Abstract
Amyloid fibers are filamentous protein structures commonly associated with neurodegenerative diseases. Unlike disease-associated amyloids, which are the products of protein misfolding, Escherichia coli assemble membrane-anchored functional amyloid fibers called curli. Curli fibers are composed of two proteins, CsgA and CsgB. In vivo, the polymerization of the major curli subunit protein, CsgA, is dependent on CsgB-mediated nucleation. The amyloid core of CsgA features five imperfect repeats (R1–R5), and R1 and R5 govern CsgA responsiveness to CsgB nucleation and self-seeding by CsgA fibers. Here, the specificity of bacterial amyloid nucleation was probed, revealing that certain aspartic acid and glycine residues inhibit the intrinsic aggregation propensities and nucleation responsiveness of R2, R3, and R4. These residues function as “gatekeepers” to modulate CsgA polymerization efficiency and potential toxicity. A CsgA molecule lacking gatekeeper residues polymerized in vitro significantly faster than wild-type CsgA and polymerized in vivo in the absence of the nucleation machinery, resulting in mislocalized fibers. This uncontrolled polymerization was associated with cytotoxicity, suggesting that incorrectly regulated CsgA polymerization was detrimental to the cell.
Keywords: amyloid, curli, protein aggregation, nucleation, Escherichia coli
Amyloids are ordered proteinaceous fibers commonly associated with mammalian neurodegenerative diseases and prion-based encephalopathies (1). Amyloid fibers have distinct biochemical and biophysical properties, such as remarkable resistance to chemical and thermal denaturation, and specific tinctorial properties when bound to Congo red and thioflavin T (ThT) (1). The molecular basis of neurodegenerative disease development induced by amyloid propagation remains elusive, partially because of the seemingly erratic and uncontrolled nature of amyloidogenesis. An emerging focus of amyloid biosynthesis has shown that amyloids can also be an integral part of physiology found in different organisms including bacteria, fungi, and mammals (2, 3). How nature coordinates functional amyloid propagation and reduces the associated cytotoxicity is poorly understood.
Curli, a bacterially produced functional amyloid, is an important component of the extracellular matrix and is involved in bacterial community behaviors (4). Because of the amyloid properties of curli fibers (5, 6), the colonies of curli-producing Escherichia coli stain red when grown on Congo red indicator plates, which provides a convenient assay to monitor curli assembly in vivo (7). In E. coli, at least six proteins are dedicated to directing efficient curli formation. Curli fibers are composed of a major subunit CsgA and a minor subunit CsgB. CsgA remains unpolymerized until it encounters the surface-tethered nucleator CsgB, which initiates CsgA polymerization (8). CsgD is a transcriptional activator for the csgBA operon (4). CsgG, CsgE, and CsgF are nonfiber structural accessory proteins involved in secretion and stabilization of the fiber subunits and modulation of fiber assembly (6). CsgG is proposed to be the curli secretion apparatus that directs the secretion of CsgA, CsgB, and CsgF across the outer membrane (9, 10). CsgE and CsgF interact with CsgG at the outer membrane (9). CsgF is required for efficient CsgB-mediated nucleation, and CsgE is critical for CsgA, CsgB, and CsgF stability (6, 9, 10).
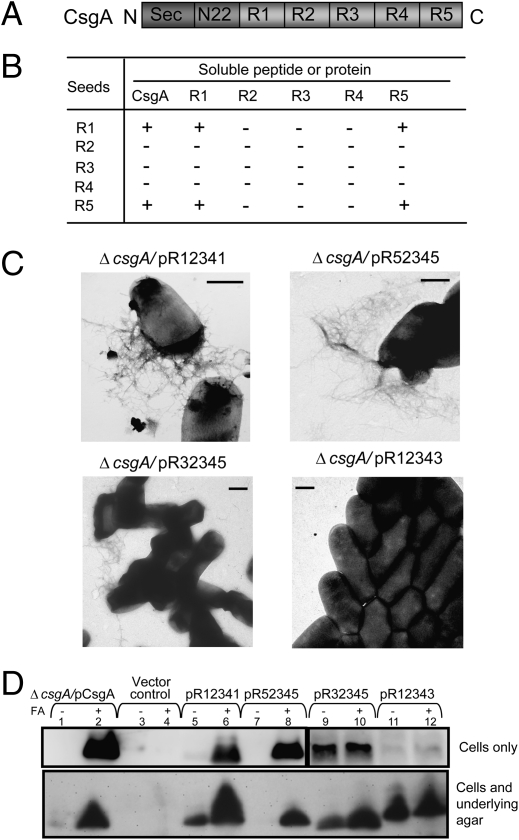
The CsgA primary amino acid sequence comprises a Sec-signal peptide (positions 1–20), a 22-residue (positions 21–42) CsgG-specific N-terminal domain, and an amyloid core region composed of five imperfect repeats (positions 43–151), each composed of 19–23 amino acids (Fig. 1A) (4). The repeats (R1, R2, R3, R4, and R5) share at least 30% identity at the amino acid level and are distinguished by the consensus sequence Ser-X5-Gln-X4-Asn-X5-Gln (11). We previously found that R1 and R5 direct CsgA to respond to CsgB-mediated nucleation and are critical for fiber elongation (12). Moreover, internally conserved Gln and Asn residues located in R1 and R5 of CsgA are required for CsgB nucleation and curli assembly (13). Because all five repeats have these critical Gln and Asn residues, it has been unclear why R1 and R5 are functionally distinct from R2, R3, and R4. We elucidated these determinants and found that certain Asp and Gly residues in R2, R3, and R4 function as gatekeeper residues, which inhibit the amyloidogenic properties of these repeating units.
