Abstract
DNA methylation might have a significant role in preventing normal differentiation in pediatric cancers. We used a genomewide method for detecting regions of CpG methylation on the basis of the increased melting temperature of methylated DNA, termed denaturation analysis of methylation differences (DAMD). Using the DAMD assay, we find common regions of cancer-specific methylation changes in primary medulloblastomas in critical developmental regulatory pathways, including Sonic hedgehog (Shh), Wingless (Wnt), retinoic acid receptor (RAR), and bone morphogenetic protein (BMP). One of the commonly methylated loci is the PTCH1-1C promoter, a negative regulator of the Shh pathway that is methylated in both primary patient samples and human medulloblastoma cell lines. Treatment with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-dC) increases the expression of PTCH1 and other methylated loci. Whereas genetic mutations in PTCH1 have previously been shown to lead to medulloblastoma, our study indicates that epigenetic silencing of PTCH1, and other critical developmental loci, by DNA methylation is a fundamental process of pediatric medulloblastoma formation. This finding warrants strong consideration for DNA demethylating agents in future clinical trials for children with this disease.
Keywords: epigenomics, cancer, PTCH1
Whereas cancer is widely viewed as a genetic disease, the role of epigenetic modifications, especially cytosine methylation in the promoter regions of genes, has been a major research focus in attempting to delineate the mechanisms leading to the formation of cancer, as well as for biomarker discovery (1). Genomic DNA of cancer cells is generally hypomethylated compared to DNA of normal cells, but displays a striking hypermethylation in the promoter regions of a subset of genes. This DNA hypermethylation has been correlated with transcriptional repression, indicating that epigenetic silencing of tumor suppressor genes may be an early step in the process of carcinogenesis (2).
In this report, we describe a genomewide DNA methylation assay that identifies CpG methylation on the basis of the biophysical property that 5-methylcytosine increases the melting temperature (Tm) of DNA. We show that this denaturation analysis of methylation differences method detects differentially methylated loci with high CpG density. Assessment of differential methylation in pediatric medulloblastomas compared to normal cerebellum DNA identifies cancer-specific methylation of genes associated with developmental processes. Of particular interest is cancer-specific methylation of the PTCH1-1C promoter, a negative regulator of the Sonic hedgehog (Shh) pathway. Whereas genetic mutations in PTCH1 have been described in human medulloblastomas, this demonstrates that epigenetic silencing by DNA methylation of PTCH1 contributes to the formation of this childhood cancer and suggests the use of DNA demethylating agents as a potential strategy for therapy.
Results
An Assay to Detect Palindrome Formation Enriches for CpG Methylation.
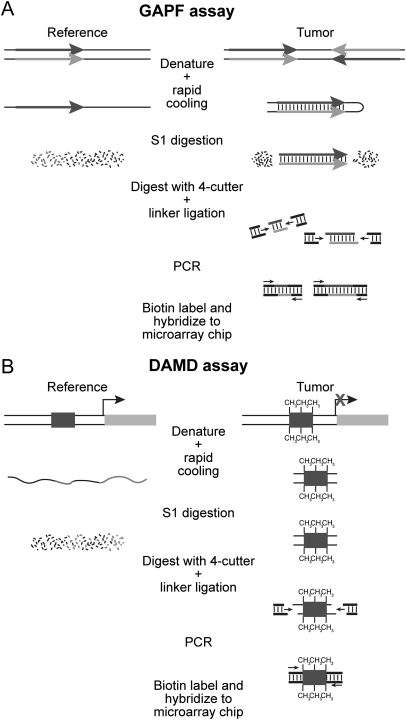
Earlier work from our laboratory focused on identifying regions of the genome susceptible to DNA palindrome formation, a rate-limiting step in gene amplification. We previously described a method to obtain a genomewide analysis of palindrome formation (GAPF) on the basis of the efficient intrastrand base pairing in large palindromic sequences (3). Palindromic sequences can rapidly anneal intramolecularly to form “snap-back” DNA under conditions that do not favor intermolecular annealing. This snap-back property was used to enrich for palindromic sequences in total genomic DNA by denaturing the DNA at 100°C in the presence of 100 mM NaCl, rapidly renaturing it by cooling, and then digesting the mixture with the single-strand-specific nuclease S1. Snap-back DNA formed from palindromes is double stranded and resistant to S1, whereas the remainder of genomic DNA is single stranded and thus is sensitive to S1 digestion (Fig. 1A). Using this assay, we have shown that de novo palindromes can form in cancers (3, 4) and that the GAPF-positive signals at the CTSK and EMC1 loci in Colo320DM cells represent DNA palindromes that define the border of an amplicon (4).
Fig. 1.
Schematic of the genomewide analysis of palindrome formation (GAPF) and denaturation analysis of methylation differences (DAMD) assays. (A) GAPF assay. Palindromic sequences can rapidly anneal intramolecularly to form “snap-back” DNA under conditions that do not favor intermolecular annealing. This snap-back property is used to enrich for palindromic sequences in total genomic DNA by denaturing the DNA at 100°C, rapidly renaturing it in the presence of 100 mM NaCl, and then digesting the mixture with the single-strand-specific nuclease S1. Snap-back DNA formed from palindromes is double stranded and resistant to S1, whereas the remainder of genomic DNA is single stranded and thus is sensitive to S1 digestion. Ligation-mediated PCR is performed, and then the DNA is labeled and hybridized to a microarray for analysis. (B) DAMD assay. DNA manipulations are the same as those performed in GAPF. During the denaturation step of the assay, the increase in the Tm caused by 5-methylcytosine renders methylated DNA resistant to denaturation compared to unmethylated DNA. After rapid cooling, the methylated DNA remains double stranded and resistant to S1 nuclease, whereas the same unmethylated sequence in the reference sample is single stranded and thus sensitive to S1 digestion.
