Abstract
We identified an autosomal recessive condition in 11 individuals in the Old Order Amish of northeastern Ohio. The syndrome was characterized by distinctive craniofacial dysmorphism, skeletal anomalies, and mental retardation. The typical craniofacial dysmorphism included brachycephaly, highly arched bushy eyebrows, synophrys, long eyelashes, low-set ears, microdontism of primary teeth, and generalized gingival hyperplasia, whereas Sprengel deformity of scapula, fusion of spine, rib abnormities, pectus excavatum, and pes planus represented skeletal anomalies. The genome-wide homozygosity mapping using six affected individuals localized the disease gene to a 3.3-Mb region on chromosome 1q23.3-q24.1. Candidate gene sequencing identified a homozygous frameshift mutation, c.139_140delAG, in the transmembrane and coiled-coil domains 1 (TMCO1) gene, as the pathogenic change in all affected members of the extended pedigree. This mutation is predicted to result in a severely truncated protein (p.Ser47Ter) of only one-fourth the original length. The TMCO1 gene product is a member of DUF841 superfamily of several eukaryotic proteins with unknown function. The gene has highly conserved amino acid sequence and is universally expressed in all human tissues examined. The high degree of conservation and the ubiquitous expression pattern in human adult and fetal tissues suggest a critical role for TMCO1. This report shows a TMCO1 sequence variant being associated with a genetic disorder in human. We propose “TMCO1 defect syndrome” as the name of this condition.
Keywords: Amish, transmembrane and coiled-coil domains 1 gene, genotyping and mapping, homozygosity, SNP arrays
Although there are a significant number of autosomal phenotypes associated with syndromic and nonsyndromic mental retardation, the identification of these genes has been relatively slow, and the underlying pathophysiology of mental retardation remains unexplained in most cases (1). The use of homozygosity mapping in consanguineous families and isolated populations is a powerful tool for revealing genetic lesions in various recessive conditions (2–6). Yet, this approach, as with conventional linkage mapping, requires that the clinical phenotype be well defined and easily recognizable. Unfortunately, mental retardation as a clinical descriptor is nonspecific unless found in association with other features. Here, we describe a unique genetic condition characterized by craniofacial dysmorphism, skeletal anomalies, and mental retardation in the Old Order Amish of northeastern Ohio. Through a genome-wide mapping study, we localized the disease gene to chromosome 1q23.3-q24.1 and identified a homozygous frameshift mutation in the transmembrane and coiled-coil domains 1 gene (TMCO1) as the cause of this autosomal recessive condition, thus we propose “TMCO1 defect syndrome” as the name for this disease.
Results
Clinical Phenotype.
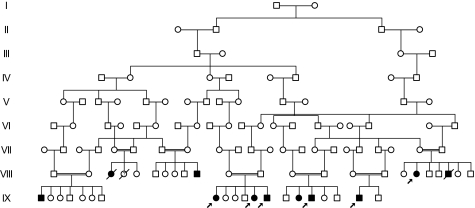
TMCO1 defect syndrome was diagnosed in 11 individuals (6 males, 5 females) ranging in age from 3 to 39 years. All 11 patients demonstrated Old Order Amish ancestry, and genealogical analyses revealed multiple lines of common descent between all parents of affected children (Fig. 1). The phenotype was not observed in the parents or 23 unaffected siblings.
Fig. 1.
Partial pedigree of the family with TMCO1 defect syndrome. Filled symbols represent affected individuals, and open symbols represent unaffected individuals. Circles and squares denote females and males, respectively. A double line identifies consanguinity. Arrows indicate affected individuals included in genetic mapping study and sequence analysis.
A history of first trimester spontaneous abortions was reported by four mothers with an overall incidence of 10 of 45 pregnancies (22%). The abnormalities in affected individuals were observed from prenatal period (Table 1). Ventriculomegaly, cleft lip and palate were noted through prenatal ultrasound in one patient, although this procedure was not routinely performed in this ethnic group. All affected individuals were born with normal birth weight for their gestational age (10 of 11 being full term), and five exhibited length exceeding the 90th percentile. Nine infants were reportedly macrocephlic at birth, probably secondary to their dysmorphic craniofacial features because their occipitofrontal circumference in fact was not significantly increased (range 3–95%, average 33%) (Table 1 and Table 2). Hypotonia and poor feeding were found in all newborns. Routine hematologic and metabolic assays were generally within normal ranges. The standard karyotype analysis was performed on at least four patients and reported as normal.