Fig. 1.
Seeding specificity of CsgA was shared by R1 and R5, and R1 and R5 can replace each other. (A) The schematic of CsgA primary structure. (B) Summary of peptides seeding specificity. +, preformed fiber seeds that promote the polymerization of soluble proteins or peptides; −, preformed seeds that did not change polymerization significantly. (C) Negative-stain EM micrographs of the csgA mutant strain containing plasmids pR12341, pR52345, pR32345, or pR12343. (Scale bar, 500 nm.) (D) Western blots of whole-cell lysates (Top) or plugs (whole cells and underlying agar; Bottom) from csgA mutant cells containing plasmids pCsgA (lanes 1 and 2), vector control (lanes 3 and 4), pR12341 (lanes 5 and 6), pR52345 (lanes 7 and 8), pR32345 (lanes 9 and 10), or pR12343 (lanes 11 and 12) grown for 48 h at 26°C on YESCA plates. Cells were treated with (+) or without (−) FA before electrophoresis as indicated. The blots were probed with anti-CsgA antibody.
Gatekeeper residues are found in many globular proteins; they act to mask aggregation-prone sequences and thereby, promote native protein folding (14–17). Herein, we explore the role of gatekeeper residues in functional amyloidogenesis. Instead of simply inhibiting aggregation, we found that gatekeeper residues actually harness the aggregation of CsgA. A CsgA mutant lacking all gatekeeper residues efficiently polymerized into amyloid fibers independently of the curli assembly proteins CsgB and CsgF, leading to uncontrolled and mislocalized fibers. Moreover, the induced expression of CsgA without gatekeeper residues dramatically decreased the viability of E. coli cells.
Results
Seeding Specificity Is Encoded in R1 and R5.
Amyloid formation is a self-propagating process (1). Preformed amyloid fibers can accelerate the polymerization of soluble subunits by a mechanism called seeding (18). Our previous results showed that R1 and R5 of CsgA are required for both CsgA–CsgB interactions and CsgA–CsgA interactions (12). Among five repeating units, only R1 and R5 polymerization can be accelerated by CsgB and CsgA (12). In vitro, peptides composed of repeating units of R1, R3, and R5 efficiently polymerize into ThT fluorescence-positive fibrous structures (19). At higher concentrations, R2 and R4 also eventually assemble into a fibrous structure that did not significantly fluoresce in the presence of ThT (19). We used these preformed fibrous materials as seeds to directly test the interaction specificity among the repeating units of CsgA. Cross-seeding and self-seeding occurred for peptides R1 and R5 only (Fig. 1B). Preformed R1 or R5 fibers promoted CsgA polymerization (Fig. 1B). These results suggest that CsgA seeding specificity is determined and shared by R1 and R5. Recent solid-state NMR and electron microscopy data of CsgA fibers are consistent with the β-helix-like structure predicted by homology modeling where R1 and R5 are positioned at the interface of adjacent fiber subunits (5, 11).
R1 and R5 Are Interchangeable In Vivo.
R1 and R5 both can seed CsgA polymerization, and both repeating units constitute the nucleation and seeding responsive domains of CsgA (Fig. 1B) (12). In addition, cross-seeding occurred between R1 and R5 (Fig. 1B). These results suggest R1 and R5 are functionally redundant. To test this hypothesis in vivo, the residues of R1 of CsgA (R12345) were replaced by the corresponding residues from R5 to form the mutant construct with the following repeating unit order: R52345. The R52345 molecule was able to complement a csgA mutant and assembled into fibers on the cell surface (Fig. 1C and Table S1 ). Similarly, a CsgA analog in which R5 was replaced by R1 (pR12341) also assembled into fibers on the cell surface (Fig. 1C). Wild-type curli fibers are SDS insoluble, and formic acid (FA) treatment is required to dissociate the monomers from the fibers (20). An abundance of R52345 and R12341 can be resolved with FA treatment, suggesting that both of these chimeras form SDS insoluble fibers similar to those formed by wild-type CsgA (Fig. 1D, lanes 1, 2, 5, 6, 7, and 8). The in vivo polymerization of R52345 and R12341 was completely defective in csgBA cells, suggesting that their polymerization was still dependent on CsgB (Table S1).