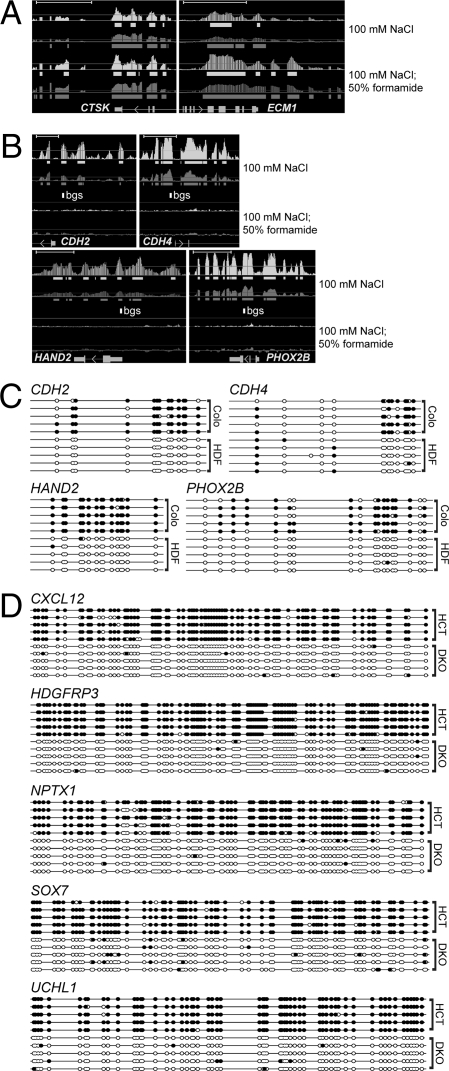
Because the presence of 5-methylcytosine increases the Tm of a DNA duplex (5–7), we recognized that a similar technique could be developed to identify regions of differential CpG methylation (Fig. 1B), and, indeed, we can detect both palindromes and regions of differential CpG methylation when we denature the DNA at 100°C in 100 mM NaCl (Fig. 2). To optimize the GAPF assay for the detection of palindromes, we added 50% formamide to the denaturation step. Formamide will decrease the Tm of duplex DNA 0.72°C for every 1% of formamide added (8) and denaturation at 100°C in 50% formamide eliminated the GAPF signals at the methylated loci and enhanced the signals at palindromes (compare Fig. 2B and 2A). Therefore, adding a high concentration of formamide, or using another stringent denaturation technique such as alkaline denaturation, will be critical for future studies using the GAPF procedure to identify palindromes.
Fig. 2.
Identification of differentially methylated genomic regions by denaturation. (A and B) Tiling array analysis of GAPF-positive regions in Colo320DM (Colo) cells compared to primary skin fibroblasts (HDF). The Upper pair of graphs is from the original GAPF method (100 mM NaCl) and the Lower pair is GAPF with the addition of formamide (100 mM NaCl, 50% formamide). Graphically displayed are P values (−10 log10, light shading) and signal [log2(signal ratio), dark shading], and the solid bars below each graph depict (−10 log10) P values >40 (= P < 0.001) and log2(signal ratio) > 1.5, where Colo > HDF. Gene names and mRNA structures (exon, large rectangle; intron, thin line; UTR, small rectangle; arrow, direction of transcription) are shown at the bottom of each panel. (Scale bars, ∼5 kb.) (A) The palindromes at CTSK and ECM1 are identified by the original GAPF assay and enhanced by the addition of 50% formamide. (B) Nonpalindromic loci showing a positive signal with the original GAPF assay that disappear with the addition of 50% formamide. The open rectangle labeled bgs depicts the area subjected to bisulfite genomic sequence analysis shown in C. (C) Bisulfite DNA sequence analysis of the nonpalindromic loci in B shows dense CpG methylation in Colo compared to HDF. Solid circles represent CpG methylation, and open circles depict unmodified CpG dinucleotides. (D) Bisulfite sequence analysis of DAMD-positive loci in HCT116 cells shows heavy CpG DNA methylation. Individual clones are shown for HCT116 (HCT) and DKO.
Denaturation Analysis of Methylation Differences (DAMD) Identifies Methylated Loci in the Colon Cancer Cell Line HCT116.
To determine whether the higher Tm of methylated DNA can effectively be used to identify regions of differential DNA methylation genomewide, we modified the original GAPF assay to identify a range of Tm differences by performing the denaturation under two conditions: (i) 100 mM NaCl and (ii) 100 mM NaCl with 0.5% formamide, both at 100°C. Similar to the original GAPF procedure, the denatured DNA was rapidly cooled, subjected to S1 nuclease digestion, amplified by ligation-mediated PCR, labeled, and applied to a tiling array. Because we were interested in identifying differential methylation in CpG islands, we used the Affymetrix GeneChip Human Promoter 1.0R Array consisting of >25,500 promoter regions with an average coverage from −7.5 to +2.45 kb relative to the transcriptional start site. As a test of the ability of this assay to identify differentially methylated regions, we compared the signal from the colorectal cancer cell line HCT116 to a double DNA methyltransferase knockout (DKO) derivative that was generated by disrupting DNMT1 and DNMT3b, reducing global DNA methylation ∼95% (9). We obtained positive signals (HCT116 > DKO) [log2(signal ratio) > 1.2 and P < 0.001] in the promoter regions of 805 genes, 563 from the 100-mM NaCl sample (Table S1) and 455 from the 100-mM NaCl 0.5% formamide sample (Table S2). No negative signals (DKO > HCT116) were identified.
