Abstract
To establish compatible rhizobial-legume symbioses, plant roots support bacterial infection via host-derived infection threads (ITs). Here, we report the requirement of plant flotillin-like genes (FLOTs) in Sinorhizobium meliloti infection of its host legume Medicago truncatula. Flotillins in other organisms have roles in viral pathogenesis, endocytosis, and membrane shaping. We identified seven FLOT genes in the M. truncatula genome and show that two, FLOT2 and FLOT4, are strongly up-regulated during early symbiotic events. This up-regulation depends on bacterial Nod Factor and the plant's ability to perceive Nod Factor. Microscopy data suggest that M. truncatula FLOT2 and FLOT4 localize to membrane microdomains. Upon rhizobial inoculation, FLOT4 uniquely becomes localized to the tips of elongating root hairs. Silencing FLOT2 and FLOT4 gene expression reveals a nonredundant requirement for both genes in IT initiation and nodule formation. FLOT4 is uniquely required for IT elongation, and FLOT4 localizes to IT membranes. This work reveals a critical role for plant flotillins in symbiotic bacterial infection.
Keywords: Medicago, plant membrane, Sinorhizobium, symbiosis
Symbiotic nitrogen-fixing rhizobial bacteria live in association with legume roots inside developmentally unique structures called nodules. Bacteria penetrate nodules via plant-derived infection threads (ITs). Invagination of the root hair plasma membrane during IT initiation resembles a partial endocytosis event where the membrane begins to bud but, instead of pinching off, elongates through the cell. As the IT finishes traversing the cell, bacteria release into the intercellular space. New membrane invagination and IT formation take place in the underlying cell layers. Eventually, the bacteria are released into host cells via an endocytosis-like event where they remain surrounded by a host-derived membrane (1, 2).
A bacterially produced lipochitooligosaccharide called “Nod Factor” (NF) is a species-specific rhizobial signal that is recognized by the legume host. NF promotes nodule development (3) and via signal transduction induces calcium spiking and transcriptional changes (4, 5). Forward genetic studies in rhizobial-legume systems have revealed genes required for host NF signal perception, signal transduction, and transcriptional changes (see ref. 6 and references therein). NF perception is mediated by LysM-family receptor-like kinases NFP and LYK3 (in Medicago truncatula; NFR1/NFR5 in Lotus japonicus). The NFP receptor induces calcium spiking via a signaling pathway that includes a Leucine-rich repeat receptor-like kinase and a putative ion channel. Calcium spiking appears to control activity of a calcium/calmodulin-dependent protein kinase and two downstream GRAS-family transcription factors, NSP1 and NSP2, which interact in the plant nucleus and directly bind the promoters of nodulation genes (7). NSP1 and NSP2 are necessary for all known NF-dependent transcriptional changes (5).
A second “entry” pathway (controlling bacterial entry into root hair cells) has been proposed that includes the high-stringency receptor encoded by LYK3. Null lyk3 mutants undergo a small number of cortical cell divisions and form no ITs. An additional transcriptional regulator NIN is required for nodule organogenesis and infection events but not NF-dependent transcriptional changes. Current evidence suggests that NIN may coordinate signals through both the signaling and entry pathways (8). Components of the proposed NF entry receptor pathway are currently limited to NIN and LYK3; how LYK3 signaling is perceived and translated into IT initiation is unknown.
Sinorhizobium meliloti, the nitrogen-fixing symbiont of Medicago (the alfalfa genus), is a close relative of the animal pathogen Brucella. Sinorhizobium and Brucella have common genes required for infection and invasion of their hosts (9). Given these common features, we asked whether eukaryotic host-cell factors required for infection might also be shared across kingdoms. Upon uptake into host cells, Brucella remains surrounded by a host-derived membrane, which contains the protein flotillin (10). Flotillins [also called Reggies (11)] are often used as markers for cholesterol-rich, detergent-resistant, membrane microdomains called “lipid rafts” and are now known to define a clathrin-independent, caveolin-independent endocytic pathway required for endocytosis of cholera toxin (12). By interacting with effectors that can bind actin, flotillins mediate membrane-shaping events including membrane budding, actin-mediated neuronal differentiation, and filopodia formation (13–15).
Because of the importance of flotillins in pathogenesis and membrane shaping, we wondered whether plant flotillin-like proteins might be involved in symbiotic events such as IT initiation and elongation, and the final endocytosis of bacteria into plant cells. Plant flotillin-like proteins, identified by sequence similarity to animal flotillins, are predicted to have many features of their mammalian counterparts including a similar tertiary structure, a conserved palmitoylation site at Cys-35, and a C-terminal coiled-coil domain (16). Many plant flotillin-like genes are annotated as nodulins (a generic name for a gene that is expressed uniquely in nodules) because of the early identification of a plant flotillin-like gene (GmNod53b) that is induced in soybean nodules (17). GmNod53b-like peptides have been isolated from the peribacteroid membranes in soybean and pea nodules (18, 19), and an Arabidopsis flotillin-like protein (named AtFlot1) was found in detergent-resistant membrane domains (20). These results imply that flotillins are conserved between plants and animals. The conserved subcellular localization suggests the possibility of conserved function(s). In this work, we employ a reverse genetics approach to investigate the role of the Medicago truncatula flotillin-like gene family (FLOTs) in symbiosis. We demonstrate that two family members, FLOT2 and FLOT4, are required for early symbiotic events and have nonredundant functions.
