a genome, either cellular (green for bacterial chromosome, yellow for eukaryotic chromosomes, and cyan for archaeal chromosome), plasmidic (purple), or phage (red). Two nodes are connected by an edge if they share homologous DNA (reciprocal best BLAST hit with a minimum of 1e-20 score, and 20% minimum identity). Edges are weighted by the number of shared DNA families. The layout was produced by Cytoscape, using an edge-weighted spring-embedded model, meaning that genomes sharing more DNA families are closer on the display. There are 3,207 nodes on that network. (B) Global network displaying connections between genomes (same color code) for a minimum of 95% identity. Imposing a minimum identity percentage on the definition of CHDs roughly filters for more recent sharing events. Bacterial clusters are indicated as follows: Burkholderia (1), Xanthomonas (2), Yersinia, (3) Streptococcus (4), Prochlorococcus/Synechocystis (5), Clostridium (6), Legionella (7), Rhodopseudomonas (8), and Helicobacter (9). There are 1,529 nodes on that network.
Interestingly, chromosomes, phages, and plasmids only fall together in the largest connected component of the network, indicating that there are also instances where at least some DNA vehicles of different types share the same DNA pool. This “giant connected component” (GCC) comprised 352,499 sequences of the 578,527 sequences of the dataset (60.9%). However, even this diverse connected component is internally structured by vehicle type. Clustering coefficient analyses by MCODE (11) and modularity maximization (12, 13) identified many modules in the GCC that almost always comprise one type of vehicle only (Table S1). It is thus remarkable that the various pools of shared DNA families appearing in the network (be they isolated connected components or the modules within the GCC) strongly correspond to the type of vehicle carrying the DNA. This finding indicates that DNA families are mostly carried and exchanged by the same type of DNA carriers: DNA families currently carried by plasmids, phages, or chromosomes are shared overwhelmingly between plasmids, phages, or chromosomes, respectively. This discontinuous structure is most important as it suggests the presence of divides in genetic diversity. We call these individual components genetic worlds because they have distinct core genes and apparently do not share homologous DNA with one another.
Each genetic world has a corresponding subnetwork with specific topological properties, even though they all correspond to highly clustered regions of the network (i.e., modules). It is not straightforward to compare networks of different sizes, yet it is worth noting that their topological parameters, reflecting the outcome of the DNA flow within the distinct genetic worlds, were quite different (Table S2), as exemplified by the large variation in their diameter and average shortest path, among others. Overall, the simplest biological explanation for these differences is that the gene flow in these genetic worlds followed rules and evolutionary histories varying among the different genetic worlds. Consistent with this claim, we did not detect any hidden common hierarchical organization (14) reflecting an accepted [e.g., National Center for Biotechnology Information (NCBI)] taxonomy in the largest subnetworks of each type of vehicle (Fig. S1) when we investigated the network reconstructed with sequences presenting at least 40% of sequence identity. This is either because of the methodological limit of the method that cannot account for edge weights, or, as it is in agreement with the literature (15–18), it might be explained if, within the limits of one type of vehicle, DNA molecules obey complex rules of transfers and losses. Yet, the lack of a strong underlying hierarchical structure does not mean that understanding the processes that led to its complexity is out of reach.
Processes Behind the Pattern
We followed the methodology of Dagan et al. (12) to estimate how the complex pattern of the GCC might have evolved (i.e., how the DNA families spread among the different types of vehicle). From an organismal standpoint, this is the most interesting part of the network to study, as it provides information about the overlap in DNA families between the private pool of cells and the mobile genetic elements. To understand the steps through which cellular chromosomes and phages on the one hand, and cellular chromosomes and plasmids on the other hand, came to share homologous DNA, we decomposed our global network in a series of embedded networks, based on the percentage of identity between the molecules of a given DNA family. We made the molecular clock–based assumption that families of DNA molecules with the highest percentages of identity were likely to be more recently shared than the ones with less identity. In particular, DNA molecules with 100% identity were considered most recently shared, those with more than 80% identity were somewhat less recently shared, and so on. Under this hypothesis, we reconstructed 11 networks representing the DNA pools shared between the vehicles at different percentages of identity, corresponding to rough “temporal” cross-sections of the GCC (Fig. S2).