The positive signals showed a strong correlation with regions previously known to be hypermethylated in HCT116 relative to the DKO line. For example, the TIMP3 gene is methylated in HCT116 cells and unmethylated in DKO cells (9), and our assay shows a positive signal in the TIMP3 promoter, as well as other loci known to be methylated in HCT116, such as SEZ6L (10), SFRP1 (10), SFRP5 (10), GATA4 (11), GATA5 (11), INHIBINα (11), NEURL (12), HOXD1 (12, 13), HIC1 (14), RASGRF2 (13), and CHFR (15) (Fig. S1). We confirmed CpG methylation at the DAMD-positive loci by randomly choosing five loci for bisulfite sequence analysis (CXCL12, HDGFRP3, NPTX1, SOX7, and UCHL1). All were heavily methylated in HCT116 compared to DKO (Fig. 2D). These data demonstrate that the DAMD assay can be used to identify differentially methylated loci in genomewide screens.
Comparison of DAMD to Methylated DNA Immunoprecipitation (MeDIP) and the Methyl-CpG Binding Domain (MBD).
The 805 DAMD-positive promoter regions identified represent a substantially larger number of differentially methylated promoters than identified in a study comparing the same cell types using MeDIP (13). One possibility for this finding is that the denaturation conditions used in the Jacinto study (95°C for 10 min) may not have been sufficient to completely denature these heavily methylated regions of the genome. Because the antibody to 5-methylcytosine recognizes only single-stranded DNA (16), these regions would not have been captured by the antibody.
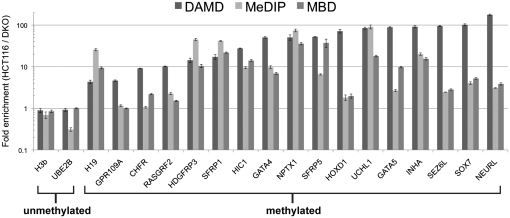
To directly compare the DAMD assay to other genomewide DNA methylation techniques, we interrogated the methylation state of 19 loci (2 unmethylated controls and 17 methylated loci) from HCT116 and DKO cells, using the MeDIP method (17) and another affinity-based method that utilizes the MBD from the human MBD2 protein (18). The methylated loci were chosen on the basis of previous validation by bisulfite sequence- or methylation-specific PCR analysis in HCT116 and DKO cells in prior publications (10–15, 19). For MeDIP, the sonicated input DNA was fully denatured by incubating at 100°C in Tris-EDTA (TE) for 10 min with rapid cooling in an ice water bath (17). Each method was performed in triplicate, and quantitative real-time PCR (qPCR) for each locus was used to determine fold enrichment of HCT116 compared to DKO. For the 2 unmethylated loci tested, all three methods did not show any enrichment (Fig. 3). For the methylated loci, however, the DAMD assay had significantly higher enrichment for the majority of loci tested compared to the other two methods (Fig. 3 and Table S3). All of the methylated loci tested contain CpG islands classified as high-CpG promoters (HCPs) (as described in ref. 20), with the exception of the H19 locus, which has an intermediate-CpG promoter (ICP).
Fig. 3.
Comparison of DAMD vs. MeDIP vs. MBD. Quantitative real-time PCR (qPCR) analysis of 19 loci (2 unmethylated, 17 methylated) depicting fold enrichment (HCT116/DKO) for each method is shown. Each method was performed in triplicate, and mean and standard deviation for the fold enrichment are displayed on a log10 scale.
Common Regions of CpG DNA Methylation in Primary Medulloblastoma Samples Detected Using DAMD.
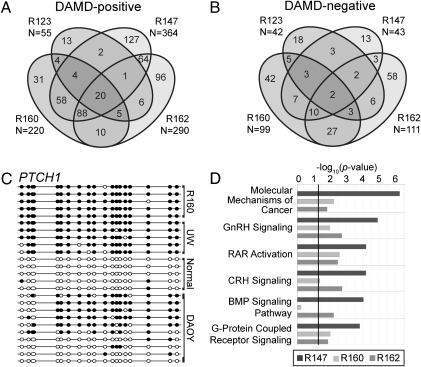
To test the hypothesis that DAMD can be used to identify cancer-specific methylation changes from patient samples, we analyzed four medulloblastoma biopsy specimens using the same two denaturation conditions as in the HCT116/DKO experiments and the Affymetrix Human Promoter array. Comparison of the medulloblastoma signals to adult normal cerebellum as a reference identified both DAMD-positive and DAMD-negative regions, some of which were shared between individual tumor samples (Fig. 4, A and B, and Table S4). Among the loci identified as DAMD-positive were members of the Notch-Hes, Sonic hedgehog (Shh), and Wingless (Wnt) pathways, pathways implicated in both normal development and the pathogenesis of medulloblastoma (21). For example, PRDM8, a negative regulator of the Notch-Hes pathway (22), AXIN2, a negative regulator of the Wnt pathway (23), and both PTCH1 and HIC1, negative regulators of the Shh pathway (24), were DAMD-positive. HIC1 is known to be frequently methylated in medulloblastoma (25) and was DAMD-positive in all four samples. PTCH1 methylation, which has not been previously identified in human medulloblastoma, was DAMD-positive in three of the samples and confirmed by bisulfite sequencing in one of the samples (Fig. 4C). Gene ontology analysis on the three tumors with the highest number of DAMD-positive loci identified the retinoic acid receptor (RAR) and bone morphogenetic protein (BMP) pathways as subject to methylation in the medulloblastomas (Fig. 4D and Table S5). Therefore, the DAMD assay identifies common regions of methylation in multiple pathways critical for cell differentiation, indicating that DNA methylation has a critical role in preventing differentiation in these pediatric cancers.