Results
Identification of Flotillin-like Genes in M. truncatula.
A search of the M. truncatula genome sequence yielded seven genomic regions with high homology (E-value < 1e−151) to the GmNod53b amino acid sequence, which we have designated FLOT1-7 (Table S1). All seven predicted ORFs have >85% identity on the nucleotide level (Fig. S1A). FLOT1-5 are located within a 30-kb region of chromosome 3 (Fig. S1B). Whereas all other plant flotillin-like proteins described thus far have a conserved predicted palmitoylation site at Cys-35 (16), FLOT4 is predicted to have a Tyr substitution at this residue (Fig. S1A). FLOT5 has no obvious translational start site and lacks a two-exon, one-intron structure. Only three FLOT genes are present in the Arabidopsis genome (16), implying an expansion of the FLOT family in M. truncatula.
FLOT2 and FLOT4 Show Nod Factor-Dependent Up-Regulation During Nodulation.
We assayed expression of M. truncatula FLOTs during nodulation by quantitative RT-PCR at times corresponding to key events in nodule development (21). FLOT2 and FLOT4 expression increases early in symbiosis with the wildtype S. meliloti strain Rm1021 (Fig. 1A). FLOT2 is up-regulated for the entire 21-day developmental time course, whereas FLOT4 expression returns to baseline by 7 days post inoculation (dpi). In contrast, neither FLOT1 nor FLOT3 expression changes during nodule development. We explored expression of FLOTs in diverse plant organs and found that FLOT4 expression is largely limited to roots and nodules, whereas FLOT2 has highest expression in flowers and green pods, and FLOT1 and FLOT3 expression was detected in all plant organs (Fig. S2). We could not detect expression of FLOT5 or FLOT7 in any plant tissue, including roots and nodules. FLOT6 expression was occasionally detected in roots and nodules at levels so low that it could not be consistently detected even in the same sample. The roles of FLOT5-7 in nodulation were therefore not explored further.
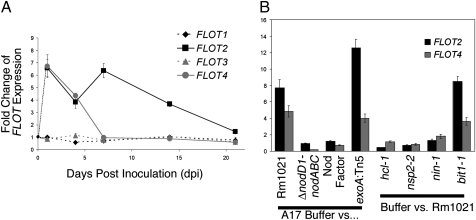
Fig. 1.
Expression of FLOTs is up-regulated during nodulation, and this regulation depends on Nod Factor. Expression of individual FLOT genes was measured by quantitative RT-PCR. Each FLOT expression level was normalized to an internal actin control. All data are the ratio of treated vs. buffer control of M. truncatula cv. Jemalong A17 plants or mutant derivatives. Each data point is an average of three replicates; error bars indicate standard error of the ratio. (A) FLOT1-4 expression in wildtype S. meliloti Rm1021 vs. buffer-treated A17 roots at 1, 4, 7 14, and 21 dpi. (B) FLOT2 and FLOT4 expression was evaluated in response to bacterial and plant mutants at 1 dpi.
A small group of plant transcripts is up-regulated in the first day of symbiosis; NF is necessary and sufficient for the majority of these early transcriptional changes (5). We asked whether NF was required for up-regulation of FLOT2 and FLOT4 at 1 dpi. An S. meliloti mutant unable to synthesize NF (ΔnodD1-nodABC) failed to up-regulate FLOT2 and FLOT4 at 1 dpi, which implies that NF is necessary, but purified NF was not sufficient for up-regulation (Fig. 1B). We wondered whether a second bacterial component such as a cell-surface polysaccharide was also necessary for FLOT induction. Mutants deficient in synthesis of succinoglycan (exoA:Tn5), lipopolysaccharide (lpsB:Tn5), cyclic β-1,2-glucan (ndvB:Tn5), and a succinoglycan overproducer (exoX:Tn5) caused up-regulation of FLOT2 and FLOT4 (Fig. 1B and Fig. S3), suggesting that known cell-surface polysaccharides are not required.
Evidence suggests that NF effects are transduced via a “signaling” pathway and a distinct “entry” pathway (6). The most downstream component of the signaling pathway, transcription factor NSP2, is required for up-regulation of FLOT2 and FLOT4 at 1 dpi (Fig. 1B). The parallel entry NF perception pathway, defined by LYK3, is also required for up-regulation of FLOT2 and FLOT4 at 1 dpi (Fig. 1B), as is the transcriptional regulator NIN, which is common to both signaling and entry pathways (Fig. 1B). However, ERN (BIT1), an ERF-like transcription factor required for up-regulation of a subset of early nodulins but not for IT initiation (22), is not required (Fig. 1B). NF perception by both signaling and entry NF receptors and signal transduction via NSP2 and NIN are required for up-regulation of FLOT2 and FLOT4 at 1 dpi.