Table 1.
Clinical features of 11 patients with TMCO1 defect syndrome
| Features | Incidence or average age |
| Prenatal and neonatal | |
| Polyhydramnios | 36% (4/11) |
| Decreased fetal movements | 36% (4/11) |
| Macrocephalic appearance | 90% (9/10) |
| Hypotonia | 100% (11/11) |
| Poor feeding | 100% (11/11) |
| Craniofacial | |
| Brachycephaly and flat face | 100% (11/11) |
| Low hairline | 100% (11/11) |
| Low-set ears | 100% (11/11) |
| Highly-arched bushy eyebrows | 100% (11/11) |
| Synophrys | 100% (11/11) |
| Orbital hypertelorism | 100% (11/11) |
| Long eyelashes | 100% (11/11) |
| Wide nose bridge | 100% (11/11) |
| Short nose with anterverted nares | 100% (11/11) |
| Oral and dental | |
| High-arched palate | 100% (11/11) |
| Cleft lip and palate | 27% (3/11) |
| Age of primary tooth eruption | 15 months (8-24) |
| Microdontism of primary teeth | 100% (9/9) |
| Generalized gingival hyperplasia | 100% (8/8) |
| Skeletal | |
| Craniosynostosis | 18% (2/11) |
| Fusion of spine | 55% (6/11) |
| Short neck | 55% (6/11) |
| Scoliosis | 64% (7/11) |
| Sprengel deformity of scapula | 80% (8/10) |
| Pectus excavatum | 82% (9/11) |
| Rib anomalies | 55% (6/11) |
| Long and hyperextensible fingers | 55% (6/11) |
| Pes planus | 100% (11/11) |
| Club feet | 27% (3/11) |
| Developmental | |
| Roll over | 11 months (5-24) |
| Sit up | 19 months (11-36) |
| Walk | 36 months (18-60) |
| Talk | 33 months (16-48) |
| Neurological | |
| Unstable gait | 100% (9/9) |
| Intention tremor | 55% (6/11) |
| Depressed deep tendon reflexes | 90% (9/10) |
| Prominence of ventricles viewed through CT or MRI | 57% (4/7) |
| Psychosocial | |
| Mental retardation with an average full scale IQ | 56 (51-61) |
| Nonverbal | 45% (5/11) |
| Sluggish speech with hoarse and loud voice | 100% (6/6) |
| Anxiety | 64% (7/11) |
| Feed themselves | 100% (10/10) |
| Dress themselves | 75% (6/8) |
| Toilet training | 75% (6/8) |
| Others | |
| Genital urinary truck abnormalities | 45% (5/11) |
| Strabismus | 45% (5/11) |
| Frequent otitis media | 73% (8/11) |
| Frequent sinusitis | 82% (9/11) |
| Mild hypertrichosis | 64% (7/11) |
| Constipation | 64% (7/11) |
Incidence is expressed as a percentage with the number of patients applied in parentheses. The numbers in parentheses behind average age are ranges.
Table 2.
