Abstract
MicroRNA (miRNAs) are negative regulators of gene expression and can function as tumor suppressors or oncogenes. Expression patterns of miRNAs and their role in the pathogenesis of hepatocellular carcinoma (HCC) are still poorly understood. We profiled miRNA expression in tissue samples (104 HCC, 90 adjacent cirrhotic livers, 21 normal livers) as well as in 35 HCC cell lines. A set of 12 miRNAs (including miR-21, miR-221/222, miR-34a, miR-519a, miR-93, miR-96, and let-7c) was linked to disease progression from normal liver through cirrhosis to full-blown HCC. miR-221/222, the most up-regulated miRNAs in tumor samples, are shown to target the CDK inhibitor p27 and to enhance cell growth in vitro. Conversely, these activities can be efficiently inhibited by an antagomiR specific for miR-221. In addition, we show, using a mouse model of liver cancer, that miR-221 overexpression stimulates growth of tumorigenic murine hepatic progenitor cells. Finally, we identified DNA damage-inducible transcript 4 (DDIT4), a modulator of mTOR pathway, as a bona fide target of miR-221. Taken together, these data reveal an important contribution for miR-221 in hepatocarcinogenesis and suggest a role for DDIT4 dysregulation in this process. Thus, the use of synthetic inhibitors of miR-221 may prove to be a promising approach to liver cancer treatment.
Keywords: hepatocarcinogenesis, microRNA, antagomiRs, mouse model, DDIT4
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in the world (1). Among the well established risk factors for HCC, chronic infection with hepatitis B (HBV) or C (HCV) virus is present in >85% of primary liver cancers (2). Early studies indicated that only a few of the protein changes detected so far can be validated at the RNA level (3). miRNAs are thought to control gene expression at the posttranscriptional level by causing mRNA degradation and/or repressing mRNA translation (4). Evidence indicating that miRNAs play an important role in various human cancers is now accumulating (5), but only a few microtranscriptome HCC profiles, involving limited numbers of samples, have been performed.
We present the miRNA expression profile of a large number of paired HCC-nontumor samples as well as liver cancer cell lines. We observed unique miRNA expression signatures that could distinguish malignant from adjacent cirrhotic tissues. We provide evidence for a major contribution of miR-221 in liver cancer cell proliferation and HCC development in vivo and identify DNA damage-inducible transcript 4 (DDIT4) as a direct target of miR-221.
Results
Twelve miRNAs Define a Signature for Liver Cancer.
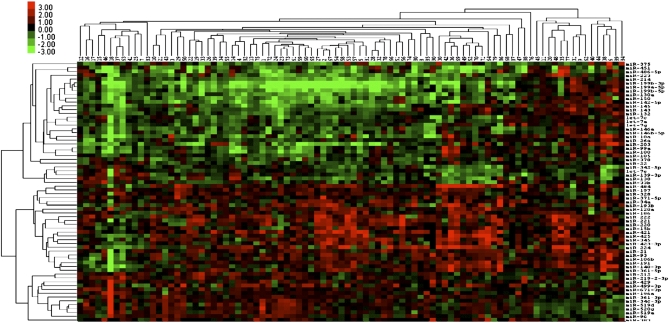
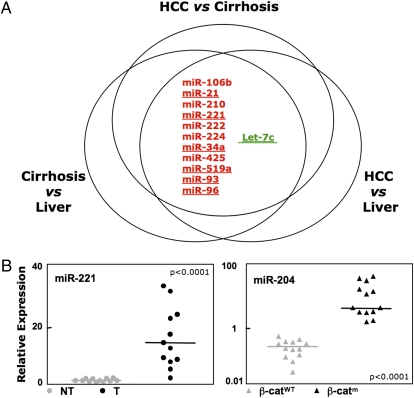
Our aim was to identify miRNAs that are significantly dysregulated during cancer progression from normal liver to full blown HCC, through the precancerous stage of cirrhosis. Thus, we profiled 21 normal livers, 104 HCC, 90 paired cirrhotic tissues (Table S1), and 35 HCC-derived cell lines. According to our strategy, we made three pairwise comparisons: (i) cirrhosis vs. normal liver, (ii) HCC vs. normal liver, and (iii) HCC tissues compared to their respective cirrhosis tissues. We hypothesized that a miRNA must be involved in liver tumorigenesis if it is consistently dysregulated through each step going from normal liver to cirrhosis and cirrhosis to HCC. miRNAs satisfying these requirements were then examined in HCC cell lines to ensure that the miRNA signature was truly a characteristic of expression in transformed liver cells per se. The heat map of the miRNAs dysregulated in the last step of progression (cirrhosis to HCC) is shown in Fig. 1. This combinatorial strategy identified the miRNAs significantly dysregulated in liver tumorigenesis: miR-106b, miR-21, miR-210, miR-221, miR-222, miR-224, miR-34a, miR-425, miR-519a, miR-93, and miR-96 were increased, and let-7c was lost during liver tumor progression (Fig. 2A and Table S2).