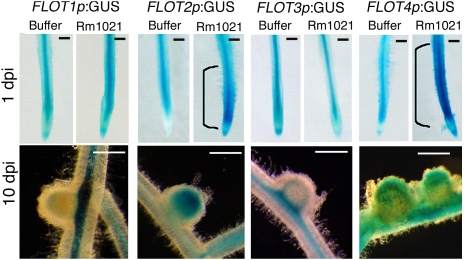
We asked whether FLOT2 and FLOT4 were up-regulated specifically in the plant cells that become infected by S. meliloti. Using transcriptional fusions consisting of FLOT promoters and the GUS reporter gene, we found that in uninoculated roots, FLOT1-4 are primarily expressed in vascular tissue (Fig. 2), and FLOT4 is also weakly expressed in elongating root hairs (Fig. S4). Upon inoculation with Rm1021, FLOT2 and FLOT4 show an expansion of expression in the root cortex in the region of elongating root hairs (Fig. 2 and Fig. S4), which will eventually become colonized by bacteria (1). In nodules, FLOT2 and FLOT4 are expressed in the infection zone (Fig. 2 Lower). In contrast, FLOT1 and FLOT3 show little change in their spatial expression or in intensity of expression in roots upon inoculation, and expression is limited to the nodule vascular tissue.
Fig. 2.
FLOT2 and FLOT4 are expressed in the root elongation zone upon inoculation and in the infection zone of nodules. A17 plants were transformed to generate hairy roots expressing FLOT1-4 promoter-GUS fusions; GUS activity is shown for the indicated strains and times. Ten transgenic lines were observed for each construct at each time point. (Scale bars: 1 mm.) Representative roots are shown.
FLOT2 and FLOT4 Localize to Membrane Microdomains and FLOT4 Becomes Polarly Localized in Response to Bacterial Signals.
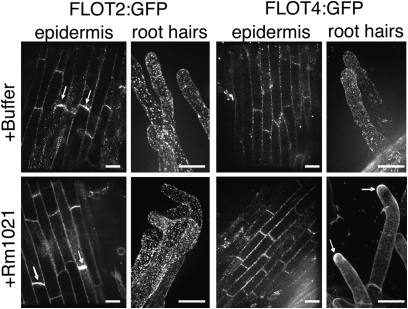
To study the localization and dynamics of FLOT2 and FLOT4 during nodulation, we constructed FLOT:GFP fusions using the FLOT genomic sequence (promoter and intron). FLOT2:GFP was not visible under control of its native promoter, so we expressed FLOT2 using the CaMV 35S promoter. In root cells, signals from both FLOT2:GFP and FLOT4:GFP appear punctate and colocalize with FM4-64, a plasma membrane marker (Fig. S5). In the absence of bacteria, FLOT4:GFP and FLOT2:GFP puncta are evenly distributed in the plasma membrane of root hair cells (Fig. 3). Upon inoculation with S. meliloti, FLOT4:GFP accumulates in the tips of elongating root hairs, whereas FLOT2:GFP puncta remain evenly distributed throughout root hair plasma membranes (Fig. 3). FLOT2:GFP is polarly localized in uninoculated and inoculated epidermal cells, whereas FLOT4:GFP is not (Fig. 3).
Fig. 3.
FLOTs localize to membrane-associated puncta and become polar after inoculation. We generated A17 hairy roots expressing 35S:FLOT2:GFP or FLOT4:GFP driven by its native promoter. Transgenic roots were visualized by using a spinning-disk confocal microscope. (Scale bars: 15 μm.) FLOT4:GFP and FLOT2:GFP are visibly punctate and evenly distributed in the membranes of uninoculated root hair cells. FLOT4:GFP puncta localize to root hair tips by 1 dpi, whereas the localization of FLOT2:GFP does not change upon inoculation. FLOT2:GFP is weakly polar in uninoculated epidermal cells (arrows) and remains polar on inoculation. At least 15 transgenic lines were observed for each treatment. Representative images are shown. Root hair images are overlays of 100 sections, taken at 0.2-μm increments.
FLOTs Are Required for Nodulation.
We tested FLOT function using RNA interference (RNAi) or artificial micro RNAs (amiRNAs) to silence FLOT expression and found that FLOTs are required for symbiosis (Fig. 4 and Fig. S6). We targeted FLOT2 and FLOT4, which are regulated during nodulation, and the constitutively expressed FLOT3 as a control [FLOT1 was also silenced and gave similar nodulation results as FLOT3 (Fig. S6)]. We confirmed efficacy of silencing by qRT-PCR at 21 dpi, a time point when no FLOTs are significantly up-regulated in inoculated hairy roots (Fig. S6C). Constructs are designated by their primary target gene (>50% reduction in expression); numbers in parentheses show genes that have partial reduction in expression due to cross silencing. The most dramatic phenotype of the FLOT-silenced lines was observed when FLOT2,3, and 4 are silenced as a group [FLOT2,3,4 RNAi (Fig. 4B); this construct also silences FLOT1 (Fig. S6 A and B)]. Roots transformed with the empty vector formed nodules 87% of the time, whereas only half of the FLOT2,3,4 RNAi roots formed nodules. Pink nodules were observed on control plants >25% of the time compared to only 2.5% for FLOT2,3,4 RNAi roots. On average, control roots formed approximately six nodules, whereas FLOT2,3,4 RNAi roots formed an average of only two nodules. FLOT2,3,4 RNAi roots also had altered morphology, including a decrease in primary root length, an increase in the number of primary and secondary lateral roots (Fig. S6A), and a reduction in root weight (Fig. S6D). These results indicate that FLOTs are required for symbiosis and normal root development.