Additional clinical features in individual patients with TMCO1 defect syndrome
| Patient | Age, yr | Wt at birth | Length at birth | OFC at birth | Wt at examination | Ht at examination | Additional clinical information |
| VII-3 | 28 | 3.884 (85%) | 55.9 (99%) | N/A | N/A | 157 (18%) | Fusion of thoracic spine and fused ribs (detailed unknown), died from sudden death at age of 28. |
| VII-10 | 30 | 3.091 (22%) | 53.3 (90%) | N/A | 79.4 (78%) | 180 (69%) | Fusion of thoracic spine, missing ribs, hydrocele, undescended right testis, optic nerve atrophy. |
| VII-18 | 39 | 3.487 (57%) | 48.3 (34%) | N/A | 101.5 (99%) | 163 (50%) | Short neck with fusion of C2 to C4 spine, obsessive-compulsive tendencies, depression. |
| VII-21 | 28 | 4.167 (90%) | 56.5 (99%) | N/A | 36.4 (<1%) | 157.5 (<1%) | Cervical spine fusion, rib deformities, hiatal hernia, right kidney and adrenal gland agenesis, died from spontaneous intestinal perforation and its complications at age of 28. |
| VIII-1 | 16 | 3.232 (29%) | 48.3 (26%) | N/A | 27.8 (<1%) | 143 (<1%) | Hydrocele, grade II vesicoureteral reflux, calyceal diverticulum, cyst in upper pole of left kidney. |
| VIII-8 | 12 | 3.317 (43%) | 53.3 (93%) | 33.5 (22%) | 52.6 (86%) | 154 (67%) | Bilateral asymmetry of ribs, attention deficit hyperactivity disorder. |
| VIII-12 | 5 | 3.430 (95%) | 44.5 (20%) | 35.5 (95%) | 20.4 (92%) | 102 (40%) | T2 to T5 spine fusion, broad or irregular ribs, partial fusion of ribs (4, 5 and 6) bilaterally, ventricular septal defect, right aortic arch, hypoplastic pituitary gland with hypopituitarism, absent infundibulum and ectopic posterior pituitary gland, left ear sensorineural hearing loss. |
| VIII-13 | 3 | 3.317 (35%) | 49.5 (43%) | 36 (54%) | 11.2 (3%) | 84 (1%) | C7 to T5 spine fusion, mutiple bifid ribs (3, 4 and 5), hypoplastic fourth rib and partial fusion of fifth rib, mild hydronephrosis within left kidney and right kidney agenesis, bladder diverticulum, left vesicoureteral reflux, left hydrocele, undescended right testis. |
| VIII-15 | 20 | 3.147 (31%) | 49.5 (54%) | 32 (3%) | 74.4 (89%) | 175 (96%) | Calcaneal valgus, obsessive-compulsive tendencies, depression. |
| VIII-16 | 18 | 4.139 (89%) | 54.6 (96%) | 34 (19%) | 64.4 (42%) | 189 (96%) | |
| VIII-19 | 5 | 2.722 (9%) | 50.2 (53%) | 32.5 (7%) | 15.9 (29%) | 109 (85%) |
Weight (Wt) at birth and at examination is expressed in kilograms, and the length and head circumference (OFC) are expressed in centimeters. The numbers in parentheses are percentiles in reference to the respective age. N/A, data are not available.
The craniofacial dysmorphism was noticeable at birth and included brachycephaly, flat face, low hairline, low-set ears, high-arched palate, and cleft lip and palate. Other characteristic dysmorphic features, such as highly arched bushy eyebrows, synophrys, orbital hypertelorism, long eyelashes, wide nose bridge, short nose with anteverted nares, and microdontism of primary teeth appeared later, generally before they reached preschool age. Generalized gingival hyperplasia usually appeared after the eruption of permanent teeth (Fig. 2, Fig. S1, and Table 1).
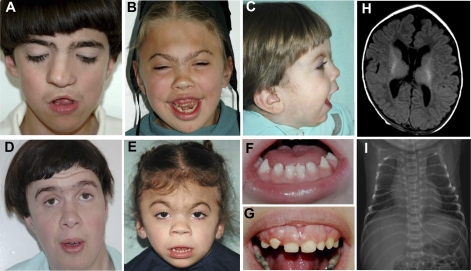
Fig. 2.
Clinical characteristics of TMCO1 defect syndrome. (A–G) Facial features of the syndrome (low hairline, brachycephaly, flat face, highly-arched bushy eyebrows, synophrys, long eyelashes, orbital hypertelorism, wide nose bridge, short nose with anterverted nares, microdontism, and generalized gingival hyperplasia) in six patients. Parental consent has been obtained for publication of these photographs. (H) Magnetic resonance imaging (MRI) of brain of a 3-month-old female, showing prominence of cerebrospinal fluid (CSF) space around the frontal lobes with extention into the anterior interhemisheric fissure. (I) Chest x-ray film of 4-day-old male showing multiple rib anomalies.