Fig. 4.
DAMD identifies common loci among primary medulloblastoma samples. (A) DAMD-positive and (B) DAMD-negative genes are depicted from four primary medulloblastoma samples (R123, R147, R160, and R162). Cerebellum from one normal individual was used as a control. The total number of DAMD-positive or DAMD-negative loci for each sample is shown, and common regions between the four samples are depicted on the Venn diagram. (C) Bisulfite sequence analysis of the PTCH1-1C DAMD-positive promoter region is shown for one of the medulloblastoma samples (R160), two human medulloblastoma cell lines [UW228 (UW) and DAOY], and the normal cerebellum control. Solid circles represent CpG methylation, and open circles depict unmodified CpG dinucleotides. (D) Gene ontology analysis of DAMD-positive loci in R147, R160, and R162 medulloblastoma samples. The top six canonical pathways (Ingenuity Pathways Analysis software) enriched are shown. The solid line represents a threshold of P < 0.05. GnRH, gonadotropin-releasing hormone; RAR, retinoic acid receptor; CRH, corticotropin-releasing hormone; BMP, bone morphogenetic protein.
In our previous study of DNA palindromes in cancer, we found common genomic regions between different medulloblastoma samples that scored as positive using the original GAPF assay (3), which we now know identifies both palindromes and regions of differential DNA methylation. To determine whether the common signals in the current analysis were due to methylation or palindromes, we performed the modified GAPF (with 50% formamide) on one of the primary medulloblastoma patient samples (R147) and used adult normal cerebellum as a control. Of the 364 DAMD-positive loci, only 4 were positive with the formamide-modified GAPF assay (AQP12A, DDT, TCEB3C, and TPSG1). Interestingly, 3 of these 4 loci map to DNA inverted repeats in the genome (26), and 1 of these (DDT) maps to a recently identified region of copy number variation (27). Thus, these results indicate that nearly all of the positive loci identified with the DAMD denaturation conditions, and with the original GAPF protocol that did not include 50% formamide, represent differential DNA methylation.
DNA Methylation Suppresses Expression of Developmental Genes in Medulloblastoma.
In a previous study, PTCH1 mRNA expression was decreased, with concomitant Shh pathway activation, in a subset of medulloblastoma patient samples. Bisulfite sequence analysis of the PTCH1-1B promoter region (CpG island 2 in Fig. 5) failed to show any CpG hypermethylation (28). The DAMD-positive signal, however, mapped to the PTCH1-1C promoter region (CpG island 1 in Fig. 5), which was not evaluated in the previous study, implying that methylation of this promoter region could be responsible for silencing the PTCH1 gene. To determine whether methylation suppresses PTCH1 expression in medulloblastoma, we screened four medulloblastoma cell lines (D283, D341, DAOY, and UW228) and found that three of the four cell lines (D283, D341, and UW228) displayed a similar level of DNA hypermethylation of the PTCH1-1C promoter region compared to the primary patient sample (Fig. 4C and Fig. S2). Interestingly, DAOY had a methylation pattern consistent with one PTCH1 allele being hypermethylated (Fig. 4C). This pattern has been observed in mouse models of medulloblastoma where the expressed PTCH1 allele harbors an inactivating mutation, whereas the wild-type PTCH1 allele is epigenetically silenced by DNA methylation (29), which suggests that a similar process occurs in human medulloblastoma.
Fig. 5.
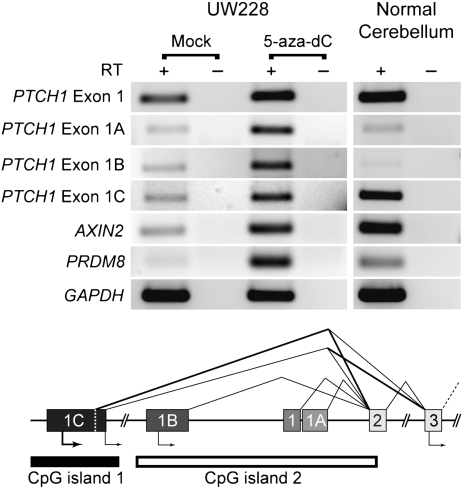
PTCH1 repression is alleviated by 5-aza-2′-deoxycytidine treatment. Reverse transcriptase PCR (RT-PCR) analysis is shown of PTCH1 isoforms, AXIN2, and PRDM8 from the human medulloblastoma cell line UW228 and normal cerebellum. UW228 was treated with either 5 μM 5-aza-2′-deoxycytidine (5-aza-dC) or vehicle alone (DMSO, Mock) for 72 h and RNA was analyzed by RT-PCR (SI Materials and Methods). −, no RT control. A schematic of the PTCH1 promoter region with alternative splicing events and CpG islands 1 (analyzed in Fig. 4C) and 2 (evaluated in ref. 28) is depicted (adapted from ref. 42).