Fig. 4.
Silencing FLOTs results in a decrease in nodule number, a decrease in infection events, and nodules that do form are nonfunctional. (A) Expression of FLOT2, FLOT3, and FLOT4 in individual hairy roots expressing the indicated RNAi or amiRNA construct was assessed by qRT-PCR, normalized to an internal actin control, and then to expression in plants transformed with the empty vector (average of at least 10 roots). Constructs are designated by their primary target gene(s) (*, >50% reduction in expression); numbers in parentheses show genes that have partial but significant (#, P < 0.05) reduction in expression due to cross silencing. (B) Nodulation phenotypes were assayed from a minimum of three biological replicates representing 50–100 plants per construct. (C) Acetylene reduction assays were performed at 21 dpi to determine the efficacy of nodules that form (14-20 plants were assayed per silenced line). (D) Hairy roots were stained for Rm1021 expressing pHemA:lacZ and stained for lacZ activity at 7 dpi to visualize infection events. Infection events and nodules were scored on ten transformed plants per amiRNA or RNAi and normalized to root weight (Fig. S6D). Between 104 (FLOT2,3,4 RNAi) and 371 (empty vector) total infection events were observed per line. (E) ITs on silenced roots (from Fig. 4C) were scored as aborting in the root hair, penetrating the hypodermal/cortical cell layers but not releasing bacteria into cortical cells, or releasing bacteria into cortical cells. Release was inferred by a blue haze in the nodule cortex. (A–E) Statistical significance was determined by pairwise two-tailed t tests comparing silenced lines to the empty vector control (*#, P < 0.05). (F–I) Abortive ITs in plants transformed with the FLOT4 amiRNA construct (F and G) compared to control ITs (H and I). (Scale bars: 15 μm.) (J) Linear regressions were conducted on plants described in A to determine whether a correlation exists between expression of FLOTs and nodule number. FLOT2 expression level has a strong linear relationship with nodule number (y = 5.1x + 2.7; P intercept = 0.005; P slope = 3 × 10−5); FLOT4 expression level has a weaker but statistically significant correlation (y = 2.0x + 5.8; P intercept = 3 × 10−8; P slope = 0.02). FLOT3 expression does correlates with nodule number (P slope = 0.6). FLOT2 and FLOT4 expression have additive effects (y = 7.1x + 0.9; P intercept = 0.003; P slope = 3 × 10−10).
Silencing different combinations of FLOTs results in varying degrees of nodulation and root morphological defects, although none as severe as FLOT2,3,4 RNAi roots (Fig. 4 A–I and Fig. S4). Silencing FLOT2 resulted in fewer nodules per plant, an increase in Nod- plants, and a decrease in plants that form pink nodules. Silencing FLOT2 resulted in a decrease in primary root length and long primary lateral roots, but no significant change in root weight. Plants silenced for FLOT4 expression were significantly less likely to form pink nodules than the empty vector; these roots had an increase in numbers of secondary lateral roots but no decrease in primary root length or root weight. The FLOT3(4) amiRNA construct silences FLOT3 as completely as FLOT2,3,4 RNAi, but these roots showed no significant nodulation defects compared to controls [this construct also targets FLOT1] (Fig. S6B). We observed shorter roots and reduced root weight in lines with the greatest reduction in FLOT3 expression. These data suggest that FLOT2 and FLOT4 play the largest role in symbiosis and that the symbiotic defects are separable from the small-root phenotype observed upon silencing FLOT2, FLOT3, and FLOT4.
FLOT2- and FLOT4-Silenced Plants Are Defective in Both Nodule Form and Function.
Nodules that form in FLOT-silenced lines are predominantly small and white, suggesting that they are unable to fix nitrogen. We found that all silenced lines [with the exception of FLOT3(4) amiRNA] had a significant decrease in their ability to reduce acetylene, a reporter for nitrogen fixation (P < 0.01; Fig. 4C).
To explore the individual contribution of each FLOT to nodule number, we used the dataset from Fig. 4A to determine whether a correlation exists between nodule number and the expression of a particular FLOT gene (each hairy root transformation event results in a unique genomic insertion of the RNAi or amiRNA construct and a different degree of gene silencing). We found that FLOT2 expression has a strong linear correlation with nodule number, whereas FLOT4 expression has a weaker but significant linear correlation with nodule number (Fig. 4J). In contrast, FLOT1 and FLOT3 expression levels have no significant correlation with nodule number (Fig. S6E and Fig. 4J). To assess whether FLOTs have additive effects, we compared the pooled expression of different combinations of FLOTs and found that the combination of FLOT2 and FLOT4 expression levels was the only grouping that increased the strength of the correlation (Fig. 4J). This is consistent with a model in which FLOT2 and FLOT4 contribute independently to nodule number.
FLOT-Silencing Results in IT Initiation and Elongation Defects.