Fig. 1.
Heat map of miRNA expression in HCC: Tree showing the miRNAs deregulated in HCC vs. cirrhotic tissues (paired t test, P < 0.01). Each column indicates the normalized log 2 ratio of the HCC/cirrhosis pairs. Clustering was performed with average linkage and uncentered correlation.
Fig. 2.
miRNA signatures in HCC. (A) Intersection of SAM analysis for three profiles. The intersection is shown among the deregulated miRNAs in the three classes of tissues: HCC, cirrhosis, and normal liver. Only the miRNA deregulated also in the HCC cell lines were considered. Red, up-regulated miRNAs; green, down-regulated miRNAs. (B) Validation of microarray by qPCR of differentially expressed miRNAs in nontumoral (NT) vs. tumoral (T) liver tissues and β-catenin mutated (β-catm) vs. wild-type (β-catwt) specimens. The relative expression fold of each miRNA is normalized to RNU6B and miR-140. P values were obtained by using a two-sided Student t test.
We then determined which of the 12 microRNAs (miRNAs) had the highest diagnostic value. Our purpose was to distinguish between the tumors (139 tumors and cell lines) and the nontumor tissues (111 cirrhotic and normal livers). We used the prediction test in BRB-ArrayTools to identify the classifier signature with the lowest misclassification error. As few as seven miRNAs could classify (15% misclassification error). The cross-validation ROC curve for the seven miRNA classifier is shown in Fig. S1A. The miRNAs used in the ROC classifier are underlined in Fig. 2A.
There were no significant differences in miRNA expression associated with p53 mutations, but we detected a signature associated with β-catenin mutation (Table S2). Among the five miRNAs up-regulated 2- to 3-fold in cases of β-catenin gene mutation was miR-34a, known previously as a p53 target, and miR-122, the miRNA most highly expressed in hepatocytes (6, 7).
miR-221/222 Target p27.
To validate the miRNA microarray data, we carried out qPCR on RNA from 12 pairs of HCC and corresponding nontumorous livers and tested four different members of the progression signature (Fig. 2B and Fig. S1B). Statistically significant differences of expression levels were observed for all miRNAs investigated. In addition, we confirmed trends observed on microarray by qPCR for four different putative β-catenin targets (Fig. 2B and Fig. S1C) (8).
We then decided to investigate the roles of miR-221 and miR-222 in detail. These miRNAs were chosen because of their consistently high levels of expression in tumors, suggesting that they act as oncomiRs in liver cancer. miR-221 and miR-222 share a high degree of homology, are neighbors in human and mouse genomes, and share common predicted targets (same seed sequence). On Northern blot analysis, each was significantly up-regulated in cancer samples and cell lines when compared to NT or normal livers (P ≤ 0.02), thus confirming qPCR data (Fig. S2 A and B). We next examined some of their putative targets for repression in human HCC vs. nontumorous livers. The cyclin-dependent kinase inhibitor p27 (Kip1-CDKN1B) and the tyrosine-kinase receptor c-kit have been identified as targets of miR-221 and miR-222 (9, 10). Because c-kit is inconsistently expressed in liver, we focused on p27. Expression levels of p27 in matched HCC-nontumor samples and liver cancer cell lines were assessed by Western blot analysis and by qPCR. We found a consistent decrease of p27 protein in samples overexpressing miR-221/222 (Fig. S2C). However, there was no difference in the mRNA expression levels of p27 by qPCR, suggesting a translational inhibition of p27 and not mRNA degradation (Fig. S3A). Finally, in transfected HeLa and 293T cells, overexpression with vectors expressing miR-221 and miR-222 was found to induce a reduction of p27 protein levels (Fig. S3B).
miR-221/222 antagomiR Inhibits Cell Growth.