To determine whether methylation suppresses PTCH1 mRNA expression, we performed RT-PCR analysis of RNA from UW228 cells treated with the DNA methyltransferase inhibitor 5-aza-2′-deoxycytidine (5-aza-dC), as well as of RNA obtained from normal adult cerebellum. When compared to normal cerebellum, the mock-treated UW228 cells demonstrated decreased expression of the PTCH1-1C isoform, which was partially restored with treatment of 5-aza-dC (SI Materials and Methods and Fig. 5). In addition, all PTCH1 isoforms were modestly increased with 5-aza-dC treatment and two other genes that were identified by DAMD as being hypermethylated in primary medulloblastoma samples and UW228, AXIN2 (two of four primary samples) and PRDM8 (all four primary samples), also showed increased expression with 5-aza-dC treatment. In summary, although genetic alterations of PTCH1 have been described in human medulloblastomas (30, 31), this is a demonstration that epigenetic suppression of PTCH1 expression by DNA methylation occurs in this disease.
Discussion
In this report, we show that an assay originally designed to identify genomic regions of tissue-specific DNA palindrome formation (3) also enriches for differential CpG DNA methylation. By modifying the denaturation conditions, the assay can be made more specific for DNA palindromes (GAPF) or enrich for different states of CpG methylation (DAMD), thereby allowing for the identification of cancer-specific genomic and epigenomic alterations.
We directly compared DAMD to MeDIP and another affinity-based purification method using a MBD protein. For the majority of the loci tested, DAMD provided a higher level of enrichment than the other two methods. It should be noted, however, that all but one of the methylated loci are classified as HCPs, and the one locus (H19) that falls in the ICP category had the lowest enrichment for DAMD (approximately fourfold). Like MeDIP and MBD, DAMD does not depend on the presence and optimal spacing of methylation-sensitive restriction enzyme recognition sites as used in the CHARM (32) and HELP (33) assays. One potential advantage for the detection of DNA methylation by DAMD is that the assay, in principle, can detect DNA methylation changes from a limited number of cancer cells in a background of normal cells or tissue following optimization of the denaturation conditions to achieve complete digestion of the unmethylated allele but not the methylated allele.
In this study, we used conditions to identify heavily methylated CpG-rich regions; however, it should be possible to “tune” DAMD to enrich for different amounts of DNA methylation at a broad range of CpG densities across the genome. By adjusting salt concentration, denaturation temperature, and formamide concentration, the assay has the potential to identify a gradient of CpG methylation densities, such as those found at ICPs and LCPs. Similarly, the addition of a high concentration of formamide allowed us to make the GAPF assay more specific for DNA palindromes by increasing the efficiency of the DNA denaturation step. It is important to note, however, that our previous study using the original GAPF procedure (3) identified both palindromes and regions of differential methylation and needs to be interpreted with these current findings in mind.
How does DAMD enrich for regions of differentially methylated DNA? The melting temperature (Tm) of DNA is primarily determined by sequence composition (34). It previously has been observed that cytosine methylation at the C-5 position increases the melting temperature (Tm) of naked DNA (5, 6). It is therefore likely that DAMD enriches for differential DNA methylation on the basis of the increase in Tm caused by methylated cytosine, and this conclusion is supported by the highly increased density of CpG methylation at DAMD-positive loci as determined by bisulfite sequence analysis. It has been hypothesized that the increase in the stability of duplex DNA caused by cytosine methylation is a result of changes in base–base stacking interactions (35). This effect of methylated cytosine on duplex DNA has previously been used to detect methylation patterns of specific loci by using denaturing gradient gel electrophoresis (36), but this technique is not amenable to genomewide studies. DAMD takes advantage of this biochemical property of methylated cytosine to detect differential DNA methylation in a genomewide assay. Although the bisulfite sequencing confirms that the DAMD-positive loci are differentially CpG methylated, we recognize that other modifications, such as the recently described 5-hydroxymethylcytosine (37, 38), might also alter the Tm and contribute to the DAMD-positive signal and protect against bisulfite modification, which will require additional studies.