Skeletal dysmorphism, such as pectus excavatum and club feet, was noted during early infancy, whereas scoliosis and long, hyperextensible fingers developed when the patients reached puberty. Fusion of the cervical or thoracic spine, rib anomalies (including fusion, hypoplasia, or broad or missing ribs), and Sprengel deformity of scapula, were usually incidental findings during routine imaging studies for other purposes. The actual incidence of these abnormalities in TMCO1 defect syndrome might be higher if systematic osseous surveys were performed in all patients (Table 1, Table 2, Fig. 2, and Fig. S1). The skeletal anomalies found in this condition mostly involve the axial skeleton, which embryologically originates from somites. There was significant divergence in patients’ stature, two of them having tall stature with height >95th percentile, whereas three exhibited short stature with height in less than the 3rd percentile. Global developmental delay was found in all affected individuals, although regression was not observed.
Neurological examination revealed depressed deep tendon reflexes and unstable gait, and intention tremor developed in some older patients. Imaging studies demonstrated mild prominence of ventricles viewed through CT or MRI (Fig. 2H). Sluggish speech with a loud and hoarse voice was found in all verbal patients. Anatomical abnormalities of the genito-urinary system were noted, including renal agenesis, hydronephrosis, vesicoureteral reflux, hypoplastic labia minora, hydrocele, and undescended testes. Other clinical features with high incidence were frequent otitis media, sinusitis, strabismus, and constipation although they were less specific (Table 1).
Genotyping and Mapping.
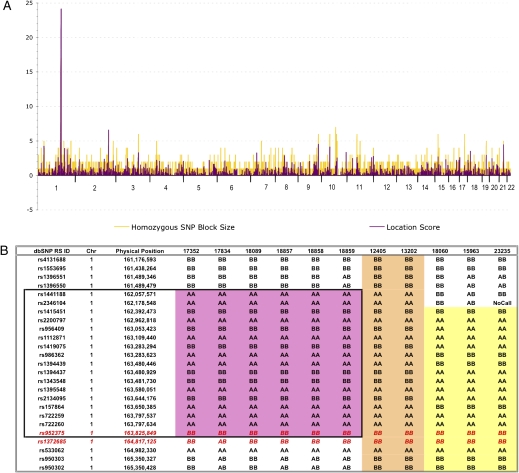
To determine the genetic basis of this syndrome, we performed a genome-wide homozygosity mapping study by using Affymetrix GeneChip Mapping 10K SNP Arrays with six affected individuals from this large consanguineous pedigree (Fig. 1). A single large, shared block of homozygous SNPs was identified on chromosome 1q23.3-q24.1 in all six patients (Fig. 3). The homozygous segment contained 17 contiguous SNPs and spanned 3.3 Mb. Examination of the minimal shared region, which was flanked by SNPs rs1396550 and rs1372685, revealed 23 known or predicted genes based on both the NCBI and Celera annotations.
Fig. 3.
Genetic mapping of TMCO1 defect syndrome found in the Old Order Amish of northeastern Ohio. (A) Autozygosity mapping using Affymetrix GeneChip 10K SNP miroarrays and six affected individuals revealed homozygous haplotype identity for 17 SNPs on chromosome 1q23.3-q24.1. (B) Genotypes for 11 affected individuals from 3 separate Amish demes reveal that the TMCO1 mutation resides on two distinct SNP haplotypes. The original six patients are displayed first (black box, pink shading). The two Somerset County, PA, patients (next two columns, tan shading) share homozygous haplotype identity with the six original patients, whereas the three Lancaster County, PA, patients are identically homozygous for a distinct haplotype surrounding TMCO1 (yellow shading). In all, the 11 genotyped patients share homozygous genotypes at a mere three SNP loci spanning no more than 1.2 Mb. TMCO1 physically maps between rs952375 and rs1372685 (red letters).
Mutation Identification and mRNA Analysis.