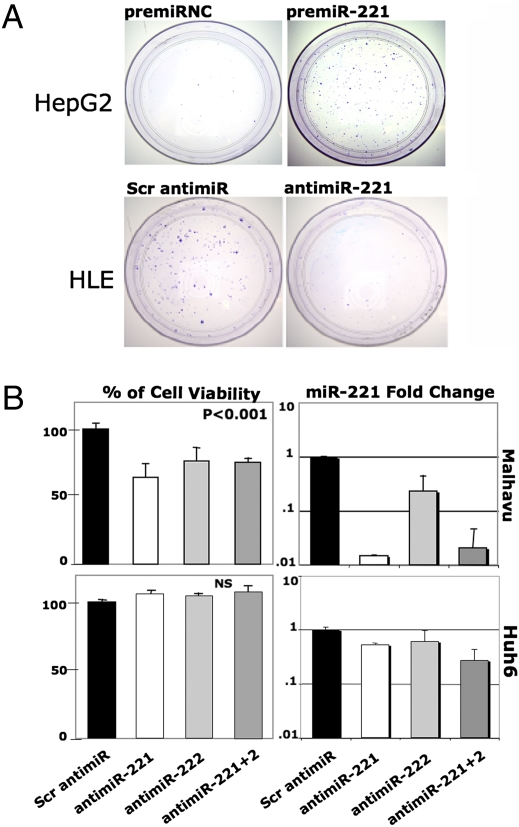
To examine the role of miR-221/222 on cell growth, we tested their effect on the transformation properties of liver cancer and other tumor-derived cell lines. After transfection of miR-221 precursor, we observed that most cell lines formed larger colonies than controls when plated at low density (Fig. 3A and Fig. S4A) (11). A few cell lines (e.g., HeLa and PLC/PRF5), however, did not display any changes, suggesting that in some contexts, an increased miR-221 level does not stimulate growth. Previous reports have indicated that miR-221 and miR-222 overexpression may cause “oncogenic addiction” (12). To address this point, LNA-modified oligonucleotides specifically designed for miR-221 (antimiR-221) and miR-222 (antimiR-222) knockdown were introduced by lipofection into liver cancer cells. Malhavu, FOCUS, and HLE cells were chosen for their high endogenous level of the miRNAs, whereas PLC/PRF5 and Huh6 were used for their reduced expression (Fig. S2B). When plated at low density, the growth of HLE was drastically inhibited (Fig. 3A), whereas the effect on other cells was more subtle. Thus, to measure the impact of synthetic inhibitors on liver cancer cell proliferation, a cell viability assay was used (Fig. S4B). As shown in Fig. 3B and Fig. S4C, treatment by antagomiRs, but not scrambled oligonucleotide, reduced cell growth in liver cancer cell lines overexpressing miR-221 and miR-222 by 35% (Malhavu) and 22% (FOCUS). By contrast, no change in proliferation was observed in treated PLC/PRF5 and Huh6 cells (Fig. 3B and Fig. S4C). Although, we cannot exclude that transfection efficiencies are different between high and low expressors, these data strongly suggest that overexpressing cells became dependent on miRNA activity. When introduced in combination, antagomiRs do not increase growth inhibition suggesting that a saturation threshold is reached in the four cell lines by a single antagomiR. A drastic reduction of miR-221 and miR-222 levels (5- to 50-fold) was observed by qPCR in high expressors, whereas a mild decrease (1.5- to 2-fold) was detected in cell lines with low expression (Fig. 3B and Fig. S4C).
Fig. 3.
miR-221 inhibition impairs tumor cell growth. (A) HepG2 and HLE cells were plated at low density after transfection by premiR negative control (NC), premiR-221 or LNA scrambled antagomiR (Scr antimiR), or LNA antimiR-221. Cells were grown for 10 days, fixed, and stained by crystal violet. (B) Malhavu (miR-221 and miR-222 high expressor) and Huh6 (miR-221 and miR-222 low expressor) liver cancer cells were transfected with antimir-221, antimiR-222, or a negative control and then plated out in 96-well plates. Cell growth was measured by MTT assay at 72 h. Viability was identical for cells treated with transfection reagent alone or scrambled antimiR. The values are the means from three separate experiments done in triplicate. P values were obtained by using a two-sided Student t test. miR-221 levels are shown as measured by qPCR 24 h after transfection of LNA oligonucleotides.
miR-221 Enhances Tumorigenesis in Vivo.