Our analysis of primary medulloblastoma samples indicates that the DAMD assay can identify biomarkers of disease. We discovered that PTCH1 can be inactivated in medulloblastoma through DNA hypermethylation. PTCH1 has previously been shown to have a number of isoforms generated by alternative use of different first exons (39–41). Interestingly, the hypermethylated CpG island identified by DAMD is adjacent to the promoter for the PTCH1-1C isoform, and expression of this isoform is abundant in normal cerebellum, although repressed in a medulloblastoma cell line. This repression can be partially alleviated by treating cells with 5-aza-dC. A previously published analysis of the function of the different PTCH1 isoforms demonstrated that the PTCH1-1B and -1C isoforms were the most potent inhibitors of the Shh pathway, acting at the level of inhibiting Smoothened and the transcription factors GLI1 and GLI2 (42). Together with our data, this suggests the expression of PTCH1-1C inhibits Shh signaling and cellular growth in normal cerebellar development, whereas in a subset of medulloblastomas PTCH1 is inactivated by transcriptional silencing through DNA methylation of the PTCH1-1C promoter. Very recently, a paper describing the response of metastatic medulloblastoma to an inhibitor of the Shh signaling pathway was reported (43). Our study indicates that an inhibitor of DNA methylation, such as 5-aza-dC, which readily crosses the blood–brain barrier, might result in the reexpression of PTCH1 and other developmental genes in a potentially large subset of medulloblastoma patients and improve existing therapies. Supporting this strategy, a recent preclinical study in a Ptch+/− mouse model of medulloblastoma and rhabdomyosarcoma treated mice with 5-aza-dC combined with the histone deacetylase inhibitor valproic acid and found that the previously hypermethylated wild-type Ptch allele was reactivated and tumor formation was efficiently prevented (44). Assessing the methylation status of PTCH1 and other developmental genes in the RAR and BMP pathways may aid in selecting which patients will benefit from this type of epigenetically targeted therapy.
Gene ontology analysis on the three tumors with the highest number of DAMD-positive loci identified members of the RAR and BMP signaling pathways (Fig. 4D and Table S5). Two tumors had methylated promoters for genes that have a normal role in the inhibition of BMP signaling (Chordin, Smurf1, Erk1/2, and Smad7). Preclinical studies have shown that isotretinoin and all-trans retinoic acid (ATRA) induce programmed cell death in a subset of medulloblastoma cells through a mechanism that involves induction of BMP2-mediated phosphorylation of p38 MAP kinase (45) and that resistant medulloblastoma cells failed to express BMP2 in response to retinoids. In that study, retinoid-resistant medulloblastoma cells could be induced to undergo apoptosis if treated with BMP-2, indicating that regulation of the BMP pathway is a central mechanism determining retinoid sensitivity or resistance in medulloblastoma. In this regard, it is very interesting that our current study shows that a subset of medulloblastomas have methylated components of the BMP signaling pathway. A phase III treatment study (ACNS0332) is currently being conducted by the Children’s Oncology Group to determine whether isotretinoin increases long-term event-free survival for high-risk medulloblastoma patients. Our study suggests that it might be informative to determine whether components of the BMP and RAR pathways are methylated in tumors that respond, or do not respond, to ATRA. In a broader context, the development of clinical assays based on DAMD may aid in early detection of disease, disease diagnosis, measurement of response to treatment, and evaluation of minimal residual disease monitoring for disease recurrence.
Materials and Methods
Ethics Statement.
Preexisting patient de-identified samples were obtained in accordance with IRB protocol.
DAMD Assay.
Genomic DNA was isolated from cells (Qiagen Blood and Cell Culture DNA kit), and a total of 2 μg of genomic DNA was used as starting material for the DAMD assay. The sample was evenly split and 1 μg was digested with KpnI (10 units) and 1 μg was digested with SbfI (10 units) for at least 8 h in a total volume of 20 μL for each digestion. Enzymes were heat inactivated at 65°C for 20 min, and digests were combined and then split between two tubes. To the 20-μL DNA mixture, 27.36 μL of water and 1.64 μL of 3 M NaCl were added. For the formamide variation of the protocol to enrich for DNA palindromes, formamide was added to a final concentration of 50% before DNA denaturation. Denaturation was performed by boiling samples in a water bath for 7 min followed by rapid cooling using an ice-water bath. S1 (Invitrogen) digestion was performed by adding 6 μL 10× S1 nuclease buffer, 4 μL 3 M NaCl, and 1 μL of S1 (diluted to 100 units/μL using S1 dilution buffer). Samples were incubated for 60 min at 37°C. S1 was inactivated by extraction with phenol followed by a phenol:chloroform extraction. DNA was ethanol precipitated in the presence of 20 μg of glycogen, and the DNA pellet was resuspended in 80 μL of 1/10 TE. The sample was divided evenly into two tubes, one subjected to digestion with MseI (40 units) and the other tube with MspI (40 units) for at least 4 h at 37°C (final volume of each digestion was 50 μL). Restriction enzymes were heat inactivated at 65°C for 20 min. Ligation-mediated PCR was performed as described (4). Amplified DNA was quantitated and 7.5 μg of DNA was fragmented: 44 μL DNA (7.5 μg total), 5 μL 10× DNase I buffer, and 1 μL DNase I (diluted to 0.017 unit in 1× DNase I buffer) for 35 min at 37°C and then heat inactivation at 95°C for 15 min. Fragmented DNA was labeled with biotin using the Affymetrix GeneChip Whole-Transcript Double-Stranded Target kit. See Table S6 for primers used in this study.
Tiling Array and Statistical Analysis.
Affymetrix Human Tiling 2.0R Arrays and 1.0R Promoter Arrays were analyzed using Tiling Array Software (v 1.1.02, Affymetrix). Raw data were scaled to a target intensity of 100 and normalized by quantile normalization. For probe analysis, a bandwidth of 250 bp was used and perfect match probes were used in a Wilcoxon rank sum two-sided test. Two independent replicates were used for sample and control unless otherwise stated. Signal and P-value thresholds are stated for each experiment. For all experiments, a maximum gap of ≤100 and minimum run of >30 bp were used. Data were visualized using the Integrated Genome Database Browser (v 5.12, Affymetrix). For the generation of gene lists, .bed files generated in the above analysis were imported into NimbleScan software (v 2.4), and a gene was denoted as positive if the region of interest mapped to −7 kb to +1.5 kb of the transcriptional start site. Gene ontology functional annotation for DAMD-positive loci from medulloblastoma samples was done using Ingenuity Pathways Analysis software (v 7.5).
