Abstract
Ebolavirus causes a severe hemorrhagic fever and is divided into five distinct species, of which Reston ebolavirus is uniquely nonpathogenic to humans. Disease caused by ebolavirus is marked by early immunosuppression of innate immune signaling events, involving silencing and sequestration of double-stranded RNA (dsRNA) by the viral protein VP35. Here we present unbound and dsRNA-bound crystal structures of the dsRNA-binding domain of Reston ebolavirus VP35. The structures show that VP35 forms an unusual, asymmetric dimer on dsRNA binding, with each of the monomers binding dsRNA in a different way: one binds the backbone whereas the other caps the terminus. Additional SAXS, DXMS, and dsRNA-binding experiments presented here support a model of cooperative dsRNA recognition in which binding of the first monomer assists binding of the next monomer of the oligomeric VP35 protein. This work illustrates how ebolavirus VP35 could mask key recognition sites of molecules such as RIG-I, MDA-5, and Dicer to silence viral dsRNA in infection.
Keywords: dsRNA, interferon, structure, virus
An enveloped, nonsegmented, negative-stranded RNA virus, ebolavirus causes severe and rapidly progressing hemorrhagic fever. Five species of ebolavirus have thus far been described: Zaire, Sudan, Côte d’Ivoire, Reston, and the proposed Bundibugyo, with each name derived from the location in which that species was first identified (1, 2). Disease from ebolavirus is marked by fever, shock, and coagulation defects with 50–90% mortality occurring 7–12 days after infection (3). However, among the five species, Reston ebolavirus appears to be uniquely nonpathogenic to humans. During 1989–1996, several outbreaks of Reston ebolavirus occurred among macaques imported from the Philippines to primate facilities in the United States (Virginia and Texas) and Italy. In early 2009, Reston ebolavirus broke out again among domesticated swine in the Philippines (4). Several animal handlers seroconverted during these outbreaks, but none of them developed hemorrhagic fever (5, 6).
Patients who develop fatal hemorrhagic fever from other species of ebolavirus display high viral titers (reaching 109 pfu/mL) and little evidence of a humoral immune response (7–9), whereas those patients that survive ebolavirus infection display rapid and robust immune responses (8). This disparity suggests that events early in viral infection determine the course of ebolavirus infection.
In general, early in infection with a pathogen, components of the innate immune system sense pathogen-associated molecular patterns (PAMPs). When PAMPs are detected, the adaptive arms of the immune system are signaled to respond. Among PAMPs, double-stranded RNA (dsRNA) is a unique product of viral infection. dsRNA is detected in the cytoplasm of an infected cell by viral dsRNA receptors such as retinoic acid–inducible gene I (RIG-I) (10) and melanoma differentiation–associated gene 5 (MDA-5) (11). Host RIG-I and MDA-5 initiate signaling cascades that activate interferon regulatory factor 3 (IRF-3), leading to the production of interferon α/β (IFN α/β), which continues the antiviral response in the infected cell and primes surrounding cells to respond. Recent studies have shown that MDA-5 is activated by dsRNA with blunt ends and that RIG-I is activated by dsRNA with a 5′ triphosphate, preferentially with blunt ends (12–15). These dsRNA structures are predicted to be present in the panhandle regions of the RNA genome of ebolavirus (16, 17) suggesting that ebolavirus would be susceptible to detection and subsequent signaling by RIG-I and MDA-5. Indeed, stimulation or overexpression of RIG-I in cell culture attenuates experimental ebolavirus replication (18).
However, ebolavirus has evolved countermeasures against innate immune signaling mechanisms, evident by the significant immunosuppression noted in natural ebolavirus infection (19, 20). In particular, a key protein of the ebolaviruses, termed VP35, may play a role in blocking critical immune signaling events early in infection. VP35 has been shown to prevent phosphorylation and dimerization of IRF-3 (21), to block induction of IFN α/β expression (21, 22), to inhibit activation of protein kinase R (PKR) (23, 24), and to serve as a suppressor of RNA silencing (25). The ability of VP35 to block IFN α/β expression has been mapped to its C-terminal dsRNA-binding domain (26, 27), also termed the IFN inhibitory domain. In this domain, three basic residues, R305, K309, and R312, as numbered in the Zaire species of ebolavirus, are critical for binding dsRNA and blocking IFN expression (26). Indeed, the functionally null Zaire VP35 mutant R312A is unable to block RIG-I activation during viral infection (28). As expected, recombinant viruses with the mutations R305A and R312A are severely attenuated and display markedly reduced growth in cell culture and murine models (29, 30). These residues lie in a basic patch on the Zaire ebolavirus VP35 dsRNA-binding domain (31), but it was previously unknown whether they directly or indirectly influence RNA binding.