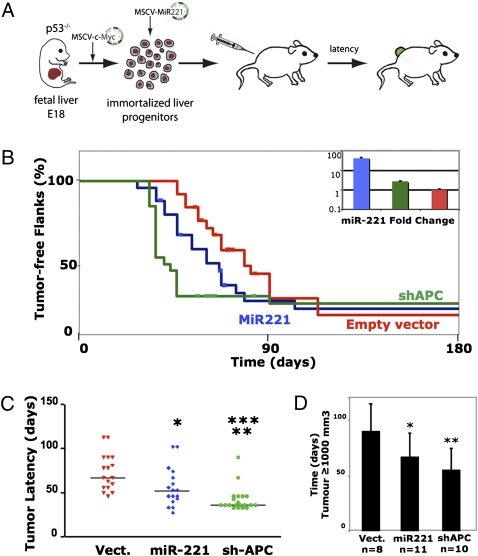
These data prompted us to investigate the role of both miRNAs in tumor development. To this aim, we used a mouse model of liver cancer (13). P53−/−;myc liver progenitors were infected with a retrovirus that overexpresses miR-221, miR-222, and sh-APC (positive control), or with an empty vector (Fig. 4A), and assayed for their ability to form tumors in vivo. Expression levels of miR-221 and miR-222 constructs were monitored and found to be >50- and 10-fold above the endogenous levels in transduced cells, respectively (Fig. 4B and Fig. S4D). Subsequently, all constructs were injected s.c. in a limited number of animals (n = 4 flanks). In this preliminary experiment, miR-222 did not accelerate tumorigenesis when compared to empty vector or even uninfected cells. We thus decided to focus on miR-221 and extended the series to a total of 24 injected flanks.
Fig. 4.
miR-221 is protumorigenic in vivo. (A) Schematic representation of the transfer protocol using immortalized p53−/−;myc liver progenitors infected by MSCV-derived retroviruses. (B) Kaplan–Meier curves showing the percentage of tumor-free injected mouse flank. Mice were injected s.c. with p53−/−;myc hepatoblasts previously infected by retroviruses expressing miR-221 or sh-APC in the PIG-MSCV vector. Each group corresponds to 24 flanks/12 mice. Mice were monitored starting at 3 weeks after injection. The histogram corresponds to miR-221 expression levels as measured by qPCR in the three types of transduced liver progenitors. (C) Average latency for tumor appearance. Significant differences of tumor onset was observed between miR-221 and empty vector (*, P = 0.0142), sh-APC and empty vector (**, P < 1 10−4), and miR-221 and sh-APC (***, P = 0.0151). (D) Time required for tumor to reach 1,000 mm3. Values are mean ± SD. Each mouse was killed when tumor reached 1,000 mm3. Significant differences were observed between miR-221- and empty vector-infected cells (*, P < 0.05) and between sh-APC and empty vector (**, P < 0.005).
In mice injected with liver progenitors carrying miR-221, the average latency for tumor apparition was decreased when compared with empty vector. This difference was statistically significant (56 ± 5 versus 73.4 ± 5 days, P = 0.0142, Student’s t test; Fig. 4 B and C). At 6 months follow-up, tumor penetrance was still incomplete although similar for all retroviral constructs (80–90% of injected flanks). We observed a trend (P = 0.0904) toward lower rates of tumor-free survival for the miR-221-injected mice when compared with empty vector-transduced hepatic progenitors (Fig. 4B). In addition, tumor progression, as measured by the time required to reach a volume of 1,000 mm3, was similar in miR-221- and sh-APC-expressing cells and significantly faster than in empty vector-transduced cells (Fig. 4D).
DDIT4 Is a Target for miR-221.
Individual miRNAs are believed to target multiple mRNAs (12). Several miR-221 targets, all of them being translationally inhibited, have been characterized in various tumor types (see above) including p27-CDKN1B, p57-CDKN1C, CKIT, and ESR1 (9, 14–16). To determine whether genes may be suppressed by miR-221 at the RNA level in HCC, we carried out an mRNA microarray analysis on 19 HCCs already screened for miRNA expression. Fifteen inversely correlated transcripts (P < 0.05 for Pearson and/or Spearman coefficients) contained putative miR-221 target sites according to the prediction software (TargetScan, PicTar-Vert, and MiRanda; Fig. S5 A and B).