Although this slicing approach may be simplistic, it unraveled connections between vehicles that were consistent with current taxonomical and empirical knowledge (Fig. 1B). The most recent network of DNA sharing among cells, plasmids, and phages (100% identity) clearly showed that different prokaryotic strains were exchanging DNA with different types of vectors (e.g., Streptococcus with phages and Yersinia, Xanthomonas, and Legionella with plasmids). Most interestingly, at 100% identity, the cellular genomes were not all connected, as their core genes are more ancient (i.e., acquired long ago) and thus more divergent. Logically, this separation of the cellular chromosomes at a high identity threshold decreases when this threshold is lowered. The cellular genomes are all connected for only identity values less than 55%. Importantly, when cellular genomes are not directly connected, but are rather in the same connected component because of their shared edges with plasmids or phages, candidate lateral gene transfers (LGTs) can be invoked and identified, as well as their likely vectors.
The problem of the evolution of the mechanisms of gene transfer in a lineage is almost philosophical and hard to deal with. In addition, we certainly underestimate the genetic diversity within plasmids and phages, as their gene pools are more transient than those of cellular entities. Yet we believed our networks could help in addressing the origin of some LGTs critically. In fact, regardless of the threshold of identity, the reconstructed networks (and their modules) continually showed preferential connections between vehicles of the same type, which indicates that when a DNA family enters a type of DNA vehicle (or a genetic world), it mainly evolves in it. Obviously, this does not imply that, over time, the genetic composition of replicating DNA vehicle is not changing, but simply that new vehicles of the same type were created through the evolution and exchange of DNA material of preexisting vehicles of that type with each other. Likewise, as long as all of the members of a cellular lineage clustered in the same taxonomically consistent group on the network, isolated from other cellular lineages, we treated this group as sharing a specific DNA pool. As we decreased the identity threshold, such lineages were logically less and less separated into isolated taxonomical clusters (Fig. S2). To be conservative, we thus ceased to estimate the relative contribution of plasmids and phages to the specific pool of each cellular lineage after a short “time period” (as roughly measured by sequence identity).
Interestingly, this conservative approach did not detect noticeable switches in the type of vehicles sharing DNA with a given lineage over long evolutionary periods, indicating that the mechanisms generating genetic diversity persists for a lineage over time, except perhaps for Burkholderia (Table S3). Our networks also provided insights into which DNA families were involved in conjugation or transduction events (Table S4). On average, approximately 97 DNA families were involved in conjugation for any temporal slice, representing between 1% and 22% of the total number of families found in all plasmids. Similarly, only approximately 26 DNA families, representing between 1% and 6% of the total number of families found in phages, were exchanged through transduction, suggesting that, in our dataset, phages were genetic couriers for a limited set of DNA families. However, for very recent events (100% identity), phages exchanged 99 families with cellular genomes, whereas plasmids exchanged only 15. An interpretation of these results could be that viral genes are more transient pools of DNA than plasmidic ones. However, transduction and conjugation rates are difficult to infer, as the possibility of sampling biases cannot be ruled out unless one knows the actual structure of genetic diversity. Importantly, at least for the networks with the highest percentage of identity (from 60% to 100%), the linkages of the GCC (overall and between phages and cellular chromosomes, or plasmid and cellular chromosomes) were caused by genes belonging to all sorts of functional categories (Fig. S3). This observation reflects that, both in the past and most recently, LGT affected a diversity of genes function, even though the proportions of functional categories being transferred seemed to vary over time (Fig. S3). In particular, we inferred a significantly higher proportion of recent LGT for the virulence genes, which suggests that anciently transferred virulence genes were either lost by the host cell lineages, or regularly affected by gene conversion with more recent virulence genes that obliterated their ancient molecular features, or that they evolved beyond recognition.
Central Role for Plasmids
In our network-based analysis of the processes of DNA exchange among various types of DNA vehicles, plasmids seemed to play a very different role than phages. For instance, at an 85% identity threshold, plasmids clustered well together, and they united 7 bacterial lineages (Xanthomonas, Prochlorococcus, Synechococcus, Streptococcus, Rhodopseudomonas, Burkholderia, and Yersinia). Interestingly, the Streptococcus lineages and the Burkholderia/Xanthomonas cluster were not directly connected, but were instead mediated by a web of plasmids. As part of a highly interconnected subnetwork, these plasmids are thus drawing on the same gene pool. The fact that 2 different—and not directly connected—bacterial clusters also share DNA with this plasmidic gene pool suggests a complex evolutionary history wherein plasmids play a central role. When the identity threshold was lowered, plasmids kept forming bridges between the various cellular chromosomes until they all eventually fell into the GCC (Fig. S2). As conjugation, in contrast with transduction, requires that both donor and recipient cells are found in proximity, our result strongly suggests that short-distance LGTs are essential in microbial evolution. This proposition is consistent with the notion that bacteria very frequently live in biofilms (19), and are not hindered by many environmental barriers as microbial cells seem to “visit” quite distant environments (20).