Peptides composed of CsgA R3 can form ThT-positive short fibrous polymers, which suggests that R3 has some amyloidogenic properties (19). In addition, the sequence similarity (70%) and identity (48%) between R3 and R5 are higher than those between R1 and R5 (52% similarity and 39% identity). We asked whether or not R3 could substitute for R1 or R5 in vivo. R32345 was somewhat defective in vivo as observed by transmission electron microscopy (TEM) from csgA/pR32345 compared with csgA/pR12341 or csgA/pR52345 (Fig. 1C and Table S1). Furthermore, R32345 remained SDS soluble and did not require FA treatment to migrate into the gel (Fig. 1D, lanes 9 and 10). The phenotype of R12343 was even more dramatic. R12343 was not detected by Western blotting of the cell pellets but instead was only detected when both cells and the underlying agar were collected. This suggests that R12343 was stably secreted but defective in curli assembly (Fig. 1D, lanes 11 and 12). Very few fibers were produced by csgA/pR12343 when analyzed by TEM (Fig. 1C and Table S1). Therefore, R3 was not able to substitute for R5 at the C terminus and was only partially capable of replacing R1 at the N terminus, suggesting that the nucleation/CsgA self-seeding response and specificity was encoded by R1 and R5.
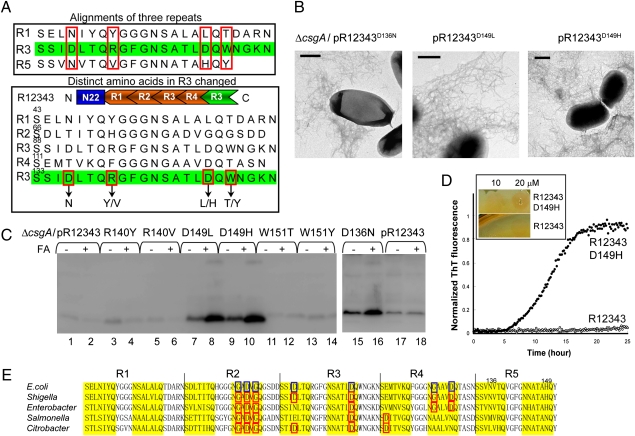
Identification of Gatekeeper Residues of R3 that Inhibit Amyloidogenic Properties.
Our results indicated that common residues found in both R1 and R5 are involved in the observed nucleation response and seeding specificity. Alternatively, R3 could contain specific residues that make it inert to the interactions critical for polymerization. Guided by this perspective, we compared R3 with R1/R5, looking for residues in R3 that were distinctive from both R1 and R5. Four residues were targeted as candidates: Asp136, Arg140, Asp149, and Trp151 in the molecule R12343 (Fig. 2A). If these residues hindered the ability of R3 to phenotypically replace R5, then changing these residues to those found in either R1 or R5 may result in a phenotype more similar to wild type. Constructs pR12343D136N, pR12343R140Y, pR12343R140V, pR12343D149L, pR12343D149H, pR12343W151T, and pR12343W151Y were engineered to test this hypothesis (Fig. 2A). Strikingly, R12343D136N, R12343D149L, and R12343D149H were able to assemble into amyloid-like fibers as measured by TEM (Fig. 2B), whereas other mutant proteins such as R12343R140Y, R12343R140V, R12343W151T, and 12343W151Y were as defective as R12343 in fiber formation in vivo as measured by TEM and Congo red binding Table S1). For R12343D136N, R12343D149L, and R12343D149H, there was more SDS insoluble than SDS soluble material detected by whole-cell Western analysis, unlike R12343R140Y, R12343R140V, R12343W151T, and 12343W151Y, which were not associated with cells (Fig. 2C). R12343R140Y, R12343R140V, R12343W151T, and 12343W151Y were stably expressed and secreted as measured by Western analysis of the cells and underlying agar (Fig. S1). In vitro polymerization of R12343D149H was dramatically faster than that of R12343 as measured by ThT fluorescence (Fig. 2D). In a CsgB-dependent manner, purified-soluble CsgA can be converted into amyloid fibers on csgA cells expressing CsgB as evidenced by TEM or Congo red staining (12). When R12343D149H and R12343 were spotted on csgA cells at 10 μM, R12343D149H assembled into fibers more readily than R12343 as indicated by stronger Congo red staining (Fig. 2D Inset).
Fig. 2.
Mutations of Asp residues of C-terminal R3 in R12343 rendered amyloidogenic properties. (A) Differences between R3 and R1/R5 amino acid sequences are indicated with red boxes at Top. The amino acid changes in C-terminal R3 of R12343 are indicated at Bottom. (B) Negative-stain EM micrographs of csgA cells containing plasmids pR12343D136N, pR12343D149L, and pR12343D149H. Cells were grown 48 h at 26°C on YESCA plates. (Scale bar, 500 nm.) (C) Western blots of whole-cell lysates from csgA cells containing constructs pR12343 (lanes 1, 2, 17, and 18), pR12343D136N (lanes 15 and 16), pR12343R140Y (lanes 3 and 4), pR12343R140V (lanes 5 and 6), pR12343D149L (lanes 7 and 8), pR12343D149H (lanes 9 and 10), pR12343W151T (lanes 11 and 12), and pR12343W151Y (lanes 13 and 14) grown 48 h at 26°C on YESCA plates. Samples were treated with (+) or without (−) FA as indicated. The blots were probed with anti-CsgA antibody. (D) The polymerization of 20-μM R12343D149H and R12343 measured by ThT fluorescence. Congo red staining of csgA (CsgB+) cells overlaid with10 μM and 20 μM R12343D149H and R12343 is shown (Inset). (E) The alignment of five repeating units of CsgA from different enteric bacteria. The consensus sequences Ser-X5-Gln and Asn-X5-Gln indicated in yellow background were predicted to form β-strands (11). The identified gatekeeper residues of CsgA from E. coli are indicated with blue boxes. The putative gatekeeper residues of CsgA from other enteric bacteria are indicated with red boxes.