In an effort to better understand the mechanism by which VP35 binds to dsRNA and blocks the IFN α/β induction pathway, we have determined the structure of the Reston ebolavirus VP35 dsRNA-binding domain (RBD) both alone and in complex with dsRNA. The dsRNA-bound crystal structure shows that Reston VP35 forms an asymmetric dimer with two unique modes of RNA binding: one monomer binds along the side of the phosphate backbone, whereas the other monomer caps the terminus of the dsRNA. The structure also illustrates that the critical third arginine, R301 in Reston (equivalent to R312 in Zaire), does not contact RNA but instead contributes to the dimer interface between the two VP35 monomers. Hence, the mutation of R301/312 abolishes IFN inhibition by disrupting the VP35 dimer interface. The dsRNA-binding studies also presented here indicate that binding of dsRNA by the two VP35s is cooperative. Furthermore, VP35 can bind the blunt-ended dsRNA observed in infection, and its binding site on RNA likely overlaps with that of RIG-I, MDA-5, and Dicer. This binding interaction between ebolavirus VP35 and dsRNA is novel and mechanistically distinct from other viral dsRNA-binding proteins.
Results
Structures of Unbound and dsRNA-Bound Reston VP35.
As it is likely that the entire VP35 molecule is required for the multistep process of IFN inhibition, we term the C-terminal domain the RNA-binding domain (RBD) rather than the IFN inhibitory domain. We have determined crystal structures of the Reston ebolavirus VP35 RNA-binding domain (residues 160–329) both alone and in complex with an 18-bp, blunt-ended dsRNA. The N-terminal residues of both the unbound and bound VP35 RBD structures are disordered: interpretable electron density begins at residue 204 in the unbound structure and 206 in the dsRNA-bound structure. Both structures reveal that the Reston VP35 RBD, like that of Zaire (31), contains two subdomains: the N-terminal half contains a four-helix bundle followed by a short fifth helix, whereas the C-terminal half contains a three-stranded mixed β sheet (a). A search for similar folds to the β sheet subdomain on the DALI server (32) yielded no significant results outside of the ebolavirus genus.
An alignment of the Reston VP35 RBD with the recently determined structure of the unbound Zaire VP35 RBD (31) shows a high degree of similarity between the two species with an overall RMSD of 0.7 Å (Fig. S1). Some flexibility is evident in residues 274–281 (Reston numbering) that form the fifth helix in the α-helical subdomain of Reston, but a loop structure in Zaire. However, outside this region there is an overall conservation of secondary structure elements and molecular architecture. Indeed, the amino acid sequences of the VP35 RNA-binding domain of all five species of ebolavirus are 85% identical, reflecting the functional importance of this region. In addition, the bound and unbound structures of the Reston VP35 RBD align with an RMSD of 0.42 Å, demonstrating that no significant conformational changes occur upon dsRNA binding.
The crystallographic asymmetric unit of the Reston VP35 RBD-dsRNA complex contains four VP35 RNA-binding domains in complex with one 18-bp dsRNA molecule, with a pseudo 2-fold symmetry axis running through the midpoint of the dsRNA (Fig. 1B). The 18-bp length of the oligo was chosen to reflect a biologically relevant length compact enough to be crystallizable, but long enough to reflect panhandle regions in the ebolavirus genome.
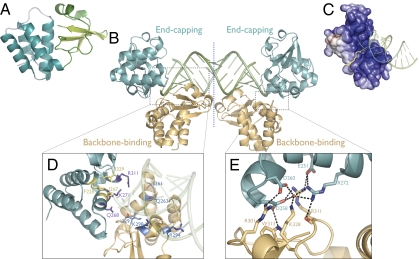
Fig. 1.
(A) Subdomain organization of the unbound VP35 RBD monomer. The α-helical subdomain is teal and the β-sheet subdomain is green. (B) Asymmetric unit of the VP35 RBD-dsRNA complex structure. Two VP35 RBDs (backbone-binding, orange; end-capping, teal) bind as an asymmetric dimer to each end of an 18-bp dsRNA oligomer (green). A pseudo 2-fold axis runs through the midpoint of the dsRNA. (C) Electrostatic surface potential of the asymmetric VP35 RBD dimer (colored continuously) highlighting the basic pocket into which blunt-ended dsRNA binds. Yellow line denotes the boundary between VP35 monomers. Positive surface is colored blue; negative surface is colored red, with limits ±10 kT/e. (D) Both the backbone-binding RBD (orange) and end-capping RBD (teal) contain polar residues (backbone-binding, blue; end-capping, purple) that interact with the dsRNA phosphate backbone. Only the end-capping RBD also uses hydrophobic residues (yellow) to form a nonpolar face that packs against the terminal bases of the dsRNA. (E) Polar residues at the dimeric interface between the backbone-binding (orange) and end-capping (teal) RBDs with possible hydrogen bonds (dashed black lines).
Dimer Assembly.
The monomeric VP35 RBD forms an asymmetric dimer in the dsRNA-complex structure, with one dimer bound to each end of the 18-bp dsRNA. Within each dimer, one VP35 RBD of the pair binds the phosphate backbone of the dsRNA (the “backbone-binding” VP35), whereas the other VP35 RBD of the pair binds terminal nucleotides of the dsRNA, capping the end of the dsRNA (the “end-capping” VP35) (Fig. 1 B and D). The dimeric VP35 RBD forms a continuous, positively charged pocket that binds the blunt-ended dsRNA (Fig. 1C).