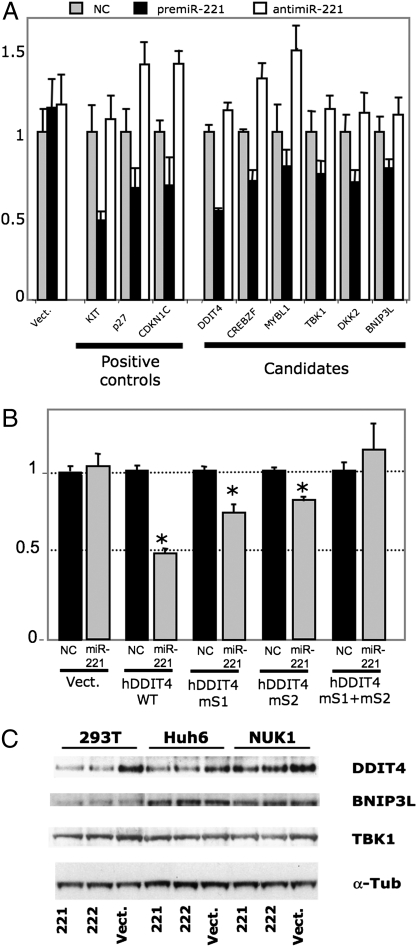
To experimentally validate that miR-221 can target these genes, 3′UTRs of 11 different genes were subcloned downstream of a Renilla reporter vector to analyze whether miR-221 can directly targets these genes (Table S3). In 293T cells, premiR-221 but not premiR negative control, is able to reduce Renilla activity for six of these constructs, indicating that this miRNA may directly target these 3′UTRs. Conversely, reporter activity was stimulated in the presence of the antimiR-221, indicating that endogenous miR-221 can directly influence targeted protein levels (Fig. 5A). The five other constructs failed to be negatively regulated by miR-221 (Fig. S6A). Two highly conserved binding sites for miR-221, subsequently named S1 and S2, are present in DDIT4 3′UTR (Fig. S6B). To ascertain the direct effect of miR-221 on DDIT4, mutant constructs targeting S1, S2, or S1+S2 were generated from human and murine DDIT4 3′UTR. Mutations that disrupted the two binding sites for miR-221 entirely restored luciferase activity (Fig. 5B and Fig. S6C), suggesting that miR-221, through binding to S1 and S2, directly regulates DDIT4 protein level. To address this point, we transfected cells from different cell lines with precursor miR-221. Western blot and qPCR experiments confirmed that miR-221 represses DDIT4 expression at both the protein and RNA levels in the different cell types (Fig. 5C and Fig. S6D). A similar decrease was observed for BNIP3L and TBK1 protein/RNA levels, although the effect of miR-221 was weaker for these two targets (Fig. 5C and Fig. S6D). Finally, we observed a significant inverse correlation between DDIT4 mRNA levels and miR-221 expression in miR-221-transduced mouse tumors (Fig. S6E), thus substantiating the relevance of this mouse model for human HCC.
Fig. 5.
DDIT4 inhibition by miR-221. (A) Luciferase activity 72 h after cotransfection of 293T cells with psiCHECK-2-UTRs vectors, premiR-221, premiR negative control (NC), and LNA antimiR-221. Data are means ± SD of four independent experiments performed in triplicate. Renilla luciferase activity was significantly decreased for 3′ UTRs of positive controls (Kit, p27, CDKN1C) and for DDIT4, CREBZF, MYBL1, TBK1, DKK2, and BNIP3L (P < 0.05, Student’s t test). (B) Luciferase activity at 72 h in 293T cells cotransfected with premiR negative control (NC, black) or premiR-221 (gray) and luciferase reporters containing the wild-type (WT) or mutated miR-221 binding sites (mS1 and mS2) of human DDIT4 3′UTR or vector (V) (*, P < 0.01, Student’s t test). (C) 293T, Huh6, and NU.K.-1 cells were transfected with vector expressing miR-221, miR-222, or empty vector, and protein levels were checked by Western blotting with the indicated antibodies. One representative of three independent experiments is shown.
Discussion
The current knowledge about miRNA expression in HCC is still rather preliminary (17–19). In this study, 104 HCC samples and their corresponding nontumorous counterparts were analyzed by using microarrays to determine miRNA expression profiles. The results clearly distinguished the tumor profile from the nonneoplastic tissue profile. Overall, liver tumorigenesis is characterized by many up-regulated and repressed miRNAs, potentially exerting oncogenic and tumor-suppressive functions, respectively.
Our analysis identified a 12-miRNA signature that defines disease progression from normal liver to full-blown tumors through liver cirrhosis. Four of them (miR-21, miR-221, miR-222, and miR-224) are commonly found to be overexpressed across previously published profiles of the HCC microtranscriptome (8, 20–22). Some belong to commonly up-regulated clusters, such as miR-106b and miR-93 at 7q22 and miR-221 and miR-222 at Xp11 (23). Interestingly, miR-34a, a target of p53, known to induce senescence and/or apoptosis in vitro, was found to be overexpressed in the progression signature, suggesting a potential versatility of miRNA activity depending on the cell type investigated (24).