We confirmed this visual interpretation of the results mathematically, by computing our networks’ centralities (i.e., indices revealing particular properties of the various nodes), to assess whether some nodes indeed occupy noteworthy positions in the topology. We used 2 classical centralities—degree and betweenness—to test which vehicles were holding together and shaping our DNA family network. Degree is the most common centrality index, and measures the number of edges connecting a given node. Betweenness quantifies the frequency with which a given node lies on the shortest path between any pair of nodes in the network. High-betweenness nodes are like reservoirs of DNA families. They appear to be “between” because they either distributed some of their DNA families to otherwise unrelated vehicles or comprise a mosaic content of DNA families originating from other vehicles. To obtain a robust estimate of these centralities, they were computed for the members of the GCC only. As different vehicles do not harbor the same number of DNA families, we corrected for the bias in genome size and kept only edges corresponding to a significant number of shared DNA families (18).
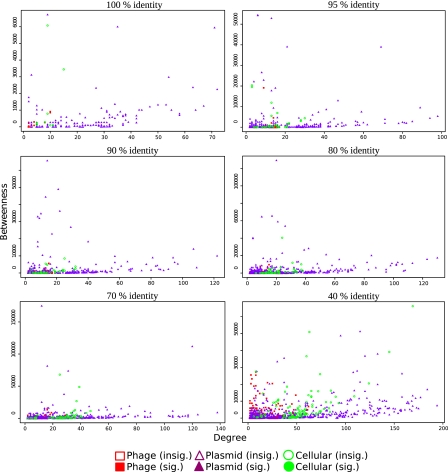
For various identity thresholds, plasmids globally displayed a much higher betweenness than phages (Fig. 2), confirming their predominant role as genetic couriers. By contrast, phages showed lower betweenness centralities, either appearing on the periphery of networks or producing their own connected components. Less surprisingly, chromosomes generally appeared to be central, particularly as the identity threshold decreased. As vehicles harboring a large number of DNA families are likely to mechanically display high betweenness centralities, we tested their significance using the methodology proposed by Lima-Mendez et al. (18). We also tested that there was no correlation between genome size and betweenness. Although most chromosomes showed significantly high values, so did a large number of plasmids.
Fig. 2.
Betweenness of nodes as function of their degree for various identity threshold networks. Cellular chromosomes are displayed as green circles, plasmids as purple triangles, and phages as red squares. When the betweenness of a node is significantly higher than expected (P < 0.05), the corresponding symbol is filled, and it is empty otherwise. Although betweenness generally increases with degree, plasmids and cellular clearly show higher betweenness values, suggesting they play a central role in the sharing of DNA. Note that scale differs among plots. There are 171 nodes for a 100% identity threshold, 342 for a 95% identity threshold, 372 for a 90% identity threshold, 509 for a 80% identity threshold, 618 for a 70% identity threshold, and 1,029 for a 40% identity threshold.