There are at least two possibilities for why R12343D136N, R12343D149L, and R12343D149H function similarly to wild-type CsgA. First, Asp residues at positions 136 and 149 in R12343 could play a negative role in nucleation and fiber elongation. By changing the Asp residues, the amyloidogenic property was improved. Alternatively, Asn and His residues at positions 136 and 149 in wild-type CsgA are critical for CsgA polymerization. To elucidate these possibilities, the Asp residue of R12343 at position 149 was changed to Arg or Lys, and curli assembly was measured. Although His, Arg, and Lys have side chains with different properties, R12343D149R and R12343D149K could assemble into fibers in vivo similarly to R12343D149H as detected by Congo red binding and Western blotting (Table S1), suggesting that at least His at position 149 is not absolutely required for fiber assembly. In addition, the Asn136 and His149 residues of CsgA are not conserved among CsgA homologs from different enteric bacteria (Fig. 2E). These observations indicate that the Asn136 and His149 residues are not essential for fiber formation. Therefore, it is plausible that the Asp residues at positions 136 and 149 in the molecule R12343 specifically impeded the amyloidogenic properties of this mutant protein and that the Asp residues prevented R3 peptide polymerization. To further explore this hypothesis, Asp residues were introduced into R5 of wild-type CsgA at positions 136 and 149. The resulting mutant CsgAN136D/H149D was stably secreted but defective in fiber assembly in vivo as detected by whole-cell Western analysis and TEM (Fig. S2 and Table S1). In addition, an R3 derivative peptide with these Asp residues changed to R1-like residues (D4N/D17L) was synthesized and found to polymerize significantly faster than the R3 peptide, supporting the idea that these Asp residues inhibit the intrinsic aggregation of R3 (Fig. S2). In summary, we designated gatekeeper residues in CsgA as those that negatively influence CsgA assembly and that substitution to other residues results in an amelioration of CsgA polymerization. Therefore, we designated the Asp residues at position 91 and 104 in R3 peptides as gatekeeper residues, because they prevented R3 from participating in critical interactions needed for polymerization.
Identification of Gatekeeper Residues in R2 and R4.
Peptides R2 and R4 have poor amyloidogenic properties and cannot efficiently self-assemble into ThT-positive amyloid fibers in vitro (19). The CsgA mutant proteins R2345 (ΔR1) and R1234 (ΔR5) were defective in curli assembly, possibly because R2 and R4 cannot function like R1 or R5 (12). To test the hypothesis that R2 and R4 also contain gatekeeper residues, we compared R4 with R1/R5, looking for residues in R4 that were different from both R1 and R5. Four residues were targeted as candidates: Thr114, Lys116, Gly123, and Asp127 in the molecule R1234 (Fig. S3A). When Gly123 or Asp127 were mutated to the corresponding residues from R5 (Asn and His, respectively), in vivo amyloidogenesis was mildly improved as detected by Congo red binding, Western analysis, and TEM (Fig. S3 and Table S1). However, when both Gly123 and Asp127 were mutated together, curli assembly was significantly improved, suggesting that Gly123 and Asp127 are gatekeeper residues in R4 (Fig. S3 and Table S1). All R1234-derivative mutants were stably expressed as shown by Western analysis of the cells and underlying agar (Fig. S3). Similarly, Asp67, Gly78, Asp80, and Gly82 (the positions of these amino acids are defined in wild-type CsgA) were targeted as potential gatekeeper residues in R2 in a molecule R2345 (Fig. S4). After Gly78, Asp80 and Gly82 were mutated to the corresponding residues from R1 (Ser, Leu, and Leu, respectively). R2345 could efficiently assemble into fibers as detected by Western analysis, Congo red binding, and TEM, suggesting that Gly78, Asp80, and Gly82 comprise the gatekeeper residues of R2 (Fig. S4). Not all Asp residues in R2 function as gatekeeper residues. When Asp67 was mutated to Glu, the resulting mutant R2345D67E was as defective as R2345, indicating that Asp67 is not a gatekeeper residue (Fig. S4). To test whether or not these gatekeeper residues impede self-polymerization of R2 or R4, we tested self-polymerization of the R4-derivative peptide with mutations of gatekeeper residues (R4G13N/D17H). When the gatekeeper residues were mutated to R5-like residues, R4G13N/D17H polymerized into amyloid fibers much more efficiently compared with the original R4 peptide as measured by ThT fluorescence Fig. S3). Therefore, the residues Gly78, Asp80, Gly82, Gly123, and Asp127 in R2 and R4 of wild-type CsgA were designated as gatekeeper residues.
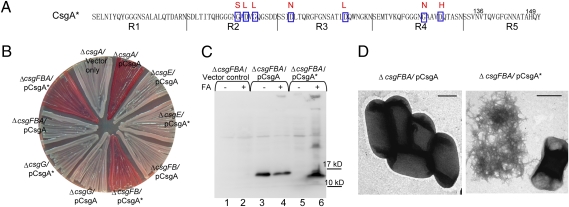
CsgA Lacking All Identified Gatekeeper Residues Polymerized in the Absence of CsgB and CsgF In Vivo.