We used lacZ-staining of S. meliloti to visualize ITs in silenced roots. FLOT-silencing results in reductions in both the number of nodules and the number of infection events [with the exception of FLOT3(4) amiRNA] (Fig. 4D). The decrease in the total number of infection events in FLOT2- and FLOT4-silenced roots indicates that the decrease in nodule number observed may in part be due to an IT initiation defect. We wondered whether the infection defects (Fig. 4D) are limited to IT initiation or whether FLOT2- and FLOT4-silenced roots also have IT elongation defects. We evaluated the progression of ITs on silenced roots and found that ITs on FLOT4-silenced roots were significantly more likely to abort in the root hair than control plants (Fig. 4E). ITs on roots silenced for FLOT4 and all FLOTs were also significantly less likely to have bacterial release into cortical cells. FLOT2- and FLOT3-silenced roots showed normal IT development (Fig. 4E). Representative images of the abortive ITs observed on FLOT4-silenced roots are shown in Fig. 4 F and G (compare with controls in Fig. 4 H and I). This finding suggests that FLOT4, but not other FLOTs, is required for normal IT elongation. It may suggest a defect in bacterial release in FLOT4-silenced plants, or that ITs fail to reach the cell layers where release normally occurs.
FLOT4 Localizes to IT Membranes.
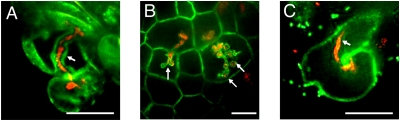
Because silencing FLOT4 results in IT elongation defects (Fig. 4 E–I), we explored whether FLOT4 localizes to ITs. In infected root hair cells, FLOT4:GFP localizes to IT membranes (Fig. 5A). A section through the infection zone of a 7-day-old nodule revealed that FLOT4 is present in IT membranes in the nodule (Fig. 5B). In contrast, FLOT2 is not present on IT membranes (Fig. 5C).
Fig. 5.
FLOT4 localizes to IT membranes. (A–C) Transgenic roots expressing FLOT4p:FLOT4:GFP or 35S:FLOT2:GFP (green) are inoculated with S. meliloti Rm1201 expressing pTryp:mCherry (red). (Scale bars: 30 μm.) (A) FLOT4:GFP localizes to IT membranes in root hair cells (stack of 50 images taken at 0.2-μm increments, arrows mark IT). (B) FLOT4:GFP localizes to IT membranes in the infection zones of maturing nodules (arrows). (C) FLOT2:GFP does not localize to IT membranes in root hair cells.
Discussion
A Requirement for FLOT2 and FLOT4 for Early Nodulation Events.
Data presented in this study indicate that FLOT2 and FLOT4 have roles in early nodulation. FLOT2 and FLOT4 are among a select group of plant genes up-regulated at 1 dpi; this up-regulation depends on both the NF signaling and entry receptor pathways, and on NIN (Fig. 1). The unexpected result that NF is insufficient for induction of FLOT2 and FLOT4 suggests that a second bacterial factor may also be required, although known surface polysaccharides are not apparently involved (Fig. S3).
FLOT4 shows a prominent accumulation in the tips of elongating root hairs upon inoculation with S. meliloti (Fig. 3). Plant proteins with similar localizations have been implicated in polar growth of root hairs and pollen tubes (23). Upon silencing FLOT4, we did not observe an obvious defect in normal root hair elongation or in curling around bacterial colonies, so it seems unlikely that FLOT4 is required for root hair elongation or directional growth. It seems more likely that this protein is involved in polar growth of the IT.
We observed that silencing FLOT2 and FLOT4 results in fewer ITs (Fig. 4D), and that silencing FLOT4 causes IT elongation defects (Fig. 4E). By analogy to flotillins in other organisms, one might predict that FLOT2 and FLOT4 have a role in the initial invagination of ITs into the root hair and that FLOT4 additionally functions in elongation of the IT. It is also possible that FLOTs coordinate IT initiation and nodule organogenesis [as has been proposed for NIN (8)] or that FLOTs are involved in endocytosis or trafficking of a signal involved in nodule organogenesis. Further studies are needed to determine whether, like their counterparts, plant flotillins are phosphorylated and interact with the actin cytoskeleton to facilitate membrane rearrangements.
FLOTs were previously isolated from soybean and pea peribacteroid membranes (18, 19); however, we were unable to detect either FLOT2:GFP or FLOT4:GFP on symbiosome membranes in M. truncatula. This may be due to limitations in the sensitivity of microscopy or to an absence of the proteins on the symbiosome membranes in this system. We found evidence that FLOTs are required for later stages of nodulation: FLOT2-silenced nodules were populated with ITs that looked similar to wild-type ITs (Fig. 4E); however, these nodules were unable to fix nitrogen (Fig. 4C), indicating a postinfection defect. Future studies are needed to determine whether FLOTs have a role in endocytosis and trafficking of bacteria.
Flotillin-like Genes Are Ubiquitous in Plants.
Possible roles for flotillin-like genes in general plant development may be suggested by the root phenotypes of FLOT-silenced plants (Figs. S4 and S5). We found that FLOT3-silenced plants had a reduction in root weight, FLOT2-silenced plants had a decrease in primary root length, and FLOT4-silenced plants had an increase in the number of secondary lateral roots (Figs. S4 and S5). These silencing phenotypes suggest that FLOTs may have a normal role in plant growth and development and that legumes may have co-opted the ancestral FLOT function for symbiosis.