As with many centrality measures, there is generally a positive correlation between degree and betweenness. Some nodes, however, showed a much higher betweenness than most nodes of the same degree. Such outliers, characterized by a low degree but a high betweenness, are especially important in any given network, as they can be seen as bridges between smaller, more connected parts of the network. Most interestingly, these bridges were almost always plasmids in our networks (Fig. 2), strongly suggesting that these vehicles, and not viruses, are key vectors in the spreading of DNA in nature. Interestingly, many of these plasmids with a remarkable betweenness—although diverse in size, G+C content, and percentage of coding genes—were already known for their phylogenetic mosaicism, the presence of mobile elements such as transposons and integrons in their genomes (resulting from recombination events), and their resistance to drugs and/or heavy metals, which likely contributed to their diffusion in various hosts and environments (Table S5). For instance, pB10 [degree (d) = 63, betweenness (b) = 2,358, in the 100% identity network] is a promiscuous IncP-1 plasmid, isolated from a waste water treatment plant. Its recombined mosaic backbone structure encompasses 5 distinct mobile genetic elements. pB10 is able to self-transfer among diverse bacterial species and confers resistance to multiple antimicrobial agents and to inorganic mercury ions, and would play a major role in rapid adaptation of bacterial communities to changing environments (21). Likewise, p1658/97 (d = 71, b = 5,933, in the 100% identity network) is a recombined mosaic plasmid from a pathogenic Escherichia coli. Its backbone codes for its replication, conjugative transfer, and stable maintenance through an active partition system plus 2 postsegregational killing systems, and it contains 19 mobile genetic elements (14% of its sequences), by which it confers antimicrobial resistance (22). Similarly, almost all the phages with noticeable betweenness in the 40% identity network [e.g., HK022 (d = 20, b = 20974.7), TM4 (d = 8, b = 27106.1), phiPV83 (d = 25, b = 20408.4), STSV1 (d = 3, b = 25507.5), SIRV2 (d = 3, b = 20342), and Corndog (d = 8, b = 25397.9)], and likely Psy315.3 in the 95% identity network (d = 9, b = 19,105), were already reported as harboring mosaic genomes (Table S5).
Lesson from the Network Robustness
Network analyses display a structure of genetic diversity and provide analytical ways to study its robustness. Typically, if the molecular data collected thus far correctly sampled our planet's genetic diversity, then any new data should fall within the already identified connected components. The structure of our network should then be robust with respect to the quantity of data investigated: as our knowledge of the genetic diversity grows, the connected component should become larger, but not more numerous. Eventually, progressively, a larger sampling could even fill the gaps between some connected components and unite some of the genetic worlds. For instance, 2 independent clusters of phages sharing a DNA pool could be linked by the discovery of a new vehicle, i.e., an environmental phage, with an intermediate genetic content. By contrast, if there are still unknown genetic worlds evolving in nature, an increasing number of new connected components should appear with the addition of new data. Eventually this number should saturate as we approach a fair sampling of the genetic diversity. Thus, by counting the number of persisting and new connected components as new data are added, it becomes possible to estimate whether our current knowledge of genetic diversity (and the lessons of evolutionary biology derived from it) is based on a representative sample of the natural diversity.
We added 45,845 environmental sequences of phages from 7 metagenomic projects, and reconstructed a global network for an identity level of 20%. Of this additional molecular data, 30.8% (14,139 sequences) fell into 31 of the 260 already existing connected components (95.7% falling into the GCC). In addition, 36.2% (16,600 sequences) created 4,533 new connected components, whereas the remaining 32.9% (15,106 sequences) were unique and thus did not appear on the network. Overall, these results demonstrate that we are presently underestimating the genetic diversity and the number of evolutionary groups. To improve our knowledge, we will need to sample in a broader diversity of environments, as the type of environments structures genetic diversity. This is at least very clear for the gene content of environmental phages (Fig. S4).
Conclusion
Metagenomics has encouraged the emergence of a more integrated and less cell-centered perspective on microbial and molecular evolution, broadening the evolutionist’s horizon. There are good reasons to include the DNA carried by mobile elements in models of the evolution of genetic biodiversity, even if it confronts us to a very different picture than the tree-like model, or the organismal-centered web-like one—the Tree of Life or Web of Life—that biologists have been progressively accustomed to think with. First, for all these entities, the genetic material is the same. DNA is a component of some phages, plasmids and chromosomes, not of any of these vehicles exclusively. Second, although this DNA is preferentially transferred (be it vertically or laterally) within a given genetic world, there is some inter-world transfer of DNA molecules occurring, leading to exchanges among different DNA vehicles (2.5% of the DNA families). This observation indicates that the changes accumulated relatively independently in the molecules of any of these worlds (i.e., the results of molecular evolution for different regimes of selective pressures and for different historical constraints) do regularly cross into another world. In principle, selected (or drifting) DNA molecules with their special adaptations can then invade and impact a new genetic world. Deciphering the rules of transitions of transfer between genetic worlds could then become a central question, prompting an integrated study of genetic evolution. In any case, the picture of the evolution of the natural genetic biodiversity should not be considered complete without the DNA molecules of any of these worlds. It implies that no general model of genetic evolution can be universally valid. Rather, many evolutionary models of the genetic biodiversity should legitimately coexist: DNA molecules change in some phages differently than they do in plasmids, or in populations of prokaryotic chromosomes. Sequencing and making trees out of the molecular data cannot hope to adequately deal with this disconnected network of genetic diversity. In the future, a plurality of evolutionary research fields will be required to understand the evolution of the various genetic worlds.