The five repeating units of CsgA have the consensus sequence Ser-X5-Gln-X4-Asn-X5-Gln. The X5 regions between Ser/Gln or Asn/Gln were predicted to comprise of β-strands in CsgA (11). Seven gatekeeper residues (Gly78, Asp80, Gly82, Asp91, Asp104, Gly123, and Asp127 for E. coli CsgA) found in R2, R3, and R4 were located in these five-residue stretches (Fig. 2E). R1 and R5 of CsgA homologs from different enteric bacteria do not have any Asp or Gly residues in the same regions (Fig. 2E). The introduction of Asp residues into R5 of wild-type CsgA at positions of Asp residues in R3 resulted in defective fiber assembly (Fig. S2), suggesting that gatekeeper residues are not tolerated at the critical end repeats. All of the CsgA homologs from different enteric bacteria have putative gatekeeper residues in R2, R3, and R4, indicating that the gatekeeper residues may play an important role in CsgA assembly. To gain some insight into the roles of gatekeeper residues in bacterial amyloid propagation, we constructed a CsgA mutant lacking all gatekeeper residues called CsgA*. In the amino acid sequence of CsgA*, all of the gatekeeper residues were replaced by the corresponding residues found in R1 or R5 (CsgAG78S/D80L/G82L/D91N/D104L/G123N/D127H) (Fig. 3A). CsgB and CsgF are required for efficient polymerization of wild-type CsgA in vivo (6, 8). Surprisingly, in the absence of CsgB and CsgF, CsgA*-expressing cells contained a deep Congo red staining pattern, which suggests that the fibers had been assembled (Fig. 3B). Western blotting of csgFBA/pCsgA cells and the underlying agar showed that wild-type CsgA remained SDS soluble in the absence of CsgB and CsgF (Fig. 3C, lanes 3 and 4), whereas CsgA* from csgFBA/pCsgA* cells was SDS insoluble (Fig. 3C, lanes 5 and 6). Many of the CsgA* fibers formed in csgFBA genetic background were mislocalized and were not tethered with cells as detected by TEM, whereas no fibers were produced by csgFBA/pCsgA cells (Fig. 3D). In both csgBA and csgFB genetic backgrounds, CsgA* formed SDS insoluble fibers and bound Congo red (Fig. 3B, Fig. S5, and Table S1). After cells were scraped off the plate, the underlying agar was visibly red, again suggesting that fibers composed of CsgA* were mislocalized (Fig. S6). Like CsgA, the stability of CsgA* was still dependent on CsgG and CsgE. In the absence of CsgG or CsgE, CsgA* was undetectable by Western blotting of cells and underlying agar and could not bind Congo red (Fig. 3B and Fig. S5). Collectively, secretion and stability of CsgA* is dependent on CsgG and CsgE. However, in contrast to wild-type CsgA, the polymerization of CsgA* in vivo does not require CsgB and CsgF. It is possible that these gatekeeper residues modulate CsgA fiber assembly by keeping the polymerization of CsgA dependent on CsgB and CsgF.
Fig. 3.
In vivo polymerization of CsgA* is independent of nucleator CsgB and CsgF. (A) Seven gatekeeper residues were changed to R1/R5-like residues (indicated above the CsgA sequence) in CsgA*. (B) A Congo red-containing YESCA plate with csgA, csgFBA, csgG, csgFB, and csgE cells transformed with the plasmids encoding CsgA or CsgA*. Cells were grown 48 h at 26°C on YESCA plates. (C) Western blot of whole cells and underlying agar (agar plugs) from csgFBA cells containing control vector (lanes 1 and 2), pCsgA (lanes 3 and 4), or pCsgA* (lanes 5 and 6). Samples were treated with (+) or without (−) FA as indicated. The blots were probed with anti-CsgA antibody. The electrophoretic mobility of wild-type CsgA was reported to be slightly slower than its molecular weight (5). CsgA* migrated slightly faster than wild-type CsgA. Similarly, CsgA* purified from the cytosol also migrated faster than CsgA, suggesting that the aberrant migration may be caused by the intrinsic properties of CsgA* compared with wild-type CsgA. (D) Negative-stain EM micrographs of csgFBA cells containing plasmids pCsgA or pCsgA*. (Scale bar, 500 nm.)
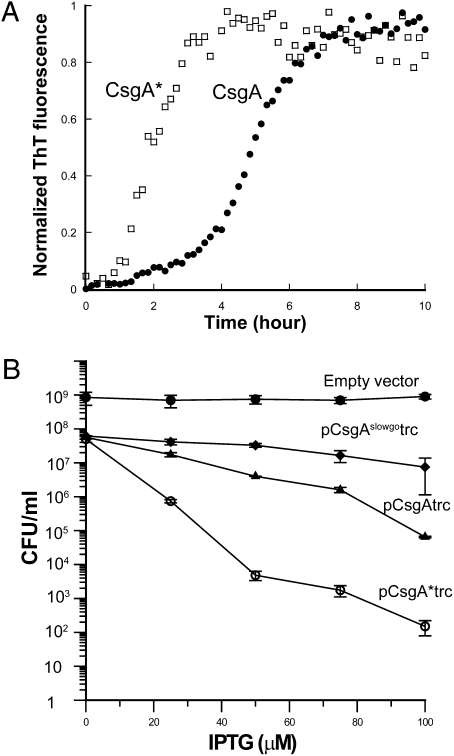
CsgA* Polymerized Faster than CsgA In Vitro and Induced Expression of CsgA* Caused E. coli to Be Less Viable.