The asymmetric Reston VP35 RBD dimeric interface is formed by a hydrogen-bonding network of residues R301, R311, W313, and K328 from the backbone-binding VP35 RBD, residues E251, D258, D260, and K272 of the end-capping VP35 RBD and a solvent molecule (Fig. 1E). The buried surface area between the two RBD monomers in the dimer interface is ≈660 Å2 (330 Å2 on each VP35). The VP35 surface buried by dsRNA binding on each monomer is larger: 860 Å2 for the dimer as a whole (520 Å2 buried by the backbone-binding interaction and 340 Å2 buried by the end-capping interaction). The greater surface buried by dsRNA binding of VP35, as compared with that generated by the two VP35s binding each other, coupled with the absence of this dimer in crystal packing of the unbound Reston VP35 RBD, suggests that the VP35 RBD does not form a dimer in the absence of dsRNA but, rather, assembles into a dimer upon RNA binding.
RNA Binding.
Binding of the two VP35s to dsRNA is mediated by polar contacts with the phosphate backbone as well as hydrophobic interactions with the terminal nucleotide bases. The backbone-binding VP35 RBD/dsRNA interface is formed by interactions with the side chains of S261, Q263, R294, K298, and the C-terminal carboxyl of I329 to the phosphate backbone of the dsRNA. By contrast, the end-capping VP35 RBD/dsRNA interface is formed by interactions of the side chains of K271 and R311 with the phosphate backbone and the side chain of Q268 with the O2’ and O3′ of the 3′ terminal ribose. Notably, residues F228, I267 and I329 of the end-capping VP35 also form a hydrophobic face that packs against the terminal RNA bases, in which the phenol ring of F228 forms a herringbone structure with the ring of the terminal nitrogen base (Fig. S2).
VP35 residues R305, K309, and R312 of the Zaire species of ebolavirus have been indicated as critical for dsRNA binding. Among these, the single point mutant R312A is sufficient to disrupt IFN suppression and to restore dsRNA PAMP recognition (28). Reston ebolavirus VP35 has a naturally occurring N-terminal truncation, and hence residues R305, K309, and R312 in the Zaire species correspond to residues R294, K298, and R301 in the numbering of the Reston species. In the Reston crystal structure, R294, K298, and R301 similarly form a positively charged surface patch in the β-sheet-loop subdomain on the VP35 RBD surface that comprises one portion of the complete dsRNA-binding site. At this site, residues R294 and K298 are positioned to directly contact the RNA phosphate backbone. Interestingly, residue R301 (the equivalent of R312 in Zaire) does not interact with dsRNA. Instead, R301 participates in VP35 RBD dimerization, in which R301 of the backbone-binding VP35 RBD hydrogen bonds to D258 and D260 on the adjacent, end-capping VP35. The R301 of the end-capping VP35 RBD is exposed to the bulk solvent and does not appear to interact with the backbone-binding VP35 RBD or the dsRNA. Thus, the critical nature of this residue does not lie in dsRNA binding but, rather, in proper VP35 RBD dimer formation in the presence of dsRNA.
Binding Assays.
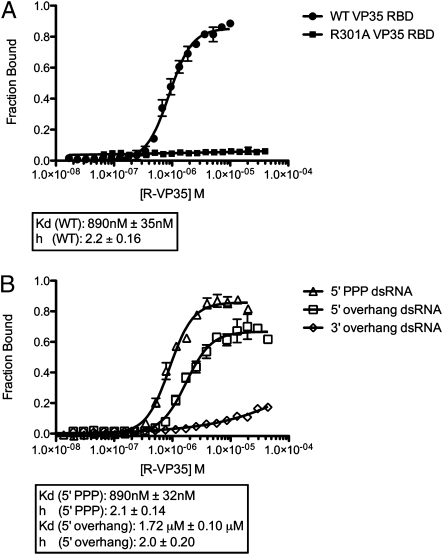
To better understand the mechanism of binding of dsRNA by VP35 in solution, we performed dual-filter dot blot binding assays using radiolabeled dsRNA corresponding to the dsRNA construct used for crystallization. These assays showed that VP35 RBD binds to dsRNA in solution with half saturation at a protein concentration of ≈1 μM (Fig. 2A). Importantly, we observe a Hill coefficient of 2.2 (±0.16 SEM), indicating that the VP35 RBD binds dsRNA in a cooperative fashion. Furthermore, the point mutation R301A, which partially disrupts the hydrogen-bonding network between the two RBDs, results in a complete loss of dsRNA binding (Fig. 2A). Hence, VP35 must assemble into a dimer to form a stable complex with dsRNA.
Fig. 2.
RNA binding studies of VP35 RBD. (A) Dual-filter dot blot experiments with blunt-ended 18bp dsRNA indicate that the wild-type Reston VP35 RBD (filled circles, n = 60) binds cooperatively, with a Hill coefficient of 2.2 ± 0.16 SEM. By contrast, the R301A point mutant of the Reston VP35 RBD (filled squares, n = 40) abrogates dsRNA binding. (B) Wild-type Reston VP35 RBD binds dsRNA with 5′ triphosphate (open triangles, n = 40) and 5′ overhangs (open squares, n = 60) with affinity and cooperativity similar to that of blunt-ended dsRNA. The VP35 RBD retains limited ability to bind dsRNA with 3′ overhangs (open diamonds, n = 60), but affinity is greatly reduced.