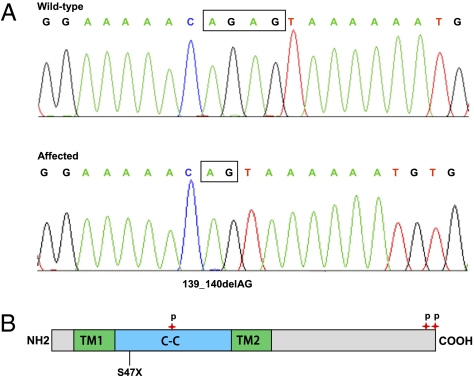
For each gene within the mapped interval, we assessed function and expression to generate a priority list for sequencing. Candidate gene sequencing was performed to screen the coding region and associated intronic splice junctions by using genomic DNA from one patient. We sequenced six genes before we identified a homozygous 2-base pair deletion (c.139_140delAG) within exon 2 of the TMCO1 gene (Fig. 4A). This frameshift variant is predicted to result in premature truncation of translation at amino acid position 47 (p.Ser47Ter, Fig. 4B). Targeted sequencing of TMCO1 exon 2 revealed that all affected individuals (n = 9) were homozygous for the mutation, their parents were heterozygous, and no unaffected siblings were homozygous for the change. We next screened 145 ethnically matched control samples from the same geographic area where the patients were found (n = 290 chromosomes) and determined that none were homozygous, whereas one was heterozygous for the c.139_140delAG mutation (estimated carrier frequency of 0.7%).
Fig. 4.
Identification of the disease-causing mutation in TMCO1 gene. (A) Sequence electropherograms showing the homozygous c.139_140delAG mutation compared with a normal control. (B) Schematic of TMCO1 protein indicating position of identified mutation and protein structure. Protein structural regions are predicted by SMART. The transmembrane (TM1 and TM2) and coiled-coil domains are shown in green and blue, respectively. The three verified phosphoserine residues (S60, S184, and S188) are indicated by red stars.
To determine the distribution of the TMCO1 mutation in other Amish populations, we screened 75 Old Order Amish from southeastern Pennsylvania for c.139_140delAG mutation. This sample included normal controls and undiagnosed patients. We found five c.139_140delAG heterozygotes for an estimated carrier frequency of 6.6%. Surprisingly, among the undiagnosed patients, we identified a single mutation homozygote. Review of his clinical history revealed a phenotype strikingly similar to the affected children from Ohio. Further testing of undiagnosed children at the Clinic for Special Children identified a total of five affected c.139_140delAG homozygotes. Review of the SNP genotype data generated previously showed that all five patients from Pennsylvania were indeed homozygous for SNPs spanning the TMCO1 locus, but the haplotypes were different (Fig. 3B). The three homozygotes from the Lancaster County Amish settlement were homozygous for a different background haplotype than the other two homozygotes, who lived in Somerset County of Pennsylvania. The Somerset County Amish haplotype matched the Ohio Amish haplotype.
To determine the mRNA expression pattern of TMCO1, human adult and fetal multiple tissue cDNA (MTC) panels were used as templates for PCR analysis. Amplification of first-strand cDNA showed ubiquitous expression in all 16 adult and 8 fetal tissues along with the control gene GAPDH (Fig. S2), with relatively higher level in the tissues of adult thymus, prostate, and testis.
Discussion
We describe here a unique autosomal recessive condition characterized by craniofacial dysmorphism, skeletal anomalies, and mental retardation in the Old Order Amish. The clinical features of this condition overlap with several previously defined syndromes such as Cornelia de Lange syndrome, chromosome 3q duplication, Smith-Magenis syndrome, and Klippel-Feil syndrome, although it is clearly distinct from these syndromes. This condition also shares numerous clinical features with some lysosomal storage diseases, but the disease does not progress as do the storage diseases. Because of the tall stature in several patients, the condition has been suspected as a Marfan-like syndrome or Sotos-like syndrome as well. It is obvious that the phenotype of this condition is fairly heterogeneous and the clinical diagnosis of the disease may be challenging, particularly before the phenotype is well documented. The unique position as a primary care facility has given us the opportunities to carefully and continually observe these patients in the description of the phenotype while we provide medical services. The value of accessible, continuous, and comprehensive care provided by such a clinic for affected children has been well demonstrated in a similar clinic previously (6). We anticipate that the current work and future translational research of this condition will not only further enhance our cost-effective medical services for these patients, but also provide an opportunity for early diagnosis through newborn screening and adequate genetic counseling for the high risk families.