To understand the biochemical distinction between CsgA* and CsgA, we purified CsgA and CsgA* and compared their polymerization kinetics in vitro using a previously described method (19). Wild-type CsgA was found to be secreted to the medium by co-overexpressing CsgA and CsgG (19). Overexpression of CsgA* resulted in cell lysis, and no CsgA* was secreted to the medium or associated with cells, which suggests that CsgA* is potentially cytotoxic. We took an alternative approach to purify mature CsgA and CsgA*. A vector (pET11d) containing CsgA or CsgA* lacking the Sec-dependent periplasmic signal sequence was transformed into the expression strain NEB C2566. Protein expression was induced by Isopropyl β-D-1-thiogalactopyranoside (IPTG), and CsgA or CsgA* was purified from cell pellets that were resuspended in guanidine hydrochloride (GdnHCl; see Materials and Methods). Wild-type CsgA polymerization followed a triphasic process with lag, rapid growth, and stationary phases, similar to previous observations (19). The polymerization lag phase for CsgA at a concentration higher than 5 μM was around ∼2 h as detected by the ThT assay (Fig. 4A) (19). In contrast to CsgA, CsgA* started polymerizing without a clear lag phase, and the total time interval for its ThT signal to reach stationary phase was 3 h compared with 7 h for wild-type CsgA (Fig. 4A). Like CsgA, CsgA* self-assembled aggregates were ordered amyloid-like fibers as evidenced by TEM (Fig. S7).
Fig. 4.
In vitro polymerization of CsgA* is faster than that of CsgA, and induced expression of CsgA* causes cells less viable. (A) The polymerization of CsgA and CsgA* at 7 μM was measured by ThT fluorescence. (B) Normalized cell suspensions of csgA containing empty vector or plasmids encoding CsgAslowgo, CsgA, or CsgA* under trc promoter were spotted on LB chloramphenicol plates in the presence of IPTG at various concentrations. Colony forming units of serial dilutions of cell suspensions were manually counted.
Although there was no obvious cytotoxicity when CsgA* was expressed under the csgBA native promoter, significant cytotoxicity was observed when CsgA* was expressed under the control of an inducible promoter (the trc promoter). To carefully compare the potential cytotoxicity of CsgA-like molecules, we measured cell viability when full-length CsgA, CsgA*, or CsgAslowgo (CsgAQ49A/N54A/Q139A/N144A) expression was induced. CsgAslowgo was previously reported to be defective in curli assembly, and its self-polymerization had a lag time two orders of magnitude greater than that of CsgA polymerization (13). The viability of csgA cells (MC4100 background) did not change significantly with the expression of CsgAslowgo (Fig. 4B). At relatively high IPTG concentration (>50 μM), expression of wild-type CsgA resulted in less viable cells compared with either CsgAslowgo or empty vector control (Fig. 4B). The induced expression of CsgA* with IPTG at 25 μM caused a marked loss of cell viability compared with wild-type CsgA (Fig. 4B). The colony-forming units of cells expressing CsgA* were approximately three orders of magnitude lower than those of cells expressing CsgA when IPTG concentration was 50 μM or higher (Fig. 4B). Expression of CsgA* resulted in similar decreased viability of various E. coli K12 cells such as C600 and BW25113. These results suggest that the presence of gatekeeper residues of CsgA decreases potential cytotoxicity.
Discussion
It was postulated that charged amino acid residues and other β-breaker residues in globular proteins could function as gatekeepers to prevent amyloid formation (16, 17, 21). Results from several different studies suggest that Pro, Asp, and Gly are among the residues least likely to be incorporated into a β-sheet structure (22, 23). Data presented here identified specific residues (gatekeeper residues) that inhibited the amyloidogenic properties of internal CsgA repeats. Our work showed that the absence of gatekeeper residues resulted in uncontrolled aggregative properties and increased potential cytotoxicity. It is possible that the CsgA amino acid sequence had evolved to balance self-polymerization and still remained under the control of the curli nucleation proteins so that polymerization could occur faithfully at the correct time and place.
Roles of Gatekeeper Residues of CsgA in Curli Biogenesis.