Dysregulation of miR-221 and miR-222 was found in the miRNA progression signature involved in liver tumorigenesis. They were previously shown to target the CDK inhibitors p27 and p57 at the protein level. Despite the impact on major players in cell cycle control, little is known about the role played in vivo by miR-221 and miR-222. The mouse model of liver cancer indicates that miR-221 stimulates the onset of tumors when compared with vector-infected cells. miR-221 also promotes tumor progression, significantly shortening the mean time to death. Such pleiotropic properties, however, appear milder when compared with the outcomes observed in sh-APC-positive control in vivo. Given the multiple potential targets of a single miRNA, such results are, however, plausible and in line with the fine-tuning effects usually observed in miRNA experiments (25).
Despite sharing the same seed as miR-221, miR-222 did not seem to accelerate tumors in preliminary experiments performed on a restricted number of mice. miR-221 and miR-222 share only three additional nucleotide identities 3′ to the seed. Previous reports have shown that residues downstream of the seed—particularly nucleotides 13–16, which all differ between miR-221 and miR-222—play a significant role in miRNA activity (26). In addition, expression levels of miR-222 in transduced hepatoblasts were substantially lower than those observed for miR-221 (Fig. S4D). Whether the absence of accelerated tumorigenesis by miR-222 is due to lower expression levels or to distinct functional properties when compared to miR-221 remains to be clarified.
For the most strongly repressed targets, mRNA destabilization usually represents the major component of repression (26). Although published targets of miR-221 belong exclusively to the translationally-repressed category, we looked for miR-221-destabilized mRNAs in HCC. According to prediction software, the expression of miR-221 was inversely correlated with a set of predicted targets in liver cancer. Among them, DDIT4 was confirmed in vitro both at the RNA and protein levels. DDIT4 is an essential regulator of the mTOR kinase through stimulation of the tuberous sclerosis tumor suppressor TSC1/2 complex and as such is considered to be a putative tumor suppressor (27). Thus, our data suggest the presence of another level of disruption of the PTEN-PI3K-AKT-mTOR axis relevant to liver tumorigenesis. Furthermore, another up-regulated member of the progression signature defined in the present report, miR-21, is known to target the PTEN tumor suppressor (28). Remarkably, PTEN and DDIT4 genes are only separated by 12 Mb on chromosome 10q, implying that the heterozygous deletion of 10q and coup-regulation of miR-21/miR-221 may act in concert in HCC. Thus, our data suggest that in liver tumors where PI3K-PTEN-AKT-mTOR mutations remain conspicuously rare, the microtranscriptome may act as an important regulator of this pathway, which is considered to be crucial for hepatocarcinogenesis (29).
In conclusion, our findings suggest that the expression levels of some miRNAs change gradually during the progression of liver disease. A subset of these miRNAs are already considered as bona fide oncomiRs (miR-21, miR-221, miR-222, miR-93, miR-96, and miR-106b), whereas some others (miR-210, miR-220, miR-224, miR-425, and miR-519a) are apparently more HCC-specific and merit further investigation. Among those miRNAs present in the progression signature, miR-221 is capable of stimulating tumor growth in vivo possibly through p27 and/or DDIT4 down-regulation.
Materials and Methods
Patients and Cell Lines.
A total of 104 patients (Table S1) and a collection of 35 hepatocellular carcinoma-derived cell lines were analyzed. The cell line collection is described in ref. 30. This study was approved by an Institutional Human Research review board (RBM 2005-019). Informed consent was obtained for each patient. Total RNA was extracted with Tri-Reagent (Euromedex) according to the manufacturer’s specifications. HCV infection status was determined by serology. p53 and β-catenin status were determined as described in ref. 31.
Microarray and Computational Analysis.
Microarray analysis was performed as described in ref. 32 and detailed in SI Materials and Methods. Two hundred fifteen mature miRNAs were studied in the expression table. Signatures were detected by using SAM (q-value <0.05) in BRB-ArrayTools (33). The microarray dataset was deposited in the ArrayExpress public database at the European Bioinformatics Institute.
Quantitative PCR and Western Blot Analysis.