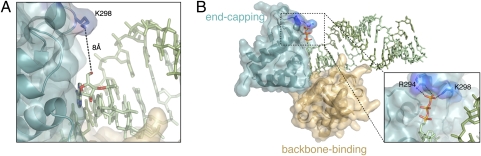
Although the nucleic acid crystallized here is blunt ended and bears 5′ hydroxyls on both ends of the duplex, examination of the structure reveals that VP35 should also be able to bind dsRNA with a 5′ triphosphate moiety, which is a key determinant of recognition by RIG-I. Specifically, the ε-amino group of K298 resides ≈8 Å from the current RNA 5′ hydroxyl, indicating that a terminal 5′ triphosphate group could fit in between (Fig. 3). Furthermore, the structure suggests that VP35 should be able to bind dsRNA with a 5′ overhang but would recognize dsRNA with a 3′ overhang less well.
Fig. 3.
Triphosphate modeled onto the 5′ end of the dsRNA in the dsRNA-dimeric VP35 RBD complex. (A) Although the dsRNA crystallized contains 5′ hydroxyls, adequate space exists to accommodate a 5′ triphosphate and VP35 indeed binds 5′ triphosphate-bearing dsRNA. (B) VP35-dsRNA complex with a triphosphate modeled onto the 5′ terminus of the dsRNA. VP35 residues R294 and K298 appear to be well positioned to hydrogen bond with a 5′ triphosphate.
Indeed, dot blot binding assays demonstrate that VP35 can bind 5′ triphosphate capped dsRNA with similar affinity and cooperativity to that of the blunt-ended dsRNA (Fig. 2B). Furthermore, dot blot binding assays using dsRNA with three nucleotide overhangs on either the 5′ or the 3′ ends show that VP35 can bind dsRNA with 5′ overhangs with similar affinity and cooperativity as blunt-ended dsRNA. By contrast, the VP35 RBD binds dsRNA with 3′ overhangs with greatly reduced affinity (Fig. 2B). These results fit well with the crystal structure observed, as the end-capping VP35 abuts the 3′ end of the dsRNA yet leaves sufficient space to accommodate either the 5′ overhang or the 5′ triphosphate.
Deuterium Exchange Mass Spectrometry.
Deuterium exchange mass spectrometry (DXMS) was performed on a construct of Zaire VP35 containing its N-terminal regions and putative coiled-coil motif in addition to the RNA-binding domain. A 10-s incubation in deuteration buffer was used to evaluate and map rapidly exchanging main-chain amides. These experiments reveal that the predicted coiled-coil motif and RNA-binding domain of VP35 exchange slowly and are likely globular. However, the N terminus and the linker region between the coiled-coil motif and the RBD exchange rapidly, reflecting a high degree of solvent accessibility and likely flexibility and disorder of these regions in solution (Fig. 4A).
Fig. 4.
Biophysical characterization of VP35. (A) Ten-second amide hydrogen–deuterium exchange map for select VP35 peptides of a near full-length Zaire VP35. Red horizontal bars indicate fast-exchanging amides; black bars indicate stretches of no exchange. (B) Molecular weight calculation of Zaire VP35 (triangles) and the Reston VP35 RBD (circles). Our Zaire VP35 expression construct encodes a 37-k Da molecule. SAXS analysis indicates a ∼100 kDa mass, consistent with the oligomeric (probably trimeric) nature of VP35 in solution. The RBD construct (here, indicated by Reston) encodes a molecule ∼20 kDa in size. SAXS indicates a ∼20–25 kDa mass, consistent with the monomeric nature of the RBD in the absence of dsRNA.
Small Angle X-Ray Scattering.
Small angle x-ray scattering (SAXS) was performed on this same Zaire VP35, as well as the truncated Reston VP35 RNA-binding domain used for crystallization (Fig. S3). Previous studies have suggested that full-length VP35 homo-oligomerizes through its N-terminal coiled-coil domain and forms trimers or tetramers (33). As expected, the Reston VP35 RNA-binding domain was found to be monomeric in solution with an estimated molecular weight of ≈20 kDa. This correlates well with size exclusion chromatography in which the VP35 RBD elutes at a size consistent with a monomeric nature (Fig. S4). The Zaire VP35 was estimated to be ≈100 kDa, indicating that the full-length VP35 is certainly oligomeric and possibly a trimer in solution (Fig. 4B).