Through genome-wide homozygosity mapping and mutational analysis in affected individuals from a consanguineous pedigree, we have identified a pathogenic sequence variant, c.139_140delAG, in the TMCO1 gene, which is associated with the disease. This alteration has not been reported as a polymorphic variant in GenBank. It cosegregates consistently with the disease phenotype, because all affected individuals are homozygous, their parents are heterozygous, and no unaffected siblings or normal controls are homozygous for the mutation. We therefore designate this condition as TMCO1 defect syndrome.
Ten of the eleven patients initially identified are from the Geauga County settlement of Ohio. One patient from Kentucky is closely related to the individuals from this settlement. The Geauga County Amish settlement originated around 1886, with initial immigrants from Holmes County, Ohio. Amish from Pennsylvania settlements later joined them, making the Geauga settlement the fourth largest Amish community with an extant population of ≈15,000. Although the history of this settlement is relatively short, genetic disorders are fairly common in the community. The carrier frequency (0.7%) is substantially lower than several other conditions recently identified in the Geauga settlement, such as hypertrophic cardiomyopathy and Charcot-Marie-Tooth disease (4, 5).
The identification of TMCO1 defect syndrome patients in the Amish populations of Pennsylvania was unexpected. Targeted sequencing of TMCO1 exon 2 for undiagnosed patients revealed five cases within two distinct Amish demes in Pennsylvania. Although several were suspected to share the same disorder, homozygosity mapping was hampered by haplotype heterogeneity. The shared haplotype for the Lancaster Amish (samples 18060, 15963, and 23235) is significantly different from the haplotype for the Somerset County, PA, and Geauga County, OH, Amish. This disparity might reflect unfortunate ancestral recombinants very near the disease gene or discrete ancestors whose genealogical ties predate emigration to the United States.
This report shows a TMCO1 sequence variant being associated with an adverse phenotype in human. The TMCO1 gene consists of seven coding exons, and the 564-bp coding region encodes a predicted protein of 188 amino acids. SMART sequence analysis predicted two transmembrane segments (amino acids 10–31 for TM1 and amino acids 90–109 for TM2) and a coiled-coil domain (amino acids 32–89) (Fig. 4B). In addition, three phosphorylation sites (phosphoserines) involved in the signaling networks across the cell cycle were verified in the previous studies (7–10) (Fig. 4B). It is predicated that the c.139_140delAG mutation would result in a severely truncated protein lacking the second transmembrane domain, the coiled-coil domain, and three potential phosphorylation sites if it escapes nonsense-mediated mRNA decay. The phosphorylation sites are essential for protein function through mediation of protein–protein interactions and signal transduction.
The function of TMCO1 is unknown. An earlier study with a GFP-tagged fusion protein suggested that HP10122, an alias for TMCO1, was localized in the endoplasmic reticulum and Golgi apparatus (11), whereas the most recent study using the similar approach with the porcine TMCO1 showed mitochondrial localization (12). Both groups have also investigated the expression patterns of TMCO1 transcript in different tissues and revealed universal expression in all tissues examined, which is consistent with our results (Fig. S2). The EST Profile Viewer of the NCBI database (UniGene Hs.31498) confirms expression of TMCO1 transcript in 42 of 45 adult tissues, including all tissue types tested by RT-PCR for this study. Moreover, mRNA expression was also noted at all developmental stages listed from embryo to adult (http://www.ncbi.nlm.nih.gov/UniGene/ESTProfileViewer.cgi?uglist=Hs.31498). Amino acid sequence comparison shows the entire sequence is highly conserved among multiple species, with 100% homology among eight mammalian TMCO1 orthologs in GenBank (Fig. S3). The extremely high degree of conservation of TMCO1 and the ubiquitous expression pattern in human adult and fetal tissues suggest a critical role for TMCO1. This speculation is supported by multiple system and organ involvement in affected individuals. The complicated pregnancy in affected individuals and high incidence of first trimester spontaneous abortions observed in these families suggests a critical role for TMCO1 in early fetal growth and development.