Because plants membranes have different compositions than animal membranes (reviewed in ref. 24), it is likely that plant membrane microdomains will have different properties than animal membrane microdomains. Plant membranes contain little cholesterol but rather a mix of sterols that vary across plant taxa. Most plants, including the model Arabidopsis, contain primarily sitosterol, campesterol, and stigmasterol. Medicago has a less common membrane composition consisting largely of spinasterol (25). Determining how the membrane microdomains defined by FLOTs are similar to and different from the membrane microdomains defined by flotillins in other plants and other organisms will give valuable insight into plant membrane microdomains and plant membrane organization.
Materials and Methods
Plant Growth and Bacterial Treatments.
Medicago truncatula Gaertner cv. Jemalong, cv. Jemalong A17 (an inbred line of Jemalong), MtNIN-1 (8), and BIT1-1 (22) were grown, inoculated, and harvested as described in ref. 26. S meliloti strain Rm1021 is a streptomycin-resistant derivative of WT field isolate SU47 (27). Bacteria were grown in liquid TY-medium or Luria–Bertani (LB) medium supplemented with appropriate antibiotics. Bacterial mutant strains are described in SI Materials and Methods. For inoculations, bacteria were grown to an OD600 of between 0.5 and 1. They were then pelleted and resuspended in 1/2× buffered nodulation medium (BNM) (28) at an OD600 of 0.05.
RNA Sample Preparation.
RNA used for 1 dpi time points [A17 inoculated with NF, or with SL44, or exoA bacterial mutants and uninoculated and inoculated NSP2-2 and HCL-1(LYK3) plant mutants] were the same RNA samples that were used in ref. 5. The RNA samples from the A17 time course were the same samples used in ref. 21, with the exception of the 21-dpi RNA samples, which were prepared by Adriana Parra-Rightmyer using methods described in ref. 21. Remaining RNA samples were isolated by using the TRIzol (Invitrogen) method described in ref. 26.
Quantitative RT-PCR.
To ensure primer specificity, primers were designed to amplify the most divergent region of each FLOT and to flank intron splice sites (Table S2). Primer pairs were tested against a cloned genomic region containing FLOT1, -2, -3, or -4 to ensure that each primer pair would only amplify its intended target. Primer pairs were also tested for specificity by using cDNA from uninoculated and inoculated M. truncatula cv. Jemalong and cv. Jemalong A17. The resulting products were sequenced to ensure that they were from a single gene. Quantitative RT-PCR, data quantification, and analysis were performed as described in ref. 5.
A. rhizogenes–Mediated Hairy Root Transformations.
Plasmids were transformed into A. rhizogenes Arqua1 (29) and selected by using the appropriate antibiotic. A. rhizogenes-mediated hairy root transformations were done according to ref. 30 with modifications (SI Materials and Methods.). After regeneration of hairy roots, plants were transferred to BNM with 0.1 μM aminoethoxyvinylglycine (AVG). Plants were flood inoculated 1 week later with 10 mL of the appropriate S. meliloti strain diluted to OD600 = 0.05 in 1/2× BNM.
β-Glucuronidase Assays.
The upstream region of each ORF (3 kb at most, less if another ORF <3 kb upstream) was amplified from BAC mth2-115c19 and cloned into pENTR D/TOPO (Invitrogen) (Table S5). LR recombination was performed with the Gateway-compatible vector pMDC163 (31). Plasmids were used to transform A. rhizogenes Arqua1. A. rhizogenes-mediated hairy root transformation was performed as described above with hygromycin selection. GUS assays were conducted as described in ref. 32.
RNAi and amiRNA Construct Design and Screen.
For RNAi constructs, 100–200 bp of either the conserved region of FLOTs or the 3′ UTR were amplified from DNA prepared from BAC mth2-115c19 (Table S3). The PCR products were first cloned into pENTR/D TOPO (Invitrogen). LR recombination was performed with the binary vector pHELLSGATE8 (33).
amiRNA constructs were designed as described in ref. 34 by using the web-based designer described in ref. 35 (http://wmd2.weigelworld.org). Details of target selection and primer design are described in SI Materials and Methods and Table S4. The final amiRNA PCR product was digested at the XhoI and XbaI sites flanking the sequence encoding the amiRNA hairpin. The resulting product was ligated into the pHELLSGATE8 vector XhoI and XbaI sites (thus removing the Gateway cassette). The result was an amiRNA construct driven by the CaMV 35S promoter with the same vector backbone as the RNA constructs.
Using A. rhizogenes to generate transgenic hairy roots, we conducted a preliminary screen of 8 RNAi and 12 amiRNA constructs to identify those that effectively silence expression of one or more FLOT (see SI Materials and Methods). ANOVA was used to identify constructs that caused significant reduction in gene expression. Pairwise two-tailed t tests were used to determine whether nodule number, percent Nod-, and percent pink nodules were significantly different in silenced lines compared to the empty vector control. Linear regressions were done by using the Microsoft Excel data analysis feature. To determine whether second- and third-order correlations existed, second- and third-order transform was performed on the data and linear regressions were conducted. Gene expression was pooled by averaging the expression of each gene in the same plant.
Plant Assays.
Acetylene reduction (36) was performed as described in ref. 37. Plants were initially grown on BNM plates as described above; at 21 dpi the entire plant was moved to test tubes for the assays. At least three uninoculated plants were assayed per construct to ensure that silencing FLOTs did not intrinsically cause an increase in ethylene production.