Building the Data Set
We downloaded 3,055,585 DNA sequences of the environmental mobilome corresponding to the phage sequences from 7 reasonable-sized metagenome projects covering various environments (Coral virome Gm00144, Human Gut Virome Gm00055, Soil virome Gm00149, Bearpaw and Octopus hot springs viromes Gm00077, Chesapeake Bay MOVE09/02 and MOVE858 Gm00053). Only 45,845 sequences larger than 300 bases were retained for further analyses (22,271 for Octopus, 10,815 for Chesapeake Bay, 8,352 for Bearpaw, 2,226 for the Coral metagenome, 1,342 for the uncultured human fecal virus, 839 for the soil metagenome). We also downloaded 50,122 phage protein sequences and 73,562 plasmid protein sequences from NCBI (http://www.ncbi.nlm.nih.gov/Entrez). Chromosomes from complete microbial genomes, corresponding to 11 of the 12 groups (excluding the E. coli/Shigella cluster, which will be discussed in detail in a separate publication) of closely related genomes described by Doolittle and Zhaxybayeva (9) were also obtained from NCBI. In addition to these genomes, we also included protein sequences from complete archaeal genomes with at least 2 close relatives within the lineage (2,605 for Pyrobaculum aerophilum str. IM2, 2,299 for Pyrobaculum arsenaticum DSM 13514, 2,149 for Pyrobaculum calidifontis JCM 11548, 1,978 for Pyrobaculum islandicum DSM 4184, 1,780 for Pyrococcus abyssi GE5, 2,125 for Pyrococcus furiosus DSM 3638, 1,955 for Pyrococcus horikoshii OT3, 1,482 for Thermoplasma acidophilum DSM 1728, and 1,499 for Thermoplasma volcanium GSS1) as well as protein sequences from the 4 smallest eukaryotic whole chromosomes (13,408 for Dictyostelium discoideum, 7,603 for Ostreococcus lucimarinus, 4,717 for Ashbya gossipy, and 8,265 Leishmania major strain Friedlin).
Definition of Homologous Families
All of the sequences (45,845 DNA sequences and 532,682 protein sequences) were compared against one another via BLAST (23) and reciprocally by using BLASTP for a protein query against a protein database, BLASTN for a nucleotide query against a nucleotide database, BLASTX for a translated nucleotide query against a protein database, and TBLASTN for a protein query against a translated nucleotide database. For each pair of sequences, all best BLAST hits with a score of 1e-20 were stored in a mySQL database. To define homologous DNA families, sequences were clustered using a single-linkage algorithm (24). In this method, a sequence is added to a cluster if it shares a reciprocal best-BLAST hit relationship with at least one of the sequences of the cluster. The DNA families so defined were called CHDs (for “cluster of homologous DNA families”). We verified that MCL clustering (25) yielded similar (92.5% identical in average) results. Additionally, sets of CHDs were clustered by the single-linkage algorithm with the added requirement that reciprocal best-BLAST hit pairs share a minimum sequence identity; 11 different sets of CHDs were produced in this manner, for various identity thresholds (100, 95, 90, 85, 80, 75, 70, 65, 60, 40, 20). Assuming a molecular clock, CHD sets produced with a higher identity threshold would represent more recently related sequences. To provide conservative estimates of the overlap between cellular genomes, plasmids and phages, any ORF of a complete cellular genome belonging to a given CHD and exactly matching a sequence from a vector known to be associated with this genome (phage or plasmid) was tagged as phage or plasmid sequences accordingly, hence considered noncellular.
Network Analyses
We built 11 networks summarizing the DNA-sharing relationships among the genomes of various DNA vehicles, according to the sets of CHDs assembled as described earlier. A network layout was produced by Cytoscape software, using an edge-weighted spring-embedded model, meaning that genomes sharing more DNA families appear closer in the display. Topological properties of these networks (e.g., diameter, radius, centralization, density, heterogeneity, average shortest path, closeness, betweenness, clustering coefficients) were estimated with the NetworkAnalyzer 2.6 Cytoscape plug-in (http://med.bioinf.mpi-inf.mpg.de/netanalyzer/help/2.6.1/index.html). Modules in the GCC of our networks were identified by the MCODE 1.3 Cytoscape plug-in (11) (default parameters) and modularity maximization (12, 13).