Supplementary Material
Acknowledgments
We thank B. Vogelstein (Johns Hopkins University) for HCT116 and DKO cell lines. We thank D. Gottschling, S. Henikoff, C. Kemp, and P. Nelson for comments on the manuscript and C. Laird for insightful comments. This study was supported by the National Institute of Arthritis and Musculoskeletal and Skin Diseases Grants AR045113 and AR045203 and by Fred Hutchinson Cancer Research Center Pilot Funding from the Early Detection and Intervention Initiative (to S.J.T.). S.J.D. is an American Society for Clinical Oncology Young Investigator and a University of Washington Child Health Research Center Scholar (National Institutes of Health Grant K12 HD43376) and was supported by National Institutes of Health Grant 2T32CA009351.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 3.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE17224).
This article contains supporting information online at www.pnas.org/cgi/content/full/0907606106/DCSupplemental.
References
- 1.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka H, Bergstrom DA, Yao MC, Tapscott SJ. Widespread and nonrandom distribution of DNA palindromes in cancer cells provides a structural platform for subsequent gene amplification. Nat Genet. 2005;37:320–327. doi: 10.1038/ng1515. [DOI] [PubMed] [Google Scholar]
- 4.Tanaka H, et al. Intrastrand annealing leads to the formation of a large DNA palindrome and determines the boundaries of genomic amplification in human cancer. Mol Cell Biol. 2007;27:1993–2002. doi: 10.1128/MCB.01313-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ehrlich M, Ehrlich K, Mayo JA. Unusual properties of the DNA from Xanthomonas phage XP-12 in which 5-methylcytosine completely replaces cytosine. Biochim Biophys Acta. 1975;395:109–119. doi: 10.1016/0005-2787(75)90149-5. [DOI] [PubMed] [Google Scholar]
- 6.Gill GE, Mazrimas JA, Bishop CC., Jr. Physical studies on synthetic DNAs containing 5-methylcytosine. Biochim Biophys Acta. 1974;335:330–348. [Google Scholar]
- 7.Szer W, Shugar D. The structure of poly-5-methylcytidylic acid and its twin-stranded complex with poly-inosinic acid. J Mol Biol. 1966;17:174–187. doi: 10.1016/s0022-2836(66)80102-x. [DOI] [PubMed] [Google Scholar]
- 8.McConaughy BL, Laird CD, McCarthy BJ. Nucleic acid reassociation in formamide. Biochemistry. 1969;8:3289–3295. doi: 10.1021/bi00836a024. [DOI] [PubMed] [Google Scholar]
- 9.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 10.Suzuki H, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 11.Akiyama Y, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schuebel KE, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–1723. doi: 10.1371/journal.pgen.0030157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jacinto FV, Ballestar E, Ropero S, Esteller M. Discovery of epigenetically silenced genes by methylated DNA immunoprecipitation in colon cancer cells. Cancer Res. 2007;67:11481–11486. doi: 10.1158/0008-5472.CAN-07-2687. [DOI] [PubMed] [Google Scholar]
- 14.Arnold CN, Goel A, Boland CR. Role of hMLH1 promoter hypermethylation in drug resistance to 5-fluorouracil in colorectal cancer cell lines. Int J Cancer. 2003;106:66–73. doi: 10.1002/ijc.11176. [DOI] [PubMed] [Google Scholar]
- 15.Toyota M, et al. Epigenetic inactivation of CHFR in human tumors. Proc Natl Acad Sci USA. 2003;100:7818–7823. doi: 10.1073/pnas.1337066100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reynaud C, et al. Monitoring of urinary excretion of modified nucleosides in cancer patients using a set of six monoclonal antibodies. Cancer Lett. 1992;61:255–262. doi: 10.1016/0304-3835(92)90296-8. [DOI] [PubMed] [Google Scholar]
- 17.Mohn F, Weber M, Schübeler D, Roloff T-C. Methylated DNA immunoprecipitation (MeDIP) Methods Mol Biol. 2009;507:55–64. doi: 10.1007/978-1-59745-522-0_5. [DOI] [PubMed] [Google Scholar]
- 18.Gebhard C, et al. Genome-wide profiling of CpG methylation identifies novel targets of aberrant hypermethylation in myeloid leukemia. Cancer Res. 2006;66:6118–6128. doi: 10.1158/0008-5472.CAN-06-0376. [DOI] [PubMed] [Google Scholar]
- 19.Zhang Y, et al. SOX7, down-regulated in colorectal cancer, induces apoptosis and inhibits proliferation of colorectal cancer cells. Cancer Lett. 2009;277:29–37. doi: 10.1016/j.canlet.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 20.Weber M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007;39:457–466. doi: 10.1038/ng1990. [DOI] [PubMed] [Google Scholar]
- 21.Gilbertson RJ. Medulloblastoma: Signalling a change in treatment. Lancet Oncol. 2004;5:209–218. doi: 10.1016/S1470-2045(04)01424-X. [DOI] [PubMed] [Google Scholar]