During the polymerization of disease-associated amyloids, the rate-limiting step is the formation of metastable protein oligomers or nuclei (18, 24). For curli assembly, amyloid nucleation is proposed to be facilitated by the CsgB protein, which adopts a β-sheet-rich secondary structure and propagates its β-sheet-rich amyloid fold onto CsgA molecules (25). This cross-nucleation mechanism separates nucleation and fiber elongation and controls curli assembly to occur at correct time and location (25). In addition, wild-type nucleation is also dependent of CsgF, an outer membrane-localized chaperone-like protein (10). CsgA polymerization in vivo is a concerted process and is under the control of the assembling factors. The gatekeeper residues of CsgA possibly provide a control mechanism for curli assembly. Self-polymerization of a CsgA mutant lacking all gatekeeper residues (CsgA*) is significantly faster than CsgA, and there is a limited lag phase before rapid polymerization (Fig. 4A). The strong aggregation propensity of CsgA* results in efficient polymerization in the extracellular environment even in the absence of CsgB and CsgF (Fig. 3 and Fig. S5). Many of the CsgA* fibers assembled in the absence of CsgB were not associated with cells and thereby, presumably may not help cells to attach to the surface. It is plausible that gatekeeper residues of CsgA modulate CsgA polymerization efficiency so that its polymerization and localization are under the control of CsgB and CsgF. In general, amyloid proteins, including functional amyloids, have less Pro and Gly residues than elastomeric proteins (26). In the E. coli proteome, 90% of 26,601 sequences predicted by TANGO (a computer algorithm for prediction of aggregating regions in unfolded polypeptide chains) to be aggregation-prone have at least one charged residue or proline at the first position on either side of the sequence, presumably to oppose aggregation (15, 16). Different gatekeeper residues seem to have varying influences on curli amyloidogenic properties. For instance, the amyloidogenic properties of R12343D149R and R12343D149K were significantly improved compared with R12343 (Table S1), although Asp, Arg, and Lys are all considered potential gatekeeper residues (15). Employing gatekeeper residues seems to be a common strategy to modulate protein aggregation.
Positional Effects of Gatekeeper Residues in CsgA.
CsgA amyloid formation is guided by both positive and negative aggregation determinants. The positive determinants are the regularly spaced Gln and Asn residues in each CsgA repeating unit that are conserved in CsgA homologs in different species. CsgA nucleation by CsgB requires the participation of specific conserved Gln and Asn residues in R1 and R5 of CsgA (13). The middle repeats (R2, R3, and R4) also have Gln and Asn residues at these conserved positions, yet these repeating units do not respond to CsgB nucleation (12). Using rationally designed mutagenesis, we elucidated negative aggregation determinants and found specific Asp and Gly residues of R2, R3, and R4 that prevented CsgB nucleation and self-assembly (Fig. 2 and Figs. S3 and Figs. S4). R2, R3, and R4 were converted to R1/R5 if these Asp and Gly residues were mutated. These Asp and Gly residues were only found in the middle repeats of CsgA homologs from different enteric bacteria, not the N-terminal or C-terminal repeating units (Fig. 2E), and our data also showed that gatekeeper residues in CsgA were not tolerated at end repeats (Fig. 2 and Figs. S2). These results suggest that the distribution of these gatekeeper residues in CsgA is not random and that the positions of N- and C-termini of CsgA are important for fiber assembly.
Potential Cytotoxicity of Functional Amyloids Was Reduced by Gatekeeper Residues.
It has been previously reported that the efficiency of amyloid formation correlates to cytotoxicity and disease development in various models (24, 27–29). Here, different CsgA derivatives with distinct polymerization properties have distinguishable cytotoxicity, which supports this argument (Fig. 4B). The induced expression of rapidly polymerizing CsgA* was more cytotoxic compared with equivalent expression of wild-type CsgA (Fig. 4B). One possibility is that the existence of gatekeeper residues in CsgA decreases the load of aggregated proteins for cellular molecular chaperones and dedicated quality-control machinery. Alternatively, gain of toxic properties of CsgA* may be distinct from wild-type CsgA. Functional amyloid propagation is a balance to maximize polymerization efficiency while minimizing potential cytotoxicity associated with amyloidogenesis.
Materials and Methods
Bacterial Growth.
Bacteria were grown at 26°C on yeast extract casamino acids (YESCA) plates to induce curli production (6). When needed, antibiotics were added to the plates in the following concentrations: kanamycin, 50 μg/mL; chloramphenicol, 25 μg/mL; and ampicillin, 100 μg/mL. Curli production was monitored using Congo red YESCA plates (6).
Bacterial Strains and Plasmids.
Curli assembly phenotypes of different strains are summarized in Table S1. Strains, plasmids, and primer sequences used in this study are listed in Table S2 and S3. The details of plasmid construction are included in SI Materials and Methods.
Purification of CsgA* and CsgA.
C-terminal His6 tagged CsgA* and CsgA without the Sec periplasmic signal sequence were purified from cell pellets. The details of the purification procedure are included in SI Materials and Methods.
Preparation of Peptides.
Peptides R3 and R4 and derivative peptides R3D4N/D17L and R4G13N/D17H were synthesized by Proteintech Group Inc. Peptides were prepared similarly as previously described (see SI Materials and Methods) (12).
ThT Assay.
ThT assays were performed and normalized as previously described (12). Seeds were prepared as previously described (see SI Materials and Methods) (19).
TEM.
A Philips CM12 transmission electron microscope was used to visualize the cells and fiber aggregates. Samples were placed on formvar-coated copper grids (Ernest F. Fullam, Inc.) for 2 min, washed with deionized water two times, and stained with 2% uranyl acetate for 90 s.
SDS/PAGE and Western Blotting.
Bacteria whole-cell lysates or cells and underlying agar (plugs) were prepared and probed for CsgA using previously described methods (6). The Western blotting of whole-cell lysate detects the proteins associated with cells, whereas Western blotting of plugs detects both proteins associated with cells and those secreted to the agar plate.
Overlay Assay.