Materials and Methods
Subjects and Clinical Assessment.
The study was approved by DDC Clinic for Special Needs Children (DDC Clinic) Institutional Review Board, and written informed consent was obtained from each participant or their legal guardian. The phenotype information was based on clinic data from 11 affected individuals, 10 of them were from Ohio and 1 from Kentucky. DDC Clinic provided primary medical services to nine of them, three from birth. As part of routine medical care, each patient’s detailed medical history and records were collected. Two patients, with the typical phenotype, were clinically diagnosed, but were deceased before their genotype was confirmed, thus their parents’ and siblings’ genotype were assessed. An additional suspected patient (VIII-4 in Fig. 1), with partial phenotypic expression at birth, expired at 2 days of age and therefore was not included in the phenotype description. Most parents and siblings of the patients also chose to participate in the study. The genealogical information obtained from the affected families was confirmed and expanded through the Swiss Anabaptist Genealogical Association (SAGA) group, James C. Hostetler database (http://www.omii.org/).
Neuropsychological Assessment.
A battery of well validated neuropsychological measures, including Wechsler Intelligence Scale WISC-R, 1991, was administrated to seven affected individuals. To reduce investigator variability and bias, the same clinical psychologist (S.T.) performed the assessments for each patient in their home environment. Four nonverbal children participated in the study, but the IQ testing results were excluded because of the unreliable results.
Genotyping and Mapping.
Total genomic DNA from whole blood was isolated by using the PUREGENE DNA Isolation Kit (Gentra Systems, Minneapolis) according to the manufacturer’s protocol. Given the founder effect in isolated populations such as the Old Order Amish, the disease gene mapping strategy focused on large genomic regions demonstrating homozygosity in all of the affected individuals. DNA samples from six patients were used for genome-wide single nucleotide polymorphism (SNP) autozygosity mapping (Fig. 1) by using the Affymetrix GeneChip Mapping 10K assay kit (Affymetrix, Santa Clara, CA) as described in refs. 2 and 3. Genotyping data were analyzed by using customized Excel spreadsheets designed to identify putative autozygous regions of the genome of the affected individuals. SNP positions came from dbSNP build 127 and NCBI build 36 of the human genome. Assuming mutation homogeneity, our analysis examined large, homozygous blocks of SNPs. Population-specific SNP allele frequencies were generated from 80 Lancaster County Amish control individuals. Cumulative two-point logarithm of odds scores for a block of homozygous SNPs were considered the “location score” for the region. These location scores provided a relative measure of the likelihood that a particular genomic region was autozygous.
Mutation Identification.
PCR primers were designed to amplify each of the seven protein-coding exons and their flanking intronic sequences of TMCO1. We designed primer sequences (Table S1) by using Primer3 software (http://frodo.wi.mit.edu/primer3).; PCR amplifications were performed by using 50 ng of genomic DNA in each reaction. The PCR products were examined on 1% agarose gels and purified for sequencing by using Qiaquick spin columns (QIAGEN, Valencia, CA). Sequencing reactions were performed by using the Big Dye terminator v3.1 cycle sequencing kit (Applied Biosystems, Foster City, CA), and the extension products were analyzed on an Applied Biosystems 310 Genetic Analyzer. The identified mutation was verified with repeat PCR amplification and sequencing in both orientations. Sample sequences were compared to the GenBank reference sequences by using Mutation Surveyor software (SoftGenetics LLC, State College, PA) for identification of sequence variants. The GenBank accession numbers of both genomic DNA and mRNA reference sequences used in this study are NT_004487 and NM_019026, respectively. We performed protein domain analysis by using the Simple Modular Architecture Research Tool (SMART; http://smart.embl-heidelberg.de). We generated amino acid sequence alignments of TMCO1 orthologs by the ClustalW2 program (www.ebi.ac.uk/Tools/clustalw2/index.html).
cDNA Amplification.