- 22.Kinameri E, et al. Prdm proto-oncogene transcription factor family expression and interaction with the Notch-Hes pathway in mouse neurogenesis. PLoS ONE. 2008;3:e3859. doi: 10.1371/journal.pone.0003859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu W, et al. Mutations in AXIN2 cause colorectal cancer with defective mismatch repair by activating beta-catenin/TCF signalling. Nat Genet. 2000;26:146–147. doi: 10.1038/79859. [DOI] [PubMed] [Google Scholar]
- 24.Briggs KJ, et al. Cooperation between the Hic1 and Ptch1 tumor suppressors in medulloblastoma. Genes Dev. 2008;22:770–785. doi: 10.1101/gad.1640908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rood BR, Zhang H, Weitman DM, Cogen PH. Hypermethylation of HIC-1 and 17p allelic loss in medulloblastoma. Cancer Res. 2002;62:3794–3797. [PubMed] [Google Scholar]
- 26.Warburton PE, Giordano J, Cheung F, Gelfand Y, Benson G. Inverted repeat structure of the human genome: The X-chromosome contains a preponderance of large, highly homologous inverted repeats that contain testes genes. Genome Res. 2004;14(10A):1861–1869. doi: 10.1101/gr.2542904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhao Y, Marotta M, Eichler EE, Eng C, Tanaka H. Linkage disequilibrium between two high-frequency deletion polymorphisms: Implications for association studies involving the glutathione-S transferase (GST) genes. PLoS Genet. 2009;5:e1000472. doi: 10.1371/journal.pgen.1000472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritchard JI, Olson JM. Methylation of PTCH1, the Patched-1 gene, in a panel of primary medulloblastomas. Cancer Genet Cytogenet. 2008;180:47–50. doi: 10.1016/j.cancergencyto.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uhmann A, et al. A model for PTCH1/Ptch1-associated tumors comprising mutational inactivation and gene silencing. Int J Oncol. 2005;27:1567–1575. [PubMed] [Google Scholar]
- 30.Pietsch T, et al. Medulloblastomas of the desmoplastic variant carry mutations of the human homologue of Drosophila patched. Cancer Res. 1997;57:2085–2088. [PubMed] [Google Scholar]
- 31.Raffel C, et al. Sporadic medulloblastomas contain PTCH mutations. Cancer Res. 1997;57:842–845. [PubMed] [Google Scholar]
- 32.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–790. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khulan B, et al. Comparative isoschizomer profiling of cytosine methylation: The HELP assay. Genome Res. 2006;16:1046–1055. doi: 10.1101/gr.5273806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Howley PM, Israel MA, Law MF, Martin MA. A rapid method for detecting and mapping homology between heterologous DNAs. Evaluation of polyomavirus genomes. J Biol Chem. 1979;254:4876–4883. [PubMed] [Google Scholar]
- 35.Aradi F. Effect of methylation on the pyrimidine-pyrimidine stacking interaction studied by (1)H NMR chemical shift. Biophys Chem. 1995;54:67–73. doi: 10.1016/0301-4622(94)00115-z. [DOI] [PubMed] [Google Scholar]
- 36.Collins M, Myers RM. Alterations in DNA helix stability due to base modifications can be evaluated using denaturing gradient gel electrophoresis. J Mol Biol. 1987;198:737–744. doi: 10.1016/0022-2836(87)90214-2. [DOI] [PubMed] [Google Scholar]
- 37.Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929–930. doi: 10.1126/science.1169786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tahiliani M, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kogerman P, et al. Alternative first exons of PTCH1 are differentially regulated in vivo and may confer different functions to the PTCH1 protein. Oncogene. 2002;21:6007–6016. doi: 10.1038/sj.onc.1205865. [DOI] [PubMed] [Google Scholar]
- 40.Nagao K, et al. Identification and characterization of multiple isoforms of a murine and human tumor suppressor, patched, having distinct first exons. Genomics. 2005;85:462–471. doi: 10.1016/j.ygeno.2004.11.014. [DOI] [PubMed] [Google Scholar]
- 41.Shimokawa T, Rahnama F, Zaphiropoulos PG. A novel first exon of the Patched1 gene is upregulated by Hedgehog signaling resulting in a protein with pathway inhibitory functions. FEBS Lett. 2004;578:157–162. doi: 10.1016/j.febslet.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 42.Shimokawa T, et al. Distinct roles of first exon variants of the tumor-suppressor Patched1 in Hedgehog signaling. Oncogene. 2007;26:4889–4896. doi: 10.1038/sj.onc.1210301. [DOI] [PubMed] [Google Scholar]
- 43.Rudin CM, et al. Treatment of medulloblastoma with hedgehog pathway inhibitor GDC-0449. N Engl J Med. 2009;361:1173–1178. doi: 10.1056/NEJMoa0902903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ecke I, et al. Antitumor effects of a combined 5-aza-2′deoxycytidine and valproic acid treatment on rhabdomyosarcoma and medulloblastoma in Ptch mutant mice. Cancer Res. 2009;69:887–895. doi: 10.1158/0008-5472.CAN-08-0946. [DOI] [PubMed] [Google Scholar]
- 45.Hallahan AR, et al. BMP-2 mediates retinoid-induced apoptosis in medulloblastoma cells through a paracrine effect. Nat Med. 2003;9:1033–1038. doi: 10.1038/nm904. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.