The presence of an underlying hierarchical structure in the GCC for an identity threshold of 40%, which included all of the cellular vehicles as well as phages and plasmids that share genetic elements with them, was tested using fitHRG (14). Analyses were stopped after 2.4/1.2/1.1 billion iterations for the cellular/plasmid/phage subnetworks, respectively, as likelihood seemed to have reached convergence. The different hierarchies sampled from the convergence zone were assembled into a majority consensus tree using consensuplot.m script under MATLAB. The network of phage metagenomes was achieved by pooling all of the data according to their source environment and counting the number of CHDs between these environments.
Evaluation of Betweenness and Degree Centralities
To identify which vehicles were most central in gene sharing, subnetworks were produced from members of the GCC for different identity thresholds using the method described by Lima-Mendez et al. (18). Briefly, for all pairs of vehicles, the probability that the pair would share at least the observed number of CHDs was calculated, given the number of CHDs in the smaller of the 2 vehicles. This probability was multiplied by the number of vehicle pairs in the GCC to get an expect value. Pairs of vehicles were connected by an unweighted edge if the expect value was less than 0.01 and unconnected otherwise. Betweenness centrality is a measure of the tendency of a node to fall along the shortest paths between other nodes. Betweenness centralities were calculated from the resulting graph for all vehicles using the Brandes algorithm (26). Larger genomes are more likely to have larger betweenness centralities, resulting from their increased degree. To assess significance of betweenness measures, we constructed a null distribution from 100 random graphs produced by shuffling genome content. Genome content was represented as a matrix with vehicles as rows and all CHDs represented in vehicles of the GCC as columns. If a given vehicle contained no members of a particular CHD, the corresponding matrix entry was 0, whereas the entry was 1 otherwise. Each random graph was produced by randomly permuting matrix entries within rows, thus producing a set of vehicles of the same size, but with random contents (18). For each set of random vehicles, a graph was constructed and betweenness centralities calculated for each vehicle. Betweenness for a given vehicle was considered significant if it exceeded 95% of the betweenness centralities calculated for the corresponding random vehicle among the graphs in the null distribution.
Testing the Evolution of LGT
For every cellular lineage, we counted the number of phage and plasmid sequences that were present in CHDs containing that lineage. This method was repeated for all identity thresholds in decreasing order, each time removing previously observed associations. This way, we counted phage and plasmid sequences associated with a given lineage at 100% identity, then only the new associations appearing at 95%, then the new ones appearing at 90%, and so on. We also computed the thresholds beyond which any given cellular lineage was no longer isolated from other lineages.
Testing the Functional Categories involved in the linkages
Taking advantage of the SEED (27) annotation repositories (ftp://ftp.theseed.org/genomes/SEED/), we assigned a functional category to every CHD containing at least one cellular or plasmidic sequence, whenever such a function was known. Phages sequences were annotated using MG-RAST (28), providing a classification of these sequences in the functional categories of the SEED. We plotted the functional distribution of the gene families involved in the linkages within the GCC at various identity thresholds. Given that this overall distribution could include vertically inherited genes in addition to laterally transferred ones, especially as the threshold of identity decreased, we also plotted the distribution of the functional categories of the gene families connecting exclusively plasmids and cellular chromosomes and connecting phages and cellular chromosomes at various identity thresholds. Most of the genes comprised in this subset were likely involved in LGT, especially at stringent identity criteria, thus offering a conservative picture of the functional categories of genes likely involved in LGT at different times.
Supplementary Material
Acknowledgments
We thank H. Le Guyader, D. Higuet, J. Deutsch, J. Shapiro, F.J. Lapointe, and Y. Boucher for comments on this work. We thank Tal Dagan, who kindly provided us with MATLAB scripts for computing modularity maximization; and Klaus Schliep for advice in statistics.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908978107/DCSupplemental.