Discussion
Multiple lines of evidence presented here demonstrate cooperative dimeric binding of dsRNA by ebolavirus VP35. Small angle x-ray scattering experiments, as well as unbound and dsRNA-bound crystal structures of VP35, illustrate that the C-terminal domain of VP35 likely dimerizes upon binding dsRNA. Accompanying RNA-binding experiments illustrate that dsRNA binding by VP35 is cooperative. Furthermore, R312 (R301 in Reston) has been previously shown to be critical for RNA binding; yet our crystal structure indicates that it does not contact dsRNA but instead is an important component of the VP35 dimeric interface. Hence, the mutation of R312/301 likely abolishes dsRNA binding and restores RIG-I signaling by disrupting VP35 dimer formation. Cooperative formation of the VP35 RBD dimer thus appears critical to the formation of a stable dsRNA complex.
The apparent flexibility of the linker in full-length VP35 between the N-terminal regions and the C-terminal dsRNA-binding domain, as evidenced by rapid deuterium exchange (Fig. 4a), would allow different monomers of the VP35 oligomer to simultaneously couple with dsRNA in different orientations. Two of these monomers would form the dimer observed in the crystal structure. A third member of the oligomer might form an additional interaction with the exposed dsRNA backbone of the complementary strand. Alternatively, the third RBD could couple with viral factors such as NP and L (34–36), or host factors usurped in infection, such as SUMO (37), dynein light chain 8 (38), IκB kinase ε (IKKε) (39), Tank-binding kinase 1 (TBK-1) (39), and protein kinase R (PKR) (24).
The RNA-binding domains of Zaire and Reston ebolavirus VP35 are 87% conserved in sequence, reflecting the functional importance of this region for the ebolavirus life cycle. Indeed, primary differences in sequence map to regions that do not take part in dsRNA binding, and it is likely that VP35s from both species will recognize dsRNA similarly (Fig. S5). Reston ebolavirus does have a reduced ability to evade cellular antiviral responses (40). However, the conservation of VP35 RBD structures between pathogenic and nonpathogenic viruses suggests that the virulence determinant lies elsewhere: in a contact of the VP35 RBD with a non-dsRNA factor, in N-terminal regions of VP35, or in another component of the virus.
This crystal structure shows that each VP35 in the dimer contacts the dsRNA exclusively through the sugar-phosphate backbone and hydrophobic faces of terminal bases, allowing the VP35 dimer to recognize dsRNA in a sequence-independent manner. Furthermore, the VP35 RBD dimer efficiently occludes the open end of the dsRNA helix, which is a crucial determinant of recognition by the enzyme Dicer and subsequent activation of host RNA silencing pathways (41, 42). The crystal structure and biophysical experiments presented here illustrate how VP35 could block the entry of viral dsRNAs into the RNA silencing pathways and function as a general suppressor of the innate immune system, as has been previously observed (25).
No structure of RIG-I or MDA-5 in complex with dsRNA is yet available. However, there is a structure available of a protein termed “laboratory of genetics and physiology 2” (LGP2) (43) in complex with dsRNA (15). LGP2 is a close structural homolog of RIG-I and MDA-5 (Fig. 5A), and similar binding surfaces are involved in dsRNA recognition by all three proteins (15) (Fig. 5A). The structure of LGP2 in complex with dsRNA shows that LGP2 and VP35 bind dsRNA using similar mechanisms. A basic surface patch of LGP2 makes charge interactions with the phosphate backbone, and an exposed loop containing hydrophobic residues packs against the terminal bases of the dsRNA. One monomer of LGP2 forms both interactions, whereas VP35 must dimerize to do so, with one VP35 monomer binding the backbone and the other binding the terminal bases. Binding sites for VP35 on dsRNA overlap with that of LGP2, and by homology, likely overlap with those of RIG-I and MDA-5. Thus, by binding and sequestering key recognition motifs on dsRNA, VP35 could block recognition and signaling by RIG-I and MDA-5 and could prevent initiation of the innate immune response (Fig. 6).
Fig. 5.
Similar modes of binding between host dsRNA receptors and VP35 RBD. (A) Superimposition of the RIG-I C-terminal domain (CTD) (pale cyan; PDB 2QFB) and MDA-5 CTD (mauve; PDB 3GA3) with the LGP2 CTD (brown; PDB 3EQT), highlighting the structural similarity among the three homologs. Conserved polar residues (blue) make key contacts with the phosphate backbone in the LGP2-dsRNA complex structure, whereas hydrophobic residues (yellow) pack against terminal nucleotide bases of the dsRNA (transparent green). (B) Electrostatic surface potential of the LGP2 CTD (colored continuously) bound to an 8-bp dsRNA oligo (green) highlighting surface charge distribution similar to that of the asymmetric VP35 dimer. Positive surface is colored blue; negative surface is colored red with limits ± 10 kT/e. (C) Schematic view of LGP2 CTD and VP35 RBD dimer, illustrating organization of polar (blue) and hydrophobic (yellow) interaction sites for dsRNA.
Fig. 6.