PCR amplification of TMCO1 cDNA was performed by using primers F117 and R462 located in exons 1 and 6 (Table S1, the base pair numbers of primers were determined according to the mRNA sequence NM_019026). The tissue distribution of TMCO1 transcript was determined by RT-PCR. Both adult and fetal human multiple tissue cDNA (MTC) panels were obtained commercially (Clontech, Mountain View, CA). PCR amplification of TMCO1 and GAPDH (positive control) sequences were performed according to the manufacturer’s protocol. Thirty PCR cycles for TMCO1 and twenty-four cycles for GAPDH were performed. The PCR products were then electrophoresed on a 1.5% agarose gel.
Supplementary Material
Acknowledgments
We thank the families for their patience and support. We appreciate the many physicians who provided outstanding and compassionate care to the children affected by the disease, particularly Dr. John Tumbush in Middlefield and many others from Akron Children’s Hospital, Rainbow Babies and Children’s Hospital, and Shriners Hospitals for Children. We are indebted to Dr. Irvin Schafer and Dr. Kurt Wegner, who helped us evaluate three patients; Dr. Guiyun Wu, who helped us review and interpret imaging studies; and Miss Carol Troyer, who helped with the data collection. We acknowledge Drs. Holmes Morton and Kevin Strauss for their continuous support throughout the project, their valuable contributions in the evaluation of patients at the Clinic for Special Children, critical reading, and suggestions to the manuscript. The study was supported in part by The Elisabeth Severance Prentiss Foundation, The Reinberger Foundation, and the Leonard Krieger Fund of the Cleveland Foundation (L2009-0078).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0908457107/DCSupplemental.
References
- 1.Raymond FL, Tarpey P. The genetics of mental retardation. Hum Mol Genet. 2006;15(Spec No 2):R110–R116. doi: 10.1093/hmg/ddl189. [DOI] [PubMed] [Google Scholar]
- 2.Puffenberger EG, et al. Mapping of sudden infant death with dysgenesis of the testes syndrome (SIDDT) by a SNP genome scan and identification of TSPYL loss of function. Proc Natl Acad Sci USA. 2004;101:11689–11694. doi: 10.1073/pnas.0401194101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Strauss KA, et al. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354:1370–1377. doi: 10.1056/NEJMoa052773. [DOI] [PubMed] [Google Scholar]
- 4.Xin B, Puffenberger EG, Tumbush J, Bockoven JR, Wang H. Homozygosity for a novel splice site mutation in the cardiac myosin-binding protein C gene causes severe neonatal hypertrophic cardiomyopathy. Am J Med Genet A. 2007;143:2662–2667. doi: 10.1002/ajmg.a.31981. [DOI] [PubMed] [Google Scholar]
- 5.Xin B, Puffenberger EG, Nye L, Wiznitzer M, Wang H. A novel mutation in the GDAP1 gene is associated with autosomal recessive Charcot-Marie-Tooth disease in an Amish family. Clin Genet. 2008;74:274–278. doi: 10.1111/j.1399-0004.2008.01018.x. [DOI] [PubMed] [Google Scholar]
- 6.Strauss KA, Puffenberger EG. Genetics, medicine, and the Plain people. Annu Rev Genomics Hum Genet. 2009;10:513–536. doi: 10.1146/annurev-genom-082908-150040. [DOI] [PubMed] [Google Scholar]
- 7.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 8.Nousiainen M, Silljé HHW, Sauer G, Nigg EA, Körner R. Phosphoproteome analysis of the human mitotic spindle. Proc Natl Acad Sci USA. 2006;103:5391–5396. doi: 10.1073/pnas.0507066103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daub H, et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Dephoure N, et al. A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwamuro S, Saeki M, Kato S. Multi-ubiquitination of a nascent membrane protein produced in a rabbit reticulocyte lysate. J Biochem. 1999;126:48–53. doi: 10.1093/oxfordjournals.jbchem.a022435. [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, et al. Molecular cloning, expression patterns and subcellular localization of porcine TMCO1 gene. Mol Biol Rep. 2009 doi: 10.1007/s11033-009-9573-8. in press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.