Quantitative real-time PCR (qPCR) was used to confirm microarray data and measure miRNA impact on putative targets. A TaqMan assay was used both for micRNA and mRNA analysis (Applied Biosystems) according to the manufacturer’s protocol (SI Materials and Methods). The TaqMan MicroRNA assays for U6 RNA and miR-140 were used to normalize the relative abundance of miRNA. Total protein extracts were prepared in Laemmli buffer, loaded onto 10% Tris·HCl-ready gel (Bio-Rad), and transferred to nitrocellulose membranes. Membranes were hybridized with antibodies against p27 (610241; BD Biosciences), DDIT4 (0638-1-AP; Proteintech Group), BNIP3L (ab8399; Abcam), TBK1 (ab40676; Abcam), tubulin (ab52866; Abcam), and actin(C-2) (sc-8432; Santa Cruz Biotechnology). The secondary alkaline phosphatase-conjugated antibody (T2191 or T2192; Applied Biosystems) was detected by using CDP-Star Western blotting detection reagents (TROPIX). Bands were quantified with ImageJ 1.38 Instrument software.
premiRs and antagomiRs Transfection.
premiRs (Ambion, Applied Biosystems) or antagomiR oligonucleotides (Eurogentec) were transfected (100 μM) using siIMPORTER (Millipore) according to the manufacturer’s procedures. Negative control premiR (Ambion) or scrambled antagomiR were transfected as controls. Chemically modified antisense oligonucleotides (antagomiR) have been used to inhibit miR expression in vitro. The sequences of antagomiR-221 and antagomiR-222 used are as follows: 5′-GAAACCCAGCAGACAATGTAGCT-3′ and 5′-GAGACCCAGTAGCCAGATGTAGCT-3′, respectively. The scrambled antimiR sequence was 5′-CAGCTGAAGTAAATACCGACCAG-3′. Five 5′ and 3′ bases of each oligonucleotides were LNA (locked nucleic acid)-modified. Transfected cells were plated at low density (2,500 cells per 10-cm plate), grown for 7–10 days, fixed, and stained by crystal violet as described in ref. 11. To detect milder effects, cell growth was monitored 96 h after transfection by the MTT [(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyltetrazolium] assay.
Mouse Model.
E18 p53−/− liver progenitors were immortalized by Myc expression as described in ref. 34. Early passage immortalized progenitors were transduced with a MSCV-based retroviral vector expressing miR-221, empty vector, or an shRNA targeting APC (35). After puromycin selection, 3 million cells per flank were injected s.c. into irradiated NCR nu/nu mice (6-8 weeks of age) 24 h after irradiation (400 rad). Injections were performed in the two flanks of each animal. Tumor volume was measured by caliper and calculated as length × width × depth × (pi/6).
Analysis of Putative miR-221 Targets.
The different 3′UTRs were amplified from human genomic DNA with the Advantage 2 PCR kit (Ozyme-Clontech). Sequences of primers are available on request. PCR products were subcloned to pGEM T-easy vector (Promega) and sequenced. 3′UTRs were then cloned into psiCHECK-2 vector (Promega) and analyzed by using the Dual-luciferase Reporter assay system (Promega). Mutant 3′UTRs were obtained by using the QuikChange site-directed mutagenesis kit according to the manufacturer’s instructions (Stratagene). Details are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Annick Harel-Bellan for helpful discussion, Colin Crist for help in luciferase assay set-up, and Solene Lebrun for technical assistance. This work was supported by La Ligue Nationale contre le Cancer (Equipe Labellisée) and Agence Nationale de la Recherche. K.M. is supported by the Robert and Theresa Lindsay Fellowship of the Watson School of Biological Sciences. C.M.C. is supported by National Institutes of Health Grant R01-CA-135030. S.V. is supported by Associazione Italiana Per La Ricerca Sul Cancro Grant IG8588.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907904107/DCSupplemental.