Model of the cooperative, dimeric assembly mechanism of the VP35 RBD on dsRNA. The interface between the dsRNA and the backbone-binding VP35 RBD is larger (540 Å2) than either the dsRNA-end-capping RBD interface (340 Å2), or the RBD-RBD interface in the dimer (330 Å2). Furthermore, the affinity for uncapped and 5′triphosphate-capped dsRNA is similar, suggesting that a 5′ triphosphate is not a major recognition signal. These results and the observed in-solution cooperativity of dsRNA binding together suggest a mechanism by which the backbone-binding VP35 forms the first and initial contact to the phosphate backbone along the side of the dsRNA (brown VP35, blue box to blue bar). The second VP35 RBD (the end-capping molecule) would then bind the larger surface now assembled by both the terminus of the RNA and the dimer interface of the backbone-binding VP35 (teal VP35, yellow box to dsRNA end). In this model, the assembly of the dimeric VP35 RBDs on the end of the dsRNA oligonucleotide masks the binding site of RIG-I and MDA-5, preventing their recognition of dsRNA and subsequent signaling.
The Reston VP35-dsRNA complex structure reveals an unusual binding interaction between the protein and the RNA that isnot typically seen in viral proteins that bind dsRNA in a sequence-independent manner, such as Tombusvirus P19 (44), Flock House virus B2 (45), or influenza virus NS1 (46, 47). Each of these viruses has evolved a unique structural mechanism for binding dsRNA, creating a palette of possibilities by which a virus may silence and sequester this pathogen-associated molecular pattern (Fig. S6).
For ebolavirus in particular, these crystal structures of the VP35 RBD alone, and in complex with dsRNA, illustrate that the functionally important RNA-binding domain of VP35 is conserved in structure, and probably function, between two species of ebolavirus with dramatically different pathogeneses in humans. More importantly, the structures presented here illustrate a novel, bimodal binding mechanism for cooperative, sequence-independent dsRNA recognition that permits ebolavirus to avoid the innate immune response and enhance virulence. The nature of the two unique modes of interaction with dsRNA achievable by any given VP35, as well as the necessity of formation of the dimer interface itself, present enticing targets for structure-based design of antivirals for human health and biodefense.
Methods
Crystallization and Data Collection.
Purified unbound and dsRNA-complexed Reston VP35 RBD (SI Methods) were screened for crystallization using a TOPAZ microfluidic system (Fluidigm), and hits were translated to hanging drop vapor diffusion. For the unbound structure, native Reston VP35 RBD was crystallized in 90 mM tripotassium citrate, 100 mM sodium acetate pH4.0, and 5% (wt/vol) PEG 6K. Se-Met containing Reston VP35 RBD was crystallized in 150 mM ammonium citrate dibasic, pH 4.0, and 3% (wt/vol) PEG 3350. For determination of the dsRNA-bound structure, native Reston VP35 RBD was complexed with an 18-bp synthetic dsRNA oligo at a 1:1.7 molar ratio. The VP35-RNA complex was crystallized in 365 mM ammonium acetate, 100 mM trisodium citrate, pH 6.5, and 18.5% PEG 4K. Crystals were harvested and cryoprotected in sequential soaks of mother liquor plus 5%, 10%, 15%, and 20% glycerol. Crystals were then flash cooled in liquid nitrogen and transported to the beam line under cryogenic conditions.
Native and Se-Met containing crystals of the unbound Reston VP35 RBD belonging to space group R32 diffracted to 2.4 Å and 3.3 Å, respectively at Beam Line 19ID of the Advanced Photon Source. Data were collected at 0.9795 Å corresponding to the Selenium K-edge. Native crystals of the dsRNA-bound Reston VP35 belonging to space group P31 diffracted to 2.1 Å at Beam Line 11–1 of the Stanford Synchrotron Radiation Laboratory.
Data Processing and Structure Determination.
For the unbound structure, data were indexed, integrated, and scaled in space group R32 using d*TREK (Rigaku). As the native and SeMet crystals were isomorphous, the structure was determined by SIR(AS) with three of six Se-atom positions identified using autoSHARP (48). An initial model was obtained using ARP/wARP (49). For the dsRNA-bound structure, data were indexed and integrated in space group P31 with HKL2000 (50) and scaled with XPREP (Bruker Inc.). Correction for anisotropic diffraction was done using the Diffraction Anisotropy Server (51). The structure was determined by molecular replacement with PHASER (52) using the unbound Reston VP35 as a search model. Refinement was carried out using PHENIX (53). TLS restraints were generated using the TLSMD server (54). Model inspection and manual rebuilding was performed using COOT (55). Final refinement statistics are listed in Table S1.
Supplementary Material
Acknowledgments
We thank Dr. Christopher Basler (Mount Sinai School of Medicine) for the gifts of Reston and Zaire VP35 cDNA, Marnie Fusco and Dafna Abelson for technical assistance, Drs. Gerald Joyce and Bianca Lam for advice on in vitro transcription, Simon Hsu for assistance with DXMS, and Beam Lines 11-1 of SSRL, 19ID of APS, and 8.3.1 and 12.3.1 of ALS for crystallographic and SAXS data collection. This is manuscript #20349 from TSRI. This work was supported by the Skaggs Institute for Chemical Biology; a Career Award in the Biomedical Sciences and an Investigators in the Pathogenesis of Infectious Diseases Award from the Burroughs Wellcome Fund; National Institutes of Health grants R21AI053423 (E.O.S.) and R01GM086701 (I.J.M.) as well as CA099835, CA118595, and AI076961; a Discovery Grant (UC10591) from the University of California IUCRP Program and BiogenIDEC (V.L.W.), Training grants 5T32AI00760 and 2T32AI007244-26 to the TSRI Department of Immunology and Microbial Science (C.R.K. and Z.A.B., respectively).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3KS4 and 3KS8).