To visualize infection events, plants were inoculated with Rm1021 expressing the lacZ expression plasmid pXLGD4 and stained for β-galactosidase activity at 7 dpi as described in ref. 38. Because of the altered lateral branching observed in some FLOT-silenced lines, we used root weight rather than root length as a measure of root size.
Protein Localization.
The genomic regions of FLOT2 and FLOT4 were amplified from BAC DNA (primer details in Table S5; see SI Materials and Methods. for construct details). For 1 dpi localization studies, plants were inoculated with Rm1021 overexpressing NodD3 from the plasmid pRmE65 (39). For IT colocalization studies, plants were inoculated with Rm1021 constitutively expressing mCherry from the plasmid pQDN03 (SI Materials and Methods).
Root segments and nodule hand sections (for imaging the infection zone) were excised and mounted in 0.1 M potassium phosphate buffer (pH 7). Spinning-disk confocal microscopy was performed on a system described in ref. 40, using a 63×/1.3 N.A. glycerol immersion objective. GFP and RFP were excited at 491 nm and 561 nm, respectively, by solid-state lasers. Z-projections of root hairs are from 100 to 200 images taken at increments of 0.2 μm (MCL NanoDrive). Stacks were processed by using ImageJ software (http://rsbweb.nih.gov/ij). Typical exposure times were 1,000 ms for GFP and 500 ms for mCherry.
Supplementary Material
Acknowledgments
We thank C. G. Starker, R. M. Mitra, and A. L. Parra-Rightmyer for the use of RNA samples, and C. G. Starker for assistance with hairy root transformations. We thank J. Griffitts for use of pJG161 and Q. A. Nguyen for assisting with GUS assays and for construction of ptrp-mCherry (pQDN03). We are grateful to E. Xiao for help with transformation experiments and for generating amiRNA constructs. We thank D. Ehrhardt for confocal microscopy expertise, R. Fisher, D. Wang and E. J. Chen for critical reading of the manuscript and fellow lab members for helpful discussions. Financial support was provided by an NSF GRFP award to C.H.H., by prior funds to S.R.L. from the Howard Hughes Medical Institute, and from the Hoover Circle Fund.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Database deposition: The sequences reported in this paper have been deposited in the GenBank database accession nos. GU224278-GU224282.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910081107/DCSupplemental.
References
- 1.Gage DJ. Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol Mol Biol Rev. 2004;68:280–300. doi: 10.1128/MMBR.68.2.280-300.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Newcomb W. A correlated light and electron microscopic study of symbiotic growth and differentiation in Pisum sativum root nodules. Can J Bot. 1976;54:2163–2186. [Google Scholar]
- 3.Debellé F, Moulin L, Mangin B, Dénarié J, Boivin C. Nod genes and Nod signals and the evolution of the Rhizobium legume symbiosis. Acta Biochim Pol. 2001;48:359–365. [PubMed] [Google Scholar]
- 4.Ehrhardt DW, Wais R, Long SR. Calcium spiking in plant root hairs responding to Rhizobium nodulation signals. Cell. 1996;85:673–681. doi: 10.1016/s0092-8674(00)81234-9. [DOI] [PubMed] [Google Scholar]
- 5.Mitra RM, Shaw SL, Long SR. Six nonnodulating plant mutants defective for Nod factor-induced transcriptional changes associated with the legume-rhizobia symbiosis. Proc Natl Acad Sci USA. 2004;101:10217–10222. doi: 10.1073/pnas.0402186101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Oldroyd GE, Downie JA. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu Rev Plant Biol. 2008;59:519–546. doi: 10.1146/annurev.arplant.59.032607.092839. [DOI] [PubMed] [Google Scholar]
- 7.Hirsch S, et al. GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell. 2009;21:545–557. doi: 10.1105/tpc.108.064501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsh JF, et al. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007;144:324–335. doi: 10.1104/pp.106.093021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batut J, Andersson SG, O'Callaghan D. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat Rev Microbiol. 2004;2:933–945. doi: 10.1038/nrmicro1044. [DOI] [PubMed] [Google Scholar]
- 10.Watarai M, Makino S, Fujii Y, Okamoto K, Shirahata T. Modulation of Brucella-induced macropinocytosis by lipid rafts mediates intracellular replication. Cell Microbiol. 2002;4:341–355. doi: 10.1046/j.1462-5822.2002.00195.x. [DOI] [PubMed] [Google Scholar]
- 11.Schulte T, Paschke KA, Laessing U, Lottspeich F, Stuermer CA. Reggie-1 and reggie-2, two cell surface proteins expressed by retinal ganglion cells during axon regeneration. Development. 1997;124:577–587. doi: 10.1242/dev.124.2.577. [DOI] [PubMed] [Google Scholar]
- 12.Glebov OO, Bright NA, Nichols BJ. Flotillin-1 defines a clathrin-independent endocytic pathway in mammalian cells. Nat Cell Biol. 2006;8:46–54. doi: 10.1038/ncb1342. [DOI] [PubMed] [Google Scholar]
- 13.Babuke T, Tikkanen R. Dissecting the molecular function of reggie/flotillin proteins. Eur J Cell Biol. 2007;86:525–532. doi: 10.1016/j.ejcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 14.Frick M, et al. Coassembly of flotillins induces formation of membrane microdomains, membrane curvature, and vesicle budding. Curr Biol. 2007;17:1151–1156. doi: 10.1016/j.cub.2007.05.078. [DOI] [PubMed] [Google Scholar]