References
- 1.Bosch FX, Ribes J, Díaz M, Cléries R. Primary liver cancer: worldwide incidence and trends. Gastroenterology. 2004;127(5, Suppl 1):S5–S16. doi: 10.1053/j.gastro.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 2.McGlynn KA, London WT. Epidemiology and natural history of hepatocellular carcinoma. Best Pract Res Clin Gastroenterol. 2005;19:3–23. doi: 10.1016/j.bpg.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 3.Kim W, et al. Comparison of proteome between hepatitis B virus- and hepatitis C virus-associated hepatocellular carcinoma. Clin Cancer Res. 2003;9:5493–5500. [PubMed] [Google Scholar]
- 4.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng S, Calin GA, Croce CM, Coukos G, Zhang L. Mechanisms of microRNA deregulation in human cancer. Cell Cycle. 2008;7:2643–2646. doi: 10.4161/cc.7.17.6597. [DOI] [PubMed] [Google Scholar]
- 6.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 7.Chang J, et al. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106–113. doi: 10.4161/rna.1.2.1066. [DOI] [PubMed] [Google Scholar]
- 8.Ladeiro Y, et al. MicroRNA profiling in hepatocellular tumors is associated with clinical features and oncogene/tumor suppressor gene mutations. Hepatology. 2008;47:1955–1963. doi: 10.1002/hep.22256. [DOI] [PubMed] [Google Scholar]
- 9.Felli N, et al. MicroRNAs 221 and 222 inhibit normal erythropoiesis and erythroleukemic cell growth via kit receptor down-modulation. Proc Natl Acad Sci USA. 2005;102:18081–18086. doi: 10.1073/pnas.0506216102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He H, et al. The role of microRNA genes in papillary thyroid carcinoma. Proc Natl Acad Sci USA. 2005;102:19075–19080. doi: 10.1073/pnas.0509603102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 12.le Sage C, et al. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zender L, et al. An oncogenomics-based in vivo RNAi screen identifies tumor suppressors in liver cancer. Cell. 2008;135:852–864. doi: 10.1016/j.cell.2008.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Galardi S, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282:23716–23724. doi: 10.1074/jbc.M701805200. [DOI] [PubMed] [Google Scholar]
- 15.Zhao JJ, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283:31079–31086. doi: 10.1074/jbc.M806041200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Fornari F, et al. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- 17.Wong QW, et al. MicroRNA-223 is commonly repressed in hepatocellular carcinoma and potentiates expression of Stathmin1. Gastroenterology. 2008;135:257–269. doi: 10.1053/j.gastro.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Jiang J, et al. Association of MicroRNA expression in hepatocellular carcinomas with hepatitis infection, cirrhosis, and patient survival. Clin Cancer Res. 2008;14:419–427. doi: 10.1158/1078-0432.CCR-07-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Varnholt H, et al. MicroRNA gene expression profile of hepatitis C virus-associated hepatocellular carcinoma. Hepatology. 2008;47:1223–1232. doi: 10.1002/hep.22158. [DOI] [PubMed] [Google Scholar]
- 20.Murakami Y, et al. Comprehensive analysis of microRNA expression patterns in hepatocellular carcinoma and non-tumorous tissues. Oncogene. 2006;25:2537–2545. doi: 10.1038/sj.onc.1209283. [DOI] [PubMed] [Google Scholar]
- 21.Li W, et al. Diagnostic and prognostic implications of microRNAs in human hepatocellular carcinoma. Int J Cancer. 2008;123:1616–1622. doi: 10.1002/ijc.23693. [DOI] [PubMed] [Google Scholar]
- 22.Ji J, et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology. 2009;50:472–480. doi: 10.1002/hep.22989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim YK, et al. Functional links between clustered microRNAs: suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 2009;37:1672–1681. doi: 10.1093/nar/gkp002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumamoto K, et al. Nutlin-3a activates p53 to both down-regulate inhibitor of growth 2 and up-regulate mir-34a, mir-34b, and mir-34c expression, and induce senescence. Cancer Res. 2008;68:3193–3203. doi: 10.1158/0008-5472.CAN-07-2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siomi H, Siomi MC. On the road to reading the RNA-interference code. Nature. 2009;457:396–404. doi: 10.1038/nature07754. [DOI] [PubMed] [Google Scholar]
- 26.Baek D, et al. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev. 2008;22:239–251. doi: 10.1101/gad.1617608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Meng F, et al. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Villanueva A, et al. Pivotal role of mTOR signaling in hepatocellular carcinoma. Gastroenterology. 2008;135:1972–1983. e1–e11. doi: 10.1053/j.gastro.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pineau P, et al. Homozygous deletion scanning in hepatobiliary tumor cell lines reveals alternative pathways for liver carcinogenesis. Hepatology. 2003;37:852–861. doi: 10.1053/jhep.2003.50138. [DOI] [PubMed] [Google Scholar]
- 31.Pineau P, et al. Chromosome instability in human hepatocellular carcinoma depends on p53 status and aflatoxin exposure. Mutat Res. 2008;653:6–13. doi: 10.1016/j.mrgentox.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simon R, Lam A. BRB-ArrayTools User Guide. Biometric Research Branch, National Cancer Institute; 2007. Version 3.2. http//linus.nci.nih.gov/brb. [Google Scholar]
- 34.Zender L, et al. Identification and validation of oncogenes in liver cancer using an integrative oncogenomic approach. Cell. 2006;125:1253–1267. doi: 10.1016/j.cell.2006.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dickins RA, et al. Probing tumor phenotypes using stable and regulated synthetic microRNA precursors. Nat Genet. 2005;37:1289–1295. doi: 10.1038/ng1651. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.