This article contains supporting information online at www.pnas.org/cgi/content/full/0910547107/DCSupplemental.
References
- 1.Sanchez A, Geisbert TW, Feldmann H. Filoviridae: Marburg and Ebola Viruses. In: Knipe DM, Howley PM, editors. Fields Virology. Philidelphia: Lippincott Williams & Wilkins; 2007. pp. 1409–1448. [Google Scholar]
- 2.Towner JS, et al. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. doi: 10.1371/journal.ppat.1000212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bwaka MA, et al. Ebola hemorrhagic fever in Kikwit, Democratic Republic of the Congo: Clinical observations in 103 patients. J Infect Dis. 1999;179(Suppl 1):S1–S7. doi: 10.1086/514308. [DOI] [PubMed] [Google Scholar]
- 4.Barrette RW, et al. Discovery of swine as a host for the Reston ebolavirus. Science. 2009;325:204–206. doi: 10.1126/science.1172705. [DOI] [PubMed] [Google Scholar]
- 5.CDC. Update: Filovirus infections among persons with occupational exposure to nonhuman primates. MMWR Morb Mortal Wkly. 1990;39:266–7. 273. [PubMed] [Google Scholar]
- 6.Editorial Team Ebola Reston virus detected pigs in the Philippines. Euro Surveill. 2009;14 pii=19105. [PubMed] [Google Scholar]
- 7.Towner JS, et al. Rapid diagnosis of Ebola hemorrhagic fever by reverse transcription-PCR in an outbreak setting and assessment of patient viral load as a predictor of outcome. J Virol. 2004;78:4330–4341. doi: 10.1128/JVI.78.8.4330-4341.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baize S, et al. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 9.Sanchez A, et al. Analysis of human peripheral blood samples from fatal and nonfatal cases of Ebola (Sudan) hemorrhagic fever: Cellular responses, virus load, and nitric oxide levels. J Virol. 2004;78:10370–10377. doi: 10.1128/JVI.78.19.10370-10377.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 11.Andrejeva J, et al. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264–17269. doi: 10.1073/pnas.0407639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmidt A, et al. 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA. 2009;106:12067–12072. doi: 10.1073/pnas.0900971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schlee M, et al. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity. 2009;31:25–34. doi: 10.1016/j.immuni.2009.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li X, et al. Structural basis of double-stranded RNA recognition by the RIG-I like receptor MDA5. Arch Biochem Biophys. 2009;488:23–33. doi: 10.1016/j.abb.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Li X, et al. The RIG-I-like receptor LGP2 recognizes the termini of double-stranded RNA. J Biol Chem. 2009;284:13881–13891. doi: 10.1074/jbc.M900818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crary SM, Towner JS, Honig JE, Shoemaker TR, Nichol ST. Analysis of the role of predicted RNA secondary structures in Ebola virus replication. Virology. 2003;306:210–218. doi: 10.1016/s0042-6822(02)00014-4. [DOI] [PubMed] [Google Scholar]
- 17.Volchkov VE, et al. Characterization of the L gene and 5′ trailer region of Ebola virus. J Gen Virol. 1999;80:355–362. doi: 10.1099/0022-1317-80-2-355. [DOI] [PubMed] [Google Scholar]
- 18.Spiropoulou CF, et al. RIG-I activation inhibits ebolavirus replication. Virology. 2009;392:11–15. doi: 10.1016/j.virol.2009.06.032. [DOI] [PubMed] [Google Scholar]
- 19.Harcourt BH, Sanchez A, Offermann MK. Ebola virus inhibits induction of genes by double-stranded RNA in endothelial cells. Virology. 1998;252:179–188. doi: 10.1006/viro.1998.9446. [DOI] [PubMed] [Google Scholar]
- 20.Harcourt BH, Sanchez A, Offermann MK. Ebola virus selectively inhibits responses to interferons, but not to interleukin-1beta, in endothelial cells. J Virol. 1999;73:3491–3496. doi: 10.1128/jvi.73.4.3491-3496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Basler CF, et al. The Ebola virus VP35 protein inhibits activation of interferon regulatory factor 3. J Virol. 2003;77:7945–7956. doi: 10.1128/JVI.77.14.7945-7956.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Basler CF, et al. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feng Z, Cerveny M, Yan Z, He B. The VP35 protein of Ebola virus inhibits the antiviral effect mediated by double-stranded RNA-dependent protein kinase PKR. J Virol. 2007;81:182–192. doi: 10.1128/JVI.01006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schümann M, Gantke T, Mühlberger E. Ebola virus VP35 antagonizes PKR activity through its C-terminal interferon inhibitory domain. J Virol. 2009;83:8993–8997. doi: 10.1128/JVI.00523-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haasnoot J, et al. The Ebola virus VP35 protein is a suppressor of RNA silencing. PLoS Pathog. 2007;3:e86. doi: 10.1371/journal.ppat.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cárdenas WB, et al. Ebola virus VP35 protein binds double-stranded RNA and inhibits alpha/beta interferon production induced by RIG-I signaling. J Virol. 