Purified proteins were spotted on the csgA cells and stained with 0.5 mg/mL Congo red solution as previously described (12).
Spotting Assay for Cell Viability.
Freshly transformed cells with plasmids pCsgAtrc, empty vector pNH3, pCsgAslowgotrc, or pCsgA*trc were scraped off the plates and normalized by absorbance at 600 nm. Cells with serial dilutions were spotted on LB chloramphenicol plates with various concentrations of IPTG to induce expression of CsgA, CsgAslowgo, or CsgA*. After overnight growth, the colony number was manually counted, and colony forming units per milliliter of cell suspension under different IPTG concentrations were calculated.
Supplementary Material
Acknowledgments
We thank the members of the Chapman Laboratory for helpful discussions and review of this manuscript. This work was supported by the National Institutes of Health Grant AI073847-02.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908714107/DCSupplemental.
References
- 1.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 2.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid—from bacteria to humans. Trends Biochem Sci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 3.Hammer ND, Wang X, McGuffie BA, Chapman MR. Amyloids: Friend or foe? J Alzheimers Dis. 2008;13:407–419. doi: 10.3233/jad-2008-13406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnhart MM, Chapman MR. Curli biogenesis and function. Annu Rev Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shewmaker F, et al. The functional curli amyloid is not based on in-register parallel beta-sheet structure. J Biol Chem. 2009;284:25065–25076. doi: 10.1074/jbc.M109.007054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman MR, et al. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science. 2002;295:851–855. doi: 10.1126/science.1067484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collinson SK, et al. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hammar M, Bian Z, Normark S. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci USA. 1996;93:6562–6566. doi: 10.1073/pnas.93.13.6562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol. 2006;59:870–881. doi: 10.1111/j.1365-2958.2005.04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nenninger AA, Robinson LS, Hultgren SJ. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci USA. 2009;106:900–905. doi: 10.1073/pnas.0812143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collinson SK, Parker JM, Hodges RS, Kay WW. Structural predictions of AgfA, the insoluble fimbrial subunit of Salmonella thin aggregative fimbriae. J Mol Biol. 1999;290:741–756. doi: 10.1006/jmbi.1999.2882. [DOI] [PubMed] [Google Scholar]
- 12.Wang X, Hammer ND, Chapman MR. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem. 2008;283:21530–21539. doi: 10.1074/jbc.M800466200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Chapman MR. Sequence determinants of bacterial amyloid formation. J Mol Biol. 2008;380:570–580. doi: 10.1016/j.jmb.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rousseau F, Serrano L, Schymkowitz JW. How evolutionary pressure against protein aggregation shaped chaperone specificity. J Mol Biol. 2006;355:1037–1047. doi: 10.1016/j.jmb.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 16.Monsellier E, Chiti F. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Otzen DE, Kristensen O, Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: A structural clue to amyloid assembly. Proc Natl Acad Sci USA. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jarrett JT, Lansbury PT., Jr Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer's disease and scrapie? Cell. 1993;73:1055–1058. doi: 10.1016/0092-8674(93)90635-4. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Smith DR, Jones JW, Chapman MR. In vitro polymerization of a functional Escherichia coli amyloid protein. J Biol Chem. 2007;282:3713–3719. doi: 10.1074/jbc.M609228200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collinson SK, Emödy L, Müller KH, Trust TJ, Kay WW. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thirumalai D, Klimov DK, Dima RI. Emerging ideas on the molecular basis of protein and peptide aggregation. Curr Opin Struct Biol. 2003;13:146–159. doi: 10.1016/s0959-440x(03)00032-0. [DOI] [PubMed] [Google Scholar]
- 22.Kallberg Y, Gustafsson M, Persson B, Thyberg J, Johansson J. Prediction of amyloid fibril-forming proteins. J Biol Chem. 2001;276:12945–12950. doi: 10.1074/jbc.M010402200. [DOI] [PubMed] [Google Scholar]
- 23.Street AG, Mayo SL. Intrinsic beta-sheet propensities result from van der Waals interactions between side chains and the local backbone. Proc Natl Acad Sci USA. 1999;96:9074–9076. doi: 10.1073/pnas.96.16.9074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selkoe DJ. Folding proteins in fatal ways. Nature. 2003;426:900–904. doi: 10.1038/nature02264. [DOI] [PubMed] [Google Scholar]
- 25.Hammer ND, Schmidt JC, Chapman MR. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci USA. 2007;104:12494–12499. doi: 10.1073/pnas.0703310104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rauscher S, Baud S, Miao M, Keeley FW, Pomès R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure. 2006;14:1667–1676. doi: 10.1016/j.str.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 27.Luheshi LM, et al. Systematic in vivo analysis of the intrinsic determinants of amyloid Beta pathogenicity. PLoS Biol. 2007;5:e290. doi: 10.1371/journal.pbio.0050290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Nostrand WE, Melchor JP, Cho HS, Greenberg SM, Rebeck GW. Pathogenic effects of D23N Iowa mutant amyloid beta -protein. J Biol Chem. 2001;276:32860–32866. doi: 10.1074/jbc.M104135200. [DOI] [PubMed] [Google Scholar]
- 29.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer's disease: Progress and problems on the road to therapeutics. Science. 2002;297:353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.