References
- 1.Whitman WB. The modern concept of the procaryote. J Bacteriol. 2009;191:2000–2005. doi: 10.1128/JB.00962-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dagan T, Martin W. Getting a better picture of microbial evolution en route to a network of genomes. Philos Trans R Soc Lond B Biol Sci. 2009;364:2187–2196. doi: 10.1098/rstb.2009.0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ragan MA, McInerney JO, Lake JA. The network of life: genome beginnings and evolution. Introduction. Philos Trans R Soc Lond B Biol Sci. 2009;364:2169–2175. doi: 10.1098/rstb.2009.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rohwer F, Thurber RV. Viruses manipulate the marine environment. Nature. 2009;459:207–212. doi: 10.1038/nature08060. [DOI] [PubMed] [Google Scholar]
- 5.Norman A, Hansen LH, Sørensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci. 2009;364:2275–2289. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugenholtz P, Tyson GW. Microbiology: metagenomics. Nature. 2008;455:481–483. doi: 10.1038/455481a. [DOI] [PubMed] [Google Scholar]
- 7.Brüssow H. The not so universal tree of life or the place of viruses in the living world. Philos Trans R Soc Lond B Biol Sci. 2009;364:2263–2274. doi: 10.1098/rstb.2009.0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tringe SG, et al. Comparative metagenomics of microbial communities. Science. 2005;308:554–557. doi: 10.1126/science.1107851. [DOI] [PubMed] [Google Scholar]
- 9.Doolittle WF, Zhaxybayeva O. On the origin of prokaryotic species. Genome Res. 2009;19:744–756. doi: 10.1101/gr.086645.108. [DOI] [PubMed] [Google Scholar]
- 10.Doolittle WF, Bapteste E. Pattern pluralism and the Tree of Life hypothesis. Proc Natl Acad Sci USA. 2007;104:2043–2049. doi: 10.1073/pnas.0610699104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. doi: 10.1186/1471-2105-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dagan T, Artzy-Randrup Y, Martin W. Modular networks and cumulative impact of lateral transfer in prokaryote genome evolution. Proc Natl Acad Sci USA. 2008;105:10039–10044. doi: 10.1073/pnas.0800679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newman ME. Finding community structure in networks using the eigenvectors of matrices. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:036104. doi: 10.1103/PhysRevE.74.036104. [DOI] [PubMed] [Google Scholar]
- 14.Clauset A, Moore C, Newman ME. Hierarchical structure and the prediction of missing links in networks. Nature. 2008;453:98–101. doi: 10.1038/nature06830. [DOI] [PubMed] [Google Scholar]
- 15.Brilli M, et al. Analysis of plasmid genes by phylogenetic profiling and visualization of homology relationships using Blast2Network. BMC Bioinformatics. 2008;9:551. doi: 10.1186/1471-2105-9-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fricke WF, et al. Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J Bacteriol. 2008;190:6779–6794. doi: 10.1128/JB.00661-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hatfull GF, Cresawn SG, Hendrix RW. Comparative genomics of the mycobacteriophages: insights into bacteriophage evolution. Res Microbiol. 2008;159:332–339. doi: 10.1016/j.resmic.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima-Mendez G, Van Helden J, Toussaint A, Leplae R. Reticulate representation of evolutionary and functional relationships between phage genomes. Mol Biol Evol. 2008;25:762–777. doi: 10.1093/molbev/msn023. [DOI] [PubMed] [Google Scholar]
- 19.Jefferson KK. What drives bacteria to produce a biofilm? FEMS Microbiol Lett. 2004;236:163–173. doi: 10.1016/j.femsle.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 20.Hooper SD, et al. A molecular study of microbe transfer between distant environments. PLoS One. 2008;3:e2607. doi: 10.1371/journal.pone.0002607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schlüter A, et al. The 64 508 bp IncP-1beta antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1beta group. Microbiology. 2003;149:3139–3153. doi: 10.1099/mic.0.26570-0. [DOI] [PubMed] [Google Scholar]
- 22.Zienkiewicz M, et al. Mosaic structure of p1658/97, a 125-kilobase plasmid harboring an active amplicon with the extended-spectrum beta-lactamase gene blaSHV-5. Antimicrob Agents Chemother. 2007;51:1164–1171. doi: 10.1128/AAC.00772-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sneath PH. The application of computers to taxonomy. J Gen Microbiol. 1957;17:201–226. doi: 10.1099/00221287-17-1-201. [DOI] [PubMed] [Google Scholar]
- 25.Enright AJ, Van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandes U. A faster algorithm for betweenness centrality. J Math Sociol. 2001;25:163–177. [Google Scholar]
- 27.Overbeek R, et al. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meyer F, et al. The metagenomics RAST server - a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics. 2008;9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.