2006;80:5168–5178. doi: 10.1128/JVI.02199-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartman AL, Towner JS, Nichol ST. A C-terminal basic amino acid motif of Zaire ebolavirus VP35 is essential for type I interferon antagonism and displays high identity with the RNA-binding domain of another interferon antagonist, the NS1 protein of influenza A virus. Virology. 2004;328:177–184. doi: 10.1016/j.virol.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 28.Hartman AL, Ling L, Nichol ST, Hibberd ML. Whole genome expression profiling reveals that inhibition of host innate immune response pathways by ebola virus can be reversed by a single amino acid change in the VP35 protein. J Virol. 2008;82:5348–5358. doi: 10.1128/JVI.00215-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hartman AL, et al. Inhibition of IRF-3 activation by VP35 is critical for the high virulence of Ebola virus. J Virol. 2008;82:2699–2704. doi: 10.1128/JVI.02344-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hartman AL, Dover JE, Towner JS, Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J Virol. 2006;80:6430–6440. doi: 10.1128/JVI.00044-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leung DW, et al. Structure of the Ebola VP35 interferon inhibitory domain. Proc Natl Acad Sci USA. 2009;106:411–416. doi: 10.1073/pnas.0807854106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reid SP, Cárdenas WB, Basler CF. Homo-oligomerization facilitates the interferon-antagonist activity of the ebolavirus VP35 protein. Virology. 2005;341:179–189. doi: 10.1016/j.virol.2005.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Boehmann Y, Enterlein S, Randolf A, Mühlberger E. A reconstituted replication and transcription system for Ebola virus Reston and comparison with Ebola virus Zaire. Virology. 2005;332:406–417. doi: 10.1016/j.virol.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 35.Johnson RF, McCarthy SE, Godlewski PJ, Harty RN. Ebola virus VP35-VP40 interaction is sufficient for packaging 3E-5E minigenome RNA into virus-like particles. J Virol. 2006;80:5135–5144. doi: 10.1128/JVI.01857-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang Y, Xu L, Sun Y, Nabel GJ. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol Cell. 2002;10:307–316. doi: 10.1016/s1097-2765(02)00588-9. [DOI] [PubMed] [Google Scholar]
- 37.Chang TH, et al. Ebola Zaire virus blocks type I interferon production by exploiting the host SUMO modification machinery. PLoS Pathog. 2009;5:e1000493. doi: 10.1371/journal.ppat.1000493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kubota T, et al. Ebolavirus VP35 interacts with the cytoplasmic dynein light chain 8. J Virol. 2009;83:6952–6956. doi: 10.1128/JVI.00480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Prins KC, Cárdenas WB, Basler CF. Ebola virus protein VP35 impairs the function of interferon regulatory factor-activating kinases IKKepsilon and TBK-1. J Virol. 2009;83:3069–3077. doi: 10.1128/JVI.01875-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kash JC, et al. Global suppression of the host antiviral response by Ebola- and Marburgviruses: Increased antagonism of the type I interferon response is associated with enhanced virulence. J Virol. 2006;80:3009–3020. doi: 10.1128/JVI.80.6.3009-3020.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–940. doi: 10.1038/nsmb1293. [DOI] [PubMed] [Google Scholar]
- 42.Lund E, Dahlberg JE. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb Symp Quant Biol. 2006;71:59–66. doi: 10.1101/sqb.2006.71.050. [DOI] [PubMed] [Google Scholar]
- 43.Miyoshi K, et al. Structure of the mouse Stat 3/5 locus: Evolution from Drosophila to zebrafish to mouse. Genomics. 2001;71:150–155. doi: 10.1006/geno.2000.6433. [DOI] [PubMed] [Google Scholar]
- 44.Baulcombe DC, Molnár A. Crystal structure of p19—a universal suppressor of RNA silencing. Trends Biochem Sci. 2004;29:279–281. doi: 10.1016/j.tibs.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 45.Chao JA, et al. Dual modes of RNA-silencing suppression by Flock House virus protein B2. Nat Struct Mol Biol. 2005;12:952–957. doi: 10.1038/nsmb1005. [DOI] [PubMed] [Google Scholar]
- 46.Bornholdt ZA, Prasad BV. X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature. 2008;456:985–988. doi: 10.1038/nature07444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cheng A, Wong SM, Yuan YA. Structural basis for dsRNA recognition by NS1 protein of influenza A virus. Cell Res. 2009;19:187–195. doi: 10.1038/cr.2008.288. [DOI] [PubMed] [Google Scholar]
- 48.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 49.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Strong M, et al. Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2006;103:8060–8065. doi: 10.1073/pnas.0602606103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Adams PD, et al. PHENIX: Building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 54.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 55.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.