- 15.Langhorst MF, et al. Reggies/flotillins regulate cytoskeletal remodeling during neuronal differentiation via CAP/ponsin and Rho GTPases. Eur J Cell Biol. 2008;87:921–931. doi: 10.1016/j.ejcb.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 16.Rivera-Milla E, Stuermer CA, Málaga-Trillo E. Ancient origin of reggie (flotillin), reggie-like, and other lipid-raft proteins: convergent evolution of the SPFH domain. Cell Mol Life Sci. 2006;63:343–357. doi: 10.1007/s00018-005-5434-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Winzer T, et al. A novel 53-kDa nodulin of the symbiosome membrane of soybean nodules, controlled by Bradyrhizobium japonicum. Mol Plant Microbe Interact. 1999;12:218–226. doi: 10.1094/MPMI.1999.12.3.218. [DOI] [PubMed] [Google Scholar]
- 18.Panter S, et al. Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Mol Plant Microbe Interact. 2000;13:325–333. doi: 10.1094/MPMI.2000.13.3.325. [DOI] [PubMed] [Google Scholar]
- 19.Saalbach G, Erik P, Wienkoop S. Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics. 2002;2:325–337. doi: 10.1002/1615-9861(200203)2:3<325::aid-prot325>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 20.Borner GH, et al. Analysis of detergent-resistant membranes in Arabidopsis. Evidence for plasma membrane lipid rafts. Plant Physiol. 2005;137:104–116. doi: 10.1104/pp.104.053041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starker CG, Parra-Colmenares AL, Smith L, Mitra RM, Long SR. Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol. 2006;140:671–680. doi: 10.1104/pp.105.072132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Middleton PH, et al. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell. 2007;19:1221–1234. doi: 10.1105/tpc.106.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang Z. Cell polarity signaling in Arabidopsis. Annu Rev Cell Dev Biol. 2008;24:551–575. doi: 10.1146/annurev.cellbio.23.090506.123233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zappel NF, Panstruga R. Heterogeneity and lateral compartmentalization of plant plasma membranes. Curr Opin Plant Biol. 2008;11:632–640. doi: 10.1016/j.pbi.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 25.Lefebvre B, et al. Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 2007;144:402–418. doi: 10.1104/pp.106.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mitra RM, Long SR. Plant and bacterial symbiotic mutants define three transcriptionally distinct stages in the development of the Medicago truncatula/Sinorhizobium meliloti symbiosis. Plant Physiol. 2004;134:595–604. doi: 10.1104/pp.103.031518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982;149:114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ehrhardt DW, Atkinson EM, Long SR. Depolarization of alfalfa root hair membrane potential by Rhizobium meliloti Nod factors. Science. 1992;256:998–1000. doi: 10.1126/science.10744524. [DOI] [PubMed] [Google Scholar]
- 29.Quandt HJ, Pühler A, Broer I. Transgenic root nodules of Vicia hirsuta: A fast and efficient system for the study of gene expression in indeterminate-type nodules. Mol Plant Microbe Interact. 1993;6:699–706. [Google Scholar]
- 30.Boisson-Dernier A, et al. Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol Plant Microbe Interact. 2001;14:695–700. doi: 10.1094/MPMI.2001.14.6.695. [DOI] [PubMed] [Google Scholar]
- 31.Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oldroyd GE, Engstrom EM, Long SR. Ethylene inhibits the Nod factor signal transduction pathway of Medicago truncatula. Plant Cell. 2001;13:1835–1849. doi: 10.1105/TPC.010193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helliwell C, Waterhouse P. Constructs and methods for high-throughput gene silencing in plants. Methods. 2003;30:289–295. doi: 10.1016/s1046-2023(03)00036-7. [DOI] [PubMed] [Google Scholar]
- 34.Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- 35.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turner GL, Gibson AH. Measurement of nitrogen fixation by indirect means. In: Bergersen FJ, editor. Methods for Evaluation Biological Nitrogen Fixation. Chichester, UK: Wiley; 1980. pp. 111–138. [Google Scholar]
- 37.Oke V, Long SR. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol Microbiol. 1999;32:837–849. doi: 10.1046/j.1365-2958.1999.01402.x. [DOI] [PubMed] [Google Scholar]
- 38.Boivin C, Camut S, Malpica CA, Truchet G, Rosenberg C. Rhizobium meliloti Genes Encoding Catabolism of Trigonelline Are Induced under Symbiotic Conditions. Plant Cell. 1990;2:1157–1170. doi: 10.1105/tpc.2.12.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fisher RF, Egelhoff TT, Mulligan JT, Long SR. Specific binding of proteins from Rhizobium meliloti cell-free extracts containing NodD to DNA sequences upstream of inducible nodulation genes. Genes Dev. 1988;2:282–293. doi: 10.1101/gad.2.3.282. [DOI] [PubMed] [Google Scholar]
- 40.Gutierrez R, Lindeboom JJ, Paredez AR, Emons AM, Ehrhardt DW. Arabidopsis cortical microtubules position cellulose synthase delivery to the plasma membrane and interact with cellulose synthase trafficking compartments. Nat Cell Biol. 2009;11:797–806. doi: 10.1038/ncb1886. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.